Abstract
Purpose: The death ligand TRAIL (tumor necrosis factor-related apoptosis inducing ligand) triggers apoptosis in a variety of cancer cells, which implies the potential for therapeutic applications. The purpose of this study was to investigate the role of the lysosomal protease cathepsin B (CB) in mediating TRAIL-induced cell death in oral squamous cell carcinoma (OSCC) cells. Methods: OSCC cell lines from primary tumor and lymph node metastasis were examined for expression of apoptosis markers by Western blots, enzyme activity assays, nuclear fragmentation assays, and FACS analysis. Gene-specific ribozymes or chemical inhibitors were used to inhibit CB or caspases in target cells. Results: TRAIL-induced activation of caspase-3, cleavage of Bid and poly–ADP-ribose polymerase, release of cytochrome c, and DNA fragmentation were blocked either by a pan-caspase inhibitor (zVAD-fmk) or a CB inhibitor (CA074Me), consistent with the involvement of TRAIL as well as CB in cell death. The primary tumor cells were more susceptible to apoptosis than their corresponding lymph node metastatic cells. Stable transfection of a ribozyme which inhibited CB expression also decreased the apoptotic process. Conclusions: We conclude that TRAIL-induced apoptotic cell death in OSCC cells is mediated through CB or through caspase activation. Our data point to a new tumor-suppressive role for CB in OSCC which is opposed to the invasion- and metastasis-promoting functions of lysosomal proteases.
Keywords: Oral cancer, Apoptosis, Cathepsin B, Caspases, TRAIL, Ribozymes
Introduction
Cytotoxic tumor therapies (chemotherapy, radiation, immunotherapy or suicide gene therapy) are primarily mediated by triggering apoptosis selectively in cancer cells (Fulda and Debatin 2004a). Activation of apoptosis has long been held responsible for the cell killing potential of some anticancer drugs. Apoptosis in response to cancer therapy proceeds through activation of the core apoptotic machinery including the extrinsic cell death receptor and the intrinsic mitochondrial signaling pathway (Herr and Debatin 2001). Stimulation of death receptors of the tumor necrosis factor (TNF) receptor superfamily such as CD95 (APO-1/Fas) or TRAIL receptors results in activation of the initiator caspase-8, which can propagate the apoptosis signal by direct cleavage of downstream effector caspases such as caspase-3 (Ashkenazi 2002). The extrinsic and the intrinsic pathways are connected through Bid, a pro-apoptotic Bcl-2 family member which is also cleaved during apoptosis by lysosomal proteases (Stoka et al. 2001). In certain cell types, caspase-8 has been shown to convert Bid from a latent to a strongly proapoptotic form, capable of inducing cytochrome c release through the action of Bax or Bak and subsequent activation of caspase-9 (Borner 2003). Cytochrome c released from the mitochondrial intermembrane space was the first mitochondrial factor actively implicated in apoptotic cell death. This release of cytochrome c into the cytosol triggers caspase-3 activation through formation of the cytochrome c/Apaf-1/caspase-9 apoptosome complex, whereas Smac/Diablo and Omi/HtrA2 promote caspase activation through neutralizing the inhibitory effects of IAPs (Van Loo et al. 2002).
TRAIL/Apo2L is a cytotoxic ligand belonging to the TNF superfamily that includes TNF-α and FasL. TRAIL engages the extrinsic apoptotic pathway by binding to its membrane-bound death receptors (DR4 and DR5), which transmit an apoptotic signal via their intracellular death domains. TRAIL rapidly triggers apoptosis in many tumor cells (Fulda and Debatin 2004b) and has a special value in cancer therapy since it has been shown to predominantly kill cancer cells while sparing normal cells (LeBlanc and Ashkenazi 2003). The underlying mechanisms for the differential sensitivity of malignant versus nonmalignant cells to TRAIL have not been defined clearly. One possible mechanism for protection of normal tissues is thought to be based on the antagonistic decoy receptors DcR1 and DcR2, which compete with DR4 and DR5 for binding with TRAIL. However, screening of various tumor and normal cell types did not reveal a consistent association between TRAIL sensitivity and TRAIL receptor expression. Hence, susceptibility for TRAIL-induced cytotoxicity may be regulated intracellularly by distinct patterns of proapoptotic and antiapoptotic factors. TRAIL appears to be a relatively safe candidate for clinical application (LeBlanc and Ashkenazi 2003). The idea to specifically target death receptors to trigger apoptosis in tumor cells is attractive for cancer therapy since death receptors have a direct link to the cell’s death machinery (Ashkenazi 2002). Also, apoptosis upon death receptor triggering is considered to occur independent of the p53 tumor suppressor gene, which is impaired in a majority of human tumors (El-Deiry 2001).
Although caspases are well established as the main players in apoptosis, other proteases such as calpains, cathepsins, and serine proteases account for alternative types of programmed cell death (Leist and Jäättelä 2001a). Lysosomal proteases can be released from the lysosomes into the cytosol, where they contribute to the apoptotic cascade upstream of mitochondria (Guicciardi et al. 2004). Many tumors express increased amounts of cathepsins, and the action of TNF is dependent on the ‘lysosomal pathway’ of apoptosis and specifically on cathepsin B (CB) (Koblinski et al. 2000; Taha et al. 2005). Recently, CB has been shown to play a dominant role in executing the apoptotic program in several tumor cell lines (Foghsgaard et al. 2001), and to be essential in different models of apoptosis (Jones et al. 1998; Canbay et al. 2003). These studies demonstrated the need for a CB-mediated proteolytic event in the apoptotic pathway triggered by TRAIL. CB inhibitors can reduce the response of cells to some apoptosis inducers, and cells deficient or downregulated in CB are more resistant to TNF-mediated apoptosis (Foghsgaard et al. 2001; Guicciardi et al. 2000). In culture, cathepsins can trigger cytochrome c release from the mitochondria into the cytosol (Stoka et al. 2001; Guicciardi et al. 2000). The fact that tumors frequently contain high levels of CB may prove useful in selectively targeting tumor cells for apoptosis induction by ligands.
In this report, we show that TRAIL-induced cell death in oral squamous cell carcinoma (OSCC) cells is reduced in the presence of CB inhibitor CA074Me or a CB-specific ribozyme. Our data support the model that TRAIL induces apoptosis through CB and is independent of caspases. We further show that TRAIL can trigger activation of CB, providing evidence for a novel CB-dependent cell death pathway in OSCC cells. Also, we found that primary OSCC cells are more susceptible to TRAIL compared to metastatic OSCC cells. Based on this CB-mediated caspase-independent apoptosis pathway, the involvement of CB in this process needs to be considered in designing molecular therapies for OSCC in the future.
Materials and methods
Cell lines and reagents
The OSCC cell lines MDA1386Tu (1386Tu) and MDA1386Ln (1386Ln) were obtained from the primary tumor and a lymph node metastasis (tumor stage T4N3B) of a 71-year-old male patient with primary hypopharynx tumor (generous gift from Peter Sacks, New York University, New York, USA) (Sacks 1996). The cells were maintained in DMEM/F12 1:1 (v/v) mix containing 10% fetal bovine serum and 0.4 μg/ml hydrocortisone at 37°C with 5% CO2. Rabbit polyclonal antibody for human CB was obtained from Athens Research and Technology (Athens, GA, USA) and HRP-conjugated goat anti-rabbit or anti-mouse antiserum were from BD Biosciences (San Diego, CA, USA). Protease inhibitor CA074, cell permeable CA074Me and z-Arg-Arg-NHMec were obtained from Peptides International (Louisville, KY, USA); G418 sulfate was from Mediatech Inc. (Herndon, VA, USA). Human recombinant TRAIL was purchased from Biosource International (Camarillo, CA, USA). Cell-permeable inhibitors z-IETD-fmk and z-VAD-fmk, rabbit polyclonal IgGs for caspase-3 (H-277), caspase-8 (H-134) and Bid (FL-195) were all from Santa Cruz Biotechnology (Santa Cruz, CA, USA). DEVD-AFC, IETD-AMC, DEVD-CHO and IETD-CHO were from Alexis (San Diego, CA, USA), anti-PARP antibody from Cell Signaling Technology (Beverly, MA, USA). Mouse monoclonal β-actin antibody, phenylmethylsulfonyl fluoride, aprotinin, leupeptin, pepstatin and MTT reduction assay kit (TOX-1 kit) were from Sigma (St. Louis, MO, USA). Mouse monoclonal anti-cytochrome C IgG was from BD Biosciences-Pharmingen (San Diego, CA, USA). Bradford protein assay kit was purchased from Bio-Rad (Hercules, CA, USA), M-PER Mammalian Protein Extraction Reagent from Pierce (Rockford, IL, USA). Protease inhibitor cocktail, In Situ Cell Death Detection Kit POD and Apoptotic DNA Ladder Kit were from Roche (Indianapolis, IN, USA), and enhanced chemiluminescence ECL substrate from Amersham Biosciences (Piscataway, NJ, USA). All protocols for the use of human cell lines in this work were approved by the Institutional Review Boards of The University of Louisville and The University of Texas at Houston.
Cell treatments
Cells were treated in triplicate wells at 37°C for 24 h with or without recombinant human TRAIL protein (0–250 ng/ml for titrations, or 100 ng/ml for all other assays) or CA074Me (0–50 μM for titrations, or 25 μM for all other assays). Pre-incubation assays were done for 3 h in the absence or presence of 10 μM zVAD-fmk and/or 25 μM CA074Me; control wells had the same volume of DMSO solvent.
Ribozyme design and expression
The design, cloning, and inhibitory effects of CB-targeted ribozymes were previously described (Wickramasinghe et al. 2005).
Cytotoxicity assay
Cells (1.5×104 cells/well in 96-well plates) were treated in triplicate with or without TRAIL protein and/or CA074Me for 24 h at 37°C; preincubation was for 3 h in the presence or absence of zVAD-fmk and/or CA074Me. Concentrations of TRAIL or inhibitors are listed in the corresponding figure legends; control assays had the same volume of DMSO. After incubation, cytotoxicity rates were determined by the MTT reduction assay (TOX-1 kit) and absorbance measurement at 570 nm (A 570). Cytotoxicity (%) was calculated using the following equation: % cytotoxicity = (C-S)/C×100, where C is the average A 570 of the untreated control and S is the average A 570 of the sample. Each experiment was performed independently at least three times in triplicate and cytotoxicities are given as means±SD.
Cathepsin B activity assay
Cells were grown in 6-well plates to 60–70% confluence, and treated for 24 h with or without TRAIL; with or without pretreatment of zVAD-fmk (10 μM) and/or CA074Me (0–50 μM), and also with CA074Me or TRAIL alone. Concentrations of TRAIL or inhibitors are listed in the corresponding figure legends; control assays had the same volume of DMSO. After treatment, the cells were rinsed twice with ice-cold PBS, treated with lysis buffer (400 mM Na-phosphate, 75 mM NaCl, 4 mM EDTA, 0.25% Triton-X 100, pH 6.0) for 1 h on ice, ultrasonicated at 40 W for 1 min (1.0 s on/0.5 s off pulses) (Model 550 Sonic Dismembrator, Fisher Scientific, Pittsburgh, PA, USA), and centrifuged at 25,000 g (10 min, 4°C) to remove cell debris. Total protein amounts were determined using to the Bradford protein assay kit. CB activity was determined fluorimetrically using the methyl-coumarylamide substrate z-Arg-Arg-NHMec at pH 6.0, as described (Wickramasinghe et al. 2005). Fluorescence was measured with an excitation wavelength of 360 nm and emission wavelength of 460 nm. The CB substrate was used in conjunction with the CB inhibitor CA074 (50 μM) in all control assays; the difference between values without and with CA074 corresponded to CB activity. One unit of enzyme activity was defined as the release of 1 μmol of product/min.
Caspase activity assay
Cell cultures were treated for 24 h with or without TRAIL and with pretreatment of zVAD-fmk and/or CA074Me. Concentrations of TRAIL or inhibitors are listed in the corresponding figure legends; control assays had the same volume of DMSO. Then cells were rinsed once and collected in cold PBS by scraping. After centrifugation, cell pellets were resuspended in caspase lysis buffer (10 mM HEPES, pH 7.4, 2 mM EDTA, 0.1% CHAPS) supplemented with protease inhibitors (5 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 10 μg/ml pepstatin A, 10 μg/ml aprotinin, and 20 μg/ml leupeptin). Freeze–thaw cell lysis cycles were performed by alternatively transferring the samples from an ethanol/dry ice bath to a 37°C water bath five times. The supernatant was collected after 20 min of centrifugation at 12 000 rpm in a cold microcentrifuge. Assays were performed in caspase buffer (10 mM PIPES, pH 7.4, 2 mM EDTA, 0.1% CHAPS, 5 mM dithiothreitol), to which 50 μM of substrate and 5 μl of protein extract were added to yield a final volume of 100 μl. Peptide substrates used for caspase-3 like (caspase-3 and -7) and caspase-8 assays were Ac-DEVD-AFC and Ac-IETD-AMC, respectively, each dissolved in dimethyl sulfoxide; the respective specific inhibitors DEVD-CHO and IETD-CHO were used in control reactions. Assays were performed in black-wall, clear bottom plates using a Spectramax Gemini XS Microplate Spectrofluorometer (Molecular Devices); reading was at 500 nm after excitation at 405 nm for AFC, and at 460 nm after excitation at 380 nm for AMC. The results were compared against AFC and AMC standard curves generated in parallel. Specific activity was expressed as units, with 1 unit defined as AFC or AMC release of 1 nmol/h/μg protein.
Protein extractions and Western blotting
Treated or control cells were rinsed with ice-cold PBS, scraped into 1 ml of PBS, and centrifuged at 4,000 rpm for 3 min. The pellets were resuspended into RIPA buffer (10 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Triton X-100, 0.1% SDS, and 1 mM EDTA) containing protease inhibitors (0.5 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and 2 μg/ml of both leupeptin and pepstatin). Then, cell extracts were sonicated as described above and cell debris was removed by centrifugation. To obtain cellular proteins for CB, cells were washed in ice-cold PBS and protein was extracted using M-PER Mammalian Protein Extraction Reagent with protease inhibitor cocktail. Proteins were quantified using the Bradford protein assay kit and compared with a γ-globulin standard curve. Equal amounts of total proteins were separated on an SDS-polyacrylamide gel and transferred onto a nitrocellulose membrane by electroblotting overnight at 20 V. Membranes were blocked in TBS-T (10 mM Tris-HCl, 150 mM NaCl, 0.25% Tween 20, pH 7.5) with 5% fat-free powdered milk at room temperature for 1 h. After rinsing membranes in TBS-T, the following primary antibodies were used: rabbit polyclonal antibodies for human cathepsin B, rabbit polyclonal IgGs for caspase-3, caspase-8, Bid, poly (ADP-ribose) polymerase (PARP), or mouse monoclonal β-actin antibody. After incubation overnight at 4°C or 1 h at room temperature, the membranes were washed four times, 10 min each, in TBS-T. Secondary antibodies used were either horseradish peroxidase-conjugated goat anti-rabbit IgG or goat anti-mouse IgG, followed by five washes with TBS-T. Bands were detected using the enhanced chemiluminescence ECL substrate. For β-actin detection, previously probed membranes were soaked in stripping buffer (70 mM Tris–HCl, pH 6.8, 2% SDS, 0.1% β-mercaptoethanol) at 60°C for 30 min, and incubated as above.
Cytochrome c release assay
Cells were collected at the indicated times and washed once in ice-cold PBS. Cell pellets were resuspended in cytosol extraction buffer, and cytosolic extracts were prepared as described previously (Nagaraj et al. 2004). Western blotting for cytochrome c was done with mouse monoclonal anti-cytochrome c IgG; the absence of intra-mitochondrial proteins was verified by blotting for mitochondrial cytochrome oxidase with mouse monoclonal anti-cytochrome oxidase IgG.
Analysis of DNA fragmentation
Apoptotic cells were detected by in situ TdT-mediated dUTP nick end labeling (TUNEL) assays using the In Situ Cell Death Detection Kit POD. Apoptotic nuclei were detected by diaminobenzidine (DAB), and the percentage of TUNEL-positive cells counted at ×400 magnification. Nucleosomal DNA fragments were detected with the Apoptotic DNA Ladder Kit after electrophoresis on 2% agarose gels for visualizations of apoptosis-indicative DNA ladders.
FACS analysis
Apoptosis detection by FACS analysis for sub-G1 DNA content was performed after staining cells with propidium iodide. Cells were trypsinized, washed and resuspended in cold PBS. After centrifugation at 1,000 rpm for 5 min, 5×105 cells were fixed with 1 ml 70% ethanol at −20°C and washed with PBS. Each pellet was resuspended in 500 μl propidium iodide buffer (20 μg/ml propidium iodide, 10 mg/ml RNAse in PBS). After incubation for 30 min in the dark at room temperature, cell fluorescence signals were determined using a FACScalibur flow cytometer (BD Biosciences) and analyzed with CellQuest software (BD Biosciences). Cells to the left of the first peak at sub-G1 (M1) contained hypodiploid DNA and were considered apoptotic.
Statistical analysis
All data represent at least three independent experiments and are expressed as the means±SD unless otherwise indicated. ANOVA was used to assess the differences between experimental groups.
Results
Sensitivity of OSCC cell lines to TRAIL and CA074Me
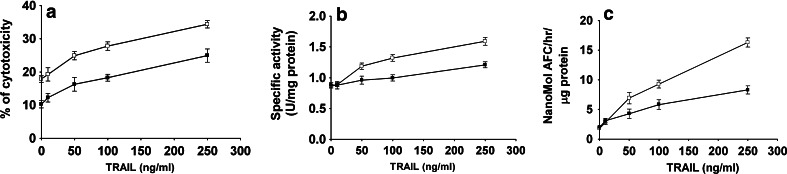
To investigate the susceptibility of OSCC cell lines to TRAIL-induced cell death, the 1386Tu and 1386Ln cell lines were cultured with 0–250 ng/ml recombinant TRAIL for 24 h, and cytotoxicity in response to increasing amounts of TRAIL was assessed by MTT assays (Fig. 1a). The 1386Tu cells were highly sensitive to TRAIL-induced cell death, whereas 1386Ln was much less affected, in agreement with our previous observations (Vigneswaran et al. 2005). Interestingly, CB and caspase-3 like activities were both increased by increasing concentration of TRAIL, and this effect was more pronounced in 1386Tu than in 1386Ln cells (Fig. 1b, 1c).
Fig. 1.
Effects of TRAIL on OSCC cells. The 1386Tu (open squares) and 1386Ln (closed squares) cells were treated with TRAIL (0–250 ng/ml) for 24 h and analyzed for a cytotoxicity, b CB, and c caspase-3 like activity. Cytotoxicity was determined by MTT assays, CB and caspase-3 like activity by using fluorogenic substrates and specific inhibitors in control assay as described in Materials and Methods. Results shown are the means±SD from three separate experiments
We also investigated whether these cell lines were sensitive to the CB inhibitor CA074Me. CA074 is a highly specific inhibitor of CB in vitro and its methylated variant CA074Me was shown to penetrate into cells more easily than the parental molecule (Buttle et al. 1992). Cytotoxicity of CA074Me for 1386Tu cells was higher than for 1386Ln cells (Fig. 2a). As expected, CB activity was diminished by increasing the concentration of CA074Me (0–50 μM) and was decreased by more than 95% at 25 μM of inhibitor (Fig. 2b). These results indicated that OSCC cells are sensitive to both TRAIL and CA074Me, and that 1386Tu cells are more susceptible than 1386Ln. The same differential cytotoxicities for Tu versus Ln cells were observed after treatment with caspase inhibitors (zVAD-fmk and zIETD-fmk) alone (data not shown).
Fig. 2.
Effects of cathepsin B inhibitor CA074Me on OSCC cells. The 1386Tu (open squares) and 1386Ln (closed squares) cells were treated with CB inhibitor CA074Me (0–50 μM) for 24 h and analyzed for a cytotoxicity, and b CB activity. Cytotoxicity was determined by MTT assays and CB activity as described in Materials and Methods. Results shown are the means±SD from three separate experiments
Inhibition of CB or caspase activities protects against TRAIL-induced nuclear changes and DNA fragmentation
TRAIL induces cell death with morphologic features typical for apoptosis, characterized by cell shrinkage, cleavage of nuclear DNA, condensation of chromatin and fragmentation of nucleus and cytoplasm with subsequent formation of apoptotic bodies. To ascertain the occurrence of such features in OSCC cells, we examined the effects of TRAIL exposure on nucleosomal DNA fragmentation in cells treated with or without the cell-permeable pan-caspase inhibitor zVAD-fmk or CA074Me (Fig. 3a). There was prominent fragmentation of DNA after treatment with TRAIL alone. With 10 μM zVAD-fmk, where no residual caspse-3 like activity was detected, DNA fragmentation was inhibited compared to TRAIL-only treated cells (Figs. 3, 4). Importantly, CA074Me also had the same effect as zVAD-fmk and inhibited DNA fragmentation even in the absence of zVAD-fmk in both Tu and Ln cells (Fig. 3a).
Fig. 3.
Effect of TRAIL on DNA fragmentation. a Nucleosomal DNA fragments in 1386Tu (Tu) or 1386Ln (Ln) cells were analyzed by gel electrophoresis after 24 h TRAIL treatment (100 ng/ml) or without TRAIL treatment. Pre-incubation for 3 h prior to TRAIL was done with zVAD-fmk (10 μM) or/and CA074Me (25 μM) inhibitors as indicated. Apoptosis was confirmed by the appearance of a ladder of oligonucleosomal DNA; DNA ladder, DNA 1 kb size marker. b Cells were treated as above, labeled with propidium iodide (20 μg/ml) after 24 h, and analyzed by flow cytometry. Apoptotic cells with sub-G1 chromosomal DNA content are expressed as a percentage of total gated cells. The data are representative of two independent experiments with similar results
Fig. 4.
TRAIL-induced nuclear TUNEL staining in OSCC cells. The 1386Tu (a) or 1386Ln (b) cells were incubated for 24 h with TRAIL (100 ng/ml), with or without pretreatment of zVAD-fmk (10 μM) or CA074Me (25 μM) or both, and photographed after TUNEL staining with DAB. Cells with nuclei showing strong chromatin condensation and nuclear fragmentation were considered apoptotic; magnification ×400. Bar diagrams show the apoptotic index scores as mean±SD from two independent experiments, each done in duplicate. The values were counted from 12 to 14 photographic fields per experiment: 1 no treatment; 2 TRAIL only; 3 TRAIL+zVAD-fmk; 4 TRAIL+CA074Me; 5 TRAIL + zVAD-fmk + CA074Me; * significantly different from TRAIL treated cells alone, P<0.05; # significantly different from control cells treated DMSO alone, P<0.05
The protective effects of zVAD-fmk and CA074Me in OSCC cells exposed to TRAIL were further quantitated by FACS analysis. Apoptotic cells with degraded DNA appear as cells with hypodiploid DNA content and were represented in so called “sub-G1” peaks to the left of the main G1 peak on the DNA histograms (Fig. 3b). A large proportion of OSCC cells treated with TRAIL exhibited a sub-G1 peak (20.5% for Tu and 16.8% for Ln), which was diminished in the presence of the pan-caspase inhibitor zVAD-fmk (5.4% for Tu and 8.0% for Ln) or the CB inhibitor CA074Me (11.2% for Tu and 12.6% for Ln). CA074Me also reversed the FACS profile of OSCC cells treated with TRAIL and reduced the amount of cells with a sub-G1 DNA-content from 20.5 to 11.2% in Tu cells and from 16.8 to 12.6% in Ln cells. Addition of zVAD-fmk (10 μM) to cells reduced the proportion of sub-G1 cells from 20.5 to 5.4% in Tu cells and from 16.8 to 8% in Ln cells (Fig. 3b). These data indicate that TRAIL generates an apoptotic signal in a caspase-independent mode, and confirm the involvement of CB in cell death induced by TRAIL.
TUNEL studies confirmed that treatment with TRAIL for 24 h was associated with extensive chromatin alterations, as 29–34% of cells showed apoptotic morphology (Fig. 4). The CB inhibitor CA074Me prevented the apoptotic morphology after 24 h treatment with or without zVAD-fmk in both cell lines. The vast majority of untreated cells displayed normal nuclear morphology, with only 2.1–3.1% of cells showing characteristics of apoptotic cells with chromatin condensation, nuclear fragmentation, and apoptotic bodies. CA074Me clearly prevented the induction of apoptosis by reducing the incidence to 9.2–12.2%. Also, the pan-caspase-inhibitor zVAD-fmk reduced the percentage of cells with apoptotic morphology to 7.3–9.2%. Both zVAD-fmk and CA074Me independently had significant protective effects against TRAIL-induced cell death (Fig. 4). The zVAD-fmk could almost completely prevent cell death, which could also be inhibited by the CA074Me, but not as efficiently as with zVAD-fmk (Figs. 3, 4).
TRAIL induces caspase activation in OSCC cells
To directly assess the role of caspase activation, we performed enzymatic caspase assays using fluorogenic caspase substrates Ac-IETD-AMC and Ac-DEVD-AFC for caspase-8 and -3 activities, respectively (Fig. 5a). TRAIL treatment significantly increased caspase-3 and -8 like activities, which were reduced in zVAD-fmk or zIETD-fmk treated cells as expected. Indeed, the initiator caspase-8 and effector caspases (caspase-3 and -7) seemed to be activated simultaneously, rendering it difficult to determine which of them initially triggered the proteolytic cascade. Since caspase-3 and -7 have the same substrate specificities in vitro, we hypothesized that caspase-7 could also be activated in the TRAIL treatment. The caspase-8 inhibitor zIETD-fmk completely inhibited caspase-8 activity. Co-treatment with zVAD-fmk and TRAIL resulted in decreased caspase-3 like and caspase-8 activity (Fig. 5a). Interestingly, inhibition of caspase-8 activity with zIETD-fmk did not completely inhibit the caspase-3 like activity. Together, these experiments demonstrated that TRAIL treatment resulted in the activation of both initiator and effector caspases.
Fig. 5.
Role of cathepsin B, caspase-3 and -8 in TRAIL–induced apoptosis. a The 1386Tu (open bars) and 1386Ln (closed bars) cells were exposed to TRAIL (100 ng/ml) after pretreatment with zVAD-fmk (10 μM), zIETD-fmk (20 μM) and/or CA074Me (25 μM) for 24 h. Cytotoxicity, CB, and caspase activities were assayed as described in Materials and Methods. Results shown are the means±SD from three separate experiments; *significantly different from TRAIL treated cells alone, P<0.05; #significantly different from control cells treated with DMSO alone, P<0.05. b Western blot analysis of 1386Tu (Tu) or 1386Ln (Ln) cell extracts after 24 h TRAIL treatment (100 ng/ml) with or without zVAD-fmk (10 μM) and/or CA074Me (25 μM). Individual blots show processing of CB (43 kDa) into active form (31 kDa); cleavage of procaspase-3 (32 kDa) into a 20 kDa species; cleavage of Bid (23 kDa) into lower mol. weight product (not shown); cytochrome c (15 kDa) release into cytosolic fraction; processing of PARP (113 kDa) into the typical 89 kDa protein and a lower molecular weight product (not shown); re-probing for β-actin as internal loading control. c Western blot analysis of 1386Tu cells after TRAIL treatment (100 ng/ml) with or without caspase-8 inhibitor zIETD-fmk (20 μM). Individual blots show cleavage of procaspase-8 (55 kDa) into a 32 kDa species; cleavage of Bid (23 kDa) into lower molecular weight product (not shown); cytochrome c (15 kDa) release into cytosolic fraction; re-probing for β-actin as internal loading control. Data are representative of two independent experiments with similar results
Activation of caspases was also monitored by Western blots (Fig. 5b). The highest caspase activity detected by enzymatic assays did not always correlate with caspase cleavage determined by Western blot analysis, which may be related to the fact that caspase cleavage products are subject to degradation by the proteasome. The proform of caspase-3 was cleaved after TRAIL treatment accompanied by an increase in caspase-3 like activity (Fig. 5a), indicating that the decrease of pro-caspase-3 probably resulted from enzyme activation. After treatment with zVAD-fmk, the active form of caspase-3 decreased, which was also confirmed by the activity assays. Moreover, treatment of OSCC cells with the caspase-8 inhibitor zIETD-fmk decreased caspase-3 like activity compared to TRAIL alone (Fig. 5a). Therefore, the data on caspase-3 activation is consistent with the model that caspase-8 acts upstream from caspase-3.
Analysis of the effects of TRAIL on caspase-8 activation, an early event associated with death receptor ligation, showed that this activation was blocked by the caspase-8 inhibitor zIETD-fmk and also by the pan-caspase inhibitor zVAD-fmk (Figs. 5a, 5c). Immunoblotting showed that TRAIL promoted the conversion of pro-caspase-8 into the processed active form (Fig. 5c), confirming involvement of caspase-8 in TRAIL-dependent cell death. The active form of caspase-3 cleaves PARP that functions downstream of caspase-3 to effect apoptosis; hence, zVAD-fmk blocked caspase -3 processing and subsequent PARP cleavage (Fig. 5b), indicating that zVAD-fmk interferes with TRAIL apoptosis. In the absence of TRAIL, CA074Me or zVAD-fmk did not induce caspase-3 activation and PARP cleavage (data not shown). Taken together, these data indicate that CA074Me inhibits apoptosis independent of caspase activation.
In some instances, lysosomal or proteasomal proteases trigger apoptosis by proteolytically activating caspases (Boya et al. 2003). Thus, we further investigated the interaction between these caspases (-3, -7 and 8) and CA074Me. We observed inhibition of activity (Fig. 5a) and also of proteolytic processing (Fig. 5b) for caspase-3, but not for caspase-8 activity (Fig. 5a). This indicated that CA074Me is not crossreactive with caspases, but that inhibition of CB is involved in lowering caspase activation further downstream in the cell death pathway. This data was also confirmed by transfection with CB-specific ribozymes into these cell lines, which showed that lowering CB activity and expression also decreased caspase-3 activity and expression (see below).
TRAIL-dependent apoptosis is mediated by Bid and involves cytosolic accumulation of cytochrome c
Bid is a pro-apoptotic protein of the BCL-2 family whose inactive form of 23 kDa is cleaved by caspases or CB, giving rise to an active fragment of ~15 kDa. Translocation of Bid to the mitochondria has been shown to engage the intrinsic apoptotic pathway after stimulus by a death receptor such as TRAIL. In order to investigate whether Bid is involved in TRAIL-induced apoptosis in OSCC cells, Western blot analysis was performed (Fig. 5b). Bid was present as the ~23 kDa proform in both untreated cell lines, whereas a decrease of this proform was observed after 24 h of TRAIL treatment, suggesting that the inactive form was processed to its active subunit. This TRAIL-induced Bid processing was inhibited by treatment with either zVAD-fmk or CA074Me, and these inhibitory effects were similar for both reagents (Fig. 5b). Also, addition of zIETD-fmk prevented Bid processing consistent with caspase-8-dependent Bid cleavage (Fig. 5c). Inhibition of caspase-8 cleavage, Bid cleavage and cytochrome c release by zIETD-fmk (Fig. 5c) all suggest that caspase-8 effects are active upstream of mitochondria effects. These inhibitor studies show that CB and caspase-8 directly or indirectly participate in activation of Bid in TRAIL-induced apoptosis (Fig. 5a, c). Previously, it was demonstrated that Bid is cleaved by lysosomal proteases in an in vitro system combining mitochondria and cytosol (Stoka et al. 2001). Thus, in analogy to the cleavage by caspase-8, a Bid species is created that can potentially trigger cytochrome c release from mitochondria. It was also demonstrated that Bid cleavage was in fact the major event leading to cathepsin-mediated cytochrome c release, since cytosol from Bid-deficient mice was nonfunctional in this assay (Stoka et al. 2001). Our data also support the model that both CB and caspase-8 can mediate a TRAIL-dependent apoptotic pathway in OSCC cells by activating Bid upstream of mitochondria.
In order to determine whether mitochondria are involved in the apoptotic process induced by TRAIL, the release of cytochrome c was measured by Western blots, with or without pan-caspase inhibitor zVAD-fmk added prior to TRAIL (Fig. 5b). Cytochrome c release was inhibited by zVAD-fmk, indicating that caspases are partially responsible. This effect was accompanied by cleavage of caspase-3 and also of PARP, a known substrate for caspase-3. Exposure to TRAIL induced more cytochrome c release in Ln than Tu cells, but inhibition of apoptosis was stronger in Tu than Ln cells (Fig. 5b). Co-treatment of OSCC cells with TRAIL and CA074Me also resulted in partial inhibition of cytochrome c release and PARP cleavage, but this effect was not as strong as for cells cotreated with TRAIL and zVAD-fmk.
TRAIL increases expression and activity of CB
It has been suggested that CB plays an essential role in TNF-mediated apoptosis (Leist and Jaattela 2001b), and has been proposed as a therapeutic target in cancer therapy of tumors with elevated levels of cysteine proteases (Foghsgaard et al. 2001). CB is synthesized as an inactive pro-enzyme (43 kD) which is processed into active 25 kDa or 31 kDa species. In our cell lines, the levels of inactive pro-enzyme were very small, and the active 31 kDa product was the predominant form. An increase in the cellular amount of active 31 kDa CB was observed after TRAIL treatment (Fig. 5b).
Because TRAIL appears to initiate apoptotic signaling by activating caspases, we sought to determine whether TRAIL could directly increase CB activation through caspase-8. We observed that the pan-caspase inhibitor zVAD-fmk also inhibited CB (Fig. 5a), in agreement with previously published data showing that not only CB, but also other cathepsins (L, V, S, K, F, X), as well as papain and legumain, can be inhibited by common caspase-specific inhibitors (Rozman-Pungercar et al. 2003). The effect of CA074Me on cell death was interpreted as specific CB effect after determining the relevant concentration required for inactivation of CB. In OSCC cells, 25 μM CA074Me fully inhibited the extractable CB activity after 24 h treatment, indicating that any effects of higher concentrations may be unrelated to CB.
In a related set of OSCC cell lines, we previously confirmed the inhibitory effect of CA074 on TRAIL-induced DNA fragmentation (Vigneswaran et al. 2005). Concentrations of CA074Me in the range of 25 μM, as used here, were necessary to elicit inhibitory effects (Foghsgaard et al.2001; Varghese et al. 2001). We also tested the effect of CA074Me on TRAIL-induced caspase activity in vitro (Fig. 5a), and confirmed that CA074Me has a CB-independent target that plays a role in cell death, as recently described (Mihalik et al. 2004). Furthermore, the effects of CA074Me on CB, Bid, and cytochrome c release indicated that the target of CA074Me was functionally located upstream to the lysosomal breakdown and the mitochondria. These results strongly imply that CA074Me inhibited cell death by inactivation of CB, and that this is a necessary yet not very efficient effect of CA074Me to inhibit apoptosis.
Inhibition of CB expression suppresses apoptosis in OSCC cells
The observation that the CB inhibitor CA074Me was able to reduce TRAIL-induced apoptosis in OSCC cells suggested an involvement of CB in this event. As independent alternative to CA074Me, a vector expressing a CB-specific ribozyme was stably transfected into the OSCC target cells. This ribozyme (RzCB) had documented inhibitory effects on CB RNA, protein expression and activity (Wickramasinghe et al. 2005). RzCB-transfected 1386Tu and 1386Ln cells showed decreased apoptosis when compared to vector control transfected cells (Figs. 6, 7). After treatment with TRAIL, both Tu and Ln vector-transfected control cells clearly had fragmented DNA, whereas the RzCB-transfected cells showed decreased DNA fragmentation compared to vector controls (Fig. 6a). Also FACS analysis showed that both Tu and Ln vector control cells treated with TRAIL exhibited sub-G1 percentages of apoptotic cells of 13.5 and 12.9%, respectively; these were diminished to 8.4 and 9.1%, respectively, when transfected with RzCB (Fig. 6b). TUNEL staining also showed that the Tu cells were more apoptotic (27.3%) compared to Ln cells (21.2%) based on the percentage of TUNEL-positive nuclei (Fig. 7).
Fig. 6.
Effects of cathepsin B ribozyme on DNA fragmentation. a Nucleosomal DNA fragments in 1386Tu (Tu) or 1386Ln (Ln) cells (vector- or CB ribozyme-transfected) with or without TRAIL (100 ng/ml) treatment for 24 h were analyzed by gel electrophoresis. Apoptosis was confirmed by the appearance of a ladder of oligonucleosomal DNA; DNA ladder, DNA 1 kb size marker. b Cells were treated as above, labeled with propidium iodide, and analyzed by flow cytometry. Apoptotic cells with sub-G1 chromosomal DNA content are expressed as a percentage of total gated cells. The data are representative of two independent experiments with similar results
Fig. 7.
Effects of cathepsin B ribozyme on TRAIL-induced nuclear TUNEL staining. The 1386Tu (a) or 1386Ln (b) cells (vector- or CB ribozyme-transfected) were incubated for 24 h with or without TRAIL (100 ng/ml) and photographed after TUNEL staining with DAB. Cells with nuclei showing strong chromatin condensation and nuclear fragmentation were considered apoptotic; magnification x400. Bar diagrams show the apoptotic index scores as mean±SD from two independent experiments, each in duplicate. The values were counted from 12 to 14 photographic fields per experiment: 1 vector, 2 RzCB, 3 vector + TRAIL, 4 RzCB + TRAIL; *significantly different from vector + TRAIL treated cells alone, P<0.05; #significantly different from control cells, P<0.05
Immunoblot analysis demonstrated efficient downregulation of CB at both the protein as well as activity level by the ribozyme, whereas wild-type levels of CB were quite high in comparison (Fig. 8a, b). We previously observed decreases in CB activity by at least 50–60% compared to wild-type cells (Wickramasinghe et al. 2005). With TRAIL treatment (100 ng/ml for 24 h), these cells showed more CB activity and expression which did not affect the inhibition of apoptosis. Clearly, inhibition of CB by RzCB had similar effects on apoptosis as inhibition by CA074Me, thus independently pointing to a novel role of CB in OSCC apoptosis. The weaker effects seen for the ribozyme compared to CA074Me can be explained by the different nature of the two inhibitors. The chemical inhibitor CA074Me was titrated to an optimal concentration leading to an almost complete (≥90%) inhibition of CB protease activity (Fig. 2b). On the other hand, inhibitory activity of a ribozyme mainly depends on its intrinsic expression level, cleavage site accessibility, and turnover of ribozyme RNA, all of which are determined by the ribozyme design and expression construct. Our ribozyme led to less than complete (~60–80%) inhibition of CB expression (Fig. 8a, b) (Wickramasinghe et al. 2005), thus leaving some residual CB protease to still be active.
Fig. 8.
Inhibition of TRAIL-induced apoptosis events by cathepsin B ribozymes. a Western blot analysis of 1386Tu (Tu) or 1386Ln (Ln) cells (vector- or CB ribozyme-transfected) with or without TRAIL treatment (100 ng/ml) for 24 . Individual blots show cleavage of pro-CB (43 kDa) into active form (31 kDa); cleavage of procaspase-3 (32 kDa) into a 20 kDa species; cleavage of Bid (23 kDa) into lower mol. weight product (not shown); cytochrome c (15 kDa) release into cytosolic fraction; processing of PARP (113 kDa) into the typical 89 kDa protein and a lower molecular weight product (not shown); re probing for β-actin as internal loading control. Data are representative of at least two independent experiments with similar results. b CB activity after the same treatment of 1386Tu (open bars) or 1386Ln (closed bars) cells; c caspase-3 like activity after the same treatment of 1386Tu (open bars) or 1386Ln (closed bars) cells. Activities were analyzed as described in Materials and Methods; means ± SD of three independent experiments are shown, each in duplicate; *significantly different from vector-transfected TRAIL treated cells alone, P<0.05; #significantly different from control cells, P<0.05
Interestingly, TRAIL treatment of vector control cells resulted in increased caspase-3 like activity compared to untreated cells, and such increase was lower in RzCB-transfected cells (Fig. 8c). Also, the cleaved 20 kDa caspase-3 band was less prominent in TRAIL-treated RzCB cells compared to vector control cells (Fig. 8a). TRAIL treatment in vector control cells also induced the loss of full-length Bid, release of cytochrome c, and PARP cleavage (Fig. 8a), whereas the RzCB-transfected cells showed much less effect on those downstream apoptosis markers. These cumulative data point to the presence and involvement of CB upstream of the effector caspases and Bid cleavage events in the cell death pathway.
Discussion
Tumor cell killing by anticancer therapy is primarily mediated by triggering apoptosis selectively in cancer cells. Intrinsic or extrinsic routes for effector caspase activation are frequently the most rapid and efficient; however, if any of these routes is blocked, the cell can activate other mechanisms (Lockshin and Zakeri 2004). Until recently, cathepsins were believed to be primarily involved in nonselective intracellular protein degradation in lysosomes, and their function outside lysosomes was ignored because of their inactivity at neutral pH (Castino et al. 2003). Outside lysosomes, cathepsin activity is regulated by their endogenous inhibitors, the cystatins, which function as threshold inhibitors similar to IAPs, the endogenous inhibitors of caspases (Tenev et al. 2005). Release of lysosomal enzymes to the cytosol precedes lysosomal breakdown and loss of acidity, and moderate lysosomal damage, sufficient only to slightly overcome the inhibitory potential of cystatins, can lead to apoptosis (Lieuallen et al. 2001). TRAIL signaling may involve such lysosomal permeabilization with release of lysosomal cathepsins into the cytosol, where they can activate Bid resulting in mitochondrial damage (Stoka et al. 2001).
The tumor-promoting effect of cathepsins contrasts their effects in mediating pro-apoptotic signals triggered by TNF (Taha et al. 2005; Foghsgaard et al. 2001; Guicciardi et al. 2001). CB is released from the lysosomes into the cytosol in response to TNF (Taha et al. 2005; Foghsgaard et al. 2001; Guicciardi et al. 2000), and contributes to downstream apoptotic events such as cytochrome c release and activation of executioner caspases (Stoka et al. 2001; Guicciardi et al. 2000). This, and the fact that tumors frequently contain high levels of cathepsins, may prove useful in selectively targeting tumor cells for apoptosis induction (Vigneswaran et al. 2005).
There is evidence for a major role for lysosomal cathepsins in apoptosis pathways especially when initiator caspase activity is suppressed (Leist and Jaattela 2001b). Leakage of CB into the cytosol resulted in the activation of effector caspases as well as induction of the nuclear morphology associated with apoptotic cells (Vancompernolle et al. 1998). The nearly complete prevention of TRAIL-induced cell death with CB inhibitors suggests that activation of CB represents an early step in this process.
The mediating factors and activation mechanisms of the apoptotic pathways remain to be identified, and may vary with the type of cells and the applied death stimulus (Herr and Debatin 2001; Wyllie and Golstein 2001). In some systems, CB has been reported to contribute to apoptosis by induction of mitochondrial membrane permeabilization, possibly via cleavage of Bid, thereby acting upstream to the caspase cascade (Stoka et al. 2001; Boya et al. 2003). Others have found that CB can act as an effector protease downstream to caspases (Foghsgaard et al. 2001; Jones et al. 1998). Loss of mitochondrial membrane potential may contribute to this process as a secondary event. We previously reported that hypoxia-mediated apoptosis occurs via two independent pathways in OSCC cell lines, and documented the release of cytochrome c from mitochondria, PARP cleavage, DNA fragmentation and upregulation of caspases (-3, -8, -9 and -10) during the cell death process (Nagaraj et al. 2004).
Our studies provide evidence of a participation of CB in the TRAIL signaling pathway, as apoptotic markers induced with TRAIL were significantly suppressed by the CB inhibitor CA074Me, in line with our previous report (Vigneswaran et al. 2005). Since some caspase inhibitors used in apoptotic studies also inhibit CB, its role remains obscure, as also pointed out previously (Turk et al. 2002a). Nevertheless, we showed that both chemical as well as RNA inhibitors of CB blocked TRAIL-induced apoptosis in OSCC cells, in support of a study in which a CB inhibitor caused the reduction of the number and volume of tumors developed in rat liver (Van Noorden et al. 1998). Our studies also showed that Bid can play a role as the lysosomal protease target, as previously observed (Stoka et al. 2001). Incubation of OSCC cells with the pan-caspase inhibitor zVAD-fmk prevented morphological signs of apoptosis, indicating that caspase activation is also required in this model for apoptosis progression, as suggested previously (Schotte et al. 1999). It was also reported that CB could weakly process the precursors of caspases -2, -3, -6, -7, and -14, as was observed here for caspase-3 and -7 (Vancompernolle et al. 1998).
Our results support a model in which caspase-8 activation and subsequent activation of Bid are necessary for lysosomal permeabilization and release of CB. This conclusion is based on the observation that treatment with the pan-caspase inhibitor zVAD-fmk inhibited expression of TRAIL-induced late apoptosis markers. Moreover, both CA074Me and ribozymes targeted at CB also inhibited these events in TRAIL-treated cells. Thus, lowering expression and activity of CB by two independent mechanisms decreased activation of the apoptotic cascade. The inhibitor CA074Me is considered to be highly selective for CB (Mihalik et al. 2004), as is the designed ribozyme, and both showed that CB is the non-caspase protease contributing to TRAIL-mediated apoptosis in the OSCC cells. The observed non-specific inhibition of CB by zVAD-fmk may complicate the interpretation of results obtained with caspase inhibitors as observed previously (Schotte et al. 1999). In other systems, including lymphocytes exposed to glucocorticoids, inhibition of CB or proteasomal proteases interfered with cell death or apoptosis more effectively than did inhibition of caspase-3 (Distelhorst 2002).
When isolated mitochondria were incubated with purified CB in the presence of cytolic extracts, a substantial release of cytochrome c was detected, suggesting that cytosolic factor(s) activated by CB mediated mitochondrial dysfunction (Guicciardi et al. 2000, 2004). Bid is cleaved and translocated to the mitochondria following lysosomal disruption by lysosomotropic agents (Cirman et al. 2004), and lysosomal extracts cleave Bid in vitro to generate a fragment with cytochrome c-releasing activity (Stoka et al. 2001). Thus, CB seems to play a key role in lysosome-mediated Bid cleavage, since Bid cleavage after TRAIL treatment was indeed diminished in the presence of either CA074Me or RzCB, confirming the role of CB in tumor cell death pathways (Foghsgaard et al. 2001). It is possible that different cathepsins may take a similar role as CB in other circumstances, since there is evidence of a dominant role of cathepsin D in other systems (Johansson et al. 2003; Heinrich et al. 2004). This might be due to the ratios of different cathepsins and their cystatin inhibitors/activators in different cell types. Currently, the proposed chain of events mediated by CB is as follows: activated caspase -8 induces CB release from lysosomes; CB in turn activates cytosolic Bid that triggers cytochrome c release and subsequent apoptosis (Fig. 9; Turk et al. 2002b).
Fig. 9.
Model for TRAIL induced apoptotic pathway with involvement of cathepsin B. TRAIL binding to DR4 or DR5 stimulates recruitment and activation of caspase-8 which directly, or indirectly through activation by CB, cleaves Bid. The cleaved Bid is translocated from the cytosol to the mitochondria where it promotes cytochrome c release. Cleavage of Bid, release of cytochrome c and activation of downstream executioner caspases promotes cell death pathways
In conclusion, this is to our knowledge the first report directly investigating a CB-dependent apoptosis mechanism in OSCC cells, thus providing mechanistic insight into CB-dependent TRAIL resistance and suggesting novel strategies for reversing TRAIL resistance in CB-expressing tumors. The role of CB in initiation and execution of the apoptotic program has become clear in several models, and the existence of a ‘lysosomal pathway of apoptosis’ is now generally accepted. Thus, current strategies for prevention of tumor malignancy by CB inhibition need to consider two opposing pathways. On one hand, CB inhibition should lead to diminished invasiveness, and thus diminished tumor spread and metastasis. On the other hand, CB inhibition may lead to enrichment of otherwise apoptosis-prone tumor cells, possibly leading to selective survival of highly malignant sub-populations of cells. Future studies need to further define the regulatory events and cellular factors that control these two apparently opposing functions of CB in OSCC progression.
Acknowledgements
This work was supported by NIH grant DE13150 and by Philip Morris USA Inc. and Philip Morris International (W.Z.), and by a postdoctoral fellowship award from the University of Louisville Brown Cancer Center (N.N.). The cell lines 1386Tu and 1386Ln were kind gifts from Dr. P. Sacks, New York University, New York, NY. We thank Dr. Samuel R. Wellhausen (Brown Cancer Center, University of Louisville) for technical assistance in the FACS analyses.
Abbreviations
- AFC
7-Amino-4-trifluoromethylcoumarin
- AMC
7-Amino-4-methylcoumarin
- Bid
BH3 interacting domain death agonist
- CA074
l-trans-epoxy-succinyl-Ile-Pro-OH
- CA074Me
l-trans-epoxy-succinyl-Ile-Pro-Methyl Ester
- CB
Cathepsin B
- CHAPS
3-[(3-Cholamidopropyl) dimethylammonio]-1-propanesulfonate hydrate
- DAB
Diaminobenzidine
- DEVD-AFC
N-acetyl-Asp-Glu-Val-Asp-AFC
- DEVD-CHO
N-acetyl-Asp-Glu-Val-Asp-CHO
- ECL
Enhanced chemiluminescence
- FACS
Fluorescence activated cell sorting
- HEPES
N-(2-Hydroxyethyl) piperazine-N’-(2-ethanesulfonic acid) hemisodium salt
- IAP
Inhibitor of apoptosis protein
- IETD-AMC
N-acetyl-Ile-Glu-Thr-Asp-AMC
- IETD-CHO
N-acetyl-Ile-Glu-Thr-Asp-CHO
- MTT
1-(4,5-Dimethylthiazol-2-yl)-3,5-diphenylformazan
- OSCC
Oral squamous cell carcinoma
- PARP
Poly (ADP-ribose) polymerase
- PIPES
Piperazine-1, 4-bis (2-ethanesulfonic acid)
- TRAIL
Tumor necrosis factor (TNF)-related apoptosis-inducing ligand
- TUNEL
TdT-mediated dUTP nick end labeling
- z-Arg-Arg-NHMec
N-CBZ-l-arginyl-l-arginine7-(4-methylcoumarin)
- z-IETD-fmk
z-Ile-Glu-Thr-Asp-fluoromethylketone
- z-VAD-fmk
z-Val-Ala-Asp-fluoromethylketone
References
- Ashkenazi A (2002) Targeting death and decoy receptors of the tumour-necrosis factor superfamily. Nat Rev Cancer 2:420–430 [DOI] [PubMed] [Google Scholar]
- Borner C (2003) The Bcl-2 protein family: sensors and checkpoints for life-or-death decisions. Mol Immunol 39:615–647 [DOI] [PubMed] [Google Scholar]
- Boya P, Gonzalez-Polo RA, Poncet D, Andreau K, Vieira HL, Roumier T, Perfettini JL, Kroemer G (2003) Mitochondrial membrane permeabilization is a critical step of lysosome-initiated apoptosis induced by hydroxychloroquine. Oncogene 22:3927–3936 [DOI] [PubMed] [Google Scholar]
- Buttle DJ, Murata M, Knight CG, Barrett AJ (1992) CA074 methyl ester: a proinhibitor for intracellular cathepsin B. Arch Biochem Biophys 299:377–380 [DOI] [PubMed] [Google Scholar]
- Canbay A, Guicciardi ME, Higuchi H, Feldstein A, Bronk SF, Rydzewski R, Taniai M, Gores GJ (2003) Cathepsin B inactivation attenuates hepatic injury and fibrosis during cholestasis. J Clin Invest 112:152–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castino R, Demoz M, Isidoro C (2003) Destination ‘lysosome’: a target organelle for tumour cell killing?. J Mol Recognit 16:337–348 [DOI] [PubMed] [Google Scholar]
- Cirman T, Oresic K, Droga-Mazovec G, Turk V, Reed JC, Myers RM, Salvesen GS, Turk B (2004) Selective disruption of lysosomes in HeLa cells triggers apoptosis mediated by cleavage of Bid by multiple papain-like lysosomal cathepsins. J Biol Chem 279:3578–3587 [DOI] [PubMed] [Google Scholar]
- Distelhorst CW (2002) Recent insights into the mechanism of glucocorticosteroid- induced apoptosis. Cell Death Differ 9:6–19 [DOI] [PubMed] [Google Scholar]
- El-Deiry WS (2001) Insights into cancer therapeutic design based on p53 and TRAIL receptor signaling. Cell Death Differ 8:1066–1075 [DOI] [PubMed] [Google Scholar]
- Foghsgaard L, Wissing D, Mauch D, Lademann U, Bastholm L, Boes M, Elling F, Leist M, Jaattela M (2001) Cathepsin B acts as a dominant execution protease in tumor cell apoptosis induced by tumor necrosis factor. J Cell Biol 153:999–1010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulda S, Debatin KM (2004a) Apoptosis signaling in tumor therapy. Ann N Y Acad Sci 1028:150–156 [DOI] [PubMed] [Google Scholar]
- Fulda S, Debatin KM (2004b) Signaling through death receptors in cancer therapy. Curr Opin Pharmacol 4:327–332 [DOI] [PubMed] [Google Scholar]
- Guicciardi ME, Deussing J, Miyoshi H, Bronk SF, Svingen PA, Peters C, Kaufmann SH, Gores GJ (2000) Cathepsin B contributes to TNF-alpha-mediated hepatocyte apoptosis by promoting mitochondrial release of cytochrome c. J Clin Invest 106:1127–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guicciardi ME, Miyoshi H, Bronk SF, Gores GJ (2001) Cathepsin B knockout mice are resistant to tumor necrosis factor-alpha-mediated hepatocyte apoptosis and liver injury: implications for therapeutic applications. Am J Pathol 159:2045–2054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guicciardi ME, Leist M, Gores G (2004) Lysosomes in cell death. Oncogene 23:2881–2890 [DOI] [PubMed] [Google Scholar]
- Heinrich M, Neumeyer J, Jakob M, Hallas C, Tchikov V, Winoto-Morbach S, Wickel M, Schneider-Brachert W, Trauzold A, Hethke A, Schutze S (2004) Cathepsin D links TNF-induced acid sphingomyelinase to Bid-mediated caspase-9 and -3 activation. Cell Death Differ 11:550–563 [DOI] [PubMed] [Google Scholar]
- Herr I, Debatin KM (2001) Cellular stress response and apoptosis in cancer therapy. Blood 98:2603–2614 [DOI] [PubMed] [Google Scholar]
- Johansson AC, Steen H, Ollinger K, Roberg K (2003) Cathepsin D mediates cytochrome c release and caspase activation in human fibroblast apoptosis induced by staurosporine. Cell Death Differ 10:1253–1259 [DOI] [PubMed] [Google Scholar]
- Jones B, Roberts PJ, Faubion WA, Kominami E, Gores GJ (1998) Cystatin A expression reduces bile salt-induced apoptosis in a rat hepatoma cell line. Am J Physiol 275:G723–G730 [DOI] [PubMed] [Google Scholar]
- Koblinski JE, Ahram M, Sloane BF (2000) Unraveling the role of proteases in cancer. Clin Chim Acta 291:113–135 [DOI] [PubMed] [Google Scholar]
- LeBlanc HN, Ashkenazi A (2003) Apo-2L/TRAIL and its death and decoy receptors. Cell Death Differ 10:66–75 [DOI] [PubMed] [Google Scholar]
- Leist M, Jäättelä M (2001a) Four deaths and a funeral: from caspases to alternative mechanisms. Nat Rev Mol Cell Biol 2:589–598 [DOI] [PubMed] [Google Scholar]
- Leist M, Jaattela M (2001b) Triggering of apoptosis by cathepsins. Cell Death Differ 8:324–326 [DOI] [PubMed] [Google Scholar]
- Lieuallen K, Pennacchio LA, Park M, Myers RM, Lennon GG (2001) Cystatin B-deficient mice have increased expression of apoptosis and glial activation genes. Hum Mol Genet 10:1867–1871 [DOI] [PubMed] [Google Scholar]
- Lockshin RA, Zakeri Z (2004). Caspase-independent cell death?. Oncogene 23:2766–2773 [DOI] [PubMed] [Google Scholar]
- Mihalik R, Imre G, Petak I, Szende B, Kopper L (2004) Cathepsin B-independent abrogation of cell death by CA-074-OMe upstream of lysosomal breakdown. Cell Death Differ 11:1357–1360 [DOI] [PubMed] [Google Scholar]
- Nagaraj NS, Vigneswaran N, Zacharias W (2004) Hypoxia-mediated apoptosis in oral carcinoma cells occurs via two independent pathways. BMC Mol Cancer 3(38):1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozman-Pungercar J, Kopitar-Jerala N, Bogyo M, Turk D, Vasiljeva O, Stefe I, Vandenabeele P, Bromme D, Puizdar V, Fonovic M, Trstenjak-Prebanda M, Dolenc I, Turk V, Turk B (2003) Inhibition of papain-like cysteine proteases and legumain by caspase-specific inhibitors: when reaction mechanism is more important than specificity. Cell Death Differ 10:881–888 [DOI] [PubMed] [Google Scholar]
- Sacks PG (1996) Cell, tissue and organ culture as in vitro models to study the biology of squamous cell carcinomas of the head and neck. Cancer Metast Rev 15:27–51 [DOI] [PubMed] [Google Scholar]
- Schotte P, Declercq W, Van Huffel S, Vandenabeele P, Beyaert R (1999). Non-specific effects of methyl ketone peptide inhibitors of caspases. FEBS Lett 442:117–121 [DOI] [PubMed] [Google Scholar]
- Stoka V, Turk B, Schendel SL, Kim TH, Cirman T, Snipas SJ, Ellerby LM, Bredesen D, Freeze H, Abrahamson M, Bromme D, Krajewski S, Reed JC, Yin XM, Turk V, Salvesen GS (2001) Lysosomal protease pathways to apoptosis. Cleavage of bid, not pro-caspases, is the most likely route. J Biol Chem 276:3149–3157 [DOI] [PubMed] [Google Scholar]
- Taha TA, Kitatani K, Bielawski J, Cho W, Hannun YA, Obeid LM (2005) TNF induces the loss of sphingosine kinase-1 by a cathepsin B dependent mechanism. J Biol Chem 280:17196–17202 [DOI] [PubMed] [Google Scholar]
- Tenev T, Zachariou A, Wilson R, Ditzel M, Meier P (2005) IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat Cell Biol 7:70–77 [DOI] [PubMed] [Google Scholar]
- Turk B, Turk D, Salvesen GS (2002a) Regulating cysteine protease activity: essential role of protease inhibitors as guardians and regulators. Curr Pharm Design 8:1623–1637 [DOI] [PubMed] [Google Scholar]
- Turk B, Stoka V, Rozman-Pungercar J, Cirman T, Droga-Mazovec G, Oresic K, Turk V (2002b) Apoptotic pathways: involvement of lysosomal proteases. Biol Chem 383:1035–1044 [DOI] [PubMed] [Google Scholar]
- Vancompernolle K, Van Herreweghe F, Pynaert G, Van de Craen M, De Vos K, Totty N, Sterling A, Fiers W, Vandenabeele P, Grooten J (1998). Atractyloside-induced release of cathepsin B, a protease with caspase-processing activity. FEBS Lett 438:150–158 [DOI] [PubMed] [Google Scholar]
- Van Loo G, Saelens X, van Gurp M, MacFarlane M, Martin SJ, Vandenabeele P (2002) The role of mitochondrial factors in apoptosis: a Russian roulette with more than one bullet. Cell Death Differ 9:1031–1042 [DOI] [PubMed] [Google Scholar]
- Van Noorden CJ, Jonges TG, Van Marle J, Bissell ER, Griffini P, Jans M, Snel J, Smith RE (1998) Heterogeneous suppression of experimentally induced colon cancer metastasis in rat liver lobes by inhibition of extracellular cathepsin B. Clin Exp Metast 16:159–167 [DOI] [PubMed] [Google Scholar]
- Varghese J, Radhika G, Sarin A (2001) The role of calpain in caspase activation during etoposide induced apoptosis in T cells. Eur J Immunol 31:2035–2041 [DOI] [PubMed] [Google Scholar]
- Vigneswaran N, Wu J, Nagaraj N, Adler-Storthz K, Zacharias W (2005) Differential susceptibility of metastatic and primary oral cancer cells to TRAIL-induced apoptosis. Int J Oncol 26:103–112 [PubMed] [Google Scholar]
- Wickramasinghe NS, Nagaraj NS, Vigneswaran N, Zacharias W (2005) Cathepsin B promotes both motility and invasiveness of oral carcinoma cells. Arch Biochem Biophys 436:187–195 [DOI] [PubMed] [Google Scholar]
- Wyllie AH, Golstein P (2001) More than one way to go. Proc Natl Acad Sci USA 98:11–13 [DOI] [PMC free article] [PubMed] [Google Scholar]