Abstract
The association of anti-EGFR to gemcitabine and oxaliplatin (GEMOX) chemotherapy did not improve survival in biliary tract carcinoma (BTC) patients. Multiple mechanisms might be involved in the resistance to anti-EGFR. Here, we explored the mutation profile of EGFR extracellular domain (ECD), of tyrosine kinase domain (TKD), and its amplification status. EGFR mutational status of exons 12, 18–21 was analyzed in 57 tumors by Sanger sequencing. EGFR amplification was evaluated in 37 tumors by Fluorescent In Situ Hybridization (FISH). Kaplan-Meier curves were calculated using the log-rank test. Six patients had mutations in exon 12 of EGFR ECD and 7 in EGFR TKD. Neither EGFR ECD nor TKD mutations affected progression free survival (PFS) or overall survival (OS) in the entire population. In the panitumumab plus GEMOX (P-GEMOX) arm, ECD mutated patients had a worse OS, while EGFR TKD mutated patients had a trend towards shorter PFS and OS. Overall, the presence of mutations in EGFR or in its transducers did not affect PFS or OS, while the extrahepatic cholangiocarcinoma (ECC) mutated patients had a worse prognosis compared to WT. Nineteen out of 37 tumors were EGFR amplified, but the amplification did not correlate with survival. ECC EGFR amplified patients had improved OS, whereas the amplification significantly correlated with poor PFS (p = 0.03) in gallbladder carcinoma patients. The high molecular heterogeneity is a predominant feature of BTC: the alterations found in this work seem to have a prognostic impact rather than a predictive role towards anti-EGFR therapy.
Introduction
Different strategies aimed at inhibiting EGFR with small molecules (erlotinib and gefitinib) or with monoclonal antibodies (cetuximab and panitumumab) have been developed over the years in many cancer types [1–6]. Panitumumab (Vectibix, Amgen), a fully human antibody directed against EGFR, was initially approved in KRAS wild type (WT) metastatic colorectal cancer (mCRC) patients refractory to previous chemotherapy [7, 8]. In biliary tract carcinoma (BTC), preclinical evidence of antitumor activity [9] and the lack of compelling therapies suggested that the combination of standard chemotherapy and EGFR inhibitors could be an attractive option to improve patient outcome [10, 11]. The randomized, open-label, phase II Vecti-BIL trial compared the efficacy of gemcitabine and oxaliplatin (GEMOX) chemotherapy with or without panitumumab (P) in KRAS WT advanced BTC (Clinical Gov Identifier NCT01389414). The study, which enrolled and stratified intrahepatic cholangiocarcinoma (ICC) and extrahepatic cholangiocarcinoma (ECC) including gallbladder carcinoma (GBC), revealed that the addition of panitumumab to the standard chemotherapy did not improve progression free survival (PFS), which was 5.3 months in experimental arm and 4.4 months in control arm. No differences were observed in overall survival (OS), being of 9.9 with GEMOX and 10.2 months with P-GEMOX [12]. Hence, we concluded that KRAS WT status was not sufficient to select patients who can achieve tumor response to anti-EGFR therapies. Over the years, the phenomenon of resistance to anti-EGFR therapies has been deeply studied, in particular in CRC. The panel of potential drivers of resistance was expanded and KRAS exons 3–4, in addition to exon 2, NRAS, PIK3CA, and BRAF analyses were introduced in the clinical practice [13–15]. Thus, we retrospectively analyzed the mutational status of these genes in patients enrolled in the Vecti-BIL study and we found that the presence of these mutations did not affect the response to treatments. Recently, new mechanisms of resistance to anti-EGFR antibodies have been recognized in mutations of EGFR exon 12 of the extracellular domain (ECD); in CRC it was demonstrated that they prevented the correct binding of anti-EGFR, reducing their activity [16]. Moreover, even if controversial, EGFR amplification seemed to be a predictive marker of prognosis and response to the anti-EGFR therapies in CRC [15, 17]. EGFR amplification was also described in BTC [18, 19], but its prognostic role is unknown. Overall, in both arms of the Vecti-BIL trial, there was a broad range of PFS and OS: in the experimental arm, PFS ranged from 1.1 to 21.3 months and OS from 2.7 to 34.9 months, while in the control arm PFS ranged between 1.1 to 15.4 months, and OS between 1.1 and 31.7 months.
Here, we extended the molecular analyses to the EGFR ECD and TKD mutation profiling, and to the EGFR amplification status to explain these differences, and to correlate them to the arm of treatment.
Materials and methods
Patients
The Vecti-BIL trial (ClinicalTrials.gov Identifier: NCT01389414) enrolled 89 BTC patients selected for the absence of KRAS exon 2 mutations. Forty-five patients were assigned to receive GEMOX in association with panitumumab (ARM-A) and 44 patients GEMOX alone (ARM-B). All patients enrolled in the study have signed the Independent Ethical Committees (IEC) informed consent, which provided the authorization to perform molecular analyses of EGFR status and of its principle transducers on archival tumor tissues. The study was performed in accordance with the Declaration of Helsinki and was approved by the “Comitato Etico San Luigi Gonzaga di Orbassano”. Of 89 patients enrolled in the study, 57 had formalin fixed paraffin embedded tissues (FFPE blocks or tissues slides) available for molecular analyses.
Mutational analysis
For all the specimens, the tumor areas, identified by a pathologist, were scraped from two tissue slides (5 micron thick). DNA was then extracted by using Qiamp DNA FFPE mini kit (Qiagen s.r.l. Milano, Italy) following manufacturers’ instruction. Briefly, For DNA extraction, tissues were deparaffinized by xylene, rehydrated by ethanol, lysed with appropriate buffer and proteinase K at 56°C for one hour and subsequently for another hour at 90°C. Samples were then transferred on columns, washed with different buffers and then eluted in nuclease free water. DNA quantity was evaluated by Nanodrop (ThermoScientific Italia, Monza, Italy). The integrity of DNA was tested by a specific PCR for the housekeeping gene GAPDH. The EGFR exon 12 was amplified using nested PCR [16]. Exons 18–21 of EGFR were amplified by nested PCR using primers and conditions already described [20]. PCR products were purified using The Wizard SV Gel and PCR Clean-Up System kit (Promega Italia, Milano Italy). Each exon was sequenced using the BigDye Terminator Cycle sequence following the PE Applied Biosystem strategy and Applied Biosystems ABI PRISM3100 DNA Sequencer (Applied Biosystem, Forster City, CA). Sense and antisense sequences were obtained by using forward and reverse internal primers, respectively. All mutations were confirmed by performing two independent PCR amplifications.
Fluorescent in situ hybridization (FISH) analysis
EGFR gene status evaluation was performed by FISH on 3-μm thick tissue sections. Dual-color FISH assay was performed using LSI EGFR/CEP7 probes (Vysis, Abbott Laboratories, USA) following the manufacturer’s instructions already described [21]. Fluorescent in situ hybridization signals were evaluated with a Zeiss Axioscope (Carl Zeiss SPA, Italy). Tumors with ≥4 copies of EGFR or gene amplification in ≥40% of cells were classified as FISH+.
Statistical analysis
KaplanMeier survival curves were calculated using the Graph Pad Prism 6 software. Molecular findings (mutational status of exon 12, 18–21 of EGFR, its transducers KRAS, NRAS, BRAF, PIK3CA,and FISH) were correlated with PFS an OS in the entire study population and separately in the two arms of treatment. The Cox proportional hazards regression model was used to identify prognostic factors. PFS and OS were calculated using Kaplan-Meier estimation and examined by log-rank test. The association between the radiological response and molecular findings was analyzed using the two tailed Chi-Square test (C.I. 95%). A p-value less than 0.05 was considered as statistically significant.
Results
Mutational status of EGFR extracellular domain (ECD) and tyrosine kinase domain (TKD)
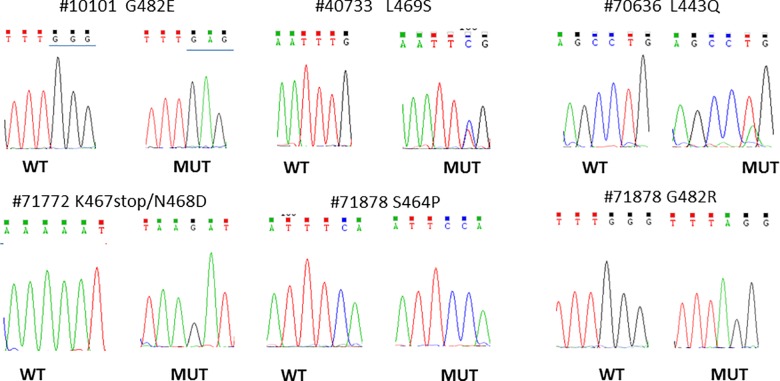
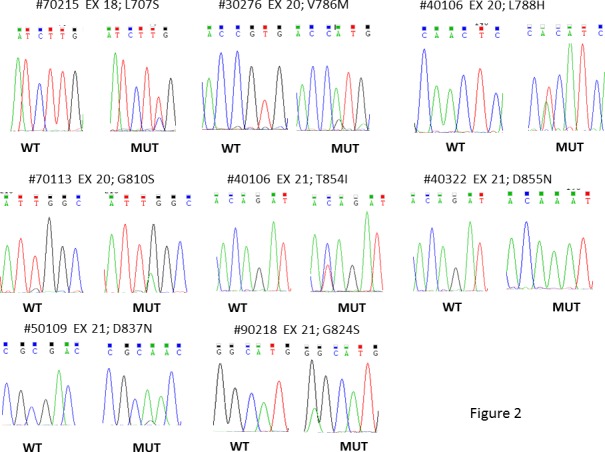
Six out of 57 patients (10.5%) displayed mutations in EGFR ECD (exon 12). Four patients belonged to the ARM-A and two patients to the ARM-B. Fig 1shows the electropherograms of EGFR ECD (exon 12) mutated samples. In the EGFR TKD, we found mutations of exons 18–21 in 7 out of 57 samples (12.3%). In detail, exon 18 was mutated in one patient, exon 20 was mutated in 3 patients, and exon 21 was mutated in 4 patients; no mutations were found in exon 19. Fig 2showed the electropherograms of EGFR TKD. S1 Table summarizes the mutations found, some of which have already been described in literature. In our previous work we described 3 patients with BRAF V600E mutations, 2 with NRAS mutations (A146S and Q61R), and 2 with PIK3CA E545K mutations [12]. One patient, the 71772, which harbored a PIK3CA mutation, was also mutated in EGFR ECD. Overall, we found that 18 out of 57 patients (31.6%) had mutations in EGFR or in its transducers. The mutations were homogeneously distributed between the two arms of treatment (8 patients in P-GEMOX arm, and 10 patients in GEMOX arm). Nine out of 18 mutated tumors (50%) were ICC, while 3 were ECC (16.7%), and 6 were GBC (33.3%).
Fig 1. Representative electropherograms of EGFR ECD mutated samples.
Fig 2. Representative electropherograms of EGFR TKD mutated samples.
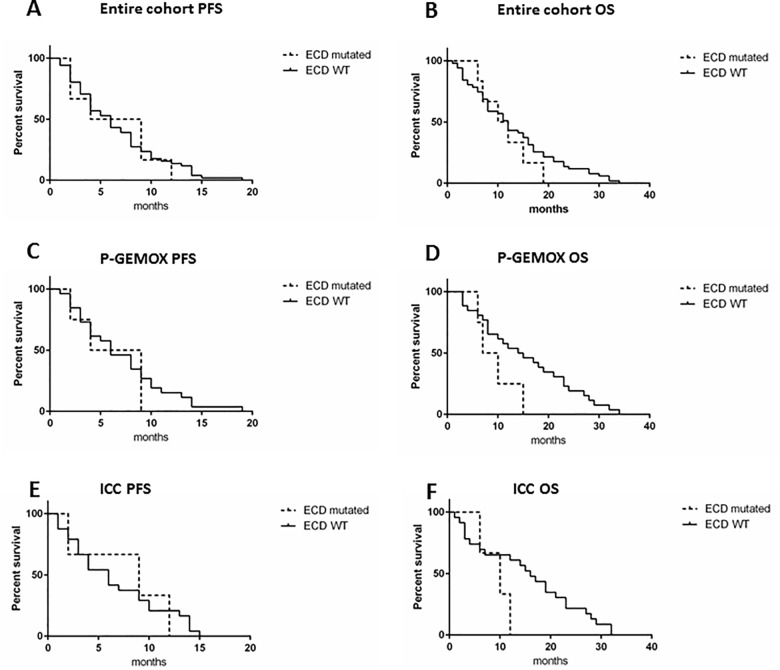
EGFR ECD mutations are associated with a worsening survival in subgroups
Analyzing all the 57 patients, independently from the treatment, we found that the presence of EGFR ECD mutations did not significantly affect PFS (p = 0.81) and OS (p = 0.44) (Fig 3A and 3B). In P-GEMOX arm, patients with EGFR ECD mutations had worse OS, even if not significantly (p = 0.1), while the PFS was not affected (p = 0.6) (Fig 3C and 3D). In the ICC subgroup, EGFR ECD mutated patients had impaired OS compared to WT patients (p = 0.14), but similar PFS (p = 0.92) (Fig 3E and 3F). Due to the small number of mutated patients, the analysis was not conducted for the GEMOX arm and for the other site subgroups.
Fig 3. KaplanMeier survival curves in ECD mutated vs WT patients.
A-B) association between the presence of ECD mutations and PFS and OS respectively, in the entire cohort of the Vecti-BIL study. C-D) association between the presence of ECD mutations and PFS and OS respectively, in P-GEMOX treated patients. E-F) association between the presence of ECD mutations and PFS and OS respectively, in ICC patients.
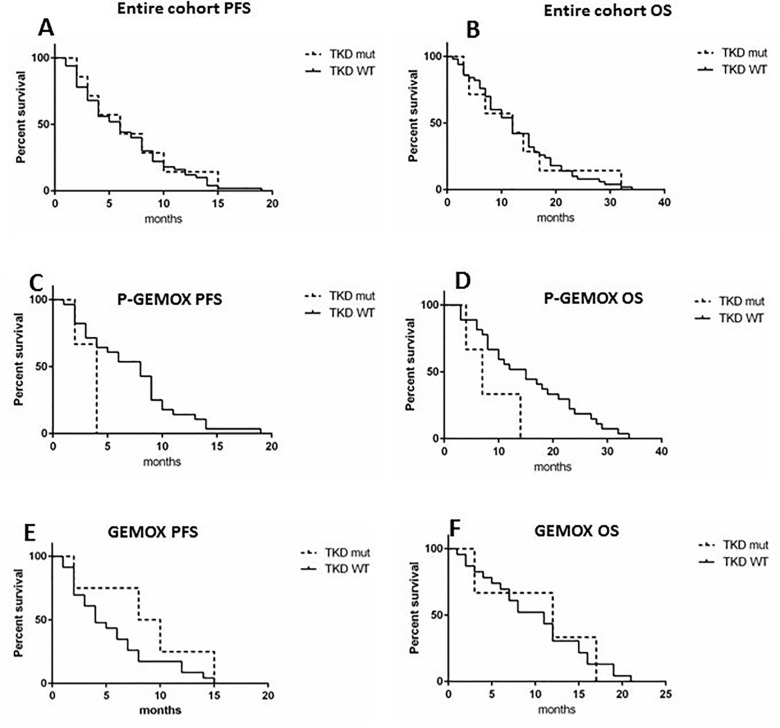
EGFR TKD mutations have a predictive negative role in P-GEMOX treated patients
The survival analysis of EGFR TKD mutated versus WT patients in the entire cohort of the study did not reveal any difference in terms of PFS (p = 0.75) and OS (p = 1.0) (Fig 4A and 4B). Even though, by analyzing P-GEMOX treated patients, we found that EGFR TKD mutations caused a trend towards decrease of PFS (p = 0.06) and OS (p = 0.06) (Fig 4C and 4D). On the contrary, EGFR TKD mutated patients treated with GEMOX had a better PFS (p = 0.2), while no differences in OS was observed (p = 0.9) (Fig 4E and 4F). For GBC patients, the presence of EGFR TKD mutations did not affect the PFS or OS (data not shown). Due to the small number of mutated patients, the analysis was not conducted for the other site subgroups.
Fig 4. Kaplan-Meier survival curves in TKD mutated vs WT patients.
A-B) Association between the presence of TKD mutations and PFS and OS, respectively, in the entire cohort of the Vecti-BIL study. C-D) Association between the presence of TKD mutations and PFS and OS, respectively, in P-GEMOX treated patients. E-F) association between the presence of TKD mutations and PFS and OS, respectively, in GEMOX treated patients.
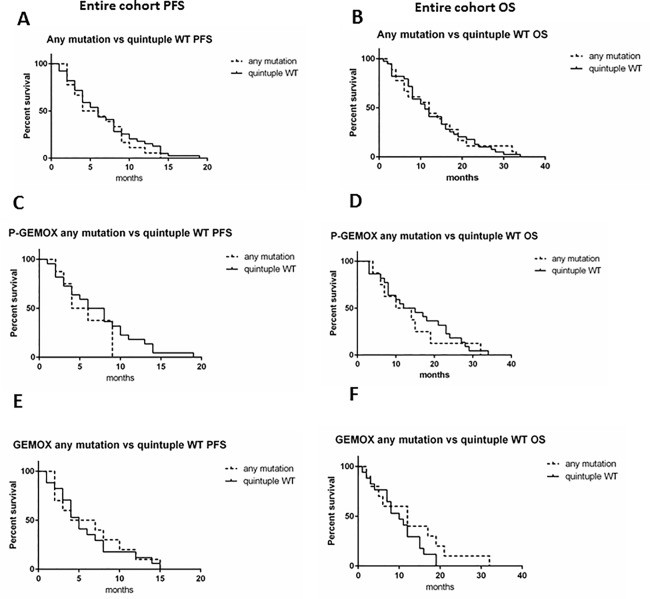
Wild-type status of EGFR and its transducers is not associated with efficacy of panitumumab
We analyzed if the presence of any mutation affecting EGFR and its main transducers (KRAS, NRAS, BRAF, PIK3CA) could influence survival in BTC patients. As shown in Fig 5A and 5B, PFS and OS were not impaired by the presence of mutations in EGFR and/or its transducers in the entire cohort (p = 0.49 and p = 0.93, respectively) as well as considering the two arms of treatment (Fig 5C–5F) (P-GEMOX; PFS: p = 0.29; OS: p = 0.6. GEMOX; PFS: p = 0.76; OS: p = 0.19). Then, we stratified patients according to the site subgroup and we found that ECC patients harboring mutations had a lower survival rates, even if not significantly, compared to WT patients (PFS: p = 0.12; OS: p = 0.17) (S1C and S1D Fig). No differences in terms of PFS and OS were evidenced in ICC (p = 0.57 and p = 0.25, respectively, S1A and S1B Fig) and GBC patients (p = 0.79 and p = 0.87, respectively, S1E and S1F Fig).
Fig 5. Kaplan-Meier survival curves in EGFR or its transducers mutated vs WT patients.
A-B) association between the presence of any mutation and PFS and OS, respectively, in the entire cohort of the Vecti-BIL study. C-D) association between the presence of any mutation and PFS and OS, respectively, in P-GEMOX treated patients. E-F) association between the presence of any mutation and PFS and OS, respectively, in GEMOX treated patients. Quintuple WT (absence of EGFR, KRAS, NRAS, BRAF, or PIK3CA mutations).
EGFR amplification does not affect patient survival
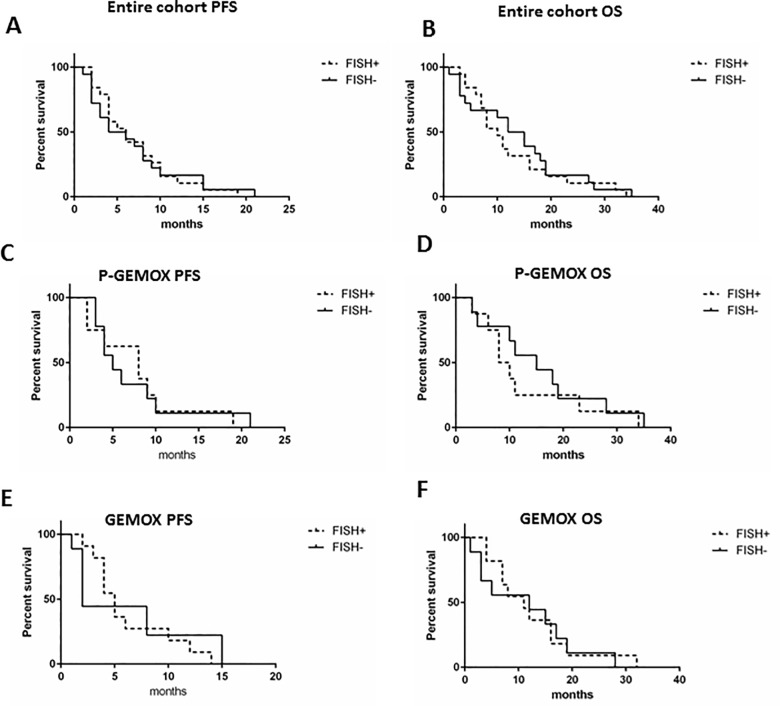
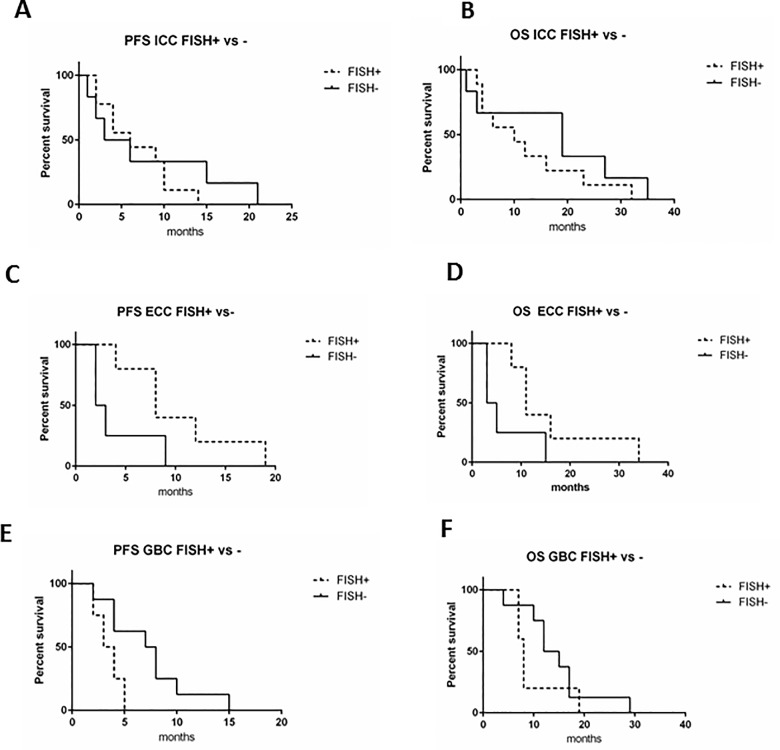
FISH analysis was conducted in 37 tumor samples. Due to the inadequate tissue preservation, the other tumor samples resulted unsuitable for this test. The analysis revealed that 19 tumors (51.4%) had amplification of EGFR (FISH+), while the remaining 18 were classified as negative (FISH-). Among the FISH+ tumors, 8 belonged to ARM-A and 11 to ARM-B of treatment. Nine out of 19 FISH+ tumors were ICC, five were ECC and five GBC. Considering all the 37 patients analyzed, PFS and OS were comparable in EGFR FISH+ and EGFR FISH- patients (PFS: p = 0.98; OS: p = 0.59) (Fig 6A and 6B). Similar results were obtained analyzing the two arms of treatment separately. Fig 6C and 6D shows the PFS and OS survival curves of P-GEMOX treated patients (p = 0.97 and p = 0.45, respectively) and Fig 6E and 6F represents the PFS and OS survival curves of GEMOX-treated patients (p = 0.68 and p = 0.46, respectively). Further, we stratified patients according to the site subgroup; as shown in Fig 7, ECC EGFR FISH+ patients had a better PFS (p = 0.07) and OS (p = 0.08) compared to FISH- patients (Fig 7C and 7D), even if not significantly. On the contrary, in GBC patients, FISH+ was associated to a PFS significantly worse (p = 0.03) and an OS slightly reduced (p = 0.23, Fig 7E and 7F). Finally, in ICC, FISH+ did not affect PFS (p = 0.54) and OS (p = 0.33, Fig 7A and 7B).
Fig 6. Kaplan-Meier survival curves in FISH+ or FISH- patients.
A-B) Association between EGFR amplification (FISH+) and PFS and OS, respectively, in the entire cohort of the Vecti-BIL study. C-D) association between EGFR amplification (FISH+) and PFS and OS, respectively, in P-GEMOX treated patients. E-F) association between EGFR amplification (FISH+) and PFS and OS, respectively, in GEMOX patients.
Fig 7. Kaplan-Meier survival curves in FISH+ versus FISH- patients.
A-B) Association between EGFR amplification (FISH+) and PFS and OS, respectively, in ICC patients. C-D) Association EGFR amplification (FISH+) and PFS and OS, respectively, in ECC patients. E-F) Association between EGFR amplification (FISH+) and PFS and OS, respectively, in GBC patients.
Discussion
The role of EGFR in cholangiocarcinogenesis is well documented. Its high expression as well as the low percentage of mutations in its downstream effectors suggested that it could be a suitable target for molecular therapy. Even though, these premises did not reflect the success obtained in mCRC and now the scientific community agreed about the marginal role of anti-EGFR therapies in BTC. The lack of efficacy obtained by the addition of panitumumab to standard chemotherapy demonstrated in the Vecti-BIL trial confirmed previous results available in literature [22–24]. The difference in median PFS and OS obtained in the two arms of treatment were not statistically different, but each arm displayed a broad range of PFS and OS, suggesting that both prognostic and predictive factors may have affected the results. The predefined collection of tumor tissues of patients enrolled in the prospective trial allowed a successive investigation of the role of EGFR mutational status, both in ECD and TKD, and the EGFR amplification. We demonstrated that 6 patients harbored mutations in EGFR ECD, none of them previously described. Four mutated patients were enrolled in the P-GEMOX arm; their PFS ranged from 2 to 9 months and the OS between 5.7 to 15.2 months. The survival curves showed that EGFR ECD mutations may have a predictive negative role in patients treated with panitumumab, but considering the worst survival of ICC patients having EGFR ECD mutations as compared to WT, the impact of EGFR ECD mutations may be rather prognostic, at least in this site subgroup.
Seven out of 57 patients harbored mutations in the exons 18–21 of EGFR, coding for the TKD: of them, two were novel mutations, while the others were already described in literature in other cancer types [25–28]. In particular, the V786M is described as high sensitive to gefitinib treatment in bronchioloalveolar adenocarcinoma [29]. Interestingly, none of EGFR TKD mutated patients had a tumor response, even if the correlation is not statistically significant (p = 0.07, data not shown). Even if not affecting the survival in the entire cohort, EGFR TKD mutated patients treated with P-GEMOX had a worse prognosis compared to WT patients.
Although the biological impact of each of the molecular alterations investigated may be different, we have considered all the mutations affecting EGFR, NRAS, KRAS, BRAF, and PIK3CA as potentially involved in the resistance to anti-EGFR therapies in BTC. For this reason, we evaluated the effect of treatments in patients without any of the mutations, referred to as “quintuple negative” population. This population does not seem to have different prognosis, and received similar benefit with both GEMOX and P-GEMOX as compared with patients with any of the mutations. By subgroup analysis, ECC quintuple negative patients have a trend towards a better survival than mutated patients, whereas in other site subgroups, mutation analysis revealed no prognostic impact.
Thirty-seven patients had tumor sample available for EGFR amplification analysis by FISH. More than half of the tested samples resulted FISH+. EGFR was amplified in all site subgroups (ICC, ECC, and GBC). Overall, EGFR amplification did not correlate with PFS or OS. According to the site of the disease, we found that EGFR amplification significantly correlated with poor PFS in GBC patients, while in ECC there was a trend towards a better survival of FISH+. Of note, 18 out of 19 EGFR amplified patients had lymph-node infiltration, suggesting a more advanced disease. This finding is enforced by a work on gastroesophageal cancers, in which EGFR amplification seemed to correlate with worse prognosis and lymph node metastasis [30]. However, the role of EGFR amplification as prognostic biomarker remains controversial. In two studies in mCRC, authors demonstrated that EGFR amplification was associated with longer PFS and OS [31, 32]; otherwise, in another work, EGFR amplification was not associated to prognosis [17].
The EGFR protein expression was not evaluated in our case series; previous studies demonstrated that EGFR is expressed in about 60% of BTC, ranging from 54% to 65%, and that the EGFR positivity does not affect the clinical outcome, independently from the treatment and site of origin [9, 23, 24, 33]. In contrast, Yang and collaborators evaluated the EGFR expression in 175 BTC, demonstrating that EGFR expression is a negative prognostic factor in ICC, but not in ECC [19, 34]. Therefore, the role of EGFR expression remains controversial, mainly due to the high heterogeneity of tumors analyzed.
The Vecti-BIL study included all KRAS exon 2 WT patients, but no other molecular stratification was performed at the time of enrollment, yielding to a highly heterogeneous population both in terms of site subgroups and of molecular alterations. Even if patients were selected for the absence of KRAS mutation, the antitumor effect of panitumumab was demonstrated neither in the overall population, nor in the molecular subgroups in which EGFR pathway was deemed suitable for effective inhibition. Most of the genetic alterations found seem to have a prognostic significance rather than a predictive role towards the anti-EGFR therapy. A high molecular heterogeneity seems to be a predominant feature of BTC, preventing the identification of well-defined subgroup of patients, which could benefit from targeted therapies. Due to the small cohort of patients analyzed, we can only speculate about prognostic or predictive role of EGFR status; nevertheless, the meta-analysis conducted by Chen and collaborators [35], which pooled four studies to examine the efficacy of anti-EGFR therapies combined to standard chemotherapy, revealed promising results of EGFR-targeted therapy in increasing the survival rate (PFS) of advanced BTC patients. These data keep the door open to the anti-EGFR therapies, provided that an accurate selection of patients is carried out. Moreover, it is highly recommended the identification of other biomarkers able to select a priori subgroups of patients who could benefit from these treatments. One of the suggested predictive biomarkers of response/resistance to anti-EGFR therapies is the expression of EGFR ligands; Luraghi and collaborators demonstrated that NRG1 is a marker of resistance to cetuximab, while EREG and AREG expression does not confer resistance to anti-EGFR therapies in mCRC xenospheres [36]. Further, another proposed mechanism is the presence of a specific oncogenic variant of EGFR, the EGFRvIII, which is constitutively active, without the requirement of ligand binding; in fact, it has been demonstrated that anti-EGFR antibodies are less effective in the presence of this variant in glioblastoma multiforme [37]. EGFR promoter methylation could be involved in the lack of response to anti-EGFR antibodies; in the work of Scartozzi, it has been demonstrated that mCRC patients treated with cetuximab displaying EGFR promoter methylation are less responder compared to unmethylated patients, with a significant worse prognosis [38]. This is only an overview of the main mechanisms involved in pathogenesis and drug resistance in BTC, evidencing the complexity of EGFR pathway and the major points of failure of anti-EGFR therapies. In fact, EGFR activation could be sustained also by a defective ubiquitination and subsequent internalization, which can cause a prolonged activation as well as a transactivation through COX-2/PgE2 signaling, as shown by Yoon and collaborators in BTC cell lines [39].
Since BTCs are rare disease, great efforts have been made in recent years in conducting prospective studies to investigate the efficacy of new therapies. Standard of care has been identified in advanced disease [40, 41] and a variety of targeted therapies has been tested so far [11, 12, 42–44]. The inclusion of patients with primary cancers arising in different part of the biliary tree is a compromise generally accepted in clinical trials. However, with the advent of new technologies as next-generation sequencing, it has become evident that mutational profiling of BTC varies within the tumor location. As a consequence, a neglected genetic heterogeneity can result in findings difficult to interpret. Thus, it is desirable that the genomic landscape of BTC will take the place of pathologic criteria based on the site of origin in future trials [45].
Supporting information
A-B) association between the presence of any mutations and PFS and OS, respectively, in ICC patients. C-D) association between the presence of any mutation and PFS and OS, respectively, in ECC patients. E-F) association between the presence of any mutation and PFS and OS, respectively, in GBC patients.
(TIF)
(DOCX)
Data Availability
All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was supported by a 5 per Mille 2010 Ministero della Salute, Fondazione Piemontese per la Ricerca sul Cancro (FPRC)—Onlus grant and a Associazione Italiana Ricerca sul Cancro–AIRC 5X1000 2010-Ministry of Health, FPO grant to MA. FL received a grant from Università di Torino anno 2014 (Fondo per la ricerca locale (Linea B), LEOF_RIC_LOC_14_01). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Goss G, Ferry D, Wierzbicki R, Laurie SA, Thompson J, Biesma B, et al. Randomized phase II study of gefitinib compared with placebo in chemotherapy-naive patients with advanced non-small-cell lung cancer and poor performance status. J Clin Oncol. 2009;27(13):2253–60. Epub 2009/03/16. doi: 10.1200/JCO.2008.18.4408 ; PubMed Central PMCID: PMCPMC4886538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qi WX, Fu S, Zhang Q, Guo XM. Anti-epidermal-growth-factor-receptor agents and complete responses in the treatment of advanced non-small-cell lung cancer: a meta-analysis of 17 phase III randomized controlled trials. Curr Med Res Opin. 2015;31(1):25–33. Epub 2014/10/30. doi: 10.1185/03007995.2014.978448 . [DOI] [PubMed] [Google Scholar]
- 3.Zhao N, Zhang XC, Yan HH, Yang JJ, Wu YL. Efficacy of epidermal growth factor receptor inhibitors versus chemotherapy as second-line treatment in advanced non-small-cell lung cancer with wild-type EGFR: a meta-analysis of randomized controlled clinical trials. Lung Cancer. 2014;85(1):66–73. Epub 2014/04/13. doi: 10.1016/j.lungcan.2014.03.026 . [DOI] [PubMed] [Google Scholar]
- 4.Lee CK, Davies L, Wu YL, Mitsudomi T, Inoue A, Rosell R, et al. Gefitinib or Erlotinib vs Chemotherapy for EGFR Mutation-Positive Lung Cancer: Individual Patient Data Meta-Analysis of Overall Survival. J Natl Cancer Inst. 2017;109(6). doi: 10.1093/jnci/djw279 . [DOI] [PubMed] [Google Scholar]
- 5.Nie F, Shen J, Tong JL, Xu XT, Zhu MM, Ran ZH. Meta-analysis: the efficacy and safety of monoclonal antibody targeted to epidermal growth factor receptor in the treatment of patients with metastatic colorectal cancer. J Dig Dis. 2009;10(4):247–57. doi: 10.1111/j.1751-2980.2009.00393.x . [DOI] [PubMed] [Google Scholar]
- 6.Jiang Z, Li C, Li F, Wang X. EGFR gene copy number as a prognostic marker in colorectal cancer patients treated with cetuximab or panitumumab: a systematic review and meta analysis. PLoS One. 2013;8(2):e56205 Epub 2013/02/18. doi: 10.1371/journal.pone.0056205 ; PubMed Central PMCID: PMCPMC3575344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amado RG, Wolf M, Peeters M, Van Cutsem E, Siena S, Freeman DJ, et al. Wild-type KRAS is required for panitumumab efficacy in patients with metastatic colorectal cancer. J Clin Oncol. 2008;26(10):1626–34. Epub 2008/03/03. doi: 10.1200/JCO.2007.14.7116 . [DOI] [PubMed] [Google Scholar]
- 8.Kennecke H, Chen L, Blanke CD, Cheung WY, Schaff K, Speers C. Panitumumab monotherapy compared with cetuximab and irinotecan combination therapy in patients with previously treated KRAS wild-type metastatic colorectal cancer. Curr Oncol. 2013;20(6):326–32. doi: 10.3747/co.20.1600 ; PubMed Central PMCID: PMCPMC3851344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pignochino Y, Sarotto I, Peraldo-Neia C, Penachioni JY, Cavalloni G, Migliardi G, et al. Targeting EGFR/HER2 pathways enhances the antiproliferative effect of gemcitabine in biliary tract and gallbladder carcinomas. BMC Cancer. 2010;10:631 doi: 10.1186/1471-2407-10-631 ; PubMed Central PMCID: PMCPMC3000850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gruenberger B, Schueller J, Heubrandtner U, Wrba F, Tamandl D, Kaczirek K, et al. Cetuximab, gemcitabine, and oxaliplatin in patients with unresectable advanced or metastatic biliary tract cancer: a phase 2 study. Lancet Oncol. 2010;11(12):1142–8. doi: 10.1016/S1470-2045(10)70247-3 . [DOI] [PubMed] [Google Scholar]
- 11.Lee J, Park SH, Chang HM, Kim JS, Choi HJ, Lee MA, et al. Gemcitabine and oxaliplatin with or without erlotinib in advanced biliary-tract cancer: a multicentre, open-label, randomised, phase 3 study. Lancet Oncol. 2012;13(2):181–8. doi: 10.1016/S1470-2045(11)70301-1 . [DOI] [PubMed] [Google Scholar]
- 12.Leone F, Marino D, Cereda S, Filippi R, Belli C, Spadi R, et al. Panitumumab in combination with gemcitabine and oxaliplatin does not prolong survival in wild-type KRAS advanced biliary tract cancer: A randomized phase 2 trial (Vecti-BIL study). Cancer. 2015. doi: 10.1002/cncr.29778 . [DOI] [PubMed] [Google Scholar]
- 13.Diaz LA, Williams RT, Wu J, Kinde I, Hecht JR, Berlin J, et al. The molecular evolution of acquired resistance to targeted EGFR blockade in colorectal cancers. Nature. 2012;486(7404):537–40. doi: 10.1038/nature11219 ; PubMed Central PMCID: PMCPMC3436069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.De Roock W, Claes B, Bernasconi D, De Schutter J, Biesmans B, Fountzilas G, et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: a retrospective consortium analysis. Lancet Oncol. 2010;11(8):753–62. Epub 2010/07/08. doi: 10.1016/S1470-2045(10)70130-3 . [DOI] [PubMed] [Google Scholar]
- 15.Ålgars A, Sundström J, Lintunen M, Jokilehto T, Kytölä S, Kaare M, et al. EGFR gene copy number predicts response to anti-EGFR treatment in RAS wild type and RAS/BRAF/PIK3CA wild type metastatic colorectal cancer. Int J Cancer. 2017;140(4):922–9. Epub 2016/11/23. doi: 10.1002/ijc.30507 . [DOI] [PubMed] [Google Scholar]
- 16.Arena S, Bellosillo B, Siravegna G, Martínez A, Cañadas I, Lazzari L, et al. Emergence of Multiple EGFR Extracellular Mutations during Cetuximab Treatment in Colorectal Cancer. Clin Cancer Res. 2015;21(9):2157–66. Epub 2015/01/26. doi: 10.1158/1078-0432.CCR-14-2821 . [DOI] [PubMed] [Google Scholar]
- 17.Italiano A, Follana P, Caroli FX, Badetti JL, Benchimol D, Garnier G, et al. Cetuximab shows activity in colorectal cancer patients with tumors for which FISH analysis does not detect an increase in EGFR gene copy number. Ann Surg Oncol. 2008;15(2):649–54. Epub 2007/11/07. doi: 10.1245/s10434-007-9667-2 . [DOI] [PubMed] [Google Scholar]
- 18.Nakazawa K, Dobashi Y, Suzuki S, Fujii H, Takeda Y, Ooi A. Amplification and overexpression of c-erbB-2, epidermal growth factor receptor, and c-met in biliary tract cancers. J Pathol. 2005;206(3):356–65. doi: 10.1002/path.1779 . [DOI] [PubMed] [Google Scholar]
- 19.Yang X, Wang W, Wang C, Wang L, Yang M, Qi M, et al. Characterization of EGFR family gene aberrations in cholangiocarcinoma. Oncol Rep. 2014;32(2):700–8. Epub 2014/06/13. doi: 10.3892/or.2014.3261 . [DOI] [PubMed] [Google Scholar]
- 20.Leone F, Cavalloni G, Pignochino Y, Sarotto I, Ferraris R, Piacibello W, et al. Somatic mutations of epidermal growth factor receptor in bile duct and gallbladder carcinoma. Clin Cancer Res. 2006;12(6):1680–5. doi: 10.1158/1078-0432.CCR-05-1692 . [DOI] [PubMed] [Google Scholar]
- 21.Frattini M, Saletti P, Romagnani E, Martin V, Molinari F, Ghisletta M, et al. PTEN loss of expression predicts cetuximab efficacy in metastatic colorectal cancer patients. Br J Cancer. 2007;97(8):1139–45. Epub 2007/10/16. doi: 10.1038/sj.bjc.6604009 ; PubMed Central PMCID: PMCPMC2360431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sohal DP, Mykulowycz K, Uehara T, Teitelbaum UR, Damjanov N, Giantonio BJ, et al. A phase II trial of gemcitabine, irinotecan and panitumumab in advanced cholangiocarcinoma. Ann Oncol. 2013;24(12):3061–5. Epub 2013/10/20. doi: 10.1093/annonc/mdt416 . [DOI] [PubMed] [Google Scholar]
- 23.Chen JS, Hsu C, Chiang NJ, Tsai CS, Tsou HH, Huang SF, et al. A KRAS mutation status-stratified randomized phase II trial of gemcitabine and oxaliplatin alone or in combination with cetuximab in advanced biliary tract cancer. Ann Oncol. 2015;26(5):943–9. doi: 10.1093/annonc/mdv035 . [DOI] [PubMed] [Google Scholar]
- 24.Malka D, Cervera P, Foulon S, Trarbach T, de la Fouchardière C, Boucher E, et al. Gemcitabine and oxaliplatin with or without cetuximab in advanced biliary-tract cancer (BINGO): a randomised, open-label, non-comparative phase 2 trial. Lancet Oncol. 2014;15(8):819–28. doi: 10.1016/S1470-2045(14)70212-8 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Metzger B, Chambeau L, Begon DY, Faber C, Kayser J, Berchem G, et al. The human epidermal growth factor receptor (EGFR) gene in European patients with advanced colorectal cancer harbors infrequent mutations in its tyrosine kinase domain. BMC Med Genet. 2011;12:144 Epub 2011/10/25. doi: 10.1186/1471-2350-12-144 ; PubMed Central PMCID: PMCPMC3215960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu M, Zhang W, Shan L, Song J, Shang D, Ying J, et al. Mutation status of somatic EGFR and KRAS genes in Chinese patients with prostate cancer (PCa). Virchows Arch. 2014;464(5):575–81. Epub 2014/03/05. doi: 10.1007/s00428-014-1566-x . [DOI] [PubMed] [Google Scholar]
- 27.Boldrini L, Alì G, Gisfredi S, Ursino S, Baldini E, Melfi F, et al. Epidermal growth factor receptor and K-RAS mutations in 411 lung adenocarcinoma: a population-based prospective study. Oncol Rep. 2009;22(4):683–91. . [DOI] [PubMed] [Google Scholar]
- 28.Locatelli-Sanchez M, Couraud S, Arpin D, Riou R, Bringuier PP, Souquet PJ. Routine EGFR molecular analysis in non-small-cell lung cancer patients is feasible: exons 18–21 sequencing results of 753 patients and subsequent clinical outcomes. Lung. 2013;191(5):491–9. Epub 2013/06/09. doi: 10.1007/s00408-013-9482-4 . [DOI] [PubMed] [Google Scholar]
- 29.Ludovini V, Gori S, Pistola L, Della Fazia MA, Piobbico D, Servillo G, et al. Long-lasting complete remission with tyrosine kinase inhibitor in bronchioloalveolar carcinoma with a so far unknown EGFR mutation. J Thorac Oncol. 2008;3(4):452–3. doi: 10.1097/JTO.0b013e318169e341 . [DOI] [PubMed] [Google Scholar]
- 30.Liao JB, Lee HP, Fu HT, Lee HS. Assessment of EGFR and ERBB2 (HER2) in Gastric and Gastroesophageal Carcinomas: EGFR Amplification is Associated With a Worse Prognosis in Early Stage and Well to Moderately Differentiated Carcinoma. Appl Immunohistochem Mol Morphol. 2016. Epub 2016/10/07. doi: 10.1097/PAI.0000000000000437 . [DOI] [PubMed] [Google Scholar]
- 31.Yang J, Li S, Wang B, Wu Y, Chen Z, Lv M, et al. Potential biomarkers for anti-EGFR therapy in metastatic colorectal cancer. Tumour Biol. 2016;37(9):11645–55. Epub 2016/07/16. doi: 10.1007/s13277-016-5140-9 . [DOI] [PubMed] [Google Scholar]
- 32.Laurent-Puig P, Cayre A, Manceau G, Buc E, Bachet JB, Lecomte T, et al. Analysis of PTEN, BRAF, and EGFR status in determining benefit from cetuximab therapy in wild-type KRAS metastatic colon cancer. J Clin Oncol. 2009;27(35):5924–30. Epub 2009/11/02. doi: 10.1200/JCO.2008.21.6796 . [DOI] [PubMed] [Google Scholar]
- 33.Harder J, Waiz O, Otto F, Geissler M, Olschewski M, Weinhold B, et al. EGFR and HER2 expression in advanced biliary tract cancer. World J Gastroenterol. 2009;15(36):4511–7. ; PubMed Central PMCID: PMCPMC2751995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yoshikawa D, Ojima H, Iwasaki M, Hiraoka N, Kosuge T, Kasai S, et al. Clinicopathological and prognostic significance of EGFR, VEGF, and HER2 expression in cholangiocarcinoma. Br J Cancer. 2008;98(2):418–25. doi: 10.1038/sj.bjc.6604129 ; PubMed Central PMCID: PMCPMC2361442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen L, Chen C, Yen Y, Tam KW. Chemotherapy for advanced biliary tract carcinoma: A meta-analysis of randomized controlled trials. Medicine (Baltimore). 2016;95(33):e4584 doi: 10.1097/MD.0000000000004584 ; PubMed Central PMCID: PMCPMC5370815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luraghi P, Bigatto V, Cipriano E, Reato G, Orzan F, Sassi F, et al. A molecularly annotated model of patient-derived colon cancer stem-like cells to assess genetic and non-genetic mechanisms of resistance to anti-EGFR therapy. Clin Cancer Res. 2017. Epub 2017/10/03. doi: 10.1158/1078-0432.CCR-17-2151 . [DOI] [PubMed] [Google Scholar]
- 37.Gajadhar AS, Bogdanovic E, Muñoz DM, Guha A. In situ analysis of mutant EGFRs prevalent in glioblastoma multiforme reveals aberrant dimerization, activation, and differential response to anti-EGFR targeted therapy. Mol Cancer Res. 2012;10(3):428–40. Epub 2012/01/09. doi: 10.1158/1541-7786.MCR-11-0531 . [DOI] [PubMed] [Google Scholar]
- 38.Scartozzi M, Bearzi I, Mandolesi A, Giampieri R, Faloppi L, Galizia E, et al. Epidermal growth factor receptor (EGFR) gene promoter methylation and cetuximab treatment in colorectal cancer patients. Br J Cancer. 2011;104(11):1786–90. Epub 2011/05/10. doi: 10.1038/bjc.2011.161 ; PubMed Central PMCID: PMCPMC3111171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoon JH, Gwak GY, Lee HS, Bronk SF, Werneburg NW, Gores GJ. Enhanced epidermal growth factor receptor activation in human cholangiocarcinoma cells. J Hepatol. 2004;41(5):808–14. doi: 10.1016/j.jhep.2004.07.016 . [DOI] [PubMed] [Google Scholar]
- 40.Valle JW, Furuse J, Jitlal M, Beare S, Mizuno N, Wasan H, et al. Cisplatin and gemcitabine for advanced biliary tract cancer: a meta-analysis of two randomised trials. Ann Oncol. 2014;25(2):391–8. doi: 10.1093/annonc/mdt540 . [DOI] [PubMed] [Google Scholar]
- 41.Sharma A, Dwary AD, Mohanti BK, Deo SV, Pal S, Sreenivas V, et al. Best supportive care compared with chemotherapy for unresectable gall bladder cancer: a randomized controlled study. J Clin Oncol. 2010;28(30):4581–6. Epub 2010/09/20. doi: 10.1200/JCO.2010.29.3605 . [DOI] [PubMed] [Google Scholar]
- 42.Valle JW, Wasan H, Lopes A, Backen AC, Palmer DH, Morris K, et al. Cediranib or placebo in combination with cisplatin and gemcitabine chemotherapy for patients with advanced biliary tract cancer (ABC-03): a randomised phase 2 trial. Lancet Oncol. 2015;16(8):967–78. Epub 2015/07/12. doi: 10.1016/S1470-2045(15)00139-4 ; PubMed Central PMCID: PMCPMC4648082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Santoro A, Gebbia V, Pressiani T, Testa A, Personeni N, Arrivas Bajardi E, et al. A randomized, multicenter, phase II study of vandetanib monotherapy versus vandetanib in combination with gemcitabine versus gemcitabine plus placebo in subjects with advanced biliary tract cancer: the VanGogh study. Ann Oncol. 2015;26(3):542–7. Epub 2014/12/23. doi: 10.1093/annonc/mdu576 . [DOI] [PubMed] [Google Scholar]
- 44.Kim ST, Jang KT, Lee J, Jang HM, Choi HJ, Jang HL, et al. Molecular Subgroup Analysis of Clinical Outcomes in a Phase 3 Study of Gemcitabine and Oxaliplatin with or without Erlotinib in Advanced Biliary Tract Cancer. Transl Oncol. 2015;8(1):40–6. doi: 10.1016/j.tranon.2014.12.003 ; PubMed Central PMCID: PMCPMC4350637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jain A, Javle M. Molecular profiling of biliary tract cancer: a target rich disease. J Gastrointest Oncol. 2016;7(5):797–803. doi: 10.21037/jgo.2016.09.01 ; PubMed Central PMCID: PMCPMC5056250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A-B) association between the presence of any mutations and PFS and OS, respectively, in ICC patients. C-D) association between the presence of any mutation and PFS and OS, respectively, in ECC patients. E-F) association between the presence of any mutation and PFS and OS, respectively, in GBC patients.
(TIF)
(DOCX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files.