ABSTRACT
An incomplete understanding of native human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV) envelope glycoproteins (Envs) impedes the development of structural models of Env and vaccine design. This shortcoming is due in part to the low number of Env trimers on virus particles. For SIV, this low expression level can be counteracted by truncating the cytoplasmic tail (CT) of Env. CT truncation has been shown to increase Env incorporation into the virion and is commonly used in vaccine and imaging studies, but its effects on viral antigenicity have not been fully elucidated. To study the effects of a CT truncation of Env in viruses in similar genetic contexts, we introduced stop codons into the CT of a SIVsmE660 molecular clone and two neutralizing antibody (NAb) escape variants. These viruses shared 98% sequence identity in Env but were characterized as either tier 1 (sensitive to neutralization), tier 2 (moderately resistant to neutralization), or tier 3 (resistant to neutralization). However, the introduction of premature stop codons in Env at position Q741/Q742 converted all three transfection-derived viruses to a tier 3-like phenotype, and these viruses were uniformly resistant to neutralization by sera from infected macaques and monoclonal antibodies (MAbs). These changes in neutralization sensitivity were not accompanied by an increase in either the virion Env content of infection-derived viruses or the infectivity of transfection-derived viruses in human cells, suggesting that CT mutations may result in global changes to the Env conformation. Our results demonstrate that some CT truncations can affect viral antigenicity and, as such, may not be suitable surrogate models of native HIV/SIV Env.
IMPORTANCE Modifications to the SIV envelope protein (Env) are commonly used in structural and vaccine studies to stabilize and increase the expression of Env, often without consideration of effects on antigenicity. One such widespread modification is the truncation of the Env C-terminal tail. Here, we studied the effects of a particular cytoplasmic tail truncation in three SIVsm strains that have highly similar Env sequences but exhibit different sensitivities to neutralizing antibodies. After truncation of the Env CT, these viruses were all very resistant to neutralization by sera from infected macaques and monoclonal antibodies. The viruses with a truncated Env CT also did not exhibit the desired and typical increase in Env expression. These results underscore the importance of carefully evaluating the use of truncated Env as a model in HIV/SIV vaccine and imaging studies and of the continued need to find better models of native Env that contain fewer modifications.
KEYWORDS: macaques, neutralizing antibodies, simian immunodeficiency virus, viral envelope
INTRODUCTION
The human immunodeficiency virus (HIV)/simian immunodeficiency virus (SIV) envelope glycoprotein (Env) initiates virus entry into cells and is the primary target for HIV vaccine development. HIV/SIV Env is a homotrimer of noncovalently bound gp120 and gp41 subunits. The highly glycosylated gp120 subunit mediates binding to receptors on target cells, while the membrane-anchored gp41 subunit initiates virus-cell fusion. Compared to other retroviruses, the HIV/SIV gp41 subunits have unusually long cytoplasmic tails (CTs) of around 150 amino acids (1). The CTs of HIV/SIV gp41 have been implicated in several important functional activities during the HIV/SIV life cycle, including Env endocytosis, intracellular trafficking, and the incorporation of Env into viral particles (1, 2). Although the biological functions of the HIV/SIV CT have not been fully characterized, several studies revealed that a truncation of the SIV gp41 CT increased Env incorporation into the virion but did not prevent the Env function in virus-cell fusion (3–7). Hence, many HIV vaccine candidates, such as the Merck candidate, RV144 trials, and the SOSIP antigen series, have a truncation at the gp41 membrane-proximal external region (MPER) of Env immunogens (8–11).
Both HIV and SIV particles express a relatively low number of Env trimers on their surface, around 7 to 16 per virion (6, 12–14), which makes studies of the Env structure more difficult. Therefore, groups performing cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET) to study the molecular structure of SIV Env use virus preparations with gp41 CT truncations. These groups assume that the gp41 truncation does not affect the structure of the extracellular domain of Env (13–19). However, the effects of the gp41 CT truncation on Env structure and the neutralization sensitivity of the virus are still not fully understood. A recent study revealed that truncation of the gp41 CT altered the binding of neutralizing antibodies (NAbs) to HIV Env, suggesting that gp41 can affect HIV/SIV Env structure and antigenicity (20).
In this paper, we use SIV as a model to study the effects of a gp41 CT truncation on the antigenicity of HIV/SIV Env. We introduced stop codons into the gp41 CTs of several SIV isolates and compared their sensitivities to NAbs from SIV-infected rhesus macaques. Like HIV, SIV strains have a range of sensitivities to NAbs (21). We previously isolated three SIVsmE660 clones that share 98% amino acid sequence identity in Env but have distinct neutralization sensitivity phenotypes (22), representative of either tier 1 (sensitive), tier 2 (moderately resistant), or tier 3 (resistant) variants. We found that the introduction of the gp41 CT truncation increased resistance to neutralizing antibodies in all of the SIV isolates without increasing Env incorporation into virions. These results suggest that the gp41 CT plays a role in maintaining the structure and antigenicity of the extracellular domain of HIV/SIV Env proteins.
RESULTS
SIVsmE660 clones have high sequence similarity but different sensitivities to neutralization.
The three parental SIVsmE660 clones used in this study were selected because they represent a range of neutralization sensitivity phenotypes while having 98% sequence identity in the env region. Amino acid differences were confined to Env, as shown in Table 1, and were located in the gp120 domains C1, V1, V2, and V4. The tier 1 SIVsmE660 FL14 clone was isolated from a SIVsmE660 virus stock as previously described (22). The tier 2 clone H807-16w-6 and the tier 3 clone H807-24w-4 are escape mutants isolated from a macaque infected with the tier 1 clone. The env sequences of the tier 2 and tier 3 clones were cloned from plasma collected at 16 and 24 weeks postinfection, respectively. The three clones were designated tier 1 (sensitive to neutralization), tier 2 (moderately sensitive to neutralization), or tier 3 (resistant to neutralization) clones. These designations are based on the tier system used to classify HIV-1 variants according to their neutralization sensitivity (23), using chronic-stage sera from a panel of rhesus macaques infected with uncloned SIVsmE660 (22) or cloned SIVsmE543-3, SIVmac251, or SIVmac239. We used heat map hierarchical clustering, which was used previously for the tiered categorization of the neutralization sensitivity of HIV-1 isolates, to assess the neutralization sensitivities of these SIVs, and we selected one virus representative each of tier 1, tier 2, and tier 3 (22).
TABLE 1.
Env amino acid differences of the three wild-type SIVsmE660 strainsa
| Tier | Amino acid |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| C1 at position: |
V1 at position: |
V2 at position: |
V4 at position: |
||||||||
| 45 | 70 | 134 | 139 | 140 | 184 | 185 | 420 | 425 | 429 | 432 | |
| 1 | T | S | T | A | A | R | I | K | K | T | R |
| 2 | A | N | I | A | S | R | I | K | R | M | Q |
| 3 | A | N | T | — | A | K | M | N | K | T | Q |
All of the amino acid differences among the three strains are shown. The locations of these differences are labeled by residue number as well as the domain in which they are found. C1 corresponds to constant region 1, and variable loops 1, 2, and 4 are labeled V1, V2, and V4, respectively. Residues that differ from the parental E660-FL14 tier 1 clone are in boldface type, and deletions are shown as a dash.
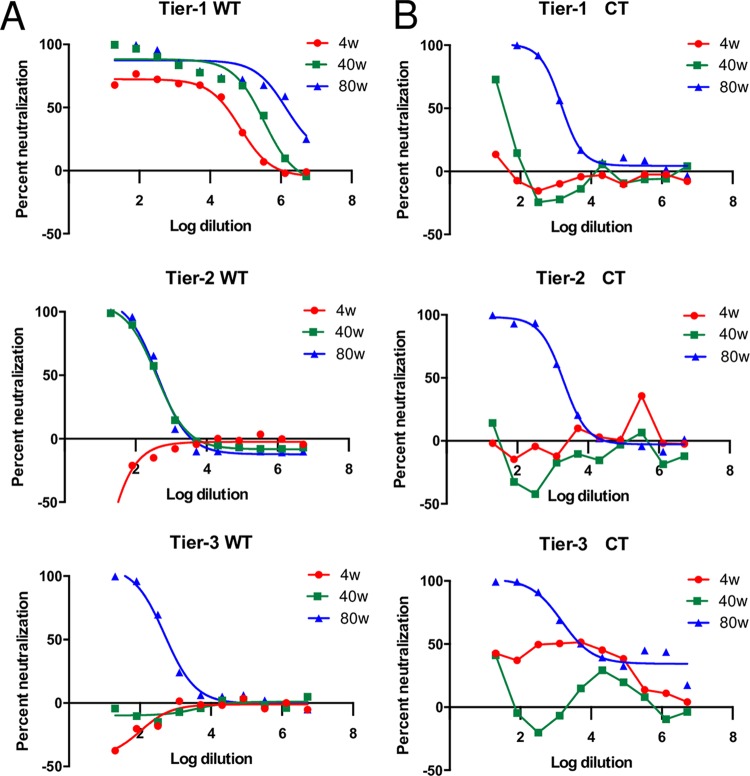
To confirm the differential sensitivities of the parental clones, we performed a TZM-bl neutralization assay (24), using sera collected at 4, 40, and 80 weeks postinfection from an unrelated SIVsmE660-infected rhesus macaque inoculated with the tier 1 virus (Fig. 1A). We previously reported that sera collected at later stages of infection have higher neutralization capabilities than do early-stage sera, which can neutralize only very sensitive strains (22). Indeed, the tier 1 virus was sensitive to rhesus sera collected at all three time points. The tier 2 virus had an intermediate phenotype and could be neutralized by sera collected at 40 and 80 weeks postinfection but not by serum collected at 4 weeks postinfection. The tier 3 virus was more resistant to neutralization and could be neutralized only by late-stage serum collected at 80 weeks postinfection.
FIG 1.
Differential neutralization sensitivities of the wild-type and CT-truncated SIVsmE660 clones. (A) Serially diluted sera of a rhesus macaque infected with a SIVsmE660 tier 1 virus collected at different time points, 4 weeks postinfection (4w), 40 weeks postinfection, and 80 weeks postinfection, were tested in duplicate against the tier 1, tier 2, and tier 3 viral clones in a TZM-bl neutralization assay. The percentages of neutralization by sera at different dilutions are shown. (B) Virus clones with truncated cytoplasmic tails are less sensitive to neutralization by rhesus sera. A neutralization assay was performed on tier 1, tier 2, and tier 3 mutant viral clones in duplicate in a TZM-bl neutralization assay as described above for panel A. The percentages of neutralization by sera at different dilutions are shown.
Truncation of the cytoplasmic tail decreases sensitivity to neutralization by rhesus sera.
We introduced a truncation in the CTs of the three viruses. The truncation was introduced at position Q741/Q742, the location analogous to that of the premature stop codon found in a SIVmac clone passaged in human cell lines, which served as the original evidence that truncation of the SIV cytoplasmic tail is an adaptation to replication in human cells (25). In the genetic context of SIVmac239, the introduction of the stop codon at this site was shown to increase Env incorporation into virions (4, 6).
Using the same neutralization assay as the one described above for the wild-type viruses, we found that truncation of the CT resulted in decreases in the sensitivities of the three transfection-derived viruses to neutralization by rhesus sera (Fig. 1B). Complete neutralization was achieved only with the late-stage sera collected at 80 weeks postinfection at a low dilution for all three mutant clones. Even at the lowest dilution, the sera collected at 4 weeks postinfection could not achieve 50% neutralization of the tier 1 ΔCT virus, whereas the same sera were able to neutralize the wild-type tier 1 virus at a 50% inhibitory dose (ID50) value of 28,900 (Table 2). The sera collected at 40 and 80 weeks postinfection also needed much lower dilutions to achieve 50% neutralization of the tier 1 ΔCT virus, with ID50 values of 65 and 1,538, respectively, compared to the ID50 values for the wild-type tier 1 virus of 211,000 and 1,360,000, respectively. Differences between the wild-type and mutant tier 2 viruses were observed with neutralization by serum from 40 weeks postinfection. The mutant could not be neutralized by serum from this time point, but the wild-type virus was neutralized with an ID50 value of 380. Like the wild-type clone, the tier 3 ΔCT virus could not be neutralized by sera collected at either 4 or 40 weeks postinfection but could be neutralized by the serum collected at 80 weeks postinfection at an ID50 value of 4,920.
TABLE 2.
Neutralizing antibody ID50 titers of rhesus sera against the wild-type and mutant clonesa
| Tier | Clone | ID50 titer |
||
|---|---|---|---|---|
| 4 wk postinfection | 40 wk postinfection | 80 wk postinfection | ||
| 1 | wt | 28,900 | 211,000 | 1,360,000 |
| ΔCT | <20 | 65 | 1,540 | |
| 2 | wt | <20 | 380 | 406 |
| ΔCT | <20 | <20 | 1,870 | |
| 3 | wt | <20 | <20 | 568 |
| ΔCT | <20 | <20 | 4,920 | |
ID50 titers were calculated with nonlinear regression by using PRISM5 and are expressed as the highest dilution of sera that resulted in a 50% reduction of RLU compared with the virus control. The sera used were collected at 4 weeks, 40 weeks, and 80 weeks postinfection from a rhesus macaque infected with the SIVsmE660 tier 1 virus. wt, wild type.
Comparison of the sensitivities of the parental and mutant clones to neutralization by monoclonal antibodies.
In addition to using polyclonal sera to assess neutralization sensitivity, we performed neutralization assays with MAbs that target different Env epitopes. The three SIV Env-specific MAbs selected for the neutralization assay were isolated and characterized as previously described (26). Antibodies ITS09.03, ITS52, and ITS01 target the variable loop 2 (V2), V3, or CD4-binding-site (CD4bs) region of Env, respectively, and here, they are labeled according to the region that they target. These epitopes are analogous to the sites on HIV Env that are major targets of broadly neutralizing antibodies (bNAbs).
The transfection-derived wild-type viruses demonstrated differential sensitivities to neutralization by the MAbs (Fig. 2). The 50% inhibitory concentrations (IC50s) for neutralization of the wild-type tier 1 virus by the V2, V3, and CD4bs MAbs were 0.045 μg/ml, 0.016 μg/ml, and 0.028 μg/ml, respectively (Table 3). Higher concentrations of the V2, V3, and CD4bs MAbs were required to reach 50% neutralization of the tier 2 virus, with values of 5.260 μg/ml, 0.065 μg/ml, and 0.160 μg/ml, respectively. The tier 3 virus could not be effectively neutralized by any of the three MAbs. As observed with neutralization by sera, truncation of the cytoplasmic tail decreased sensitivity to neutralization by MAbs. At the highest concentration tested, none of the MAbs could achieve 50% neutralization of any of the three mutant viruses.
FIG 2.
IC50 titers of monoclonal antibodies against the wild-type and mutant clones. Monoclonal antibodies targeting the variable loop 2 (V2), V3, and CD4bs regions of Env were tested in duplicate against the viral clones in a TZM-bl neutralization assay. IC50 titers were calculated with nonlinear regression by using PRISM5 and are expressed as the lowest concentration of antibody that causes a 50% reduction in RLU compared with the virus control. A titer of 50 μg/ml is plotted for data points where an IC50 was not reached at the highest MAb concentration tested.
TABLE 3.
Neutralizing antibody IC50 titers against the wild-type and mutant clones with monoclonal antibodies targeting the V2, V3, and CD4bs regions
| Tier | Clone | IC50 (μg/ml) |
||
|---|---|---|---|---|
| V2 | V3 | CD4bs | ||
| 1 | wt | 0.045 | 0.016 | 0.028 |
| ΔCT | >50 | >50 | >50 | |
| 2 | wt | 5.260 | 0.065 | 0.160 |
| ΔCT | >50 | >50 | >50 | |
| 3 | wt | >50 | >50 | >50 |
| ΔCT | >50 | >50 | >50 | |
Truncation of the cytoplasmic tails does not increase the number of trimers on the virion.
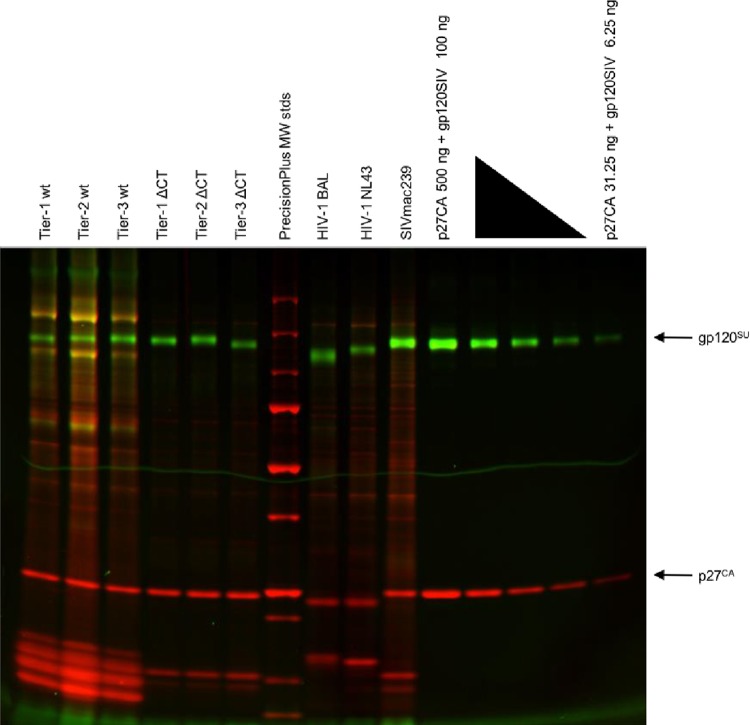
To investigate the underlying mechanism for this increase in neutralizing antibody resistance of the CT-truncated viruses, we evaluated the virion Env content. We performed dual-fluorescence imaging of an SDS-PAGE gel of virus proteins produced from infection of A66-R5 cells to quantify the virion Env content, as previously described (27, 28). To ensure that this preparation would not affect the neutralization phenotype, we tested the A66-R5-derived wild-type viruses in a TZM-bl neutralization assay. Indeed, the A66-R5-derived wild-type viruses retained their neutralization sensitivities (data not shown). Viral particles were collected after ultracentrifugation of the A66-R5 cell infection supernatant and analyzed by SDS-PAGE. The gp120 glycoproteins were then stained with green fluorescence, while a red fluorescent whole-protein stain was used to stain for p27 (Fig. 3). A standard curve, produced from the serial dilution of purified gp120 and p27 protein standards, was used to quantify the amount of Gag p27 and Env gp120 in each of the virus preparations. From the calculated Gag/Env ratios, we could approximate the number of trimers per virion, based on the estimate of an average of 1,400 Gag molecules per virion (Table 4) (13).
FIG 3.
Quantification of virion Env content using dual-fluorescence imaging of virus proteins. A66-R5-derived test viruses, SupT1-R5-derived control viruses, and a series of protein standards were run on an SDS-PAGE gel. The gel was stained with the Pro-Q Emerald 300 glycoprotein dye to detect gp120 and with the SYPRO ruby dye to detect total protein, including p27. MW, molecular weight.
TABLE 4.
Estimation of the number of Env trimers per viriona
| Virus | Gag/Env ratio | Estimated no. of trimers |
|---|---|---|
| Tier 1 wt/A66-R5 | 74 | 6 |
| Tier 2 wt/A66-R5 | 55 | 8 |
| Tier 3 wt/A66-R5 | 36 | 13 |
| Tier 1 ΔCT/A66-R5 | 32 | 15 |
| Tier 2 ΔCT/A66-R5 | 33 | 14 |
| Tier 3 ΔCT/A66-R5 | 89 | 5 |
| HIV-1 BAL/SupT1-R5 | 27 | 17 |
| HIV-1 NL43/SupT1-R5 | 65 | 7 |
| SIVmac239/SupT1-R5 | 13 | 34 |
Gag p27/Env gp120 ratios were calculated, and the numbers of virion-associated trimers were approximated based on an estimate of 1,400 Gag molecules per virion.
The numbers of trimers on the wild-type and mutant virions were not significantly different. The wild-type viruses had a range of 6 to 13 trimers/virion, while the CT-truncated mutants had a range of 5 to 15 trimers/virion. Additionally, neutralization sensitivity did not seem to correlate with the Env content for the tested SIVsmE660 viruses. For example, the wild-type tier 3 virus had only 7 more trimers than did the neutralization-sensitive wild-type tier 1 virus. Furthermore, The tier 3 mutant virus had the lowest Env content, with 5 trimers/virion, but had a resistant phenotype. Well-characterized control preparations of the HIV-1 BAL and NL43 strains, as well as a CT-truncated SIVmac239 preparation, were used as quality controls, and results from this experiment agree with previously reported observations. Thus, despite previous reports that truncation of the CT can increase the virion Env contents of viruses produced from infected cultures of human cells, the introduction of a CT truncation into SIVsmE660 clones did not increase the average numbers of trimers on the virion.
Infectivity of parental and mutant clones in rhesus and human cells.
Premature stop codons in the SIV CT were first identified in SIV strains that were passaged in human cell lines, providing the original evidence that a truncation of the SIV CT enhances replication in human cells (25, 29). However, in those studies, passage of the truncated form of SIV in rhesus cells resulted in a reversion to the full-length CT, which indicates that truncation of the SIV CT may reduce viral fitness in rhesus cells.
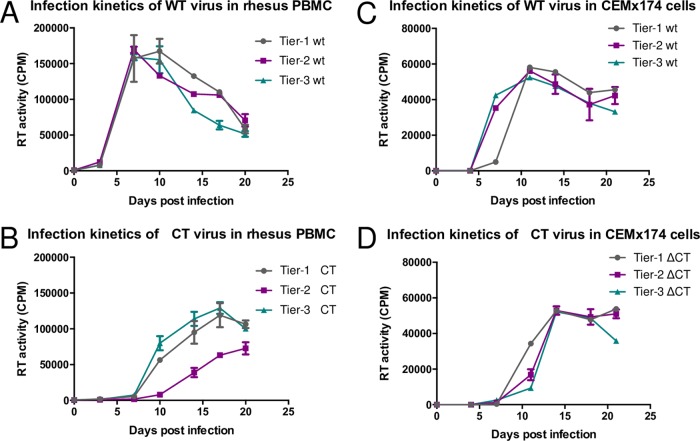
Because of this evidence that a CT truncation can differentially affect infectivity in human and rhesus cells, we compared replications of the wild-type and mutant viruses in phytohemagglutinin (PHA)-stimulated rhesus peripheral blood mononuclear cells (PBMCs) and the human cell line CEMx174. For replication assays in both cell types, the viral input was normalized by the 50% tissue culture infective dose (TCID50) values quantified for TZM-bl cells. In rhesus PBMCs, the tier 1, tier 2, and tier 3 wild-type viruses replicated similarly (Fig. 4A). Each virus reached peak viral production at around days 7 to 10 postinfection, with similar values for virus production at each time point. Compared to the wild-type viruses, the three mutant viruses had delayed kinetics and lower values for virus production in rhesus PBMCs (Fig. 4B). The tier 1 and tier 3 mutants did not reach peak virus production until day 17, and the highest observed value for tier 3 virus production was not observed until day 20. The values of these peaks were lower than those for the wild-type clones, with a 1.5-fold decrease for tier 1 mutant production, a 2.4-fold decrease for tier 2 mutant production, and a 1.2-fold decrease for tier 3 mutant production. These results show that the cytoplasmic tail truncation affected the replication of our viral clones in rhesus PBMCs.
FIG 4.
Kinetics of rhesus PBMC and CEMx174 infection with wild-type (A and C) and mutant (B and D) clones. PBMCs were stimulated with 10% IL-2 and PHA for 72 h and then infected with 10,000 TCID50 of each virus in duplicate. CEMx174 cells were infected with 1,000 TCID50 of each virus in duplicate. Viral supernatants were collected every 3 days, and virus production was quantified by RT activity.
The human cell line that we selected for use in our replication assays is the CEMx174 cell line, a hybrid of human B cell and T cell lines. The wild-type and mutant viruses had similar values for peak virus production in this cell line. However, the mutant viruses reached peak virus production slightly later, at day 14 (Fig. 4D), than did the wild-type viruses, which peaked at day 11 (Fig. 4C). Therefore, truncation of the CT did not appear to enhance the infectivity of our virus clones in human cells but instead seemed to delay the replication kinetics.
DISCUSSION
Modifications to the cytoplasmic tail of SIV Env are commonly used for both structural and vaccine studies (13–19). These modifications are used to increase Env incorporation into the virion, as the Env expression level is low on native SIV particles (6, 12–14), but it is not clear how such modifications affect viral Env antigenicity. In this study, we investigated the effects of a common modification, truncation of the CT, on the neutralization sensitivities of three SIV clones. The three clones were selected for this study because they were all derived from the same parental strain but represent different tier levels of neutralization sensitivity. However, we found that truncation of the cytoplasmic tail caused all three clones to become uniformly resistant to neutralization by both rhesus sera and monoclonal neutralizing antibodies. Furthermore, in contrast to data from previous reports, CT truncation in these strains did not lead to a higher level of Env incorporation into the virions or increased infectivity in human cells.
Previous studies of the neutralization sensitivity of viruses with truncated cytoplasmic tails have yielded mixed results. For several CD4-independent and macrophage-tropic SIVmac strains, truncation of the Env CT is associated with increased sensitivity to neutralization (30–32). Other studies of tail-truncated versions of the glycan mutant SIVmac239-M5 and macrophage-tropic isolate SIVmac316 showed that the truncation of the Env CT decreased virus sensitivity to neutralizing antibodies (7, 33). Unlike previous reports that compared different viral strains (7, 30, 32), we clarified the effects of the Env CT truncation in this study by using three viruses that have high genetic similarity while having disparate neutralization sensitivities. In all of these viruses, truncation of the CT results in decreased sensitivity to neutralization by rhesus sera and MAbs. Rhesus sera collected at different time points differentially neutralized the tier 1, tier 2, and tier 3 wild-type clones. However, when these serum samples were used to test the neutralization sensitivities of the CT-truncated clones, the mutant viruses were highly resistant to neutralization. Similar results were observed with neutralization by MAbs targeting different epitopes; the truncated mutants could not be neutralized by any of the MAbs tested.
The sites of truncation differ among these previous studies of the effect of cytoplasmic tail truncation, but the difference in sites does not account for the observed differences in observed neutralization sensitivities. The site of our truncation, Q741/Q742 in the Env sequence of the parental clone, is analogous to the location of the premature stop codons first identified in SIVmac239 produced from infected human cells (25, 29, 34). Paradoxically, truncation at this site was previously described as being associated with both increased sensitivity (30, 32) as well as decreased sensitivity (33).
Other factors that could contribute to the decreased neutralization sensitivity of cytoplasmic tail-truncated mutants are increased virion Env content and increased infectivity in human cells. Several groups have shown increased Env content with a cytoplasmic tail truncation at the site that we chose (4, 6, 33) as well as at other sites along the tail (5, 7, 12–14, 35). However, when we performed glycoprotein staining to quantify the Env content of our truncated virions, we did not see an increase in the number of Env trimers per virion compared to the wild-type strains. Although the level of Env incorporation can be cell line dependent, we do not think that this factor interfered with our results. Similar SupT1-derived cell lines were used to produce our test viruses as well as the tail-truncated SIVmac239 control, which had an increased number of trimers compared to the wild type. Thus, the level of Env expression does not seem to contribute to the decreased neutralization sensitivity of the CT-truncated viruses in our studies.
We also compared the infectivities of the wild-type and mutant viruses in human cells, as the truncation of the Env CT was first identified as an adaptation to enhance virus replication in human cells. Previously reported evidence indicates that truncation of the Env CT can increase the “inherent infectivity” of viruses in human cells, independent of increases in Env expression levels (7). However, when we used the same human cell line to test the infectivity of the truncated SIVsmE660 viruses, there was no increase in infectivity compared to the wild type.
Our results demonstrate that the decrease in the neutralization sensitivity of our CT-truncated viruses cannot be explained by an increased Env content on the virion or enhanced viral infectivity. One hypothesis that requires further investigation is that a cytoplasmic tail truncation induces a structural change in the extracellular domain of Env so that neutralizing antibodies can no longer effectively bind or neutralize the virus. Previous evidence suggests that some mutations in the cytoplasmic tail of SIV can affect the structure of the external domain of Env (30, 36, 37). Our observation that the truncated viruses cannot be neutralized by several MAbs that target different epitopes, as well as their decreased infectivity in both human and rhesus cells, may indicate a global structural change of the extracellular domain as a result of the CT truncation of Env.
From this study, as well as the work of previous groups, we suspect that the effects of cytoplasmic tail truncations are strain dependent. The observed neutralization sensitivity, Env content, and infectivity of tail-truncated mutants likely depend on the viral genetic background. We were able to control for this strain dependence by studying three genetically similar SIVsmE660 viruses with different neutralization sensitivities. We found that truncation of the cytoplasmic tail of these viruses confers resistance to antibody-mediated neutralization without increased Env contents on the virion or increased infectivity in human cells. These results suggest that modifications of the cytoplasmic tail can have major effects on the antigenicity of the extracellular domain of Envs of some SIVs. Therefore, viruses that contain these modifications should be carefully characterized before use in imaging and vaccine studies because the structure of trimers on the virions of some viruses containing truncated CT domains may not accurately reflect the structure of the corresponding trimers composed of Env subunits with a full-length CT.
MATERIALS AND METHODS
Viruses.
The three SIVsmE660 infectious molecular clones were isolated and constructed as described previously (22). Briefly, sera from chronically infected rhesus macaques inoculated with uncloned SIVsmE660, SIVsmE543-3, SIVmac251, or SIVmac239 were used to characterize the neutralization resistance of SIVsmE660 clones as tier 1 (sensitive to most sera), tier 2 (intermediate), or tier 3 (resistant). We used heat map hierarchical clustering, which was used previously for tier categorization of neutralization sensitivities of HIV-1 isolates, to assess the neutralization sensitivities of these SIVs and selected one virus representative each of tier 1, tier 2, and tier 3. Tier 1 SIVsmE660 clone FL14 was isolated from SIVsmE660 stock virus. The tier 2 SIVsmE660 H807-16w-6 and tier 3 SIVsmE660 H807-24w-4 env sequences were cloned from the plasma of a rhesus macaque infected with the tier 1 SIVsmE660 clone FL14 at 16 and 24 weeks postinfection, respectively.
Env cytoplasmic tail truncation.
To truncate the Env CT domain, two stop codons were introduced into the SIV clones at positions Q741 and Q742 (amino acid numbering based on E660-FL14 Env [GenBank accession number AFM75723]) by PCR mutagenesis with the following set of primers: forward primer 5′-CCTCCCGCTTATGTTTAGTAGATCCCTATCCAC-3′ and reverse primer 5′-GTGGATAGGGATCTACTAAACATAAGCGGGAGG-3′. The presence of the mutations was confirmed by sequencing the Env region of the mutant clones.
Virus stock preparation.
Virus stocks were produced by transfecting 293T cells with full-length wild-type and mutant molecular clones. 293T cells were maintained in Gibco GlutaMAX Dulbecco's modified Eagle medium (DMEM) plus 10% fetal bovine serum (FBS), 100 U/ml penicillin, and 100 μg/ml streptomycin and transfected with 10 μg plasmid using FuGENE 6 transfection reagent (Roche Diagnostics, Indianapolis, IN). Virus stocks were collected from the supernatant of transfected cells after 48 h and filtered with a 0.22-μm filter. The TCID50 values of virus stocks were determined on TZM-bl cells (38) and calculated by the Reed-Muench method (39).
Neutralization assay and serum samples.
The sensitivities of the SIV clones to neutralizing antibodies were evaluated by using the TZM-bl neutralization assay as previously described (24, 40). The serum samples used to compare the neutralizing antibody sensitivities of viruses were collected 4, 40, and 80 weeks after infection from an unrelated rhesus macaque inoculated with SIVsmE660-FL14. The MAbs used in our TZM-bl assay, ITS09.03, ITS52, and ITS01, which target the variable loop 2 (V2), V3, and CD4bs regions of Env, respectively, were provided by Mario Roederer and were previously characterized (26). Briefly, serially diluted, heat-inactivated serum samples or serially diluted MAbs were mixed with 100 TCID50 of transfection-derived viruses and incubated at 37°C for 90 min. After incubation, 104 TZM-bl cells, with DEAE-dextran at a final concentration of 12.5 μg/ml, were added to each well. As controls, cells were cultured alone to measure background fluorescence, and cells were cultured with virus alone to measure the maximum fluorescence of each virus without neutralization. After 40 h of culture at 5% CO2 and 37°C, luciferase activity was measured with a luciferase assay kit (Promega) and read on a Mithras LB940 instrument (Berthold Technologies). The average relative luminescence units (RLU) of the cell controls were subtracted as background. The ID50 and IC50 were calculated with nonlinear regression by using PRISM5. The ID50 is expressed as the highest dilution of sera that resulted in a 50% reduction of RLU compared with the virus control. The IC50 is expressed as the lowest concentration of MAbs that resulted in a 50% reduction of RLU compared with the virus control.
Quantification of Env trimers on the virion.
Dual-color fluorescence imaging of an SDS-PAGE gel of viral proteins was used to quantify virion p27 capsid (CA) and gp120 contents as previously described (27, 28). Briefly, virus stocks of the wild-type and mutant viruses were produced by transfection of 293T cells and used to infect A66-R5 cells (provided by Jim Hoxie, University of Pennsylvania, Philadelphia, PA, USA), a derivative of SupT1 cells that are engineered to express CCR5. The infection-derived test viruses, well-characterized HIV-1 and SIVmac239 control viruses, and a series of gp120 and p27 protein standards were purified and resolved by SDS-PAGE. The gels were stained with two fluorescent dyes: SYPRO ruby (red fluorescence), a total protein stain that detects p27, and Pro-Q Emerald 300 (green fluorescence), a glycoprotein stain that detects gp120. The fluorescence intensities of the gp120 and p27 bands were analyzed by using the VersaDoc 3000 imaging system (Bio-Rad Laboratories). The virion gp27/gp120 ratios were estimated by using the standard curve from the protein standards.
Replication assays.
The infectivities of wild-type and CT-truncated SIV clones were evaluated on rhesus PBMCs and CEMx174 cells. Rhesus PBMCs were isolated from an uninfected macaque and were cultured in complete RPMI 1640 medium (containing 10% FBS, 100 U/ml penicillin, 100 μg/ml streptomycin, 1% HEPES, and 1% l-glutamine) supplemented with 10% interleukin-2 (IL-2) and stimulated with 2 μg/ml PHA for 72 h. After washing with Hanks' balanced salt solution (HBSS), 106 activated PBMCs were infected with 104 TCID50 of the transfection-derived viruses at 37°C for 90 min. The infected PBMCs were washed and cultured in complete RPMI 1640 medium containing 10% IL-2.
CEMx174 cells, a hybrid human T cell/B cell line, were maintained in complete RPMI 1640 medium. The cells were washed with HBSS, and 0.5 × 106 cells were infected with 103 TCID50 of transfection-derived virus at 37°C for 90 min. The infected CEMx174 cells were then washed with HBSS and cultured in complete RPMI 1640 medium. Virus production was monitored by quantifying the reverse transcriptase (RT) activity of the supernatant collected at 3-day intervals. RT values were quantified with a phosphorimaging plate (Fujifilm, Japan).
ACKNOWLEDGMENTS
This work was supported in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. government.
REFERENCES
- 1.Tedbury PR, Freed EO. 2015. The cytoplasmic tail of retroviral envelope glycoproteins. Prog Mol Biol Transl Sci 129:253–284. doi: 10.1016/bs.pmbts.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Postler TS, Desrosiers RC. 2012. The tale of the long tail: the cytoplasmic domain of HIV-1 gp41. J Virol 87:2–15. doi: 10.1128/JVI.02053-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Johnston PB, Dubay JW, Hunter E. 1993. Truncations of the simian immunodeficiency virus transmembrane protein confer expanded virus host range by removing a block to virus entry into cells. J Virol 67:3077–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zingler K, Littman DR. 1993. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein increases Env incorporation into particles and fusogenicity and infectivity. J Virol 67:2824–2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Manrique JM, Celma CC, Affranchino JL, Hunter E, Gonzalez SA. 2001. Small variations in the length of the cytoplasmic domain of the simian immunodeficiency virus transmembrane protein drastically affect envelope incorporation and virus entry. AIDS Res Hum Retroviruses 17:1615–1624. doi: 10.1089/088922201753342022. [DOI] [PubMed] [Google Scholar]
- 6.Yuste E, Reeves JD, Doms RW, Desrosiers RC. 2004. Modulation of Env content in virions of simian immunodeficiency virus: correlation with cell surface expression and virion infectivity. J Virol 78:6775–6785. doi: 10.1128/JVI.78.13.6775-6785.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yuste E, Johnson W, Pavlakis GN, Desrosiers RC. 2005. Virion envelope content, infectivity, and neutralization sensitivity of simian immunodeficiency virus. J Virol 79:12455–12463. doi: 10.1128/JVI.79.19.12455-12463.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pugach P, Ozorowski G, Cupo A, Ringe R, Yasmeen A, de Val N, Derking R, Kim HJ, Korzun J, Golabek M, de Los Reyes K, Ketas TJ, Julien JP, Burton DR, Wilson IA, Sanders RW, Klasse PJ, Ward AB, Moore JP. 2015. A native-like SOSIP.664 trimer based on an HIV-1 subtype B env gene. J Virol 89:3380–3395. doi: 10.1128/JVI.03473-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klasse PJ, Depetris RS, Pejchal R, Julien J-P, Khayat R, Lee JH, Marozsan AJ, Cupo A, Cocco N, Korzun J, Yasmeen A, Ward AB, Wilson IA, Sanders RW, Moore JP. 2013. Influences on trimerization and aggregation of soluble, cleaved HIV-1 SOSIP envelope glycoprotein. J Virol 87:9873–9885. doi: 10.1128/JVI.01226-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanders RW, van Gils MJ, Derking R, Sok D, Ketas TJ, Burger JA, Ozorowski G, Cupo A, Simonich C, Goo L, Arendt H, Kim HJ, Lee JH, Pugach P, Williams M, Debnath G, Moldt B, van Breemen MJ, Isik G, Medina-Ramírez M, Back JW, Koff WC, Julien J-P, Rakasz EG, Seaman MS, Guttman M, Lee KK, Klasse PJ, LaBranche C, Schief WR, Wilson IA, Overbaugh J, Burton DR, Ward AB, Montefiori DC, Dean H, Moore JP. 2015. HIV-1 neutralizing antibodies induced by native-like envelope trimers. Science 349:aac4223. doi: 10.1126/science.aac4223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rerks-Ngarm S, Pitisuttithum P, Nitayaphan S, Kaewkungwal J, Chiu J, Paris R, Premsri N, Namwat C, de Souza M, Adams E, Benenson M, Gurunathan S, Tartaglia J, McNeil JG, Francis DP, Stablein D, Birx DL, Chunsuttiwat S, Khamboonruang C, Thongcharoen P, Robb ML, Michael NL, Kunasol P, Kim JH, MOPH-TAVEG Investigators . 2009. Vaccination with ALVAC and AIDSVAX to prevent HIV-1 infection in Thailand. N Engl J Med 361:2209–2220. doi: 10.1056/NEJMoa0908492. [DOI] [PubMed] [Google Scholar]
- 12.Chertova E, Bess JW Jr, Crise BJ, Sowder IR, Schaden TM, Hilburn JM, Hoxie JA, Benveniste RE, Lifson JD, Henderson LE, Arthur LO. 2002. Envelope glycoprotein incorporation, not shedding of surface envelope glycoprotein (gp120/SU), is the primary determinant of SU content of purified human immunodeficiency virus type 1 and simian immunodeficiency virus. J Virol 76:5315–5325. doi: 10.1128/JVI.76.11.5315-5325.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhu P, Chertova E, Bess J Jr, Lifson JD, Arthur LO, Liu J, Taylor KA, Roux KH. 2003. Electron tomography analysis of envelope glycoprotein trimers on HIV and simian immunodeficiency virus virions. Proc Natl Acad Sci U S A 100:15812–15817. doi: 10.1073/pnas.2634931100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhu P, Liu J, Bess J Jr, Chertova E, Lifson JD, Grise H, Ofek GA, Taylor KA, Roux KH. 2006. Distribution and three-dimensional structure of AIDS virus envelope spikes. Nature 441:847–852. doi: 10.1038/nature04817. [DOI] [PubMed] [Google Scholar]
- 15.Zanetti G, Briggs JA, Grunewald K, Sattentau QJ, Fuller SD. 2006. Cryo-electron tomographic structure of an immunodeficiency virus envelope complex in situ. PLoS Pathog 2:e83. doi: 10.1371/journal.ppat.0020083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.White TA, Bartesaghi A, Borgnia MJ, Meyerson JR, de la Cruz MJ, Bess JW, Nandwani R, Hoxie JA, Lifson JD, Milne JL, Subramaniam S. 2010. Molecular architectures of trimeric SIV and HIV-1 envelope glycoproteins on intact viruses: strain-dependent variation in quaternary structure. PLoS Pathog 6:e1001249. doi: 10.1371/journal.ppat.1001249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.White TA, Bartesaghi A, Borgnia MJ, de la Cruz MJ, Nandwani R, Hoxie JA, Bess JW, Lifson JD, Milne JL, Subramaniam S. 2011. Three-dimensional structures of soluble CD4-bound states of trimeric simian immunodeficiency virus envelope glycoproteins determined by using cryo-electron tomography. J Virol 85:12114–12123. doi: 10.1128/JVI.05297-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu G, Liu J, Roux KH, Taylor KA. 2017. Structure of simian immunodeficiency virus envelope spikes bound with CD4 and monoclonal antibody 36D5. J Virol 91:e00134-. doi: 10.1128/JVI.00134-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sougrat R, Bartesaghi A, Lifson JD, Bennett AE, Bess JW, Zabransky DJ, Subramaniam S. 2007. Electron tomography of the contact between T cells and SIV/HIV-1: implications for viral entry. PLoS Pathog 3:e63. doi: 10.1371/journal.ppat.0030063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen J, Kovacs JM, Peng H, Rits-Volloch S, Lu J, Park D, Zablowsky E, Seaman MS, Chen B. 2015. Effect of the cytoplasmic domain on antigenic characteristics of HIV-1 envelope glycoprotein. Science 349:191–195. doi: 10.1126/science.aaa9804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopker M, Easlick J, Sterrett S, Decker JM, Barbian H, Learn G, Keele BF, Robinson JE, Li H, Hahn BH, Shaw GM, Bar KJ. 2013. Heterogeneity in neutralization sensitivities of viruses comprising the simian immunodeficiency virus SIVsmE660 isolate and vaccine challenge stock. J Virol 87:5477–5492. doi: 10.1128/JVI.03419-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu F, Ourmanov I, Kuwata T, Goeken R, Brown CR, Buckler-White A, Iyengar R, Plishka R, Aoki ST, Hirsch VM. 2012. Sequential evolution and escape from neutralization of simian immunodeficiency virus SIVsmE660 clones in rhesus macaques. J Virol 86:8835–8847. doi: 10.1128/JVI.00923-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Seaman MS, Janes H, Hawkins N, Grandpre LE, Devoy C, Giri A, Coffey RT, Harris L, Wood B, Daniels MG, Bhattacharya T, Lapedes A, Polonis VR, McCutchan FE, Gilbert PB, Self SG, Korber BT, Montefiori DC, Mascola JR. 2010. Tiered categorization of a diverse panel of HIV-1 Env pseudoviruses for assessment of neutralizing antibodies. J Virol 84:1439–1452. doi: 10.1128/JVI.02108-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Montefiori DC. 2004. Evaluating neutralizing antibodies against HIV, SIV, and SHIV in luciferase reporter gene assays. Curr Protoc Immunol Chapter 12:Unit 12.11. doi: 10.1002/0471142735.im1211s64. [DOI] [PubMed] [Google Scholar]
- 25.Hirsch VM, Edmondson P, Murphey-Corb M, Arbeille B, Johnson PR, Mullins JI. 1989. SIV adaptation to human cells. Nature 341:573–574. doi: 10.1038/341573a0. [DOI] [PubMed] [Google Scholar]
- 26.Mason RD, Welles HC, Adams C, Chakrabarti BK, Gorman J, Zhou T, Nguyen R, O'Dell S, Lusvarghi S, Bewley CA, Li H, Shaw GM, Sheng Z, Shapiro L, Wyatt R, Kwong PD, Mascola JR, Roederer M. 2016. Targeted isolation of antibodies directed against major sites of SIV Env vulnerability. PLoS Pathog 12:e1005537. doi: 10.1371/journal.ppat.1005537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Louder MK, Sambor A, Chertova E, Hunte T, Barrett S, Ojong F, Sanders-Buell E, Zolla-Pazner S, McCutchan FE, Roser JD, Gabuzda D, Lifson JD, Mascola JR. 2005. HIV-1 envelope pseudotyped viral vectors and infectious molecular clones expressing the same envelope glycoprotein have a similar neutralization phenotype, but culture in peripheral blood mononuclear cells is associated with decreased neutralization sensitivity. Virology 339:226–238. doi: 10.1016/j.virol.2005.06.003. [DOI] [PubMed] [Google Scholar]
- 28.Chertova E, Chertov O, Coren LV, Roser JD, Trubey CM, Bess JW, Sowder RC, Barsov E, Hood BL, Fisher RJ, Nagashima K, Conrads TP, Veenstra TD, Lifson JD, Ott DE. 2006. Proteomic and biochemical analysis of purified human immunodeficiency virus type 1 produced from infected monocyte-derived macrophages. J Virol 80:9039–9052. doi: 10.1128/JVI.01013-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kodama T, Wooley DP, Naidu YM, Kestler HW, Daniel MD, Li Y, Desrosiers RC. 1989. Significance of premature stop codons in Env of simian immunodeficiency virus. J Virol 63:4709–4714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vzorov AN, Gernert KM, Compans RW. 2005. Multiple domains of the SIV Env protein determine virus replication efficiency and neutralization sensitivity. Virology 332:89–101. doi: 10.1016/j.virol.2004.10.044. [DOI] [PubMed] [Google Scholar]
- 31.Bonavia A, Bullock BT, Gisselman KM, Margulies BJ, Clements JE. 2005. A single amino acid change and truncated TM are sufficient for simian immunodeficiency virus to enter cells using CCR5 in a CD4-independent pathway. Virology 341:12–23. doi: 10.1016/j.virol.2005.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Puffer BA, Pohlmann S, Edinger AL, Carlin D, Sanchez MD, Reitter J, Watry DD, Fox HS, Desrosiers RC, Doms RW. 2002. CD4 independence of simian immunodeficiency virus Envs is associated with macrophage tropism, neutralization sensitivity, and attenuated pathogenicity. J Virol 76:2595–2605. doi: 10.1128/JVI.76.6.2595-2605.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kuwata T, Takaki K, Enomoto I, Kazuhisa Y, Matsushita S. 2013. Increased infectivity in human cells and resistance to antibody-mediated neutralization by truncation of the SIV gp41 cytoplasmic tail. Front Microbiol 4:117. doi: 10.3389/fmicb.2013.00117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chakrabarti L, Guyader M, Alizon M, Daniel MD, Desrosiers RC, Tiollais P, Sonigo P. 1987. Sequence of simian immunodeficiency virus from macaque and its relationship to other human and simian retroviruses. Nature 328:543–547. doi: 10.1038/328543a0. [DOI] [PubMed] [Google Scholar]
- 35.Vzorov AN, Compans RW. 1996. Assembly and release of SIV Env proteins with full-length or truncated cytoplasmic domains. Virology 221:22–33. doi: 10.1006/viro.1996.0349. [DOI] [PubMed] [Google Scholar]
- 36.Vzorov AN, Compans RW. 2000. Effect of the cytoplasmic domain of the simian immunodeficiency virus envelope protein on incorporation of heterologous envelope proteins and sensitivity to neutralization. J Virol 74:8219–8225. doi: 10.1128/JVI.74.18.8219-8225.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Spies CP, Ritter GD Jr, Mulligan MJ, Compans RW. 1994. Truncation of the cytoplasmic domain of the simian immunodeficiency virus envelope glycoprotein alters the conformation of the external domain. J Virol 68:585–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wei X, Decker JM, Liu H, Zhang Z, Arani RB, Kilby JM, Saag MS, Wu X, Shaw GM, Kappes JC. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob Agents Chemother 46:1896–1905. doi: 10.1128/AAC.46.6.1896-1905.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Reed LJ, Muench H. 1938. A simple method of estimating fifty per cent endpoints. Am J Epidemiol 27:493–497. doi: 10.1093/oxfordjournals.aje.a118408. [DOI] [Google Scholar]
- 40.Ourmanov I, Kuwata T, Goeken R, Goldstein S, Iyengar R, Buckler-White A, Lafont B, Hirsch VM. 2009. Improved survival in rhesus macaques immunized with modified vaccinia virus Ankara recombinants expressing simian immunodeficiency virus envelope correlates with reduction in memory CD4+ T-cell loss and higher titers of neutralizing antibody. J Virol 83:5388–5400. doi: 10.1128/JVI.02598-08. [DOI] [PMC free article] [PubMed] [Google Scholar]