Abstract
The cyclic lipodepsipeptides WAP-8294A are antibiotics with potent activity against methicillin-resistant Staphylococcus aureus (MRSA). One member of this family, WAP-8294A2 (Lotilibcin), was in clinical trials due to its high activity and distinct chemistry. However, WAP-8294A compounds are produced in a very low yield by Lysobacter and only under very stringent conditions. Improving WAP-8294A yield has become very critical for research and application of these anti-MRSA compounds. Here, we report a strategy to increase WAP-8294A production. We first used the CRISPR/dCas9 system to increase the expression of five cotranscribed genes (orf1–5) in the WAP gene cluster, by fusing the omega subunit of RNA polymerase with dCas9 that targets the operon’s promoter region. This led to the transcription of the genes increased by 5–48 folds in strain dCas9-ω3. We then refactored four putative self-protection genes (orf6, orf 7, orf 9 and orf10) by reorganizing them into an operon under the control of a strong Lysobacter promoter, PHSAF. The refactored operon was introduced into strain dCas9-ω3, and the transcription of the self-protection genes increased by 20–60 folds in the resultant engineered strains. The yield of the three main WAP-8294A compounds, WAP-8294A1, WAP-8294A2, and WAP-8294A4, increased by 6, 4, and 9 folds, respectively, in the engineered strains. The data also showed that the yield increase of WAP-8294A compounds was mainly due to the increase of the extracellular distribution. WAP-8294A2 exhibited potent (MIC 0.2–0.8 μg/mL) and specific activity against S. aureus among a battery of clinically relevant Gram-positive pathogens (54 isolates).
Keywords: Lysobacter, natural products, antibiotics, WAP-8294A, CRISPR/dCas9, gene refactoring
Graphical Abstract
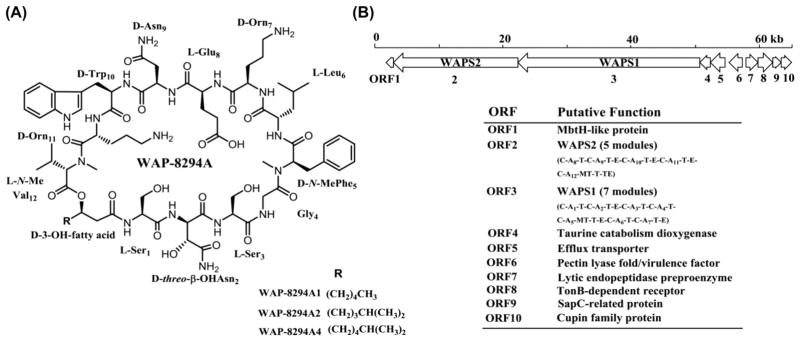
Lysobacter is a genus of Gram-negative gliding bacteria that were classified in 1978.1 Although it is a relatively new genus of bacteria, Lysobacter have emerged as new biocontrol agents against plant diseases because many species in the genus are prolific producers of bioactive natural products. These natural products often show interesting features in both their chemical structures and biological activities, such as the antifungal antibiotics HSAF and the potent anti-MRSA WAP-8294A compounds from L. enzymogenes OH11,2,3 and the phenazine-type antibiotics from L. antibioticus OH13.4 WAP-8294A compounds are a group of cyclic lipodepsipeptides consisting of 19 members, with WAP-8294A1, WAP-8294A2 and WAP-8294A4 being the main products. Among them, WAP-8294A2 (Lotilibcin) reached clinical studies (Figure 1A).5,6 These three compounds together with WAP-8294Ax8, WAP-8294Ax9 and WAP-8294Ax13 are active against MRSA in vitro.6 The cyclic lipodepsipeptides are biosynthesized by two giant nonribosomal peptide synthetases (NRPS), WAPS1 and WAPS2, and several tailoring enzymes.3 A number of putative transporters and regulators are also present in the biosynthetic gene cluster. However, L. enzymogenes OH11 only produces a minute amount of WAP-8294A compounds, which has hampered their structure-and-activity studies and therapeutic applications. The very low yield might be a mechanism of self-protection for L. enzymogenes OH11, considering the extremely potent activity of WAP-8294A compounds. The mode of WAP-8294A’s action is not clear, but the antibiotic activity of these compounds is selective to Gram-positive bacteria, such as Staphylococcus aureus. As Gram-negative species, Lysobacter are not sensitive to WAP-8294A treatment, most probably due to the outer membranes (OM) special to Gram-negative bacteria. The naturally low OM-permeability of Gram-negative bacteria provides the cell with an effective permeability barrier against external noxious agents, including antibiotics.7,8 However, L. enzymogenes OH11 is the producer of WAP-8294A compounds, and its outer membranes are no longer a barrier for the action of the antibiotics. Thus, it is reasonable to assume that if we increase expression of the genes related to self-protection (such as the efflux pumps in the gene cluster), in conjunction with overexpression of the key biosynthetic genes, the productivity of WAP-8294A in strain OH11 could be improved.
Figure 1.
Chemical structure of the main WAP-8294A compounds (A) and the WAP biosynthetic gene cluster (B) from L. enzymogenes OH11.
So far, very little genetic engineering has been conducted in Lysobacter species due to the limited knowledge about the biology of this relatively new group of bacteria and very few genetic tools available to them. The CRISPR/Cas9 system, as a novel genome editing tool, has been employed in numerous organisms including bacteria.9–12 The system contains three important components, Cas9 which is an RNA-guided double-stranded DNA nuclease, CRISPR RNA (crRNA) and trans-activating crRNA (tracrRNA).9,13 A 20–30 bp region in the crRNA that is complementary to the target DNA in the host is called the spacer.13,14 Guided by the spacer sequence, the system recognizes the NGG protospacer-adjacent motif (PAM) on the chromosomes and binds to the complementary region, followed by a double strand break at the target sequence induced by Cas9.15 This break usually triggers an automatic gene repair mechanism known as error-prone nonhomologous ending joining (NHEJ) which introduces loss-function mutations to the target gene in the repaired host chromosome.9,13,14,16 In addition, this system is an efficient tool for gene activation and repression without disrupting the host genome.17 Mutations in the catalytic domains of Cas9 abolish its cleavage activity as a nuclease but retain DNA binding ability,14 which was referred as “dead” Cas9 (dCas9).17 The CRISPR/dCas9 system was exploited to engineer a programmable transcription regulator. Using GFP report gene in E. coli and β-galactosidase gene in Streptococcus pneumonia, Bikard et al. showed that the system could achieve the transcription repression by targeting dCas9 to promoter regions or open reading frames, presumably through preventing the binding of RNA polymerase or blocking transcription elongation.17 dCas9 could also function as transcription activator by fusing with the omega subunit of RNA polymerase and directing it to promoter regions of weakly expressed genes. The idea was to use the omega subunit to recruit RNA polymerase to the promoter and thus to enhance expression.17 They tested ten spacers that targeted positions on both DNA strands with different distances from the promoter of GFP report gene and found this strategy could strongly activate the gene expression.17
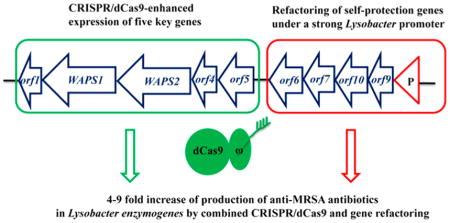
In this study, we applied the CRISPR/dCas9 system to Lysobacter for the first time to enhance the expression of the WAP-8294A production genes. We also refactored four putative self-protection genes into a single operon and put them under the control of a strong Lysobacter promoter. The results demonstrate that the combined use of CRISPR/dCas9-mediated gene expression and refactoring of self-protection gene is an effective approach to yield improvement of the anti-MRSA antibiotics WAP-8294A in L. enzymogenes OH11.
RESULTS AND DISCUSSION
Transcription Activation of WAP Core Genes Using CRISPR/dCas9-ω
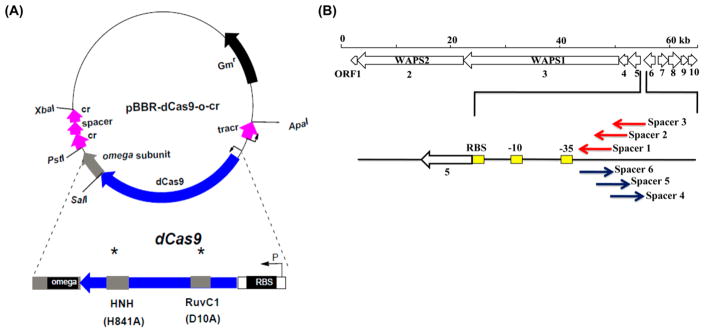
To enhance the expression of WAP-8294A biosynthetic genes, we adopted the CRISPR/dCas9 system to L. enzymogenes OH11. The strategy was based on a previously reported system,17 which contains an inactivated Cas9 gene (dCas9) that is fused with the omega subunit of RNA polymerase (Figure S1). Five core genes (orf1–orf5, including the two NRPS genes, WAPS1 and WAPS2) in the WAP cluster forms an operon, sharing a putative promoter located at the upstream of orf5 (Figure 1B). Previously, it had been shown in other bacteria that dCas9-ω fusion enhanced gene expression when it bound to a sequence with an optimal distance from the promoter, and the activation level depends on the optimal distance away from the −35 promoter element.17 Because the dCas9-ω fusion had never been tested in any Lysobacter species, no information for optimal distance was available. Therefore, six spacers (each followed by a NGG PAM) with various distances from the −35 promoter element were designed, in order to find an optimal spacer. Spacers 1–3 targeted the coding strand and spacers 4–6 targeted the noncoding strand (Figure 2 and Figure S2). All constructs were introduced into L. enzymogenes OH11 and confirmed by PCR (Figure S3 and S4). Spacer 1-containing strain was barely able to grow after it was incubated in 1/10 TSB medium, so this strain was not used in the subsequent study.
Figure 2.
Construction of a gene activation vector in L. enzymogenes OH11 using the CRISPR/dCas9-ωsystem. (A) CRISPR/dCas9-ωbased gene activation system. cr, CRISPR RNA; tracr, trans-activating crRNA. (B) Position of the six spacers used in the CRISPR/dCas9-ωsystem.
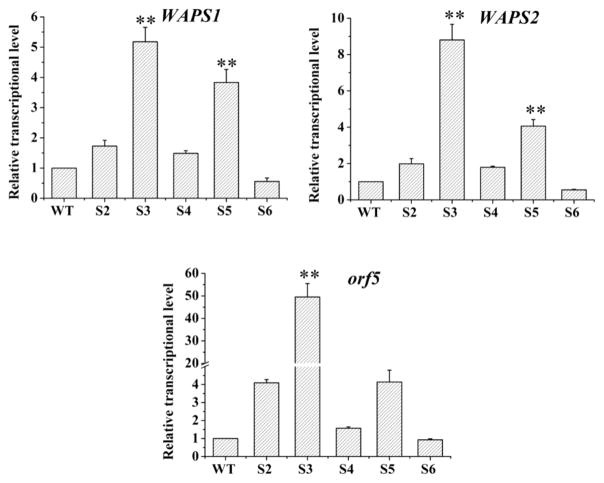
The transcription level of three representative genes, WAPS1, WAPS2 and orf5, was examined using qRT-PCR. For WAPS1 gene, four (spacers 2, 3, 4, 5) of the five strains gave a higher expression level (1.5–5.2 fold) than the wild type (WT), and one strain (spacer 6) had a lower expression level (0.5 fold) than the wild type (Figure 3). For WAPS2 gene, four (spacers 2, 3, 4, 5) of the five strains gave a higher expression level (1.8–8.8 fold) than the wild type, and one strain (spacer 6) had a lower expression level (0.6 fold) than the wild type (Figure 3). A similar pattern was observed for orf5 gene, the same four strains (spacers 2, 3, 4, 5) gave a higher expression level (1.6–48.8 fold) than the wild type, and one strain (spacer 6) had a slightly lower level (0.9) fold than the wild type (Figure 3). In all cases, the spacer-3 containing strain showed the highest expression for WAPS1, WAPS2 and orf5. Together, the results indicate that spacer-3 is likely located at an optimal position for the CRISPR/dCas9-ω system to enhance the WAP gene expression. Targeting this location of the operon’s promoter could have made the dCas9-fused omega subunit to occupy a proper position for interaction with the RNA polymerase complex.17 This interaction in turn led to a strong expression of the target genes in L. enzymogenes OH11.
Figure 3.
Transcriptional activation of WAP genes by the CRISPR/dCas9-ω system in L. enzymogenes OH11. Transcriptional levels of three key genes (WAPS1, WAPS2 and orf5) for WAP-8294A compounds biosynthesis and export were measured. WT, wild type strain; S2, S3, S4, S5, S6, the strains targeted by spacer 2, spacer 3, spacer 4, spacer 5 and spacer 6, respectively. Data are presented as averages of three independent experiments each conducted in triplicate. *, P < 0.05; **, P < 0.01 (see Table S4).
Refactoring of Self-Protection Genes under the Control of a Strong Lysobacter Promoter PHSAF
Self-protection is an important requirement for the antibiotic-producers, which includs the efflux of antibiotics, antibiotics inactivation and modification of the antibiotic targets.18 In WAP gene cluster, half (five) of the genes are predicted to function in self-protection, including three transport-related genes (orf5, orf 9 and orf10) and two resistance genes (orf6 and orf 7).3 This unusually high portion of self-protection-related genes in the gene cluster seems to imply that WAP-8294A compounds could be very toxic to the producer Lysobacter itself. The previous result had shown that a disruption of orf6 led to loss of antibacterial activity and WAP-8294A production,3 which supports the notion that self-protection is essential for Lysobacter to produce these compounds. Among the self-protection related genes, orf5 encodes an efflux transporter, orf6 encodes a protein homologous to pectin lyase fold/virulence factor that is related to peptidases, the product of orf 7 belongs to a family of bacterial extracellular metallo endopeptidases, orf 9 encodes a SapC-related protein that is related to peptide transport,19,20 and the product of orf10 belongs to the cupin superfamily with diverse functions.21 We therefore suspected that an increase of transcription of these genes may lead to an increase of WAP-8294A yield due to an enhanced self-protection. The transcription of orf5 had been significantly increased using the CRISPR/dCas9 system (Figure 3), and in this part of work we focused on the other four genes. In the native WAP gene cluster, orf6, orf 7, orf 9 and orf10 appear to belong to three different transcriptinal units (Figure 1B). So we tried to refactor the genes by rearranging them into a single operon under the control of the same promoter, PHSAF, which is a strong promoter controlling HSAF production in L. enzymogenes OH11 (Figure 4A).22 The constructed plasmid pHmgA-P::WAP containing this operon (Figure S5) was introduced into the wild type, as well as the spacer-3 containing dCas9-ω3 strain, and the resulted engineered strains (WT/refactored or dCas9-ω3/refactored) were examined for gene expression and compound production (Figure 4B and Figure S6).
Figure 4.
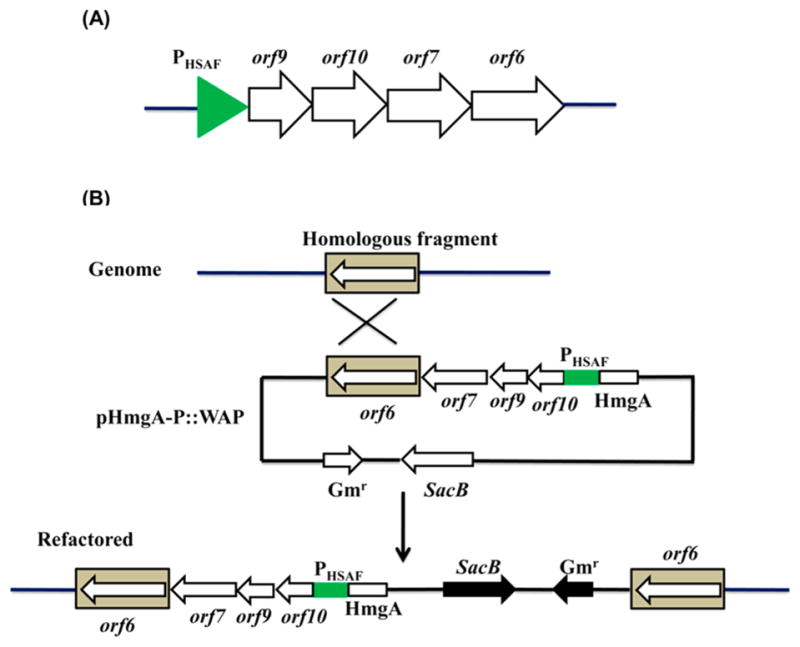
Refactoring of four self-protection genes in the WAP gene cluster. (A) Reconstruction of orf6, orf 7, orf 9 and orf10 of the WAP gene cluster into a single operon under the control of a strong Lysobacter promoter, PHSAF.22 (B) Homologous recombination of the plasmid with the WAP locus in the genome of Lysobacter enzymogens OH11.
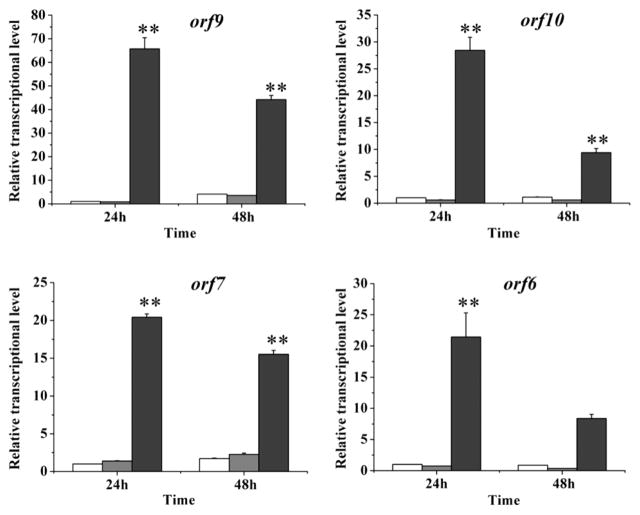
qRT-PCR results showed that the transcription of orf 9, orf10, orf 7 and orf6 increased by 65, 28, 20, and 21 fold when compared to the wild type strain, while the transcription of these genes did not change much in strain dCas9-ω3 (Figure 5). The result showed that promoter PHSAF successfully enhanced the expression of the four refactored genes (orf6, orf 7, orf 9 and orf10) in the engineered strain. To make sure the introduction of the refactored genes and the CRISPR/dCas9 system did not affect the growth of the host, we examined the growth rate of the Lysbobacter strains. The results showed that all strains had a similar growth rate, with strain dCas9-ω3 and strain dCas9-ω3/refactored slightly lower than the others (Figure S7).
Figure 5.
Transcriptional enhancement of the WAP genes in the engineered strains. Transcriptional level of orf 9, orf10, orf 7 and orf6 was measured in WT and the engineered strains. The white columns represent the gene transcriptional level in WT, the gray columns represent the gene transcriptional level in strain dCas9-ω3, and the dark gray columns represent the gene transcriptional level in strain dCas9-ω3/refactored. Data are presented as averages of three independent experiments each conducted in triplicate. **, P < 0.01 (see Table S5).
Yield Improvement of the WAP-8294A Compounds in Engineered Strains
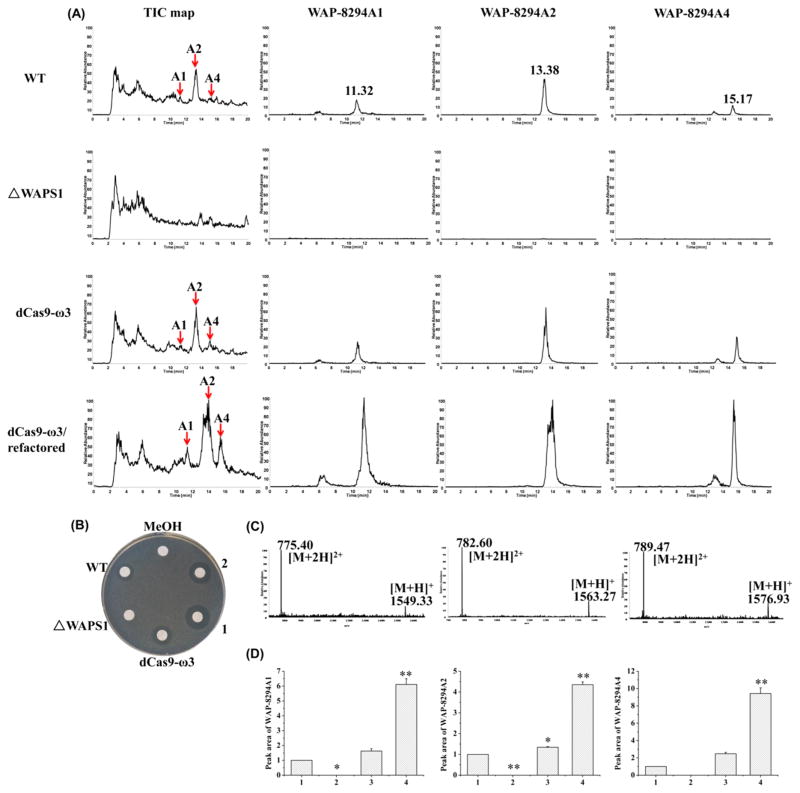
LC–MS was performed to determine the production of WAP-8294A compounds. In the extracts from the wild type, WAP-8294A1 had a retention time of 11.32 min with m/z of 1549.33 for [M + H]+ and 775.40 for [M + 2H]2+, WAP-8294A2 had a retention time of 13.38 min with m/z of 1563.27 for [M + H]+ and 782.60 for [M + 2H]2+, and WAP-8294A4 had a retention time of 15.17 min with m/z of 1576.93 for [M + H]+ and 789.47 for [M + 2H]2+ (Figure 6A and 6C). These three compounds, WAP-8294A1, WAP-8294A2 and WAP-8294A4, were also clearly produced in strain dCas9-ω3 and strain dCas9-ω3/refactored (Figure 6A). Strain ΔWAPS1 (disruption mutant of WAPS1, a gene encoding nonribosomal peptide synthetase) was used as a negative control.3 None of the WAP-8294A compounds was detected in this strain.
Figure 6.
Analysis of WAP-8294A compounds by LC–MS. (A) The yield of WAP-8294A1, WAP-8294A2 and WAP-8294A4 in WT and engineered strains. TIC, Total Ions Chromatograph. (B) Antibacterial activity test against B. subtilis. MeOH, methanol; 1, 2, two strains of dCas9-ω3/refactored. (C) The m/z of WAP-8294A1, WAP-8294A2 and WAP-8294A4. (D) Peak area of WAP-8294A compounds in WT and engineered strains. 1, WT; 2, ΔWAPS1; 3, dCas9-ω3; 4, dCas9-ω3/refactored. Data are presented as averages of three independent experiments. *, P < 0.05; **, P < 0.01 (see Table S6).
Next, the WAP-8294A extracts were tested for antibacterial activity to see if the engineered strains possess enhanced activity. The results showed that the extracts from strain dCas9-ω3/refactored exhibited clearly better activity against Bacillus subtilis than that from the wild type or strain dCas9-ω3 (Figure 6B). To quantify the yield of each of the three compounds, the peak areas of each of the extracted ions corresponding to WAP-8294A1, WAP-8294A2 and WAP-8294A4 were calculated (Figure 6A and 6D). A cyclic peptide (lysobactin) was used as external standard for quantification (Figure S8). WAP-8294A1 yield in strain dCas9-ω3 and strain dCas9-ω3/refactored increased 1.6 and 6 fold, respectively, compared to that in the wild type, WAP-8294A2 yield in strain dCas9-ω3 and strain dCas9-ω3/refactored increased 1.4 and 4 fold, respectively, compared to that in the wild type, and WAP-8294A4 yield in strain dCas9-ω3 and strain dCas9-ω3/refactored increased 2 and 9 fold, respectively, compared to that in the wild type. In all cases, the production of WAP-8294A compounds increased slightly in the strain containing the CRISPR/dCas9-ω3 system. When the CRISPR/dCas9-ω3 system was combined with the refactored genes, the yield of the compounds was improved significantly. The results demonstrate that an enhanced expression of the genes for WAP-8294A biosynthesis and self-protection in L. enzymogenes OH11 can lead to an enhanced production of the antibiotic compounds. However, the yield of WAP-8294A compounds in WT/refactored strain was almost the same as those in WT (Figure S9), indicating that only overexpressing the self-protection genes was not sufficient for the observed yield improvement of WAP-8294A compounds. In addition to the three main compounds, the minor compounds (WAP-8294Ax8, WAP-8294Ax9, and WAP-8294Ax13) seemed to become clearly detectable in the engineered strain, dCas9-ω3/refactored. WAP-8294Ax8 and WAP-8294Ax9 have the same mass as WAP-8294A1, and there were additional peaks around 6 min retention time when the m/z of WAP-8294A1 was extracted (Figure 6A). Similarly, WAP-8294Ax13 has the same as mass as WAP-8294A4, and there was an additional peak around 13 min retention time when the m/z of WAP-8294A4 was extracted (Figure 6A). The results showed that the yield of the minor compounds (WAP-8294Ax8/9/13) increased in the engineered strain in the similar trend as the main compounds (WAP-8294A1/2/4).
Distribution of WAP-8294A Compounds in the Culture
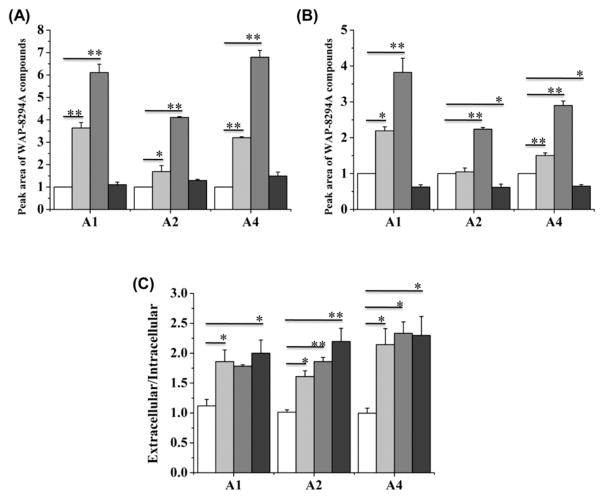
In the WAP gene cluster, orf5, orf 9 and orf10 may be related to the transport of the antibiotic compounds from inside Lysobacter cells to the outside environment, which is a common way of self-protection. Thus, the enhanced expression of these genes may change the distribution of extracellular and intracellular WAP-8294A compounds. We separately extracted the extracellular and intracellular WAP-8294A compounds and quantified them separately. All three WAP compounds, WAP-8294A1, WAP-8294A2 and WAP-8294A4, could be detected in the supernatant and in the cells (Figure S10). In the wild type, the extracellular and intracellular WAP-8294A compounds appeared to have an almost equal distribution (Figure 7C and Figure S11). In strain dCas9-ω3, the amount of extracellular WAP-8294A1, WAP-8294A2 and WAP-8294A4 increased by 3.6, 1.7, and 3.2 fold, respectively, when compared to the wild type (Figure 7A and Figure S11). In strain dCas9-ω3/refactored, the amount of extracellular WAP-8294A1, WAP-8294A2 and WAP-8294A4 increased by 6.1, 4.1, and 6.8 fold, respectively, when compared to the wild type (Figure 7A and Figure S11). In strain WT/refactored, the amount of extracellular WAP-8294A1, WAP-8294A2 and WAP-8294A4 increased only slightly, when compared to the wild type (Figure 7A and Figure S11). In contrast to the significant increase of the extracellular WAP-8294A compounds, the change of the intracellular compounds appeared much smaller. In strain dCas9-ω3, the amount of intracellular WAP-8294A1, WAP-8294A2 and WAP-8294A4 increased by 2.2, 1.2, and 1.5 fold, respectively, when compared to the wild type (Figure 7B and Figure S11). In strain dCas9-ω3/refactored, the amount of intracellular WAP-8294A1, WAP-8294A2 and WAP-8294A4 increased by 3.8, 2.2, and 2.9 fold, respectively, when compared to the wild type (Figure 7B and Figure S11). Interestingly, in strain WT/refactored, the amount of intracellular WAP-8294A1, WAP-8294A2 and WAP-8294A4 decreased slightly, when compared to the wild type (Figure 7B and Figure S11). In strain dCas9-ω3, the extracellular/intracellular ratio of the WAP-8294A compounds is clearly higher than that of WT (Figure 7C), suggesting that the efflux pump encoded by orf5 that was overexpressed in strain dCas9-ω3 is involved in export of WAP-8294A compounds. In addition, the extracellular/intracellular ratio of the WAP-8294A compounds in strain WT/refactored is also higher than that of WT (Figure 7C), indicating that orf 9 and orf10 may also contribute to the WAP-8294A export. Together, the results show that, while the extracellular and intracellular compounds distributed almost equally in the wild type, the enhanced expression of the biosynthetic genes and self-protection genes not only increased the overall yield of the WAP-8294A compounds, but also changed the distribution of the compounds. The increased extracellular distribution is the main reason for the overall increased yield of these compounds in strain dCas9-ω3 and strain dCas9-ω3/refactored.
Figure 7.
Peak area of extracellular WAP-8294A compounds (A) and intracellular WAP-8294A compounds (B). (C) The peak area ratio of extracellular WAP-8294A compounds versus intracellular WAP-8294A compounds. The white columns represent the compounds in WT, the light gray columns represent the compounds in dCas9-ω3, the gray columns represent the compounds in dCas9-ω3/refactored, and the dark gray columns represent the compounds in WT/refactored. A1, A2 and A4 represent WAP-8294A1, WAP-8294A2 and WAP-8294A4, respectively. Data are presented as averages of three independent experiments. *, P < 0.05; **, P < 0.01 (see Tables S7–S9).
Test of WAP-8294A2 Efficacy
To test the efficacy of the compounds, WAP-8294A2, the most abundant member in the family, was prepared from the Lysobacter culture. A group of clinically relevant Gram-positive pathogens, including Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Bacillus cereus, Listeria monocytogenes, Streptococcus pyogenes, Streptococcus agalactiae, and Streptococcus pneumoniae, were used in the test of WAP-8294A2 efficacy. The results demonstrated that the activity of WAP-8294A2 is rather potent and specific for S. aureus, with MICs in the range of 0.2–0.8 μg/mL against 10 isolates, including MRSA and daptomycin-resistant isolates (Table S3). Strong activity, with MICs in the range of 0.8–1.6 μg/mL, was also observed in the test against 5 isolates of S. epidermidis. The MICs increased to 3.2–6.4 μg/mL for 5 isolates of L. monocytogenes and >12.8 μg/mL for 2 isolates of B. cereus (but with 0.4 μg/mL for one undefined Bacillus isolate) (Table S3). High MIC results, 25.6–51.2 μg/mL and above, were noted for all other species (a total of 31 isolates, including vancomycin-resistant Enterococci, VRE, and daptomycin-resistant isolates).
Summary and Concluding Remarks
The results from this study show that the combined use of CRISPR/dCas9-ω3 and self-protection gene refactoring is an effective approach for yield improvement of this group of very potent anti-MRSA WAP-8294A compounds in L. enzymogenes OH11. As a relatively new genus of environmental bacteria, Lysobacter species have not been subjected to genetic manipulations by synthetic biology methods commonly used in other organisms. In this work, we achieved 5–48 fold increase in gene transcription levels upon introducing the CRISPR/dCas9-ω3 system and 20–65 fold increase upon refactoring self-protection genes in the WAP gene cluster. The resultant strains produced a 4–9 fold higher amount of WAP-8294A compounds than the starting strain. WAP-8294A2, the most abundant compound produced by the strains, exhibited potent and selective activity against S. aureus among a panel of clinically relevant pathogens of Gram-positive bacteria.
The method described in this work should be generally applicable to other species in genus Lysobacter, which exhibit a great potential as new sources of bioactive compounds.23 So far, at least five Lysobacter genomes have been reported, and most of the genomes contain multiple (12–16) gene clusters for natural product biosynthesis.24,25 Many of the gene clusters are silent, and other clusters produce compounds in a tiny amount and/or only under very special conditions. The development of the combined use of the CRISPR/dCas9-ω system and the strategy of gene refactoring in L. enzymogenes may provide an effective way to exploitation of the large number of unexplored gene clusters in other Lysobacter species.
METHODS
Bacterial Strains, Plasmids and Growth Conditions
Bacterial strains and plasmids used in this study are shown in Table S1. Luria–Bertani (LB) broth medium and 1/10 Tryptic Soy Broth (TSB) were used for the growth of L. enzymogenes OH11 (CGMCC No. 1978).26 Fermentation medium (2.5% glucose, 2% soy-bean flour, 0.4% soybean oil, 0.25% NaCl, 0.5% CaCO3; pH 7.2) was used for production of WAP-8294A compounds and RNA extraction. Vector pBBR1-MCS5 (Gmr) and pHmgA-P were used for gene expression in L. enzymogenes OH11.22,27 Plasmid pWJ66 that contains crRNA and dCas9 was purchased from Addgene (Cambridge, MA). B. subtilis was used as the indicator bacterium of WAP-8294A production. Escherichia coli strain XL-1 Blue was cultured at 37 °C in LB medium supplemented with gentamicin (50 μg/mL) to propagate plasmids.
DNA Manipulation, Sequencing and Lysobacter Transformation
Chromsomal DNA and plasmids were isolated from L. enzymogenes OH11 or E. coli according to the standard techniques.28 DNA sequencing was performed by Eurofins MWG Operon LLC. Database searching and sequence analysis were performed using the online program PSI-BLAST.29 Plasmids were transformed into L. enzymogenes OH11 by electroporation using Eppendorf Eporator (Eppendorf North America, Inc.). After electroporation, the cells were allowed to incubate in 700 μL LB medium at 30 °C for 3 h, then spread all cells on LB plates (containing gentamicin) at 30 °C for 72 h.
Primers and PCR
All primers used in this study are listed in Table S2. PCRs were carried out using Phusion High-Fidelity DNA polymerase (Thermo Scientific) or TaqDNA polymerase (Thermo Scientific). For Phusion DNA polymerase, an initial denaturation at 98 °C for 30s was followed by 30 cycles of amplification (98 °C for 10 s, 60 °C for 15 s, 72 °C for 1 min), and additional 5 min at 72 °C. For Taq DNA polymerase, an initial denaturation at 95 °C for 3 min was followed by 30 cycles of amplification (95 °C for 30 s, 60 °C for 30 s, 72 °C for 1 min), and additional 10 min at 72 °C. Considering different DNA templates and primers, the annealing temperature and the elongation time were changed in some cases.
Generation of Strains dCas9-ω from L. enzymogenes OH11
DNA fragments, including subunit ω, dCas9-tracr, and cr-repeat-cr, were amplified by PCR using plasmid pWJ66 as the template (Figure S1).17 The primers used for the PCR were listed in Table S2. The spacer region of cr-spacer-cr fragments was a 30-nt DNA that was designed to bind six different positions in the promoter region of the WAP operon (upstream of orf5) (Figure S2). These guide RNAs, each containing a different 30-nt spacer (spacers 1–6), were generated by separate PCR to produce cr-spacer and spacer-cr using primers in Table S2. Then cr-spacer and spacer-cr were used as the template for overlapping PCR to obtain complete cr-spacer-cr. The fragments of ω, dCas9-tracr, and cr-spacer-cr were cloned into plasmid pBBR1-MCS5 (Gmr), to generate six pB-dCas9-o-cr constructs, which were confirmed by PCR or double digestion (Figure S3). The constructs were transformed into L. enzymogenes OH11 by electroporation, the transformants (dCas9-ω1-dCas9-ω6) were selected on the LB plates (25 μg/mL gentamicin) and verified by PCR using primers listed in Table S2 (Figure S4).
Generation of Strains dCas9-ω3/Refactored from Strain dCas9-ω3
The orf 7 DNA fragment containing its putative ribosome binding sequence (RBS) and the entire open reading frame (ORF) was amplified using primers ORF7-up and ORF7-down. Then the amplified DNA fragment was inserted into the BamHI/XhoI sites of pHmgA-P,22 giving pHmgA-P::WAP1. The orf6 DNA fragment containing its RBS and ORF was amplified using primers ORF6-up and ORF6-down, and the fragment was inserted into the XhoI site of pHmgA-P::WAP1 to generate pHmgA-P::WAP2. The DNA fragment containing both orf 9 and orf10, which are translationally coupled, was amplified using primers ORF9–10-up and ORF9–10-down, and the fragment was inserted into the BamHI site of pHmgA-P::WAP2 to generate pHmgA-P::WAP. Subsequently, construct pHmgA-P::WAP was verified (Figure S5) and introduced into the wild type or strain dCas9-ω3 (containing spacer-3) by electroporation. The resulted transformants (WT/refactored or dCas9-ω3/refactored) were selected on the LB plates (100 μg/mL gentamicin) and verified by PCR using primers listed in Table S2 (Figure S6).
Growth Comparison between the WT and Engineered Strains
Strains WT, dCas9-ω3, dCas9-ω3/refactored and WT/refactored were grown in 3 mL LB medium at 30 °C for overnight, and a fraction (1%) of the cultures was transferred to 25 mL 1/10 TSB medium, 30 °C with shaking. The growth curve was calculated based on OD600 values of each of the cultures for every 2 h of growth.
RNA Isolation and qRT-PCR
RNA was isolated from WT, dCas9-ω and dCas9-ω3/refactored that were grown in fermentation medium for 24 h and 48h, as described previously.30 RNA was treated with DNase I (Thermo Scientific) to remove contaminated DNA, and then it was reverse transcribed to complementary DNA by using SuperScript II RT reagent kit (Invitrogen). qRT-PCR was carried out in CFX Connect Real-Time PCR Detection System (BIO-RAD Laboratories, Inc.) using PowerUp SYBR Green Master Mix (Applied Biosystems) with primers listed in Table S2. The conditions are used as follows: 50 °C for 2 min, 95 °C for 2 min, followed by 40 cycles of 95 °C for 15 s, 60 °C for 1 min. The 16S rRNA was used as an internal control. The relative transcriptional levels of tested genes were normalized to 16S rRNA and determined by using the 2−ΔΔCT method.31 The values were presented as fold change in comparison with the relative expression levels for each gene at the first test time point in the wild-type strain. Data are presented as the averages of three independent experiments conducted in triplicate, and the statistical analyses are included in Tables S4 and S5.
Extraction and Preparation of WAP-8294A Compounds
Various strains of L. enzymogenes OH11 were grown in 3 mL 1/10 TSB for 24 h, and a fraction (1%) of the culture was transferred to 3 mL fermentation medium. The cultures were incubated at 30 °C with shaking at 200 rpm for 72 h. To extract total WAP compounds, the 3 mL culture broth was collected, and the pH of the broth was adjusted to 2.5 with 37% HCl. The treated broth was then extracted with n-butanol/ethyl acetate (1/1, vol) containing 0.05% TFA. The organic phase was dried in the air, and the residues were redissolved in 200 μL methanol containing 0.05% TFA. To detect the distribution of WAP-8294A compounds, the 3 mL culture was centrifuged (12 000 rpm, 5 min), and the supernatant and the cell pellet were separated. WAP-8294A compounds were extracted from the supernatant using the same method above. To extract WAP-8294A compounds in the cells, the pellet was put into −20 °C freezer for 1 h and then incubated into 30 °C water bath for 10 min. This was repeated three times to disrupt the cells. Then the cell pellet was treated with 3 mL n-butanol/ethyl acetate (1/1, vol) containing 0.05% TFA. The organic phase was dried in the air, and the residues were redissolved in 200 μL methanol containing 0.05% TFA. A 20 μL aliquot of each extracts was analyzed by LC–MS. Water/0.05% FA (solvent A) and acetonitrile/methanol (1/1, vol)/0.05% FA (solvent B) were used as the mobile phases with a flow rate of 1.0 mL/min. The HPLC program was as follows: 57% B in A in the first 5 min, 57–100% B in 5–32 min, 100% B in 32–40 min, back to 57% B at 41 min, and maintained to 48 min. The metabolites were detected at 280 nm on a UV detector. To quantify WAP-8294A compounds, pure lysobactin (20 μg/mL), which is also a cyclic peptide like WAP-8294A compounds, but is not produced by L. enzymogenes OH11, but by a different Lysobacter species (Lysobacter sp. ATCC53042), was used as internal control for LC–MS analyses. The similar amount of lysobactin was recovered and detected in different samples during the LC–MS analyses (Figure S8 and Figure S12). The statistical analyses of yield changes in various strains are included in Tables S6–S9. For preparation of WAP-8294A2, the fermentation culture was scaled up and the compound was prepared as described previously.3
Antibacterial Activity Test
B. subtilis was used as the indicator organism for anti-Gram-positive bacteria activity of WAP-8294A compounds.3 B. subtilis was incubated in 3 mL LB medium at 37 °C for 6 h with shaking. After the OD600 value was measured, the culture was diluted using LB medium to OD600 = 0.3. The culture was mixed with LB agar (1:1000 ratio), and the mixture was poured into a plate for solidification. Sterilized small round filter-papers were placed on the surface of the LB plate, and aliquots (5 μL) of various L. enzymogenes OH11 extracts were added onto each of the filter-papers. Methanol was used as negative control. The plates were incubated at 37 °C until clear inhibition zones appeared.
The efficacy of WAP-8294A2 was tested against a panel of clinically relevant Gram-positive pathogens, including Staphylococcus aureus, Staphylococcus epidermidis, Enterococcus faecalis, Bacillus cereus, Listeria monocytogenes, Streptococcus pyogenes, Streptococcus agalactiae, and Streptococcus pneumoniae. The isolates used in these studies represent a mixture of ATCC strains and clinical isolates from the University of Nebraska Medical Center.32 All organisms were grown on blood agar at 37 °C and subcultured two times from frozen stock before testing. The broth based microdilution method was used to generate MICs. Initially, 2-fold dilutions of WAP-8294A2 were prepared in Mueller–Hinton broth (with or without 3% lysed horse blood) and tested at final concentrations ranging from 12.8–0.1 μg/mL. Wells were inoculated with each isolate to yield a final concentration of 5.0 × 105 organisms/mL, and trays were incubated at 37 °C for 18–20 h. As a control for growth, each isolate was additionally tested in a well that contained media only. MICs were visually interpreted as the lowest concentration of WAP-8294A2 that inhibited growth of the organism. Depending on the results of initial studies, the isolates were further tested using higher (51.2–0.4 μg/mL) or lower (1.6–0.125 μg/mL) WAP-8294A2 concentrations. All testing was performed in duplicate.
Supplementary Material
Acknowledgments
We thank Drs. Ronald Cerny and Kurt Wulser for technical assistance. We are grateful to Dr. Weeks and Dr. Jiang (Department of Biochemistry, University of Nebraska-Lincoln) for providing the plasmid containing sgRNA and cas9. This work was supported in part by NSFC (31329005, 31701839), the NIH (R01AI097260), and a UNL Revision Award and a UNL Faculty Seed Grant.
Footnotes
Author Contributions
L.D., F.L. and P.F. conceived and designed experiments. L.Y., S.W. and P.F. carried out experiments. L.D., L.Y., S.W. and P.F. analyzed data. L.D. and L.Y. wrote the manuscript.
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acssynbio.7b00293.
Details of primers, plasmid constructs, efficacy of WAP-8294A2, generation and verification of various strains, and quantification of metabolites (PDF)
References
- 1.Christensen P, Cook FD. Lysobacter, a new genus of non-fruiting, gliding bacteria with a high base ratio. Int J Syst Bacteriol. 1978;28:367–393. [Google Scholar]
- 2.Lou L, Qian G, Xie Y, Hang J, Chen H, Zaleta-Rivera K, Li Y, Shen Y, Dussault PH, Liu F, Du L. Biosynthesis of HSAF, a tetramic acid-containing macrolactam from Lysobacter enzymogenes. J Am Chem Soc. 2011;133:643–645. doi: 10.1021/ja105732c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W, Li Y, Qian G, Wang Y, Chen H, Li YZ, Liu F, Shen Y, Du L. Identification and characterization of the anti-methicillin-resistant Staphylococcus aureus WAP-8294A2 biosynthetic gene cluster from Lysobacter enzymogenes OH11. Antimicrob Agents Chemother. 2011;55:5581–5589. doi: 10.1128/AAC.05370-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao Y, Qian G, Ye Y, Wright S, Chen H, Shen Y, Liu F, Du L. Heterocyclic aromatic N-oxidation in the biosynthesis of phenazine antibiotics from Lysobacter antibioticus. Org Lett. 2016;18:2495–2498. doi: 10.1021/acs.orglett.6b01089. [DOI] [PubMed] [Google Scholar]
- 5.Pirri G, Giuliani A, Nicoletto SF, Pizzuto L, Rinaldi AC. Lipopeptides as anti-infectives: a practical perspective. Cent Eur J Biol. 2009;4:258–273. [Google Scholar]
- 6.Kato A, Nakaya S, Kokubo N, Aiba Y, Ohashi Y, Hirata H, Fujii K, Harada K. A new anti-MRSA antibiotic complex, WAP-8294A I. taxonomy, isolation and biological activities. J Antibiot. 1998;51:929–935. doi: 10.7164/antibiotics.51.929. [DOI] [PubMed] [Google Scholar]
- 7.Leive L. The barrier function of the gram-negative envelope. Ann N Y Acad Sci. 1974;235:109–129. doi: 10.1111/j.1749-6632.1974.tb43261.x. [DOI] [PubMed] [Google Scholar]
- 8.Vaara M. Agents that increase the permeability of the outer-membrane. Microbiol Rev. 1992;56:395–411. doi: 10.1128/mr.56.3.395-411.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang W, Bikard D, Cox D, Zhang F, Marraffini LA. CRISPR-assisted editing of bacterial genomes. Nat Biotechnol. 2013;31:233–239. doi: 10.1038/nbt.2508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM. Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res. 2013;41:4336–4343. doi: 10.1093/nar/gkt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schuster M, Schweizer G, Reissmann S, Kahmann R. Genome editing in Ustilago maydis using the CRISPR–Cas system. Fungal Genet Biol. 2016;89:3–9. doi: 10.1016/j.fgb.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 12.Tong Y, Charusanti P, Zhang L, Weber T, Lee SY. CRISPR-Cas9 based engineering of actinomycetal genomes. ACS Synth Biol. 2015;4:1020–1029. doi: 10.1021/acssynbio.5b00038. [DOI] [PubMed] [Google Scholar]
- 13.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;39:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kim H, Kim JS. A guide to genome engineering with programmable nucleases. Nat Rev Genet. 2014;15:321–334. doi: 10.1038/nrg3686. [DOI] [PubMed] [Google Scholar]
- 17.Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 2013;41:7429–7437. doi: 10.1093/nar/gkt520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hopwood DA. How do antibiotic-producing bacteria ensure their self-resistance before antibiotic biosynthesis incapacitates them? Mol Microbiol. 2007;63:937–940. doi: 10.1111/j.1365-2958.2006.05584.x. [DOI] [PubMed] [Google Scholar]
- 19.Parra-Lopez C, Baer MT, Groisman EA. Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 1993;12:4053–4062. doi: 10.1002/j.1460-2075.1993.tb06089.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Thompson SA, Shedd OL, Ray KC, Beins MH, Jorgensen JP, Blaser MJ. Campylobacter fetus surface layer proteins are transported by a type I secretion system. J Bacteriol. 1998;180:6450–6458. doi: 10.1128/jb.180.24.6450-6458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uberto R, Moomaw EW. Protein similarity networks reveal relationships among sequence, structure, and function with the cupin superfamily. PLoS One. 2013;8:e74477. doi: 10.1371/journal.pone.0074477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang Y, Qian G, Liu F, Li YZ, Shen Y, Du L. Facile method for site-specific gene integration in Lysobacter enzymogenes for yield improvement of the anti-MRSA antibiotics WAP-8294A and the antifungal antibiotic HSAF. ACS Synth Biol. 2013;2:670–678. doi: 10.1021/sb4000806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xie Y, Wright S, Shen Y, Du L. Bioactive natural products from Lysobacter. Nat Prod Rep. 2012;29:1277–1287. doi: 10.1039/c2np20064c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Panthee S, Hamamoto H, Paudel A, Sekimizu K. Lysobacter species: a potential source of novel antibiotics. Arch Microbiol. 2016;198:839–845. doi: 10.1007/s00203-016-1278-5. [DOI] [PubMed] [Google Scholar]
- 25.de Bruijn I, Cheng X, de Jager V, Expósito RG, Watrous J, Patel N, Postma J, Dorrestein PC, Kobayashi D, Raaijmakers JM. Comparative genomics and metabolic profiling of the genus Lysobacter. BMC Genomics. 2015;16:991. doi: 10.1186/s12864-015-2191-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Qian G, Hu B, Jiang Y, Liu F. Identification and characterization of Lysobacter enzymogenes as a biological control agent against some fungal pathogens. Agric Sci China. 2009;8:68–75. [Google Scholar]
- 27.Kovach ME, Elzer PH, Hill DS, Robertson GT, Farris MA, Roop RM, 2nd, Peterson KM. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene. 1995;166:175–176. doi: 10.1016/0378-1119(95)00584-1. [DOI] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 29.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu G, Tian Y, Yang H, Tan H. A pathway-specific transcriptional regulatory gene for nikkomycin biosynthesis in Streptomyces ansochromogenes that also influences colony development. Mol Microbiol. 2005;55:1855–1866. doi: 10.1111/j.1365-2958.2005.04512.x. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta DeltaC(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Fey PD, Endres JL, Yajjala VK, Widhelm TJ, Boissy RJ, Bose JL, Bayles KW. A genetic resource for rapid and comprehensive phenotype screening of non-essential Staphylococcus aureus genes. mBio. 2013;4:e00537–12. doi: 10.1128/mBio.00537-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.