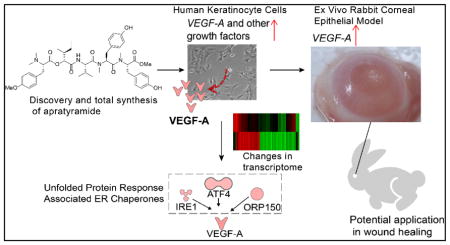
Abstract
A novel linear depsipeptide enriched with tyrosine derived moieties, termed apratyramide, was isolated from an apratoxin-producing cyanobacterium. The structure was determined using a combination of NMR spectroscopy, mass spectrometry and chiral analysis of the acid hydrolyzate and confirmed by total synthesis. Apratyramide up-regulated multiple growth factors at the transcript level in human keratinocyte (HaCaT) cells and induced the secretion of vascular endothelial growth factor A (VEGF-A) from HaCaT cells, suggesting the compound’s potential wound-healing properties through growth factor induction. Transcriptome analysis and sequential validation supported the hypothesis and indicated its mode of action (MOA) through the unfolded protein response (UPR) pathway, which is functionally related to wound healing and angiogenesis. The conditioned medium of HaCaT cells treated with apratyramide induced angiogenesis in vitro. An ex vivo rabbit corneal epithelial model was applied to confirm the VEGF-A induction in this wound-healing model.
Graphical Abstract
INTRODUCTION
Wound healing is a complex biological process and consists of a series of events including inflammation, cell proliferation, tissue granulation and remodeling of scar tissue, which involves the coordinated efforts of several cell types, such as keratinocytes, fibroblasts, endothelial cells, macrophages, and platelets.1–3 A wide variety of growth factors and cytokines are involved in each stage of the wound healing process: platelet-derived growth factors (PDGFs), vascular endothelial growth factors (VEGFs), basic fibroblast growth factors (bFGFs), and granulocyte-macrophage colony stimulating factor (GM-CSF), and many studies have shed light on the crucial roles of these growth factors on initiating and facilitating wound healing.2,4 Dysregulation of these growth factors could delay wound closure and result in chronic wounds (e.g., diabetic foot ulcers [DFUs], pressure ulcers [PUs], and chronic venous leg ulcers [VLUs]), which represent a major healthcare burden in the US.5,6
Despite many efforts that have been spent on the development of growth factors as therapeutic agents, to date, this field has been disappointing. There is only one Federal Drug Administration (FDA) approved growth factor on the market for the treatment of DFUs: recombinant platelet-derived growth factor, rhPDGF-BB, Becaplermin.7 There are also other growth factors under clinical trials, including VEGF, bFGF and GM-CSF.7 One of the limitations for topical administration of exogenous growth factors is low absorption due to the protein nature of these growth factors.8–10 Therefore, an alternative therapeutic method aimed at stimulating the production and secretion of endogenous growth factors from wound tissue by small molecules might be more promising.
VEGF is one of the most potent angiogenic growth factors and promotes all steps in the angiogenic cascade.11 In the VEGF family, VEGF-A is the best studied angiogenic growth factor regulating both physiological and disease processes such as tumor growth, psoriasis and wound healing.12–14 VEGF-A is produced by keratinocytes, fibroblast smooth muscle cells, platelets, neutrophils, and macrophages during wound healing, andkeratinocytes are thought to be a major source of VEGF-A after injury.15–17 VEGF-A stimulates angiogenesis by acting on endothelial cells in the wound sites.18 It has been found that VEGF-A gene expression is up-regulated in the skin after wounding.16 Furthermore, the altered expression pattern of VEGF mRNA during skin repair in genetically diabetic (db/db) mice suggested that the impairment in VEGF synthesis and release at the wound site might contribute to chronic wounds.16 In agreement with these observations, many in vitro and in vivo studies have shown that administration of VEGF-A topically or by gene transfer accelerates experimental wound healing through stimulation of angiogenesis, re-epithelialization, collagen deposition, and synthesis and maturation of extracellular matrix.19–22 Therefore, the above information strongly suggests the therapeutic application of VEGF-A inducers in the treatment of chronic wounds.
The use of natural products for the treatment of wounds and injuries is as old as civilization. Since ancient times, people have recognized the healing properties of herbs, honey, leaves, oil, etc.23 So far, some but not all of the active components of these natural wound healers that are identified fall into several structural classes: vitamins24–26, terpenes or terpenoids27–30, polyphenols31–34, and alkaloids35,36. These compounds enhance wound healing through various mechanisms, including promoting skin cells proliferation and migration, angiogenesis, collagen synthesis, as well as exerting anti-inflammatory and antiseptic activities.37 In addition to these traditional natural sources, marine organisms are becoming a rich source for new drugs.For example, pseudopterosins are a series of a diterpene-pentoseglycoside compounds from gorgonian corals that enhance wound healing through anti-inflammation.38,39
Marine cyanobacteria produce various secondary metabolites which belong to peptides, polyketides or hybrid of peptide-polyketides.40 Despite the fact that marine cyanobacteria produce compounds with a broad spectrum of biological activities, including anticancer, antimicrobial, protease inhibitory, immunomodulatory, neuromodulatory properties, and considered a valuable source for medicinal therapeutic use, they have not yet been linked to activities associated with wound healing, to the best of our knowledge. We report a novel linear depsipeptide isolated from marine cyanobacteria as a growth factor inducer with potential wound healing properties. This study provides new insights into the role of small peptides in wound healing and broadens the spectrum of activities of compounds from marine cyanobacteria.
RESULTS AND DISCUSSION
Isolation and structure determination
Five different Guamanian collections of freeze dried apratoxins-producing Moorea bouillonii were collected from Fingers Reef, Guam. 41 Each were individually extracted with CH2Cl2 and MeOH (2:1) followed by solvent partitioning, silica chromatography and reversed-phase HPLC purification to yield optically active compound 1 as a minor metabolite (2.0 mg, [α]D20 – 101.9 (c 0.59, MeOH)).
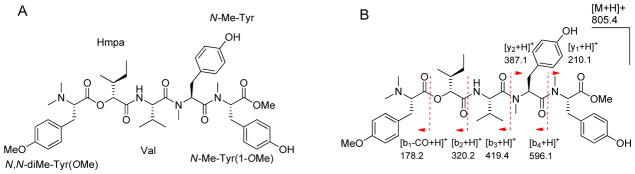
The HRESIMS of 1 in the positive mode showed a molecular ion peak at m/z 805.4388 [M+H]+, suggesting a molecular formula of C44H60N4O10 with seventeen degrees of unsaturation. The 1H NMR spectrum of 1 (Figure S1) displayed characteristic peptide signals for several α-protons (δH 3.4 – 5.3), an exchangeable proton of amide (δH 8.08), four N-methyls (δH 2.2 – 2.8) and two O-methyls (δH 3.5 – 3.7). A signal at δC 75.09 in 13C NMR spectrum (Figure S2) corresponding to a typical oxygenated sp3 carbon suggested the presence of a hydroxy acid in addition to amino acids. Following the interpretation of 1D and 2D NMR experiments, 1H and 13C NMR signals (Supporting Information Table S1) were assignable into five partial structures: three modified tyrosines [N-Me-Tyr, N-Me-Tyr(1-OMe) and N,N-diMe-Tyr(OMe)], one proteinogenic amino acid (Val) and one α-hydroxy acid moiety [2-hydroxy-3-methylpentanoic acid (Hmpa)] (Figure 1A). The sequence of these units was established on the basis of HMBC and NOESY correlations (Supplemental information Table S1) and was further verified by ESIMS fragmentation (Figure 1B). Due to its structural enrichment of tyrosine units as well as its origin from Apra Harbor, we named this compound apratyramide (1). The absolute configuration of 1 was determined by enantioselective HPLC analysis and comparison with authentic standards.
Figure 1.
A) Structure of apratyramide (1). B) ESIMS fragmentation pattern of 1.
Total synthesis of apratyramide
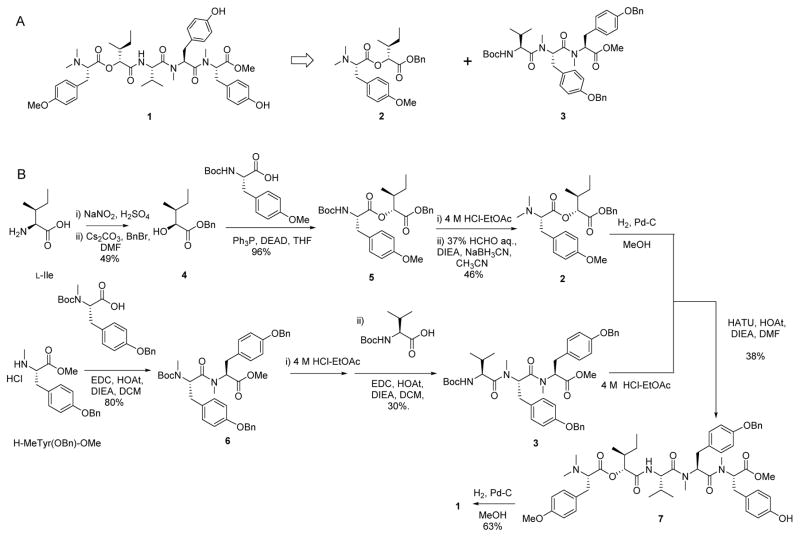
Owing to the limited supply of apratyramide from nature, we performed the total synthesis in order to obtain more material for biological evaluation. Following a retrosynthetic analysis of apratyramide (Scheme 1A), a convergent synthesis (Scheme 1B) was conducted by obtaining two building blocks: an ester and a tripeptide. To construct the ester, two commercially available amino acids were obtained as starting materials. The α-hydroxy carboxylic acid 4 was prepared from L-isoleucine by published protocol.42 The esterification of N-Boc-O-Me-tyrosine with acid 4 provided ester 5 in 96% yield by Mitsunobu reaction (Ph3P/DEAD).43 The Boc group of ester 5 was removed using 4 M HCl in ethyl acetate. Then, the desired N,N-dimethylated amino ester 2 was formed by a reductive alkylation reaction of the free amine in 5 using a mixture of aq. HCHO and NaBH3CN.44 The end acid group of ester 2 was liberated by hydrogenation with Pd/C/H2 in MeOH. The tripeptide 3 was constructed smoothly by sequential coupling of N-Boc-N-Me-tyrosine(OBn) with methyl ester of N-Me-tyrosine(OBn), then with N-Boc-valine using the coupling system EDCI/HOAt.45 The Boc group in 3 was cleaved by 4 M HCl in ethyl acetate to obtain acetate free amine, which was then coupled with the free acid in ester 2 to provide precursor 7 by coupling combination HATU/HOAt in DMF. Finally, the hydrogenation of 7 by Pd/C/H2 in MeOH gave final product 1 in 63% yield. The NMR spectra for natural and synthetic apratyramide (1) were identical (Supporting information Figure S7–8). Comparison of optical activity and HR-MS of natural and synthetic 1 further confirmed the structural assignment for the natural product.
Scheme 1.
A) Retrosynthetic analysis of 1. B) Convergent synthesis of 1.
Apratyramide Induces Transcription of VEGF-A in HCT116 Cells
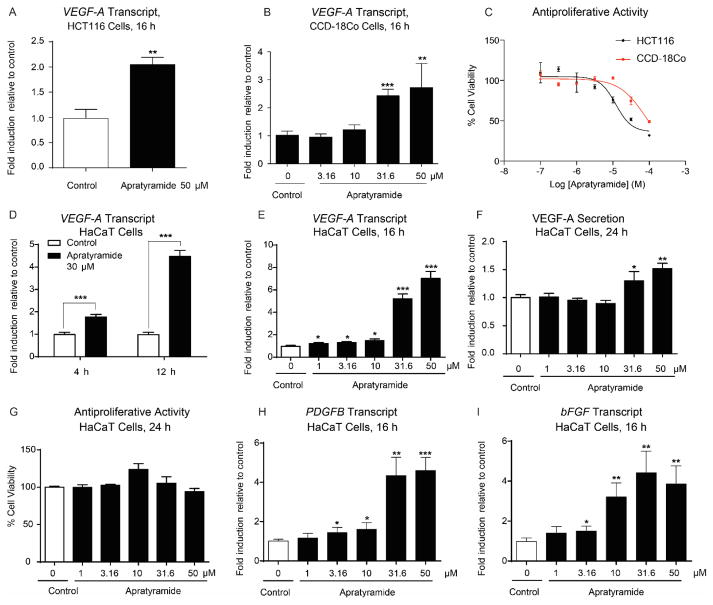
In our search for modulators of VEGF-A and angiogenesis from marine cyanobacteria, we previously identified apratoxins from the same cyanobacterium as potent inhibitors by preventing cotranslational translocation of VEGF-A and other secreted proteins.46,47 Apratyramide (1), however, had the opposite effect. Using human colon cancer HCT116 cells, we observed that 1 up-regulated VEGF-A, while displaying minimal cytotoxicity (Figures 2A,C). Fifty micro-molar apratyramide doubled VEGF-A transcript level in HCT116 cells (Figure 2A). Apratyramide (1) also exerted a similar effect in the corresponding normal colon cells (CCD-18Co) with negligible effects on cell viability (Figures 2B,C). Therefore, we aimed to further explore apratyramide’s therapeutic applications where VEGF-A upregulation might be beneficial.
Figure 2.
Apratyramide (1) is an inducer of VEGF-A and other growth factors. A) Transcript level of VEGF-A in HCT116 (human colon cancer) cells, 16 h. B) Transcript level of VEGF-A in CCD-18Co (human normal colon) cells, 16 h. C) Antiproliferative effect of 1 on HCT116 and CCD-18Co cells, 48 h. D) Transcript level of VEGF-A in HaCaT cells after 4 h and 12 h treatment with 30 μM of 1. E) Transcript level of VEGF-A in HaCaT cells after 16 h. F) Level of VEGF-A secretion from HaCaT after 24 h. G) Antiproliferative activity of 1 on HaCaT cells, 24 h. H) Transcript level of PDGFB in HaCaT cells, 16 h. I) Transcript level of bFGF in HaCaT cells, 16 h. Data are presented as mean + SD, *P < 0.05, **P < 0.01, ***P < 0.001, compared to control using unpaired t test (n = 3).
Apratyramide Induces Transcription, Secretion of VEGF-A and transcription of Other Wound-healing Related Growth Factors in HaCaT Cells
Since VEGF-A inducers are considered promising therapeutic agents for the treatment of chronic wounds, we proposed that 1 may also induce VEGF-A in normal cell types that are involved in the wound healing process. Thus, we logically moved on to a commonly used in vitro wound healing model: human keratinocyte cells (HaCaT). As expected, VEGF-A mRNA level was induced 1.7- fold after 4 h treatment with 30 μM of 1 and a greater induction effect was observed after 12 h (Figure 2D). After 16 h, 50 μM of 1 increased VEGF-A transcript level by 7-fold, while 31.6 μM of 1 led to a 5-fold increase (Figure 2E). Accordingly, 50 μM of 1 induced a 1.5-fold increase of VEGF-A secretion from HaCaT cells after 24 h, and 31.6 μM of 1 induced a 1.3-fold increase without causing cytotoxicity (Figures 2F,G). Around ninety-percent cell viability was observed at 50 μM of 1 after 24 h (Figure 2G).
Multiple growth factors and cytokines are involved in a complex integration of signals for regulating wound healing processes.7 This information prompted us to test whether 1 also induces other growth factors which might work cooperatively with VEGF-A during wound healing. RT-qPCR data indicated that PDGFB and bFGF were all stimulated by 1 (Figures 2H,I). Importantly, rhPDGF-BB is the only FDA-approved growth factor on the market for the treatment of DFUs.7
Transcriptome Profiling and Ingenuity Pathway Analysis Indicate that UPR Plays a Role in Mechanisms of Apratyramide-induced VEGF-A
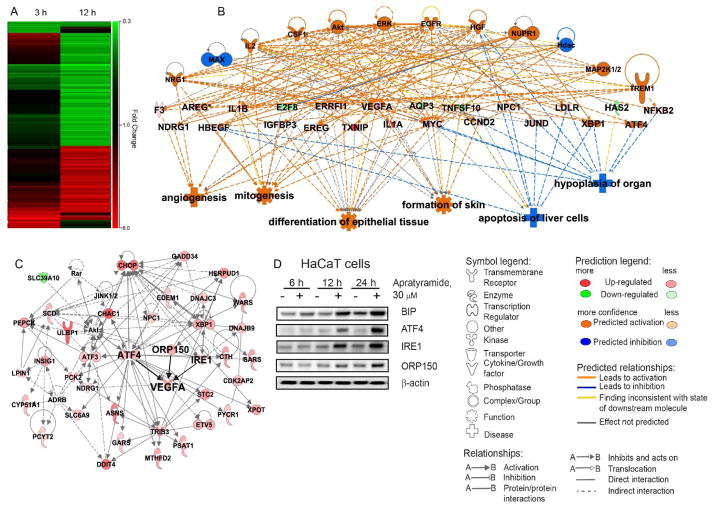
To further elucidate the mode of action of apratyramide through which multiple growth factors are induced, we performed microarray profiling using the Affymetrix GeneChip® Human Transcriptome Array 2.0 and determined global changes in transcript levels in HaCaT cells treated with apratyramide. Comparative analysis identified 371 differentially expressed genes after 12 h treatment with 30 μM of 1 (P < 0.05, FDR corrected, fold change >1.5 or <0.67) (Figure 3A). Consistent with our previous data, VEGF-A appeared to be one of the most up-regulated genes (Table 1). To examine the molecular functions and genetic networks, the 12 h microarray data was analyzed using Ingenuity Pathways Analysis (IPA) (Figures 3B,C). In accordance with our hypothesis, the global changes of transcript levels are associated with increased downstream phenotypic effects including angiogenesis, mitogenesis, differentiation of epithelial tissue and formation of skin, and decreased effects such as apoptosis of liver cells and hypoplasia of organs (Figure 3B). IPA analysis of 371 microarray hits indicated the unfolded protein response (UPR) (Figure S21) as the top canonical pathway with a p-value of 1.45 × 10−16. The IPA also elucidated that the 371 hits were most closely related to a molecular network associated with the function of cellular compromise and cellular maintenance (Figure 3C). The network contains molecular components from UPR pathway (ATF4, INSIG1, CHOP, DNAJC3, PP1R15A, JINK1/2), NRF2-mediated oxidative stress response signaling (ATF4, DNAJC3, JINK1/2, Akt, HERPUD1, DNAJB9) as well as glucocorticoid receptor signaling (ADRB, Akt, JINK1/2, PEPCK, PCK2).
Figure 3.
Ingenuity Pathway Analysis (IPA) for transcriptome profiling of apratyramide (1). A) Heat map for transcript changes after 3 h and 12 h treatment with 30 μM 1. Red indicates up-regulation. Green indicates down-regulation. B) Top regulator effect network. C) Top related molecular network associated with the function of cellular compromise and cellular maintenance. D) Validation of selected hits from transcriptome profiling using immunoblot analysis.
Table 1.
Selected groups of top up- and down-regulated genes after 12 h treatment with 30 μM apratyramide (1).a
| Up-regulated genes, FDR correction, 12 h | Down-regulated genes, FDR correction, 12 h | |
|---|---|---|
| Growth factor | AREG, VEGFA, HBEGF, EREG | |
| Cytokine | IL1A, IL1B, MYDGF | SCGB1A1, CXCL1, CCL2, TNFSF10 |
| Transcription regulator | DDIT3, CREB3L2, XBP1, ETV5, ATF4, JUND, ZNF165, ETV4, ATF3, MYC, NFE2L1, KLF6, SQSTM1, DAP, PREB, NFKB2, FOXE1, MEF2D, MAGED1, IRF2BP2, BHLHE40 | GMNN, UHRF1, GTF2I, VGLL1, DLX5, E2F8, SMAD6, ID3, GRHL3 |
| G-protein coupled receptor | GPR1 | P2RY2 |
| MicroRNA | MIR-3143, MIR-554, MIR-548 |
The P values obtained were controlled for multiple testing (false discovery rate) using the Benjamini-Hochberg method. Differentially expressed genes were then ranked by the P values, genes with P < 0.05 (with FDR correction) and fold change >1.5 or <0.67 were considered as differentially expressed genes at a statistically significant level, which were grouped based on functions. Selected groups are presented above. See Supporting Information Table S2 for a full list of top up- and down-regulated genes.
Cytoprotective Roles of UPR and Its Modulatory Effects on Growth Factors
The unfolded protein response (UPR) pathway is a cytoprotective signaling cascade in response to endoplasmic reticulum (ER) stress in cells. UPR coordinates multiple signaling pathways and controls various physiologies in cells and the whole organism including liver development, plasma cell differentiation,48 bone development,49,50 plasma cell differentiation,51,52 normal pancreatic homeostasis53 and placental development and embryonic viability.54 Importantly, UPR is activated after skin injury, suggesting the protective roles of UPR in rescuing wound injuries.55 Therefore, intervening in ER stress and modulating signaling components of UPR would provide promising therapeutics for the treatment of chronic wounds.56
Interestingly, studies have unveiled the modulatory effects of the UPR on VEGF-A. The UPR contributes to the transcriptional, protein processing and transportation of VEGF-A in the ER through activation of ATF4, IRE1 and ORP150, which were all up-regulated by apratyramide at transcript and protein levels (Figure 3Figures 3C,D).57–59 These findings suggest that apratyramide may induce VEGF-A through the UPR pathway. Besides up-regulating VEGF-A, the UPR has also been reported to enhance angiogenesis by up-regulating a number of other pro-angiogenic factors like FGFs, PDGFs and IL-8.60 These observations are also in accordance with our microarray and RT-qPCR results, indicating that many other pro-angiogenic factors (e.g., bFGF, PDGFB, HB-EGF) were also up-regulated after treatment with apratyramide (Figure 2, Table 1, Supporting Information Table S2). Collectively, improving ER homeostasis by activating the UPR pathway independent of ER stress may be a promising tool to accelerate wound closure, including diabetes-associated chronic wounds closure55 and apratyramide has the promising attributes to be one such therapeutic agent.
A closer investigation of these individual molecular components of UPR further enabled us to identify several lines of evidence demonstrating that they also directly attribute to angiogenesis, wound healing and VEGF-A up-regulation dependent or independent of UPR.54,61–63
ATF4 is an important transcription factor in the UPR signaling cascade which activates VEGF-A at both transcript and protein levels.61–63 ATF4 promotes bone angiogenesis by increasing VEGF expression and release in the bone environment.63 It has also been reported that ATF4 expression was induced in smooth muscle cells after arteries injury in rats and its overexpression further enhanced the expression of VEGF-A by an interaction between ATF4 and a recognition element located in the VEGF-A gene.62 The microarray data as well as immunoblot analysis suggested that ATF4 is activated by apratyramide at both the mRNA and protein level, which subsequently leads to the induction of the transcription of a number of its downstream molecular components: CHOP, SLC6A9, CHAC1, ATF3, SARS, WARS and others (Figure 3).
IRE1 is also an ER-located transmembrane protein, which play an essential role in physiological processes including angiogenesis, placental development and embryonic viability.54,64 It has been shown that VEGF-A expression in the placenta is partially dependent on IRE1.54 Another recent study identified the deficiency of IRE1 in type 2 diabetic db/db mice and that cell therapies using direct IRE1 gene transfer significantly accelerated cutaneous wound healing in diabetic mice through enhancing angiogenesis.64 These findings strongly suggested the therapeutic strategy for diabetic wound healing by enhancing IRE1 activity. In addition, IRE1 deletion resulted in elevation of microRNAs, while the supply of IRE1 promoted the angiogenic potential of diabetic (bone marrow–derived progenitor cells) BMPCs through modulating miRNA biogenesis.64 Accordingly, we also observed down-regulation of several microRNAs after 12 h treatment with apratyramide (Table 1, Supporting Information Table S2).
ORP150 is an ER chaperone, the expression of which is regulated by UPR. Overexpression of the ORP150 gene by adenovirus vectors accelerated wound healing by modulating intracellular VEGF transport.59 This observation implied that ORP150 was involved in skin wound healing.
Conditioned Medium from HaCaT Cell Culture treated with Apratyramide Induced Angiogenesis In Vitro
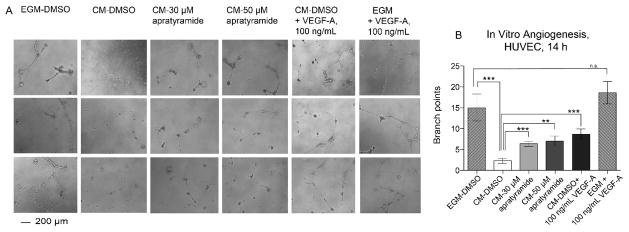
During wound healing, VEGF-A is secreted to stimulate angiogenesis, which primarily acts on endothelial cells in the wound area.18 Thus, we questioned whether the increased VEGF-A secreted from HaCaT cells induced by 1 could enhance angiogenesis. Here, conditioned medium (CM) was collected from the HaCaT cell culture with or without the presence of 1 at 24 h, when we previously detected an increase of VEGF-A secretion. We incubated human endothelial cells (HUVECs) with the obtained CM and monitored their angiogenic activity using an in vitro angiogenesis assay (Figures 4A,B). As a positive control, the complete growth medium (EGM) containing 2% FBS and bovine brain extract (BBE) enabled HUVECs to form tube-like structures from individual cells after 14 h. The CM from HaCaT cell culture (DMEM, 10% FBS) alone, however, had little effect on angiogenesis as most HUVECs remained as individual cells. This is possibly due to a lack of growth supplement required for angiogenesis in endothelial cells in the DMEM. The decreased angiogenesis in the CM-DMSO group was rescued by the treatment with 30 or 50 μM of 1 in HaCaT culture, indicated by an increase of tube-like structure formation. Similar to apratyramide’s effect, VEGF-A protein, 100 ng/mL, also induced angiogenesis in vitro. In contrast, the addition of VEGF-A at 100 ng/mL to the complete growth medium did not significantly further induce angiogenesis, probably due to the sufficient amount of angiogenic factors in the BBE. The above results demonstrated that apraytramide indirectly enhanced angiogenesis potentially through an induction of VEGF-A secreted from HaCaT cells.
Figure 4.
Conditioned medium from HaCaT cell culture treated with apratyramide (1) induced angiogenesis in vitro. A) Conditioned medium (CM) from HaCaT culture with the presence of compound 1 or solvent control DMSO (0.3 %), 24 h, induced angiogenesis in vitro, determined by matrigel assay using HUVECs (scale bar 200 μm), 14 h. VEGF-A protein, 100 ng/mL was used as positive control. Complete growth medium (EGM, Lonza) for HUVEC was also used as a positive control B) Branch point counting was used as quantification method. Three random microscope view-fields were counted and the number of branch points was averaged for each well. Error bars indicate mean + SEM of eight replicates from two independent experiments. P-values were calculated relative to control (CM-DMSO) using unpaired t test (n = 8), **P < 0.01, ***P < 0.001.
Apratyramide Induces VEGF-A in a Rabbit Corneal Epithelial Ex Vivo Model
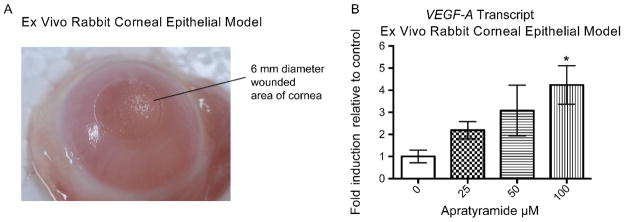
In order to evaluate apratyramide (1) in a more physiological context, we tested it in an ex vivo rabbit corneal epithelial model, a validated wound healing model.65,66 The fresh rabbit eyes were obtained and wounds were induced on the center of the cornea by a laser (Figure 5A). After that, eyeballs were trimmed to collect cornea tissues which were then cultured in medium with or without the presence of 1. Eighteen-hour later, total RNA was collected from cornea tissues and subjected for RT-qPCR analysis. Consistent with the effect in vitro, we have detected a dose-dependent increase of VEGF-A mRNA in the cornea after treatment with 1 (Figure 5B).
Figure 5.
Apratyramide induced VEGF-A in a rabbit corneal epithelial ex vivo model. Data are presented as mean + SEM, *P < 0.05.
CONCLUSIONS
We have discovered a novel linear depsipeptide, apratyramide, from marine cyanobacteria as a growth factor inducer which has the potential to rescue chronic wounds and accelerate wound healing processes by inducing growth factor secretion in the wound area. Apratyramide was isolated through a standard procedure, followed by total structural determination through a combined analysis of NMR and MS spectra as well as chiral analysis of the acid hydrolyzate. We conducted the total synthesis of apratyramide through a convergent synthetic strategy which provided us more material for further biological evaluation. We performed a series of in vitro cell-based assays to elucidate the biological activity as well as mechanism of action, although the direct biological target remains elusive. Apratyramide induced both transcription and secretion of VEGF-A in human keratinocyte (HaCaT) cells, evident by RT-qPCR and AlphaLISA analysis. Other wound-healing related growth factors were also found to be induced at the transcriptional level, including PDGFB and bFGF. Transcriptome profiling using HaCaT cells identified 371 differentially expressed genes after 12 h treatment with apratyramide. Importantly, VEGF-A and other growth factors were up-regulated, showing consistency with our previous in vitro data and supporting our hypothesis of the potential wound healing properties of apratyramide through growth factor modulation. IPA indicated that apratyramide induced growth factors through or partially through the UPR pathway mediated by ATF4, IRE1 and ORP150, three molecular components in the UPR pathway that are functionally related to wound healing and angiogenesis. Most importantly, the promising effects in a phenotypic assay and molecular changes in the ex vivo model warrant further studies. We have also elucidated new mechanisms for the unexplored wound healing bioactivities of marine cyanobacterial compounds.
MATERIALS AND METHODS
Details of experimental procedures are provided in the Supporting Information.
Supplementary Material
Acknowledgments
Research was supported in part by the National Institutes of Health, NCI grant R01CA172310, and the Debbie and Sylvia DeSantis Chair professorship (H.L.). We thank Y. Zhang from the Gene Expression and Genotyping Core of the Interdisciplinary Center for Biotechnology Research in UF for the assistance in transcriptome profiling. We also thank A. Riva from the Bioinformatics Core of the Interdisciplinary Center for Biotechnology Research for assistance in transcriptome data analysis and heat map generation. We thank the support from the China Scholarship Council (W.Z.) to work in the H.L. lab.
Footnotes
W.C., cell-based assays, IPA analysis, ex vivo rabbit experiment, experimental design, and manuscript writing; S.S., RT-qPCR experiment of rabbit corneal samples; D.J.G., supervised ex vivo rabbit studies. L.A.S., S.M., R.R., natural product chemistry; W.Z., total synthesis; Q.Y.C., S.D., scale-up total synthesis. V.J.P., sample collections and identification and edited manuscript; H.L., designed and supervised experiments and edited manuscript.
Supplementary methods, including isolation, configurational analysis, biological assays (cell viability, VEGF-A secretion, immunoblot analysis, angiogenesis, RT-qPCR, transcriptome profiling, ex vivo organ culture and mRNA measurements), total synthesis, supplementary references, Tables, Figures and NMR spectra
References
- 1.Clark RA. Cutaneous tissue repair: basic biologic considerations. I J Am Acad Dermatol. 1985;13:701–725. doi: 10.1016/s0190-9622(85)70213-7. [DOI] [PubMed] [Google Scholar]
- 2.Greaves NS, Ashcroft KJ, Baguneid M, Bayat A. Current understanding of molecular and cellular mechanisms in fibroplasia and angiogenesis during acute wound healing. J Dermatol Sci. 2013;72:206–217. doi: 10.1016/j.jdermsci.2013.07.008. [DOI] [PubMed] [Google Scholar]
- 3.Goren I, Müller E, Schiefelbein D, Gutwein P, Seitz O, Pfeilschifter J, Frank S. Akt1 controls insulin-driven VEGF biosynthesis from keratinocytes: implications for normal and diabetes-impaired skin repair in mice. J Invest Dermatol. 2009;129:752–764. doi: 10.1038/jid.2008.230. [DOI] [PubMed] [Google Scholar]
- 4.Barrientos S, Stojadinovic O, Golinko MS, Brem H, Tomic-Canic M. Growth factors and cytokines in wound healing. Wound Repair Regen. 2008;16:585–601. doi: 10.1111/j.1524-475X.2008.00410.x. [DOI] [PubMed] [Google Scholar]
- 5.Falanga V. Wound healing and its impairment in the diabetic foot. Lancet. 2005;366:1736–1743. doi: 10.1016/S0140-6736(05)67700-8. [DOI] [PubMed] [Google Scholar]
- 6.Brem H, Tomic-Canic M. Cellular and molecular basis of wound healing in diabetes. J Clin Invest. 2007;117:1219. doi: 10.1172/JCI32169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrientos S, Brem H, Stojadinovic O, Tomic-Canic M. Clinical application of growth factors and cytokines in wound healing. Wound Repair Regen. 2014;22:569–578. doi: 10.1111/wrr.12205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dou C, Lay F, Ansari AM, Rees DJ, Ahmed AK, Kovbasnjuk O, Matsangos AE, Du J, Hosseini SM, Steenbergen C, Fox-Talbot K, Tabor AT, Williams JA, Liu L, Marti GP, Harmon JW. Strengthening the skin with topical delivery of keratinocyte growth factor-1 using a novel DNA plasmid. Mol Ther. 2014;22:752–761. doi: 10.1038/mt.2014.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kryger Z, Zhang F, Dogan T, Cheng C, Lineaweaver WC, Buncke HJ. The effects of VEGF on survival of a random flap in the rat: examination of various routes of administration. Br J Plast Surg. 2000;53:234–239. doi: 10.1054/bjps.1999.3315. [DOI] [PubMed] [Google Scholar]
- 10.Andrews SN, Jeong E, Prausnitz MR. Transdermal delivery of molecules is limited by full epidermis, not just stratum corneum. Pharm Res. 2013;30:1099–1109. doi: 10.1007/s11095-012-0946-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ferrara N. Role of vascular endothelial growth factor in the regulation of angiogenesis. Kidney Int. 1999;56:794–814. doi: 10.1046/j.1523-1755.1999.00610.x. [DOI] [PubMed] [Google Scholar]
- 12.Ferrara N, Hillan KJ, Gerber HP, Novotny W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat Rev Drug Discov. 2004;3:391–400. doi: 10.1038/nrd1381. [DOI] [PubMed] [Google Scholar]
- 13.McColl BK, Stacker SA, Achen MG. Molecular regulation of the VEGF family - Inducers of angiogenesis and lymphangiogenesis. Apmis. 2004;112:463–480. doi: 10.1111/j.1600-0463.2004.apm11207-0807.x. [DOI] [PubMed] [Google Scholar]
- 14.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473:298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rossiter H, Barresi C, Pammer J, Rendl M, Haigh J, Wagner EF, Tschachler E. Loss of vascular endothelial growth factor A activity in murine epidermal keratinocytes delays wound healing and inhibits tumor formation. Cancer Res. 2004;64:3508–3516. doi: 10.1158/0008-5472.CAN-03-2581. [DOI] [PubMed] [Google Scholar]
- 16.Frank S, Hübner G, Breier G, Longaker MT, Greenhalgh DG, Werner S. Regulation of vascular endothelial growth factor expression in cultured keratinocytes. Implications for normal and impaired wound healing. J Biol Chem. 1995;270:12607–12613. doi: 10.1074/jbc.270.21.12607. [DOI] [PubMed] [Google Scholar]
- 17.Brown LF, Yeo K, Berse B, Yeo TK, Senger DR, Dvorak HF, Van De Water L. Expression of vascular permeability factor (vascular endothelial growth factor) by epidermal keratinocytes during wound healing. J Exp Med. 1992;176:1375–1379. doi: 10.1084/jem.176.5.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao P, Kodra A, Tomic-Canic M, Golinko MS, Ehrlich HP, Brem H. The Role of Vascular Endothelial Growth Factor in Wound Healing. J Surg Res. 2009;153:347–358. doi: 10.1016/j.jss.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Di Peppe SR, Mangoni A, Zambruno G, Spinetti G, Melillo G, Napolitano M, Capogrossi MC. Adenovirus-mediated VEGF(165) gene transfer enhances wound healing by promoting angiogenesis in CD1 diabetic mice. Gene Ther. 2002;9:1271. doi: 10.1038/sj.gt.3301798. [DOI] [PubMed] [Google Scholar]
- 20.Galeano M, Deodato B, Altavilla D, Cucinotta D, Arsic N, Marini H, Torre V, Giacca M, Squadrito F. Adeno-associated viral vector-mediated human vascular endothelial growth factor gene transfer stimulates angiogenesis and wound healing in the genetically diabetic mouse. Diabetologia. 2003;46:546–555. doi: 10.1007/s00125-003-1064-1. [DOI] [PubMed] [Google Scholar]
- 21.Michaels J, Dobryansky M, Galiano RD, Bhatt KA, Ashinoff R, Ceradini DJ, Gurtner GC. Topical vascular endothelial growth factor reverses delayed wound healing secondary to angiogenesis inhibitor administration. Wound Repair Regen. 2005;13:506–512. doi: 10.1111/j.1067-1927.2005.00071.x. [DOI] [PubMed] [Google Scholar]
- 22.Brem H, Kodra A, Golinko MS, Entero H, Stojadinovic O, Wang VM, Sheahan CM, Weinberg AD, Woo SLC, Ehrlich HP, Tomic-Canic M. Mechanism of sustained release of vascular endothelial growth factor in accelerating experimental diabetic healing. J Invest Dermatol. 2009;129:2275–2287. doi: 10.1038/jid.2009.26. [DOI] [PubMed] [Google Scholar]
- 23.Forrest RD. Early history of wound treatment. J R Soc Med. 1982;75:198–205. doi: 10.1177/014107688207500310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reichrath J, Lehmann B, Carlberg C, Varani J, Zouboulis CC. Vitamins as hormones. Horm Metab Res. 2007;39:71–84. doi: 10.1055/s-2007-958715. [DOI] [PubMed] [Google Scholar]
- 25.Lin TS, Abd Latiff A, Abd Hamid NA, Ngah W, Mazlan M. Evaluation of Topical Tocopherol Cream on Cutaneous Wound Healing in Streptozotocin-Induced Diabetic Rats. Evidence-Based Complement. Altern Med. 2012 doi: 10.1155/2012/491027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacKay D, Miller AL. Nutritional support for wound healing. Altern Med Rev. 2003;8:359–378. [PubMed] [Google Scholar]
- 27.Shim KM, Choi SH, Jeong MJ, Kang SS. Effects of aucubin on the healing of oral wounds. In Vivo (Brooklyn) 2007;21:1037–1041. [PubMed] [Google Scholar]
- 28.Sevimli-Gür C, Onbaşılar I, Atilla P, Genç R, Çakarc N, Deliloğlu-Gürhan I, Bedir E. In vitro growth stimulatory and in vivo wound healing studies on cycloartane-type saponins of Astragalus genus. J Ethnopharmacol. 2011;134:844–850. doi: 10.1016/j.jep.2011.01.030. [DOI] [PubMed] [Google Scholar]
- 29.Shukla A, Rasik AM, Jain GK, Shankar R, Kulshrestha DK, Dhawan BN. In vitro and in vivo wound healing activity of asiaticoside isolated from Centella asiatica. J Ethnopharmacol. 1999;65:1–11. doi: 10.1016/s0378-8741(98)00141-x. [DOI] [PubMed] [Google Scholar]
- 30.Moon EJ, Lee YM, Lee OH, Lee MJ, Lee SK, Chung MH, Park YI, Sung CK, Choi JS, Kim KW. A novel angiogenic factor derived from Aloe vera gel: beta-sitosterol, a plant sterol. Angiogenesis. 1999;3:117–123. doi: 10.1023/a:1009058232389. [DOI] [PubMed] [Google Scholar]
- 31.Schmidt CA, Murillo R, Bruhn T, Bringmann G, Goettert M, Heinzmann B, Brecht V, Laufer SA, Merfort I. Catechin derivatives from Parapiptadenia rigida with in vitro wound-healing properties. J Nat Prod. 2010;73:2035–2041. doi: 10.1021/np100523s. [DOI] [PubMed] [Google Scholar]
- 32.Clericuzio M, Tinello S, Burlando B, Ranzato E, Martinotti S, Cornara L, La Rocca A. Flavonoid oligoglycosides from Ophioglossum vulgatum L. Having wound healing properties. Planta Med. 2012;78:1639–1644. doi: 10.1055/s-0032-1315149. [DOI] [PubMed] [Google Scholar]
- 33.Wang R, Lechtenberg M, Sendker J, Petereit F, Deters A, Hensel A. Wound-healing plants from TCM: In vitro investigations on selected TCM plants and their influence on human dermal fibroblasts and keratinocytes. Fitoterapia. 2013;84:308–317. doi: 10.1016/j.fitote.2012.12.020. [DOI] [PubMed] [Google Scholar]
- 34.Sharifi R, Pasalar P, Kamalinejad M, Dehpour AR, Tavangar SM, Paknejad M, Mehrabani Natanzi M, Nourbakhsh M, Ahmadi Ashtiani HR, Akbari M, Rastegar H. The effect of silymarin (Silybum marianum) on human skin fibroblasts in an in vitro wound healing model. Pharm Biol. 2013;51:298–303. doi: 10.3109/13880209.2012.721789. [DOI] [PubMed] [Google Scholar]
- 35.Porras-Reyes BH, Lewis WH, Roman J, Simchowitz L, Mustoe TA. Enhancement of wound healing by the alkaloid taspine defining mechanism of action. Proc Soc Exp Biol Med. 1993;203:18–25. doi: 10.3181/00379727-203-43567. [DOI] [PubMed] [Google Scholar]
- 36.Nesterova YV, Povetieva TN, Suslov NI, Zhdanov VV, Hrichkova TY, Udut EV, Chaykovskiy AS, Gaydamovich NN, Andreeva TI, Dygai AM. Regeneratory characteristics of complex extract and isolated diterpene alkaloids of aconitum baikalense. Bull Exp Biol Med. 2012;152:439–443. doi: 10.1007/s10517-012-1548-4. [DOI] [PubMed] [Google Scholar]
- 37.Tsala DE, Amadou D, Habtemariam S. Natural wound healing and bioactive natural products. Phytopharmacology. 2013;4:532–560. [Google Scholar]
- 38.Mayer AMS, Glaser KB, Cuevas C, Jacobs RS, Kem W, Little RD, McIntosh JM, Newman DJ, Potts BC, Shuster DE. The odyssey of marine pharmaceuticals: a current pipeline perspective. Trends Pharmacol Sci. 2010;31:255–265. doi: 10.1016/j.tips.2010.02.005. [DOI] [PubMed] [Google Scholar]
- 39.Day DR, Jabaiah S, Jacobs RS, Little RD. Cyclodextrin formulation of the marine natural product pseudopterosin a uncovers optimal pharmacodynamics in proliferation studies of human umbilical vein endothelial cells. Mar Drugs. 2013;11:3258–3271. doi: 10.3390/md11093258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tan LT. Bioactive natural products from marine cyanobacteria for drug discovery. Phytochemistry. 2007;68:954–979. doi: 10.1016/j.phytochem.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 41.Luesch H, Yoshida WY, Moore RE, Paul VJ, Corbett TH. Total structure determination of apratoxin A, a potent novel cytotoxin from the marine cyanobacterium Lyngbya majuscula. J Am Chem Soc. 2001;123:5418–5423. doi: 10.1021/ja010453j. [DOI] [PubMed] [Google Scholar]
- 42.Poterała M, Plenkiewicz J. Synthesis of new chiral ionic liquids from α-hydroxycarboxylic acids. Tetrahedron Asymmetry. 2011;22:294–299. [Google Scholar]
- 43.Grab T, Bräse S. Efficient synthesis of lactate-containing depsipeptides by the mitsunobu reaction of lactates. Adv Synth Catal. 2005;347:1765–1768. [Google Scholar]
- 44.Conroy T, Guo JT, Linington RG, Hunt NH, Payne RJ. Total synthesis, stereochemical assignment, and antimalarial activity of gallinamide A. Chem- A Eur J. 2011;17:13544–13552. doi: 10.1002/chem.201102538. [DOI] [PubMed] [Google Scholar]
- 45.Chen Y, Bilban M, Foster CA, Boger DL. Solution-phase parallel synthesis of a pharmacophore library of HUN-7293 analogues: A general chemical mutagenesis approach to defining structure-function properties of naturally occurring cyclic (depsi)peptides. J Am Chem Soc. 2002;124:5431–5440. doi: 10.1021/ja020166v. [DOI] [PubMed] [Google Scholar]
- 46.Chen QY, Liu Y, Cai W, Luesch H. Improved total synthesis and biological evaluation of potent apratoxin S4 based anticancer agents with differential stability and further enhanced activity. J Med Chem. 2014;57:3011–3029. doi: 10.1021/jm4019965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen QY, Liu Y, Luesch H. Systematic chemical mutagenesis identifies a potent novel apratoxin A/E hybrid with improved in vivo antitumor activity. ACS Med Chem Lett. 2011;2:861–865. doi: 10.1021/ml200176m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reimold AM, Etkin A, Clauss I, Perkins A, Friend DS, Zhang J, Horton HF, Scott A, Orkin SH, Byrne MC, Grusby MJ, Glimcher LH. An essential role in liver development for transcription factor XBP-1. Genes Dev. 2000;14:152–157. [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang P, McGrath B, Li S, Frank A, Zambito F, Reinert J, Gannon M, Ma K, McNaughton K, Cavener DR. The PERK Eukaryotic Initiation Factor 2α Kinase Is Required for the Development of the Skeletal System, Postnatal Growth, and the Function and Viability of the Pancreas. Mol Cell Biol. 2002;22:3864–3874. doi: 10.1128/MCB.22.11.3864-3874.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yang X, Matsuda K, Bialek P, Jacquot S, Masuoka HC, Schinke T, Li L, Brancorsini S, Sassone-Corsi P, Townes TM, Hanauer A, Karsenty G. ATF4 is a substrate of RSK2 and an essential regulator of osteoblast biology: Implication for Coffin-Lowry syndrome. Cell. 2004;117:387–398. doi: 10.1016/s0092-8674(04)00344-7. [DOI] [PubMed] [Google Scholar]
- 51.Reimold aM, Iwakoshi NN, Manis J, Vallabhajosyula P, Szomolanyi-Tsuda E, Gravallese EM, Friend D, Grusby MJ, Alt F, Glimcher LH. Plasma cell differentiation requires the transcription factor XBP-1. Nature. 2001;412:300–307. doi: 10.1038/35085509. [DOI] [PubMed] [Google Scholar]
- 52.Iwakoshi NN, Lee AH, Vallabhajosyula P, Otipoby KL, Rajewsky K, Glimcher LH. Plasma cell differentiation and the unfolded protein response intersect at the transcription factor XBP-1. Nat Immunol. 2003;4:321–329. doi: 10.1038/ni907. [DOI] [PubMed] [Google Scholar]
- 53.Scheuner D, Song B, McEwen E, Liu C, Laybutt R, Gillespie P, Saunders T, Bonner-Weir S, Kaufman RJ. Translational control is required for the unfolded protein response and in vivo glucose homeostasis. Mol Cell. 2001;7:1165–1176. doi: 10.1016/s1097-2765(01)00265-9. [DOI] [PubMed] [Google Scholar]
- 54.Iwawaki T, Akai R, Yamanaka S, Kohno K. Function of IRE1 alpha in the placenta is essential for placental development and embryonic viability. Proc Natl Acad Sci. 2009;106:16657–16662. doi: 10.1073/pnas.0903775106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schürmann C, Goren I, Linke A, Pfeilschifter J, Frank S. Deregulated unfolded protein response in chronic wounds of diabetic ob/ob mice: A potential connection to inflammatory and angiogenic disorders in diabetes-impaired wound healing. Biochem Biophys Res Commun. 2014;446:195–200. doi: 10.1016/j.bbrc.2014.02.085. [DOI] [PubMed] [Google Scholar]
- 56.Lee J, Ozcan U. Unfolded protein response signaling and metabolic diseases. J Biol Chem. 2014;289:1203–1211. doi: 10.1074/jbc.R113.534743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ghosh R, Lipson KL, Sargent KE, Mercurio AM, Hunt JS, Ron D, Urano F. Transcriptional regulation of VEGF-A by the unfolded protein response pathway. PLoS One. 2010;5:e9575. doi: 10.1371/journal.pone.0009575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ozawa K, Tsukamoto Y, Hori O, Kitao Y, Yanagi H, Stern DM, Ogawa S. Regulation of tumor angiogenesis by oxygen-regulated protein 150, an inducible endoplasmic reticulum chaperone. Cancer Res. 2001;61:4206–4213. [PubMed] [Google Scholar]
- 59.Ozawa K, Kondo T, Hori O, Kitao Y, Stern DM, Eisenmenger W, Ogawa S, Ohshima T. Expression of the oxygen-regulated protein ORP150 accelerates wound healing by modulating intracellular VEGF transport. J Clin Invest. 2001;108:41–50. doi: 10.1172/JCI11772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pereira ER, Liao N, Neale GA, Hendershot LM. Transcriptional and post-transcriptional regulation of proangiogenic factors by the unfolded protein response. PLoS One. 2010;5:e12521. doi: 10.1371/journal.pone.0012521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Nathaniel Roybal C, Hunsaker LA, Barbash O, Vander Jagt DL, Abcouwer SF. The oxidative stressor arsenite activates vascular endothelial growth factor mRNA transcription by an ATF4-dependent mechanism. J Biol Chem. 2005;280:20331–20339. doi: 10.1074/jbc.M411275200. [DOI] [PubMed] [Google Scholar]
- 62.Malabanan KP, Kanellakis P, Bobik A, Khachigian LM. Activation Transcription Factor-4 Induced by Fibroblast Growth Factor-2 Regulates Regulates VEGF-A transcription in Vascular Smooth Muscle Cells. Circ Res. 2008;103:378–387. doi: 10.1161/CIRCRESAHA.107.168682. [DOI] [PubMed] [Google Scholar]
- 63.Zhu K, Jiao H, Li S, Cao H, Galson DL, Zhao Z, Zhao X, Lai Y, Fan J, Im HJ, Chen D, Xiao G. ATF4 promotes bone angiogenesis by increasing vegf expression and release in the bone environment. J Bone Miner Res. 2013;28:1870–1884. doi: 10.1002/jbmr.1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wang JM, Qiu Y, Yang ZQ, Li L, Zhang K. Inositol-requiring enzyme 1 facilitates Diabetic wound healing through modulating micrornas. Diabetes. 2017;66:177–192. doi: 10.2337/db16-0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gibson DJ, Schultz GS. A corneal scarring model. Methods Mol Biol. 2013;1037:277–298. doi: 10.1007/978-1-62703-505-7_16. [DOI] [PubMed] [Google Scholar]
- 66.Sriram S, Gibson DJ, Robinson P, Pi L, Tuli S, Lewin AS, Schultz G. Assessment of anti-scarring therapies in exvivo organ cultured rabbit corneas. Exp Eye Res. 2014;125:173–182. doi: 10.1016/j.exer.2014.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.