Abstract
The present study aimed to investigate the contents of glucosinolates (GSLs) and carotenoids in eleven varieties of Chinese cabbage in relation to the expression level of the important transcription factors. MS and HPLC analysis identified the presence of 13 GSLs (progoitrin, sinigrin, glucoalyssin, gluconapoleiferin, gluconapin, glucocochlearin, glucobrassicanapin, glucoerucin, 4-hydroxyglucobrassicin, glucobrassicin, 4-methoxyglucobrassicin, neoglucobrassicin and gluconasturtiin) and four carotenoids (lutein, zeaxanthin, α-carotene and β-carotene). GSL contents were varied among the different cabbage varieties. The total GSL content ranged from 2.7 to 57.88 μmol/g DW. The proportion of gluconapin (54%) and glucobrassicanapin (22%) was higher in all the varieties, respectively. Results documented the variation in total and individual carotenoid contents that have also been observed among different varieties; however, the total carotenoid contents ranged from 289.12 to 1001.41 mg kg−1 DW (mean 467.66). Interestingly, the proportion of lutein (66.5) and β-carotene (25.9) were higher than α-carotene (5.1) and zeaxanthin (2.5%). Consequently, the expression level of the regulatory gene, MYB28 was higher in ‘K0648’ and was directly proportional to GSL content. Similarly, the expression levels of 1-PSY were higher in ‘K0112’; however, the expression levels of 2-ZDS, 3-LCYB, 4-LCYE, 5-CHXB and 7-NCED genes showed no significant difference. In addition, the correlation between GSL and carotenoid contents and gene expression level showed moderate significant difference in each Chinese cabbage.
Keywords: Chinese cabbage, Glucosinolates, Carotenoids, HPLC, Gene expression
1. Introduction
The vegetables that belonged to the Brassicaceae family are economically important in the Korean vegetable market. Many commonly consumed vegetables such as broccoli, cabbage, cauliflower, Chinese cabbage, turnip, common radish and horseradish belonged to the Brassicaceae family. Among them, Chinese cabbage (Brassica rapa L. ssp. pekinensis) is one of the most important Brassica vegetables because of its regular consumption rate in Korea (Cartea et al., 2011, Cho et al., 1999). Several classes of secondary metabolites such as carotenoids, flavonoids, glucosinolates (GSLs), and other phytochemicals have been identified and quantified from the Chinese cabbage. Their compositions and contents depended on the cultivation conditions (Reif et al., 2013, Lee et al., 2015). The phytochemicals, especially GSLs and other functional compounds exhibit potential medical applications such as antidiabetic and anti-cancer agents; therefore, the interest toward this vegetable is increasing worldwide.
Plant pigments such as anthocyanins and carotenoids widely involved the metabolic and physiological regulation of plant metabolism (Park et al., 2012). Similarly, these pigment compounds play an important role in defensive mechanisms apart from the essential nutrients. Among the pigments, α- and β-carotene are the main backbone and intermediary metabolites for the synthesis of vitamin A. Vitamin A protects the human body from xerophthalmia, blindness, and premature death. Moreover, lutein and zeaxanthin have recently been recognized to be beneficial for eye health to the prevention of age-related macular degeneration (Krishnadev et al., 2010).
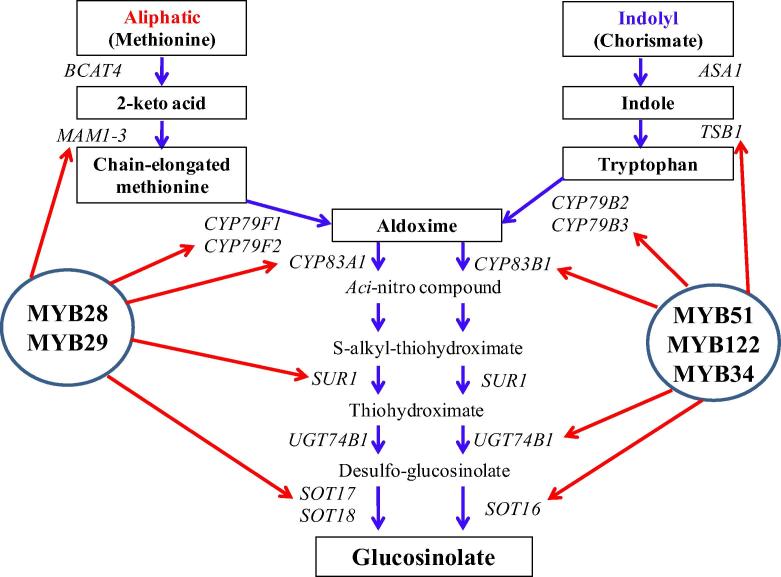
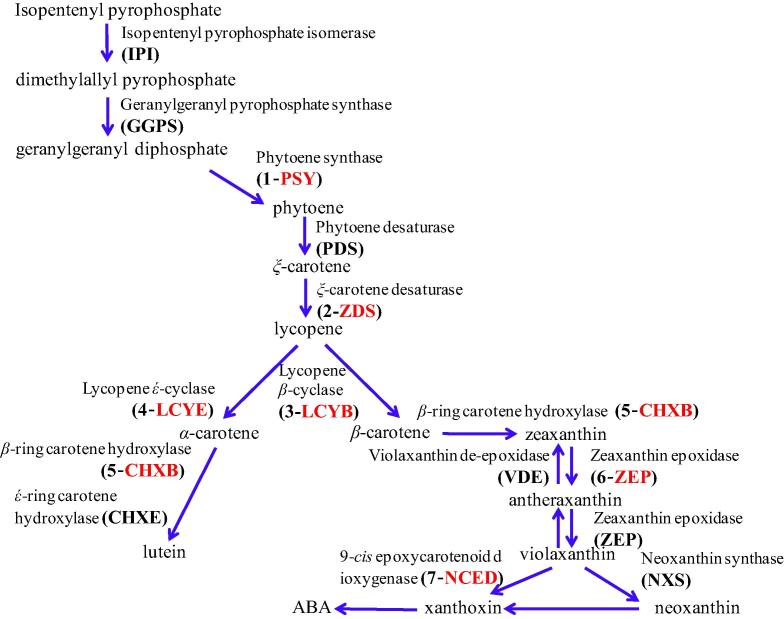
In Brassica vegetable, GSL biosynthesis is regulated by various responses. In the case of biotic and abiotic stress responses, R2R3-MYB transcription factors (TFs) are maintained the GSL biosynthesis. MYB TFs can be divided into 2 groups: MYB28, MYB76, and MYB29 control the biosynthesis of high aliphatic GSLs, while MYB51, MYB122, and MYB34, alternatively called high indolyl GSLs, can be manipulated to coordinately control the suite of enzymes that synthesize indolyl GSLs (Fig. 1) (Kim et al., 2014). Recently, Kim et al. (2013) reported GSL contents and transcription factors in different organs of Chinese cabbage. Additionally, Tuan et al. (2012) reported the carotenoid accumulation and expression of carotenogenesis gene development in Chinese cabbage (Fig. 2). However, gene expression and accumulation of GSLs and carotenoids in different varieties of Chinese cabbage have not been reported. Therefore, the objectives of this study were to quantify the gene expression and levels of GSLs and carotenoids and to discuss the correlation between them in 11 varieties of Chinese cabbage.
Figure 1.
Schematic representation of glucosinolate biosynthesis (Yatusevich et al., 2010). In this study MYB28, 29, 34, 122 were used.
Figure 2.
Carotenoid biosynthetic pathway (Tuan et al., 2012). In this study PSY, ZDS, LCYB, LCYE, CHXB, ZEP, NCED were used.
2. Materials and methods
2.1. Analysis reagents
Sinigrin and aryl sulfatase were purchased from Sigma–Aldrich. DEAE Sephadex A-25 was obtained from GE Healthcare Bio-Sciences AB (Uppsala, Sweden). Sodium acetate was provided from Samchun Pure Chemicals Co. Ltd. (Pyeongtaek, Korea). Routine reagents and chemicals used for HPLC were obtained from J.T. Baker Chemical Co. (Phillipsburg, NJ, USA).
2.2. Plant materials
Seeds of Chinese cabbage ‘CR Hakwang’ were procured from Asia seed Co. Ltd. (Seoul, Korea), ‘Chunkwang’ was obtained from Sakata Korea Seed Co. Ltd (Seoul, Korea) and ‘Bulam No. 3’, ‘BulamPlus’ were purchased from Hungnong seed Co. Ltd (Pyeongtaek, Korea). The National Agrobiodiversity Center (National Academy of Agricultural Science, Rural Development Administration, Jeonju, Korea) kindly provided seven cultivars of Chinese cabbage (‘K0015’, ‘K0112’, ‘K0416’, ‘K0461’, ‘K0588’, ‘K0648’, ‘K0651’).
2.3. Cultivation of 11 varieties of Chinese cabbage
The seeds of Chinese cabbages were sown on 72 hole-trays with ‘High’ bed soil (Punong, Gyeongju, Korea) on 28 August, 2014 at Greenhouse of the Chungnam National University (latitude, 127°35′E; longitude, 36°36′N). At 14 days after sowing (DAS), seedlings were transplanted to the individual pot (Ø 13 × 13 cm), each pot containing bed soil. 1 g of fertilizers (N-P-K, 12-10-9, Namhae Chemical Corp, Seoul, Korea) was applied on per pot in bed soil. At 49 DAS, 11 varieties of Chinese cabbage were harvested.
2.4. Extraction and desulfation of glucosinolates
Glucosinolates were extracted and desulfated by the method of Kim et al., 2007, International Standards Organization, 1992 as follows. Hundred mg of the freeze dried samples was mixed with boiling 70% (v/v) methanol for 5 min in a water bath at 70 °C. After proper mixing of the samples and centrifugation (12,000 rpm, 4 °C, 10 min), the tubes were collected very carefully, and debris free supernatant was transferred into fresh tubes for the extraction of GSLs. The crude GSLs were desulfated by the addition of 75 μL of an aryl sulfatase (E.C.3.1.6.1) solution in a mini-column packed with DEAE-Sephadex A-25 previously activated with 0.5 M sodium acetate (ca. >40 mg as dry matrix) in an anion exchange column. Aryl sulfatase (E.C.3.1.6.1) (75 μl) was transferred into the column for the complete desulfation of GSLs. After desulfation reaction for 16–18 h at room temperature, the desulfated GSLs were eluted with 0.5 ml (×3 times) of ultra-pure water. The eluates were filtered through 0.45 μm Teflon PTFE syringe filter and analyzed immediately by HPLC or stored at 4 or −20 °C until further chemical analysis.
2.5. Quantification of glucosinolates
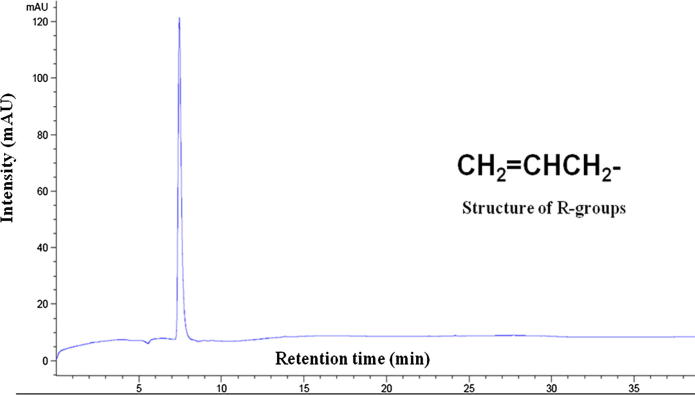
For quantitative analysis, separation of DS-GSL was conducted on a C18 column (150 × 3.0 mm i.d., 3 μm, Inertsil ODS-3; GL Sciences, Tokyo, Japan) using a Agilent 1200 Series HPLC system equipped with a diode array detector (Agilent Technologies, CA, USA). The UV–Visible detector wavelength was set at 227 nm. The elution solvent consisted of solvent A (ultra-pure water) and solvent B (acetonitrile). The samples were run for 27 min to separate entire compounds. The gradient solvent system used for the HPLC separation is mentioned in our previous report (Lee et al., 2014). Standard sinigrin was used for the quantification of individual components (Fig. 3).
Figure 3.
HPLC chromatogram and chemical structure of the standard glucosinolates.
2.6. Quantification of carotenoid by HPLC
Carotenoids were extracted and slightly modified Tuan et al. (2012) and Kim et al. (2015). Briefly, 5 ml of ethanol was mixed with 500 mg of lyophilized powder of the samples and mixed thoroughly for 30 s, further kept in the boiling water bath (75 °C) for 5 min for subsequent addition of 1.5 ml of potassium hydroxide. After incubation, the samples were kept in the ice cold condition for 10 min, to that 2.5 ml of ultra-pure water was added to suspend the compounds. Further, the crude samples were separated by the addition of an equal volume of hexane. The organic solvent layer was separated by centrifugation at 3000 rpm for 3 min. The collected upper layer of the extract was evaporated at room temperature and 1 ml of dichloromethane/methanol (50:50, v/v) was added to dissolve the collected organic layer (slightly modified from Tuan et al. (2012)).
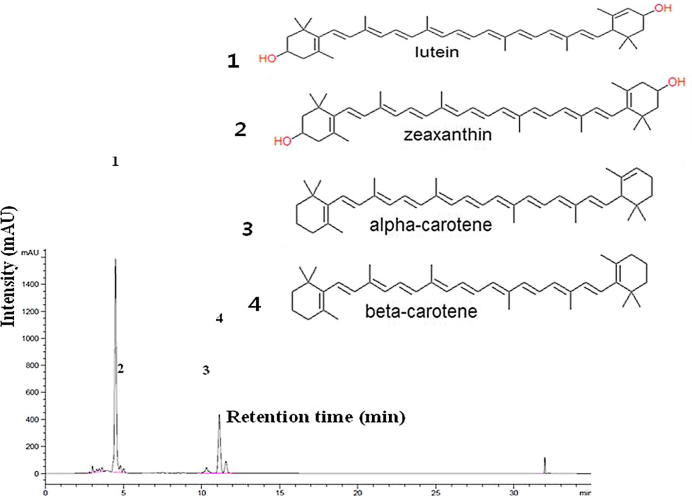
Agilent 1200 Series HPLC system with C30 column was used for the quantification of the carotenoid samples. The samples were separated by eluting with mixtures of solvents such as (A) 25:75 (v/v) water/methanol and solvent (B) 100% ethyl acetate with a flow rate of 1.0 ml/min for 35 min. The column oven temperature and the detection wavelength were maintained at 40 °C and 454 nm respectively. Individual carotenoids were quantified as mg kg−1 dry weight (DW) by comparing the peak area of standard lutein, zeaxanthin, α-carotene and β-carotene respectively (Fig. 4).
Figure 4.
HPLC chromatogram and chemical structure of the standard carotenoids.
2.7. Determination of total RNA for gene expression
2.7.1. Extraction of total RNA and cDNA synthesis
Total RNA was isolated from each development seedling using total RNA extraction Kit (GeneAll, Seoul, Korea). The concentration of total RNA was determined through the absorbance spectrum at 260:280 nm wave length by NanoVue Plus spectrophotometer (GE Healthcare Life Sciences, MA, USA). Total RNA was adjusted by the concentration by RNA free water, and the cDNA was synthesized from 1 μg of total RNA and performed on a total volume of 20 μl that contained 10 μl of 2× RT Pre-mix (SolGent, Daejeon, Korea), 50 μM (each) of oligo (dT) 20 primers (Toyobo, Osaka, Japan). The PCR reaction protocol was as follows: initial denaturation at 95 °C for 15 min, followed by 40 cycles of denaturation at 95 °C for 15 s, and then a final cycle at 72 °C for 20 s. The resulting cDNA products were used for real-time PCR analysis.
2.7.2. Quantitative determination of total RNA by real-time polymerase chain reaction (RT-PCR)
Quantitative real-time PCR was performed for transcriptional level analysis of glucosinolate biosynthesis and carotenogenesis genes from Chinese cabbage leaves using Bio-Rad CFX 96 real-time PCR system (Bio-Rad, Hercules, CA, USA). The gene-specific primers were designed for qRT-PCR as previously described (Table 1) (Kim et al., 2013, Tuan et al., 2012). A SYBR Green qRT-PCR was carried out in a 20 μl reaction volume including 1.0 μl each primer, 5 μl of 20-times diluted cDNA and 10 μl of 2× RT mix (SolGent, Daejeon, Korea) using the following conditions; 95 °C for 10 min, followed by 35 cycles of 95 °C for 10 s, annealing for 15 s at 60 °C, and then a final cycle at 72 °C for 15 s, followed by one cycle of 95 °C for 1 min, and one cycle of 55 °C for 1 min. RT-PCR results were calculated as the mean of three replicates and statistical differences between samples were evaluated by standard deviation in MS Excel 2007.
Table 1.
PCR primers for gene expression involved in GSL and carotenoid biosynthesis in Chinese cabbage.
| Name | Sequence (5′–3′) | Amplicon (base pair) | References |
|---|---|---|---|
| MYB28_RT F | ACCATACTGTCAACACGCCTCC | 219 | Gigolashvili et al., 2007, Hirai et al., 2007 |
| MYB28_RT R | CAGAAGTGACCTTAGCCGCAAC | ||
| MYB29_RT F | CTGTCTCCTCCGTGTCTCAA | 148 | |
| MYB29_RT R | CCTCGGCTGCATTGTTACTA | ||
| MYB34_RT F | ACACCGGCGACGTCGATTC | 229 | |
| MYB34_RT R | TCTAACTCCTCCATAAGGCCAACA | ||
| MYB51_RT F | GATCTCCGAAACCAGCAAATCA | 240 | |
| MYB51_RT R | GTAGTAATGAGTGGGCCACCACT | ||
| MYB122_RT F | TTGAATGATGTTGTATCTCATGATGATG | 190 | |
| MYB122_RT R | AGTTGTCAATCCCTTCAAAGGAAACA | ||
| BrPSY_RT F | GCTATCTACGTTTGGTGCAGAAGAA | 189 | Tuan et al. (2012) |
| BrPSY_RT R | AAATGGCTGAATATCGACAGGGTAT | ||
| BrZDS_RT F | CCTTCTTGTCAAAGACCACACTCAT | 160 | |
| BrZDS_RT R | AGCTAGTGAGTTCCTCAGCTTGTCA | ||
| BrLCYB_RT F | AAGATATCCAAGAGAGGATGGTTGC | 180 | |
| BrLCYB_RT R | CCACCATGTAACCTGTAGAAGGATG | ||
| BrLCYE_RT F | ATGGATGAACAGTCTAAGCTCGTTG | 185 | |
| BrLCYE_RT R | ACACCGTAGTTGTTTGTGAAAGGAA | ||
| BrCHXB_RT F | CAGAGAAAACAAGCTCTCTGGACAC | 185 | |
| BrCHXB_RT R | CATCTGCCAAGAGAATCGGTAGTAA | ||
| BrZEP_RT F | AGACTTAAGCGCCATAAGAGGAGAA | 185 | |
| BrZEP_RT R | ACTTGACATACCAAGTGCCAGAGAC | ||
| BrNCED_RT F | CACATCCTCTGTTTTGTTCACGAC | 171 | |
| BrNCED_RT R | AAGAGTTTGTTCCTGGAGTTGTTCC | ||
2.8. Statistical analysis
The Tukey’s multiple range test was performed to determine which group was significantly different at p ⩽ 0.05 among GSL and carotenoid contents and gene expression levels using SPSS statistical software (version 11.5 for Windows, SPPS Inc., Chicago, IL, USA). All the data are expressed as the mean ± the standard deviation with three replications.
The Pearson correlation (using SPSS version 11.0 for Windows SPSS Inc., Chicago, IL, USA) was used to determine relationships between GSL and carotenoid contents and their biosynthesis genes of Chinese cabbages. The Pearson’s correlation coefficients (r) and p value were used to show correlations and evaluate ∗ and ∗∗ which indicate significance at 5% and 1% levels, respectively.
3. Results and discussion
3.1. Plant growth of Chinese cabbage
The weight and water content of Chinese cabbage leaves are shown in Table 2. Water contents in 11 Chinese cabbages were 80.4%. Among 11 Chinese cabbages, some varieties (‘Chunkwang’, ‘CR Hakwang’, ‘Bulam No. 3’, ‘Bulam Plus’, ‘K0015’, ‘K0416, and ‘K0588’) look like general Chinese cabbages, but others (‘K0112’, ‘K0416’, ‘K0648’ and ‘K0651’) look like turnips.
Table 2.
Moisture content of eleven varieties of Chinese cabbage (n = 3).
| Variety | Fresh weight (g) | Dry weight (g) | Water content (%) |
|---|---|---|---|
| Chunkwang | 57.92 ± 10.57 | 9.31 ± 2.25 | 84.0 ± 1.4 |
| CR Hakwang | 35.78 ± 9.27 | 6.12 ± 1.26 | 82.7 ± 1.2 |
| Bulam No. 3 | 55.91 ± 3.21 | 7.58 ± 0.16 | 86.4 ± 0.5 |
| Bulam Plus | 44.50 ± 4.01 | 8.63 ± 3.02 | 80.7 ± 6.2 |
| K0015 | 65.01 ± 9.55 | 6.04 ± 1.53 | 90.8 ± 1.1 |
| K0112 | 25.71 ± 6.50 | 5.24 ± 1.05 | 79.2 ± 3.7 |
| K0416 | 25.75 ± 5.75 | 6.76 ± 0.55 | 73.1 ± 2.0 |
| K0461 | 50.44 ± 10.67 | 8.30 ± 4.34 | 83.4 ± 8.9 |
| K0588 | 32.61 ± 5.09 | 5.46 ± 1.05 | 83.1 ± 3.2 |
| K0648 | 15.43 ± 3.96 | 4.24 ± 0.65 | 71.8 ± 4.7 |
| K0651 | 9.46 ± 2.90 | 2.78 ± 0.17 | 69.2 ± 6.8 |
| Mean | 38.05 ± 18.17 | 6.41 ± 1.97 | 80.4 ± 6.6 |
3.2. Glucosinolate contents in Chinese cabbages
Total and individual GSLs were investigated in 11 varieties of Chinese cabbages (Table 3). The results indicated that the total GSL contents ranged from 2.70 to 57.88 μmol g−1 DW (Table 4). The total GSL levels in the different cultivars were in the following order: ‘K0416’ (57.88) > ‘K0112’ (25.99) > ‘K0461’ (12.77) > ‘K0648’ (11.86) > ‘Chunkwang’ (11.20) > ‘CR Hakwang’ (10.72) > ‘K0015’ (9.76) > ‘BulamPlus’ (8.07) > ‘Bulam No. 3’ (6.34) > ‘K0588’ (5.90) > ‘K0651’ (2.70). This result indicated that the total GSL levels varied significantly among the varieties of Chinese cabbage. Similarly, in the previous report (Lee et al., 2014), total GSL contents in sixty-two varieties of Chinese cabbage ranged from 2.83 to 48.53 μmol g−1 DW. Compared to the report of Hong et al. (2011), the contents of observed GSL were higher, especially, the proportion of gluconapin was 54%, glucobrassicanapin 22%, neoglucobrassicin, 4-methoxyglucobrassicin, glucobrassicin and progoitrin 4–6%, and other minor GSLs (glucoalyssin, sinigrin, gluconapoleiferin, 4-hydroxyglucobrassicin, glucocochlearin, gluconasturtiin, glucoerucin) less than 6%. In general the content of GSLs and the bitter taste have significant coincidence with the degradation products. Similarly, the amount of glucobrassicanapin and gluconapin in B. rapa is related to the bitterness (Padill et al., 2007) Glucobrassicin and 4-methoxyglucobrassicin represented the proportion (4.0% and 6.4%) of total GSLs in Chinese cabbage. The breakdown products from indolyl GSLs have been identified as functional compounds in various Brassica species, suggesting that they have an important role in the plant defence system. Particularly, genotype × environment interactions were a significant factor to handle the GSL levels. Plants generally respond to environmental stresses by inducing health-promoting phytochemicals as plant defense responses (Pedras et al., 2003).
Table 3.
Glucosinolates identified in Chinese cabbage.
| No.a | Trivial names | Semisystematic names of R-groupb | Compound Group | [M + H]+ (m/z)c | Response factord |
|---|---|---|---|---|---|
| 1 | Progoitrin | (2R)-2-Hydroxy-3-butenyl | Aliphatic | 310 | 1.09 |
| 2 | Sinigrin | 2-Propenyl | Aliphatic | 280 | 1.00 |
| 3 | Glucoalyssin | 5-Methylsufinylpentyl | Aliphatic | 372 | 1.07 |
| 4 | Gluconapoleiferin | 2-Hydroxy-pent-4-enyl | Aliphatic | 324 | 1.00 |
| 5 | Gluconapin | 3-Butenyl | Aliphatic | 294 | 1.11 |
| 6 | Glucocochlearine | 1-Methylpropyl | Aliphatic | 296 | 1.00 |
| 7 | 4-Hydroxyglucobrassicin | 4-Hydroxy-3-indolylmethyl | Indolyl | 385 | 0.28 |
| 8 | Glucobrassicanapin | 4-Pentenyl | Aliphatic | 308 | 1.15 |
| 9 | Glucoerucin | 4-Methylthiobutyl | Aliphatic | 342 | 1.04f |
| 10 | Glucobrassicin | 3-Indolymethyl | Indolyl | 369 | 0.29 |
| 11 | 4-Methoxyglucobrassicin | 4-Methoxy-3-indolylmethyl | Indolyl | 399 | 0.25 |
| 12 | Gluconasturtiin | 2-Phenylethyl | Aromatic | 344 | 0.95 |
| 13 | Neoglucobrassicin | 1-Methoxy-3-indolylmethyl | Indolyl | 399 | 0.20 |
No., the elution order of glucosinolates from HPLC chromatograms in Fig. 1.
R-groups in Fig. 1.
Confirmed by Yang and Quiros (2010).
According to Clarke (2010).
Table 4.
Glucosinolate contents (μmol g−1 DW) in eleven varieties of Chinese cabbage.
| No. | Trivial names | Commercial varieties |
|||||
|---|---|---|---|---|---|---|---|
| Chunkwang | CR Hakwang | Bulam No. 3 | Bulam Plus | K0015 | K0112 | ||
| 1 | Progoitrin | 0.45 ± 0.12ab | 1.20 ± 0.50a | 0.17 ± 0.08b | 0.96 ± 0.40ab | 0.34 ± 0.16ab | 0.14 ± 0.13b |
| 2 | Sinigrin | 0.04 ± 0.01ab | 0.06 ± 0.01ab | 0.05 ± 0.01ab | 0.05 ± 0.01ab | 0.07 ± 0.01a | 0.04bb |
| 3 | Glucoalyssin | 0.07 ± 0.01ab | 0.11 ± 0.02ab | 0.09 ± 0.01ab | 0.12 ± 0.02a | 0.10 ± 0.04ab | 0.11 ± 0.02ab |
| 4 | Gluconapoleiferin | 0.07 ± 0.01b | 0.44 ± 0.19ab | 0.07 ± 0.02b | 0.17 ± 0.08ab | 0.11 ± 0.04b | NDa |
| 5 | Gluconapin | 2.68 ± 1.11c | 1.04 ± 0.35c | 0.75 ± 0.61c | 1.33 ± 0.29c | 1.76 ± 0.70c | 22.64 ± 7.72b |
| 6 | Glucocochlearin | 0.07 ± 0.01a | 0.33ab | 0.06 ± 0.02a | 0.08 ± 0.04a | 0.06 ± 0.01a | 0.11ab |
| 7 | 4-Hydroxyglucobrassicin | 0.01cb | 0.01 ± 0.00c | 0.02 ± 0.00c | 0.01 ± 0.00c | 0.01 ± 0.00c | 0.08 ± 0.04a |
| 8 | Glucobrassicanapin | 4.68 ± 1.85a | 3.59 ± 1.34a | 2.98 ± 0.57a | 1.90 ± 0.44a | 5.05 ± 2.78a | 1.52 ± 1.35 |
| 9 | Glucoerucin | 0.30 ± 0.05a | 0.09 ± 0.02b | 0.15 ± 0.04ab | 0.19 ± 0.02ab | 0.13 ± 0.04b | 0.15 ± 0.07ab |
| 10 | Glucobrassicin | 0.49 ± 0.14a | 1.00 ± 0.23a | 0.46 ± 0.15a | 0.82 ± 0.43a | 0.41 ± 0.17a | 0.28 ± 0.24a |
| 11 | 4-Methoxyglucobrassicin | 1.62 ± 0.23ab | 1.87 ± 0.55a | 0.90 ± 0.34abc | 1.52 ± 0.27ab | 1.15 ± 0.51abc | 0.68 ± 0.17bc |
| 12 | Gluconasturtiin | 0.25 ± 0.06a | 0.24 ± 0.14a | 0.14 ± 0.09a | 0.12 ± 0.02a | 0.10 ± 0.02a | 0.13 ± 0.06a |
| 13 | Neoglucobrassicin | 0.48 ± 0.23a | 0.74 ± 0.30a | 0.51 ± 0.30a | 0.80 ± 0.49a | 0.47 ± 0.19a | 0.11 ± 0.04a |
| Total | 11.20 ± 3.47bc | 10.72 ± 3.57bc | 6.34 ± 0.74c | 8.07 ± 2.13c | 9.76 ± 4.04c | 25.99 ± 8.20b | |
| No. | Trivial names | Commercial varieties | |||||
| K0416 | K0461 | K0588 | K0648 | K0651 | Mean | ||
| 1 | Progoitrin | ND | 1.00 ± 0.86ab | 0.46 ± 0.28ab | 0.86 ± 0.10ab | 0.09bb | 0.57 ± 0.40 |
| 2 | Sinigrin | 0.07 ± 0.01ab | 0.05 ± 0.02ab | 0.04 ± 0.01ab | 0.06 ± 0.00ab | 0.03bb | 0.05 ± 0.01 |
| 3 | Glucoalyssin | 0.07ab | 0.10 ± 0.06ab | 0.10 ± 0.02ab | 0.05 ± 0.00ab | 0.05 ± 0.02ab | 0.09 ± 0.02 |
| 4 | Gluconapoleiferin | ND | 0.38 ± 0.38ab | 0.35 ± 0.32ab | 0.75 ± 0.38a | 0.08bb | 0.27 ± 0.23 |
| 5 | Gluconapin | 53.72 ± 9.49a | 1.71 ± 0.13c | 0.66 ± 0.36c | 1.43 ± 0.38c | 0.12 ± 0.10c | 7.98 ± 16.48 |
| 6 | Glucocochlearin | 0.04 ± 0.01a | 0.03 ± 0.01a | 0.05 ± 0.03a | 0.10 ± 0.07a | 0.13 ± 0.05a | 0.10 ± 0.08 |
| 7 | 4-Hydroxyglucobrassicin | 0.02 ± 0.01c | 0.02 ± 0.01c | 0.01 ± 0.01c | 0.04 ± 0.01abc | 0.05 ± 0.01ab | 0.03 ± 0.02 |
| 8 | Glucobrassicanapin | 2.42 ± 0.24a | 4.47 ± 0.16a | 2.19 ± 1.20a | 6.15 ± 6.19a | 0.16 ± 0.14a | 3.19 ± 1.78 |
| 9 | Glucoerucin | 0.07 ± 0.02b | 0.15 ± 0.05ab | 0.18 ± 0.01ab | 0.16 ± 0.11ab | 0.07 ± 0.03b | 0.15 ± 0.06 |
| 10 | Glucobrassicin | 0.38 ± 0.13a | 0.55 ± 0.43a | 0.41 ± 0.19a | 1.02 ± 0.44a | 0.67 ± 0.24a | 0.59 ± 0.25 |
| 11 | 4-Methoxyglucobrassicin | 0.27 ± 0.14c | 0.87 ± 0.83abc | 0.59 ± 0.07bc | 0.33 ± 0.13c | 0.65 ± 0.22bc | 0.95 ± 0.53 |
| 12 | Gluconasturtiin | 0.28 ± 0.10a | 0.29 ± 0.16a | 0.16 ± 0.08a | 0.41 ± 0.36a | 0.22 ± 0.08a | 0.21 ± 0.09 |
| 13 | Neoglucobrassicin | 0.53 ± 0.32a | 3.14 ± 4.36a | 0.70 ± 0.67a | 0.51 ± 0.43a | 0.39 ± 0.26a | 0.76 ± 0.81 |
| Total | 57.88 ± 9.48a | 12.77 ± 6.94bc | 5.90 ± 1.53c | 11.86 ± 7.63bc | 2.70 ± 0.35c | 14.84 ± 15.46 | |
Within each column, values followed by the same letters are not significantly different at p ⩽ 0.05, using Tukey’s multiple-range test (n = 3).
ND, not detected.
Detected in only one sample.
3.3. Carotenoid contents in Chinese cabbages
Carotenoid metabolites in Chinese cabbage were identified as lutein, zeaxanthin, α-carotene and β-carotene (Table 5), which was similar to the report of Pool-Zobel et al., 1997, Müller et al., 1999. The majority of carotenoids in the leaves were lutein, and β-carotene, which are essential for photosynthesis. Total carotenoid contents ranged from 289.12 to 1001.41 mg kg−1 DW (mean 467.66). Carotenoid levels in the different cultivars were in the following order: ‘K0112’ (1001.41) > ‘K0416’ (657.01) > ‘K0651’ (541.61) > ‘K0648’ (539.43) > ‘CR Hakwang’ (406.08) > ‘Bulam Plus’ (367.50) > ‘K0588’ (350.63) > ‘K0015’ (345.72) > ‘K0461’ (345.18) > ‘Bulam No. 3’ (300.58) > ‘Chunkwang’ (289.12 mg kg−1 DW). This result indicated that total carotenoid levels varied significantly among varieties of Chinese cabbage. Kopsell et al. (2007) reported the accumulation of important dietary carotenoid pigments among the genetically related Brassica species. Lutein accumulation ranging from 520 (Brassica carinata) to 870 (B. rapa), β-carotene ranging from 220 (B. carinata) to 430 (Brassica juncea) and zeaxanthin accumulation ranging from 20 (Brassica napus, B. carinata) to 40 (B. rapa) mg kg−1 DW.
Table 5.
Carotenoid contents (mg kg−1 DW) in eleven varieties of Chinese cabbage.
| No. | Trivial names | Commercial varieties |
|||||
|---|---|---|---|---|---|---|---|
| Chunkwang | CR Hakwang | Bulam No. 3 | Bulam Plus | K0015 | K0112 | ||
| 1 | Lutein | 153.17 ± 30.32d | 206.24 ± 3.92cd | 158.05 ± 19.72d | 193.41 ± 28.81cd | 161.69 ± 27.39d | 384.86 ± 16.07a |
| 2 | Zeaxanthin | 6.11 ± 0.47d | 8.24 ± 1.26bcd | 5.93 ± 1.15d | 7.60 ± 0.65d | 7.99 ± 0.46cd | 23.82 ± 2.88a |
| 3 | α-Carotene | 18.16 ± 6.57d | 28.55 ± 2.46cd | 18.67 ± 3.87d | 23.14 ± 5.25cd | 26.95 ± 6.69cd | 91.50 ± 14.88a |
| 4 | β-Carotene | 111.68 ± 40.81c | 163.05 ± 4.13c | 117.94 ± 22.64c | 143.35 ± 30.11c | 149.08 ± 40.11c | 501.23 ± 55.73a |
| Total | 289.12 ± 77.90d | 406.08 ± 8.74cd | 300.58 ± 46.14cd | 367.50 ± 64.66cd | 345.72 ± 74.45cd | 1001.41 ± 86.82a | |
| No. | Trivial names | Commercial varieties | |||||
| K0416 | K0461 | K0588 | K0648 | K0651 | Mean | ||
| 1 | Lutein | 282.24 ± 40.26bc | 167.48 ± 12.60d | 183.00 ± 21.45cd | 248.68 ± 48.12cd | 360.11 ± 76.64ab | 227.18 ± 82.26 |
| 2 | Zeaxanthin | 13.28 ± 1.88bc | 7.19 ± 1.59d | 8.55 ± 0.79bcd | 10.88 ± 3.81bcd | 13.43 ± 1.84b | 10.27 ± 5.18 |
| 3 | α-Carotene | 64.10 ± 19.86ab | 26.30 ± 2.76cd | 27.37 ± 2.39cd | 51.61 ± 20.15bc | 27.65 ± 7.56cd | 36.73 ± 22.95 |
| 4 | β-Carotene | 297.39 ± 77.80b | 144.21 ± 12.67c | 131.71 ± 16.50c | 228.27 ± 73.01bc | 140.42 ± 35.47c | 193.48 ± 115.58 |
| Total | 657.01 ± 136.87b | 345.18 ± 21.79cd | 350.63 ± 25.40cd | 539.43 ± 144.58bc | 541.61 ± 120.25bc | 467.66 ± 211.79 | |
Within each column, values followed by the same letters are not significantly different at p ⩽ 0.05, using Tukey’s multiple-range test (n = 3).
3.4. Gene expression of 4 selected glucosinolate biosynthesis-related genes
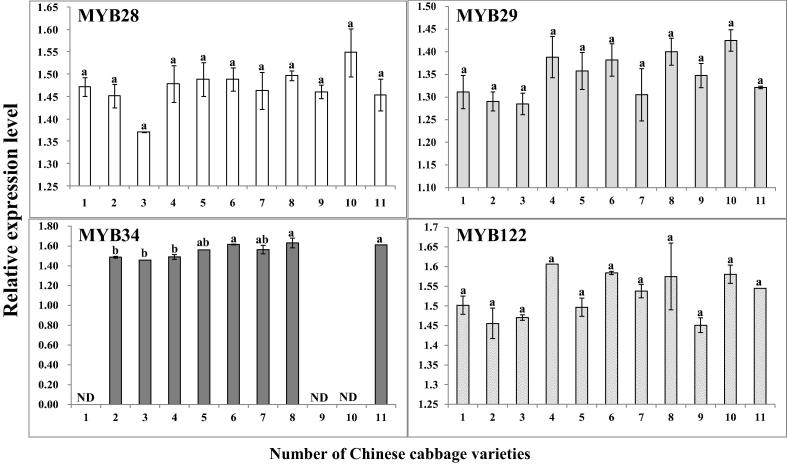
The transcription level of MYB TFs in different varieties is presented in Fig. 5. MYB28 and MYB29, involved in aliphatic GSLs showed extremely different patterns of expression among different varieties. Additionally, MYB51 was expressed only in seeds. In this study, MYB34 was not expressed in some varieties (‘Chunkwang’, ‘K0588’, ‘K0648’), but MYB51 was not expressed in all varieties. In the case of MYB122, expression level was similar to MYB28 and MYB29. However, there was little significant difference between Chinese cabbages. It was observed that, depending on the genotype, the expression level of MYB TFs for GSLs varied in Chinese cabbages. In Arabidopsis thaliana, transcription factors such as R2R3-MYB TF MYB28/HAG1 are the rate limiting step and key factor for the aliphatic GSL biosynthesis (Gigolashvili et al., 2007). Similarly, Hirai et al. (2007) described that methyl jasmonate induced the expression level of MYB29 which directly alters the aliphatic GSL levels in A. thaliana. The transcription of MYB28 in Chinese cabbages was seen in highest levels in ‘K0648’. But among 11 varieties of Chinese cabbage, transcription of MYB showed no significant difference. Likewise, the expression level of MYB29 showed a similar pattern compared with MYB28.MYB51 and MYB122 could act as activators or repressors, depending on the context, in a similar way like MYB34 (Gigolashvili et al., 2007). The transcription of MYB34 was seen in Chinese cabbage, some varieties (‘Chunkwang’, ‘K0588’, ‘K0648’) were not detected. Kim et al. (2013) indicated that transcription of MYB34-2 and MYB34-3 was only observed in seeds, young leaves, and roots.
Figure 5.
Expression of MYB transcription factors related to GSL biosynthetic pathway (see Fig. 1) in different Chinese cabbages. ND, not detected. (1) ‘Chunkwang’; (2) ‘CR Hakwang’; (3) ‘Bulam No. 3’; (4) ‘Bulam Plus’; (5) ‘K0015’; (6) ‘K0112’; (7) ‘K0416’; (8) ‘K0461’; (9) ‘K0588’; (10) ‘K0648’; (11) ‘K0651’. Within each column, values followed by the same letters are not significantly different at p ⩽ 0.05, using Tukey’s multiple-range test (n = 3).
3.5. Gene expression of seven selected carotenoid biosynthesis-related genes
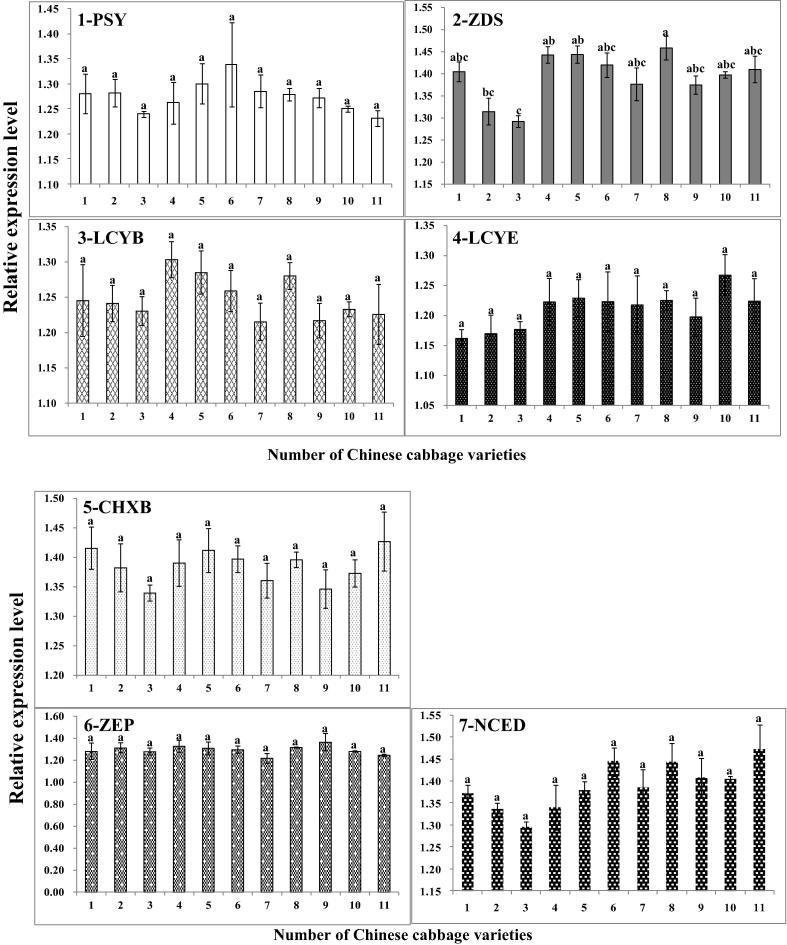
The expression levels of carotenogenesis genes were examined in 11 varieties of Chinese cabbage (Fig. 6). Begining at the carotenoid biosynthetic pathway; expression of 1-PSY was at the highest level in ‘K0112’, but among 11 varieties of Chinese cabbage, transcription of 1-PSY showed no significant difference. Except for 6-ZEP, expression pattern was similar and continued in the next five genes (2-ZDS, 3-LCYB, 4-LCYE, 5-CHXB and 7-NCED). According to the relative quantities of transcription to actin gene, carotenogenesis genes in Chinese cabbages did not show marginal difference with other varieties. Tuan et al. (2012) reported that carotenogenesis genes share a similar expression pattern during seedling development of Chinese cabbage. The transcription of 6-ZEP, related in ABA biosynthesis, was almost at same levels in the eleven varieties of Chinese cabbage.
Figure 6.
Expression of carotenogenesis genes related to the carotenoid biosynthetic pathway (see Fig. 2) in different Chinese cabbages. (1) ‘Chunkwang’; (2) ‘CR Hakwang’; (3) ‘Bulam No. 3’; (4) ‘Bulam Plus’; (5) ‘K0015’; (6) ‘K0112’; (7) ‘K0416’; (8) ‘K0461’; (9) ‘K0588’; (10) ‘K0648’; (11) ‘K0651’. Within each column, values followed by the same letters are not significantly different at p ⩽ 0.05, using Tukey’s multiple-range test (n = 3).
3.6. Correlation between second metabolites and their gene expressions
Through analysis indicated that the correlation between GSL contents and gene expression level showed little significant difference in each Chinese cabbage variety (Table 6), whereas significant differences were noted in the case of carotenoid contents and gene expression level (Table 7). In ‘K0416’ Pearson’s correlation coefficients between LCYE and total carotenoids were −1.000 (p < 0.01), and correlations found between CHXB and α-carotene, β-carotene, total carotenoids (r = −1.000; −0.999; −0.998, respectively at p < 0.05). Furthermore, weak correlations were found between LCYB and lutein in ‘Bulam No. 3’ and ‘Bulam Plus’. In ‘CR Hakwang’ and ‘K0461’, LCYE with lutein was found with weak correlations. In the case of CHXB, weak correlations were found with lutein, β-carotene, zeaxanthin in ‘Chunkwang’, ‘CR Hakwang’, ‘K0461’ respectively. Weak correlations were found between LCYB and α-carotene in ‘K0651’. No statistical correlations were observed between carotenogenesis genes and carotenoids figments in other varieties.
Table 6.
Correlation of glucosinolate contents and gene expression of the glucosinolate biosynthesis.
| Variety | Correlation | Aliphatic GSLs | Total GSLs | Variety | Correlation | Aliphatic GSLs | Total GSLs |
|---|---|---|---|---|---|---|---|
| Chunkwang | MYB28 | −0.966 | −0.984 | K0416 | MYB28 | 0.600 | 0.582 |
| p value | 0.167 | 0.114 | p value | 0.590 | 0.604 | ||
| MYB29 | −0.894 | −0.929 | MYB29 | −0.983 | −0.979 | ||
| p value | 0.295 | 0.242 | p value | 0.117 | 0.131 | ||
| CR Hakwang | MYB28 | 0.873 | 0.827 | K0461 | MYB28 | 0.651 | 0.734 |
| p value | 0.324 | 0.380 | p value | 0.549 | 0.475 | ||
| MYB29 | −0.573 | −0.643 | MYB29 | 0.734 | 0.807 | ||
| p value | 0.611 | 0.555 | p value | 0.476 | 0.402 | ||
| Bulam No. 3 | MYB28 | 0.972 | 0.995 | K0588 | MYB28 | 0.770⁎ | 0.794⁎ |
| p value | 0.152 | 0.065 | p value | 0.015 | 0.011 | ||
| MYB29 | 0.878 | 0.805 | MYB29 | 0.413 | 0.456 | ||
| p value | 0.317 | 0.404 | p value | 0.269 | 0.217 | ||
| Bulam Plus | MYB28 | 0.335 | 0.185 | K0648 | MYB28 | 0.869 | 0.913 |
| p value | 0.783 | 0.881 | p value | 0.330 | 0.267 | ||
| MYB29 | 0.517 | 0.556 | MYB29 | −0.512 | −0.425 | ||
| p value | 0.654 | 0.432 | p value | 0.658 | 0.721 | ||
| K0015 | MYB28 | −0.980 | −0.940 | K0651 | MYB28 | −0.577 | −0.406 |
| p value | 0.128 | 0.221 | p value | 0.608 | 0.733 | ||
| MYB29 | −0.423 | −0.287 | MYB29 | −0.712 | −0.835 | ||
| p value | 0.722 | 0.815 | p value | 0.496 | 0.371 | ||
| K0112 | MYB28 | −0.484 | −0.501 | ||||
| p value | 0.678 | 0.666 | |||||
| MYB29 | 0.943 | 0.937 | |||||
| p value | 0.243 | 0.228 | |||||
The data were analyzed using ANOVA.
Indicate significance at 5% levels.
Table 7.
Correlation of carotenoid contents and gene expression of the carotenoid biosynthesis.
| Variety | Correlation | Lutein | Zeaxanthin | α-Carotene | β-Carotene | Total | Variety | Correlation | Lutein | Zeaxanthin | α-Carotene | β-Carotene | Total |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chunkwang | LCYB | −0.087 | 0.314 | −0.238 | −0.236 | −0.175 | CR Hakwang | LCYB | 0.922 | 0.755 | 0.695 | 0.509 | 0.959 |
| p value | 0.945 | 0.797 | 0.847 | 0.849 | 0.888 | p value | 0.253 | 0.455 | 0.510 | 0.660 | 0.183 | ||
| LCYE | −0.851 | −0.989 | −0.760 | −0.762 | −0.801 | LCYE | 0.999⁎ | 0.403 | 0.322 | 0.828 | 0.988 | ||
| p value | 0.352 | 0.093 | 0.450 | 0.448 | 0.409 | p value | 0.028 | 0.736 | 0.791 | 0.379 | 0.098 | ||
| CHXB | −1.000⁎ | −0.930 | −0.983 | −0.984 | −0.993 | CHXB | 0.796 | −0.190 | −0.275 | 1.000⁎⁎ | 0.725 | ||
| p value | 0.019 | 0.240 | 0.117 | 0.115 | 0.076 | p value | 0.414 | 0.818 | 0.823 | 0.007 | 0.484 | ||
| Bulam No. 3 | LCYB | 1.000⁎ | 0.962 | 0.835 | 0.892 | 0.959 | Bulam Plus | LCYB | 0.999⁎ | 0.878 | 0.980 | 0.990 | 0.994 |
| p value | 0.012 | 0.176 | 0.371 | 0.299 | 0.183 | p value | 0.027 | 0.317 | 0.129 | 0.091 | 0.068 | ||
| LCYE | 0.995 | 0.987 | 0.893 | 0.938 | 0.985 | LCYE | 0.916 | 0.646 | 0.841 | 0.871 | 0.889 | ||
| p value | 0.061 | 0.102 | 0.298 | 0.226 | 0.109 | p value | 0.262 | 0.553 | 0.364 | 0.327 | 0.303 | ||
| CHXB | 0.996 | 0.939 | 0.791 | 0.855 | 0.935 | CHXB | 0.746 | 0.376 | 0.630 | 0.675 | 0.702 | ||
| p value | 0.061 | 0.223 | 0.419 | 0.347 | 0.231 | p value | 0.464 | 0.754 | 0.566 | 0.528 | 0.505 | ||
| K0015 | LCYB | 0.934 | 0.433 | 0.968 | 0.919 | 0.928 | K0112 | LCYB | 0.651 | −0.424 | 0.062 | −0.137 | −0.212 |
| p value | 0.232 | 0.715 | 0.161 | 0.258 | 0.242 | p value | 0.548 | 0.721 | 0.960 | 0.913 | 0.864 | ||
| LCYE | 0.772 | 0.122 | 0.837 | 0.745 | 0.761 | LCYE | −0.779 | −0.582 | −0.121 | −0.316 | −0.387 | ||
| p value | 0.439 | 0.922 | 0.368 | 0.465 | 0.449 | p value | 0.431 | 0.604 | 0.923 | 0.796 | 0.747 | ||
| CHXB | 0.783 | 0.140 | 0.847 | 0.756 | 0.772 | CHXB | −0.902 | −0.754 | −0.346 | −0.525 | −0.588 | ||
| p value | 0.428 | 0.911 | 0.357 | 0.454 | 0.438 | p value | 0.284 | 0.457 | 0.775 | 0.648 | 0.599 | ||
| K0416 | LCYB | −0.833 | −0.895 | −0.977 | −0.986 | −0.960 | K0461 | LCYB | 0.206 | 0.964 | −0.884 | −0.903 | −0.448 |
| p value | 0.373 | 0.294 | 0.135 | 0.105 | 0.181 | p value | 0.868 | 0.172 | 0.309 | 0.282 | 0.704 | ||
| LCYE | −0.958 | −0.726 | −0.996 | −0.991 | −1.000⁎⁎ | LCYE | 0.999⁎ | 0.543 | −0.658 | −0.689 | −0.097 | ||
| p value | 0.184 | 0.483 | 0.053 | 0.084 | 0.008 | p value | 0.022 | 0.634 | 0.543 | 0.516 | 0.938 | ||
| CHXB | −0.932 | −0.780 | −1.000⁎⁎ | −0.999⁎ | −0.998⁎ | CHXB | 0.515 | 0.998⁎ | −0.683 | −0.713 | −0.131 | ||
| p value | 0.236 | 0.431 | 0.001 | 0.031 | 0.045 | p value | 0.656 | 0.041 | 0.521 | 0.495 | 0.917 | ||
| K0588 | LCYB | 0.813 | 0.795 | 0.446 | −0.811 | 0.226 | K0648 | LCYB | −0.878 | −0.914 | −0.963 | −0.943 | −0.927 |
| p value | 0.395 | 0.415 | 0.706 | 0.397 | 0.855 | p value | 0.317 | 0.265 | 0.174 | 0.216 | 0.245 | ||
| LCYE | 0.968 | 0.960 | 0.793 | −0.546 | 0.563 | LCYE | 0.888 | 0.923 | 0.968 | 0.950 | 0.934 | ||
| p value | 0.160 | 0.180 | 0.471 | 0.633 | 0.620 | p value | 0.304 | 0.252 | 0.162 | 0.203 | 0.232 | ||
| CHXB | 0.886 | 0.871 | 0.565 | −0.723 | 0.358 | CHXB | 0.065 | −0.016 | −0.158 | −0.094 | −0.048 | ||
| p value | 0.308 | 0.327 | 0.618 | 0.485 | 0.767 | p value | 0.958 | 0.990 | 0.899 | 0.940 | 0.969 | ||
| K0651 | LCYB | 0.961 | 0.689 | 0.999⁎ | 0.991 | 0.978 | |||||||
| p value | 0.178 | 0.516 | 0.030 | 0.086 | 0.133 | ||||||||
| LCYE | 0.704 | 0.955 | 0.848 | 0.799 | 0.752 | ||||||||
| p value | 0.503 | 0.191 | 0.355 | 0.411 | 0.458 | ||||||||
| CHXB | 0.848 | 0.862 | 0.947 | 0.916 | 0.884 | ||||||||
| p value | 0.355 | 0.339 | 0.207 | 0.263 | 0.310 | ||||||||
Data were analyzed using ANOVA.
Indicates significance at 5% level, respectively.
Indicate significance at 1% level, respectively.
4. Conclusion
In summary, the expression pattern of MYB transcription factors and carotenogenesis genes involved in GSL and carotenoid biosynthesis in eleven varieties of Chinese cabbage was observed. Remarkable differences in the GSL and carotenoid contents were observed among different varieties. Chinese cabbage contained rich content of anti-carcinogenic GSLs and carotenoids for human daily consumption. Some varieties contain high levels of health promoting compounds in edible parts of Chinese cabbage, such as ‘K0112’ and ‘K0416’. The expression levels of MYB28, 29, 34, 122 and seven carotenogenesis genes were examined in eleven varieties of Chinese cabbage. Also, correlation of secondary metabolite contents and their gene expression level showed little significant difference in each Chinese cabbage. This study might be helpful in understanding GSL and carotenoid biosynthesis in Chinese cabbage. The presented results could serve as an additional source of developing a cultivation method for high functional plants. However, further study will required to investigate the biosynthesis of GSLs and carotenoids during the developmental stages of Chinese cabbage.
Financial support
This research work was supported by the research fund of Chungnam National University.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Mariadhas Valan Arasu, Email: mvalanarasu@gmail.com, mvalanarasu@ksu.edu.sa.
Sun-Ju Kim, Email: kimsunju@cnu.ac.kr.
References
- Cartea M.E., Francisco M., Soengas P., Velasco P. Phenolic compounds in Brassica vegetables. Molecules. 2011;16:251–280. doi: 10.3390/molecules16010251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho E.J., Rhee S.H., Kang K.S., Park K.Y. In vitro anticancer effect of Chinese cabbage kimchi fractions. J. Korea Soc. Food Sci. Nutr. 1999;28(6):1326–1331. [Google Scholar]
- Clarke D.B. Glucosinolates, structures and analysis in food. Anal. Methods. 2010;2:310–325. [Google Scholar]
- Gigolashvili T., Berger B., Mock H.P., Müller C., Weisshaar B., Flügge U.I. The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 2007;50:886–901. doi: 10.1111/j.1365-313X.2007.03099.x. [DOI] [PubMed] [Google Scholar]
- Hirai M.Y., Sugiyama K., Sawada Y., Tohge T., Obayashi T., Suzuki A., Araki R., Sakurai N., Suzuki H., Aoki K. Omics-based identification of Arabidopsis Myb transcription factors regulating aliphatic glucosinolate biosynthesis. Proc. Natl. Acad. Sci. USA. 2007;104:6478–6483. doi: 10.1073/pnas.0611629104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong E., Kim S.J., KIM G.H. Identification and quantitative determination of glucosinolates in seeds and edible parts of Korean Chinese cabbage. Food Chem. 2011;128:1115–1120. [Google Scholar]
- International Standards Organization (ISO) ISO 9167-1:1992 (E); Geneva, Switzerland: 1992. Rapeseed: Determination of Glucosinolates Content – Part 1: Method Using High performance Liquid Chromatography; pp. 1–9. [Google Scholar]
- Kim J.K., Chu S.M., Kim S.J., Lee D.J., Lee S.Y., Lim S.H., Ha S.H., Kweon S.J., Cho H.S. Variation of glucosinolates in vegetable crops of Brassica rapa L. ssp. pekinensis. Food Chem. 2010;119:423–428. [Google Scholar]
- Kim S.J., Kawaharada C., Jin S., Hashimoto M., Ishii G., Yamauchi H. Structural elucidation of 4-(cystein-S-yl)butyl glucosinolate from the leaves of Eruca sativa. Biosci. Biotechnol. Biochem. 2007;71:114–121. doi: 10.1271/bbb.60400. [DOI] [PubMed] [Google Scholar]
- Kim Y.B., Li X., Kim S.J., Kim H.H., Lee J.Y., Kim H.R., Park S.U. MYB transcription factors regulate glucosinolate biosynthesis in different organs of Chinese cabbage (Brassica rapa ssp. pekinensis) Molecules. 2013;18:8682–8695. doi: 10.3390/molecules18078682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y.B., Chun J.H., Kim H.R., Kim S.J., Lim Y.P., Park S.U. Variation of glucosinolate accumulation and gene expression of transcription factors at different stages of Chinese cabbage seedlings under light and dark conditions. Nat. Prod. Commun. 2014;4:533–537. [PubMed] [Google Scholar]
- Kim H.K., Chun J.H., Kim S.J. Method development and analysis of carotenoid compositions in various tomatoes. Korean J Environ. Agric. 2015;34(2):1–8. [Google Scholar]
- Kopsell D.A., McElroy J.S., Sams C.E., Kopsell D.E. Genetic variation in carotenoid concentrations among diploid and amphidiploid rapid-cycling Brassica species. HortScience. 2007;42(3):461–465. [Google Scholar]
- Krishnadev N., Meleth A.D., Chew E.Y. Nutritional supplements for age-related macular degeneration. Curr. Opin. Ophthalmol. 2010;21(3):184–189. doi: 10.1097/ICU.0b013e32833866ee. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M.K., Chun J.H., Byeon D.H., Chung S.O., Park S.U., Park S.H., Arasu M.V., Al-Dhabi N.A., Lim Y.P., Kim S.J. Variation of glucosinolates in 62 varieties of Chinese cabbage (Brassica rapa L. ssp. pekinensis) and their antioxidant activity. LWT-Food Sci. Technol. 2014;58:93–101. [Google Scholar]
- Lee D.S., Jeon D.S., Park S.G., Arasu M.V., Al-Dhabi N.A., Kim S.C., Kim S.J. Effect of cold storage on the contents of glucosinolates in Chinese cabbage (Brassica rapa L. ssp. pekinensis) South Indian J. Biol. Sci. 2015;1(1):38–42. [Google Scholar]
- Müller H., Bub A., Watzl B., Rechkemmer G. Plasma concentrations of carotenoids in healthy volunteers after intervention with carotenoid-rich foods. Eur. J. Nutr. 1999;38:35–44. doi: 10.1007/s003940050044. [DOI] [PubMed] [Google Scholar]
- Padill G., Cartea M.E., Velasco P., Haro A., Ordás A. Variation of glucosinolates in vegetable crops of Brassica rapa. Phytochemistry. 2007;68:536–545. doi: 10.1016/j.phytochem.2006.11.017. [DOI] [PubMed] [Google Scholar]
- Park W.T., Kim J.K., Park S.H., Lee S.W., Li X., Kim Y.B., Uddin R., Park N.I., Kim S.J., Park S.U. Metabolic profiling of glucosinolates, anthocyanins, carotenoids, and other secondary metabolites in kohlrabi (Brassica oleracea var. gongylodes) J. Agric. Food Chem. 2012;60:8111–8116. doi: 10.1021/jf301667j. [DOI] [PubMed] [Google Scholar]
- Pedras M.S., Chumala P.B., Suchy M. Phytoalexins from Thlaspiarvense, a wild crucifer resistant to virulent Leptosphaeria maculans: structures, syntheses and antifungal activity. Phytochemistry. 2003;64:949–956. doi: 10.1016/s0031-9422(03)00441-2. [DOI] [PubMed] [Google Scholar]
- Pool-Zobel B.L., Bub A., Müller H., Wollowski I., Rechkemmer G. Consumption of vegetable reduces genetic damage in humans: first results of a human intervention trial with carotenoid-rich foods. Carcinogenesis. 1997;18:1847–1850. doi: 10.1093/carcin/18.9.1847. [DOI] [PubMed] [Google Scholar]
- Reif C., Arrigoni E., Berger F., Baumgartner D., Nyström L. Lutein and β-carotene content of green leafy Brassica species grown under different conditions. LWT-Food Sci. Technol. 2013;53:378–381. [Google Scholar]
- Tuan P.A., Park N.I., Park W.T., Kim Y.B., Kim J.K., Lee J.H., Lee S.H., Yang T.J., Park S.U. Carotenoids accumulation and expression of carotenogenesis genes during seedling and leaf development in Chinese cabbage (Brassica rapa subsp. pekinensis) Plant Omics. 2012;5(2):143–148. [Google Scholar]
- Yang B., Quiros C.F. Survey of glucosinolate variation in leaves of Brassica rapa crops. Genet. Resour. Crop. Evol. 2010;57:1079–1089. [Google Scholar]
- Yatusevich R., Mugford S.G., Matthewman C., Gigolashvili T., Frerigmann H., Delaney S., Koprivova A., Flügge U.I., Kopriva S. Genes of primary sulfate assimilation are part of the glucosinolate biosynthetic network in Arabidopsis thaliana. Plant J. 2010;62:1–11. doi: 10.1111/j.1365-313X.2009.04118.x. [DOI] [PubMed] [Google Scholar]