Abstract
Introduction
Stroke is a multifactorial and heterogeneous disorder, correlates with heritability and considered as one of the major diseases. The prior reports performed the variable models such as genome-wide association studies (GWAS), replication, case-control, cross-sectional and meta-analysis studies and still, we lack diagnostic marker in the global world. There are limited studies were carried out in Saudi population, and we aim to investigate the molecular association of single nucleotide polymorphisms (SNPs) identified through GWAS and meta-analysis studies in stroke patients in the Saudi population.
Methods
In this case-control study, we have opted gender equality of 207 cases and 207 controls from the capital city of Saudi Arabia in King Saud University Hospital. The peripheral blood (5 ml) sample will be collected in two different vacutainers, and three mL of the coagulated blood will be used for lipid analysis (biochemical tests) and two mL will be used for DNA analysis (molecular tests). Genomic DNA will be extracted with the collected blood samples, and specific primers will be designed for the opted SNPs (SORT1-rs646218 and OLR1-rs11053646 polymorphisms) and PCR-RFLP will be performed and randomly DNA sequencing will be carried out to cross check the results.
Results
The rs646218 and rs11053646 polymorphisms were significantly associated with allele, genotype and dominant models with and without crude odds ratios (OR’s) and Multiple logistic regression analysis (p < 0.05). Correlation between lipid profile and genotypes has confirmed the significant relation between triglycerides and rs646218 and rs1105364 6polymorphisms. However, rs11053646 polymorphism was correlated with HDLC (p = 0.04). Genotypes were examined in both males' vs. males and females' vs. females in cases and control and we concluded that in rs11053646 polymorphisms with male subjects compared between cases and controls found to be associated with dominant model heterozygote genotypes (p < 0.05).
Conclusion
The results of the current study confirmed the SORT1 and OLR1 SNPs were associated in the Saudi population. The current results were in the association with the prior study results documented through GWAS and meta-analysis association. However, other ethnic population studies should be performed to rule out in the human hereditary diseases.
Keywords: Stroke, Genome-wide association studies, Meta-analysis studies, Saudi population
1. Introduction
Stroke is a common neurological disorder affecting the global population of 15 million; among them ∼5 million suffer from the permanent disability, and 5.5 million dies due to stroke-related factors (Jin et al., 2015). Etiologically, ischemic stroke (IS) is supposed to be a highly complex disease influenced by genetic and environmental issues (Qin et al., 2015). This disease has considered as a fourth commonest leading cause of death, lasts neurologic impairment, functional disability, and stroke aetiology is multifactorial and conventional forms of inheritance cannot be confirmed (Hassan and Markus, 2000). The verified risk factors as age, cigarette smoking, diabetes, hypertension, hyperlipidemia and detected comorbidities as HIV, sickle cell disease, cerebral malaria are progressively being encountered. Based on the availability, IS can be effected with the physical damage, psychological, social and cognitive functions (Benamer and Grosset, 2009, Robert and Zamzami, 2014). However, in IS, the aetiology and risk factors may vary from those for myocardial infarction (O'Donnell et al., 2010). The genetic roles in IS have been established in numerous reports entails in twins and family models (Munshi et al., 2015). The discovering of predisposing to stroke in genetic variants could be a vital step towards the expansion of enhanced diagnostic tests for stroke. Genome-wide association studies (GWAS) has recognised merely a few confirmed loci for a small amount of the heritable risk (Loci Associated with Ischaemic Stroke and Its Subtypes, 2016). Genetic epidemiology mainly involves with complex diseases such as stroke and frequently observed in the population-based studies. However, the disease is polygenic in origin with an involvement of multiple genes (Wichmann, 2005). In human diseases, GWAS were used as an influential device to identify the recognised marker and helps the classifying genetic variations that contribute to common and complex diseases (Example: Stroke or Ischemic Stroke) (Tan et al., 2014). Identification of risk through single nucleotide polymorphisms (SNPs) can be confirmed with GWAS and meta-analysis (Nalls et al., 2014). It has demonstrated various polymorphisms of different genes were strong risk locus for stroke and controversial results were adapted from global population. Many candidate genes have been studied for their potential roles in IS. However, preliminary case-control studies adopt the principal data in genetic epidemiology, and such studies are useful in originating the appropriate conclusions for genetic polymorphisms (Das et al., 2016). Meta-analysis studies are defined as a statistical tool, merging the case-control study results of different ethnic studies on the similar topic, and it has become the popular method for resolving discrepancies in genetic association studies (Lee, 2015). Many meta-analysis studies have been carried out in stroke and concluded as positive (He et al., 2017, Luo et al., 2017, Xiuju et al., 2016, Yanamala et al., 2017, Zhong et al., 2017), negative (Xia et al., 2017, Yang et al., 2017) and nominal associations (Li et al., 2017).
Sortilin (SORT1) gene has been identified through GWAS studies, and it appears on chromosome 1p13.3 regions (Hubacek, 2016). SORT1 is expressed both in neurons cum non-neuronal cells and encodes for the sorting protein that plays an important role in the uptake of lipids (Andersen et al., 1995, Arvind et al., 2014). However, Linsel-Nitschke et al. (2010) confirmed functional variants at this locus might reside within the SORT1 gene since the risk allele was associated with decreased SORT1 expression, increased plasma Low density lipoprotein-cholesterol (LDL-C) concentration, and increased coronary artery disease (CAD) risk. Earlier studies also showed the significance association with CAD risk (Buyske et al., 2012, Consortium CADG, 2011, Gudbjartsson et al., 2009, Kathiresan et al., 2009, Ozaki et al., 2002, Saade et al., 2011, Wang et al., 2011). The Oxidized Low Density Lipoprotein Receptor 1 (OLR1) gene was identified as a cell surface endocytosis receptor for oxidised low-density lipoprotein (Ox-LDL) on vascular endothelial cells (Hattori et al., 2006). The OLR1 gene consists of six exons and five introns, spans 7-kb, which located on chromosome 12p13.1-p12.3. The SNPs in the OLR1 gene have been identified, including G501C transversion on exon 4, results in an amino acidic substitution from Lysine-Asparagine at position 167. The rs11053646 polymorphism in the oxidised LDL binding domain of LOX-1 causes to decrease binding and internalisation of ox-LDL7 and has been associated with hypertension, myocardial infarction and carotid atherosclerosis and importantly, G501C SNP is statistically linked to the risk of IS. The deletion in OLR1 gene is possible while an increase in the OLR1 overexpression, plaque formation and development through the demonstration in the atherosclerosis mouse models (Au et al., 2015, Wang et al., 2011).
However, confirmed through GWAS and meta-analysis variants have little information with stroke and there are no genetic studies in Saudi Arabia, which is known to be obese as it is existent in the Arab region. The prevalence of obesity has been drastically increased due to western lifestyle and uncontrolled diet. The women are more effected with obesity compared with the males. This is the reason we have opted one-third of males (n = 69) and two-third of females (n = 138) in both the cases and controls. This present study in Saudi population aims to evaluate the potential association of SNPs identified through GWAS and meta-analysis studies in stroke patients.
2. Materials and methods
2.1. Ethical concern
Ethical approval grant has been sanctioned for this study from College of Applied Medical Sciences, King Saud University (CAMS 047-37/38).
2.2. Patient selection
In this study, 414 Saudi subjects were selected from King Saud University Hospital in a capital city of Saudi Arabia. We have recruited 420 subjects based on the inclusion and exclusion criteria. All the subjects (n = 420) were the native of the Saudi population. Patients with major cardiac, renal, hepatic, skeletal disorders and cancerous diseases (n = 6) were excluded from this study. The patients’ details such as demographic information and lipid profile parameters were included in structured proforma. Two hundred and seven healthy controls were recruiting from general Saudi population without any disorder or diseases. The patient selection details were described in our prior publications (Alharbi et al., 2014). Routinely, 5 ml of peripheral blood was collected, and 3 ml of serum sample was used for biochemical analysis and remaining 2 ml of the EDTA blood was used to separate the genomic DNA.
2.3. Biochemical assessment
A serum sample was used for analyzing the lipid profile parameters such as Total cholesterol (TC), Triglycerides (TG), High-density lipoprotein-cholesterol (HDL-C) and LDL-C, measured by automated clinical chemistry analyser (KoneLab) using the commercially available kits.
2.4. Genetic analysis and quality control
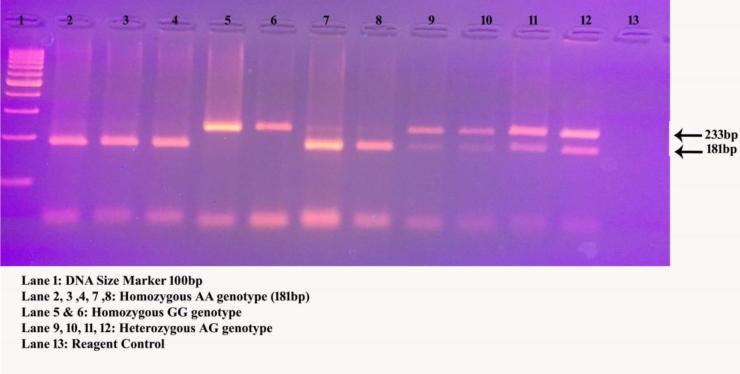
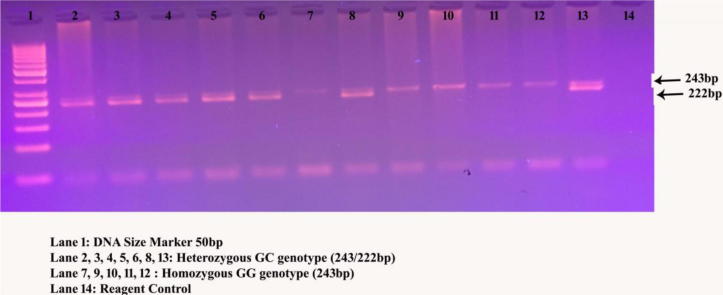
Genomic DNA was isolated with the commercial kit available from Norgen Biotech Corporation. The valuation for DNA samples were confirmed with NanoDrop spectrophotometer and stored in −80 °C. Two SNPs were selected based on the design of the study from GWAS (rs464218) and meta-analysis (rs11053646) studies. Primers were designed for Polymerase Chain Reaction-Restriction Fragment Length Polymorphism (PCR-RFLP) analysis and synthesised from Bioserve Biotechnologies Limited. Gene amplification was carried out with thermal PCR in a 25 µL reaction mixture (Alharbi et al., 2015). The PCR products were digested for 16 h for both the variants and run on 3% agarose gel. The complete PCR-RFLP details are tabulated in Table 1. The quality control was performed in both SNPs of cases and controls (Fig. 1, Fig. 2) by determining the genotypes in total call rate and minor allele frequencies, indicating ∼10% of random samples, i.e., 30 cases and 30 control samples have been performed the DNA sequencing analysis.
Table 1.
List of single nucleotide polymorphisms details involved in this study.
| Gene | rs number | Chr. Position | Forward Primer | Reverse Primer | Protein change | PCR | Enzyme | RFLP |
|---|---|---|---|---|---|---|---|---|
| SORT1 | rs464218 | 109313684 | GGTGTGCAGTGTTCTTGGAG | TGGAATTCGAAGGGACCTTTTCA | – | 233bp | HhaI | A-233bp; G-181/52bp |
| OLR1 | rs11053646 | 10160849 | GGCTCATTTAACTGGGAAAG | CCTGTCCGTCCAAGGTCATA | Lys-Asn | 243bp | FNU4HI | G-243bp; C-222/21bp |
Fig. 1.
A 3% agarose gel electrophoretograms of SORT1 digested products after HhaI restriction enzyme.
Fig. 2.
Digested OLR1 gene products run on 3% agarose gel electrophoresis with FNU4HI restriction enzyme.
2.5. Statistical analysis
Clinical characteristics of incorporated subjects were expressed as Mean ± SD. Clinical data was compared between cases and controls using Student's t-test. Genotype and allele frequencies were calculated by gene counting method, and the chi-square (X2) tests were used to identify departures from Hardy-Weinberg equilibrium (HWE). The odds ratio (OR) and 95% confidence intervals (CI/CIs) were also calculated. Multinomial logistic regression analysis was also performed within the genotype analysis incorporating age, gender, and Body mass index (BMI). Analysis of variance (ANOVA) was used to compare differences between genotypes, BMI and lipid profile parameters. Yates correction was performed in cases and controls for the rs11053646 variant. A p-value < 0.05 (two-sided) was considered to be significant. Statistical analysis was performed using the SPSS statistical package (Version 21.0, SPSS Inc., and Chicago, IL, USA).
3. Results
3.1. Baseline features
During this study process, 207 cases and 207 controls were recruited to perform the case-control study. However, the clinical details were documented in Table 2. The mean age of the cases and controls were found to be 30.6 ± 4.2, and 27.1 ± 6.1 and t-test were found to be 0.001. There was no significant difference between the groups on gender, height and HDL-C parameters (p > 0.05). However, on another side, the positive association was confirmed with weight, BMI, TC, TG and LDL-C (p < 0.05).
Table 2.
Demographic characteristics of selected cases and controls.
| S.No | Cases (n = 207) | Controls (n = 207) | P Value | |
|---|---|---|---|---|
| 1 | Gender (M:F) | 69 (33%):138 (67%) | 69 (33%):138 (67%) | 0.99 |
| 2 | Weight (kg) | 74.1 ± 12.3 | 68.7 ± 20.3 | 0.001 |
| 3 | Height (cms) | 157.7 ± 5.2 | 162.1 ± 8.1 | 0.12 |
| 4 | BMI (kg/m2) | 29.7 ± 4.3 | 25.7 ± 6.1 | 0.0001 |
| 5 | TC (mmol/L) | 5.1 ± 1.0 | 4.7 ± 1.2 | 0.009 |
| 6 | TG (mmol/L) | 1.7 ± 0.9 | 1.3 ± 0.6 | 0.0001 |
| 7 | HDL-C (mmol/L) | 0.7 ± 0.2 | 0.8 ± 0.4 | 0.13 |
| 8 | LDL-C (mmol/L) | 3.7 ± 0.9 | 2.4 ± 0.7 | 0.003 |
3.2. Genotype association
In this study, rs464218 and rs11053646 polymorphisms were carried out between the cases and controls. Both the variants appeared in HWE in both the groups. The genotype and allele frequencies of cases and controls were tabulated in Table 3. AA, AG and GG are the three genotypes were detected in the rs 464218 variant in SORT1 gene. The genotype frequencies observed in cases are 30.4% in AA, 51.2% in AG and 18.4% in GG, where as in controls, it was found to be 41.5% in AA, 44.4% in AG and 14% in GG genotypes respectively. The genotype distribution between cases and healthy controls were significantly associated with AG genotypes, dominant model and G allele [(AG Vs AA; OR = 1.57, 95%CI = 1.02–2.41; p = 0.03); (AG + GG vs AA; OR = 1.62, 95%CI = 1.08–2.43; p = 0.01) and (G vs A; OR = 1.38, 95%CI = 1.04–1.82; p = 0.02)]. The positive association was also observed when we adjusted the genotype data with age, gender and BMI i.e., with AG genotypes, Dominant model and G allele [(AG Vs AA; OR = 1.93, 95%CI = 1.19–2.65; p = 0.02); (AG + GG vs AA; OR = 1.78, 95%CI = 1.17–2.65; p = 0.03) and (G vs A; OR = 1.50, 95%CI = 1.03–2.12; p = 0.02)]. The variant model (GG vs AA) was found to be similarly associated with AG genotype (GG Vs AA; OR = 1.78, 95%CI = 1.01–3.20; p = 0.04).
Table 3.
Genotype and allele distributions of rs464218 and rs11053464 variants analyses and adjusted with potential confounders (N = 207).
| Genotype | Case (N%) | Control (N%) | X2 | Crude OR (95% CI) | P value | OR (95% CI)a | P Value |
|---|---|---|---|---|---|---|---|
| rs464218 | |||||||
| AA | 63 (30.4%) | 86 (41.5%) | Reference | ||||
| AG | 106 (51.2%) | 92 (44.4%) | 4.2 | 1.57 (1.02–2.41) | 0.03 | 1.93 (1.19–2.65) | 0.02 |
| GG | 38 (18.4%) | 29 (14.0%) | 3.8 | 1.78 (1.01–3.20) | 0.04 | 1.87 (1.22–3.41) | 0.04 |
| AG + GG Vs AA | 144 (69.6%) | 121 (58.4%) | 5.5 | 1.62 (1.08–2.43) | 0.01 | 1.78 (1.17–2.65) | 0.03 |
| AG Vs AA + GG | 106 (51.2%) | 92 (44.4%) | 1.9 | 1.31 (0.89–1.93) | 0.16 | - | - |
| GG Vs AA + AG | 38 (18.4%) | 29 (14.0%) | 1.4 | 1.38 (0.81–2.33) | 0.23 | - | - |
| A | 232 (0.56) | 264 (0.64) | Reference | ||||
| G | 182 (0.44) | 150 (0.36) | 5.1 | 1.38 (1.04–1.82) | 0.02 | 1.50 (1.03–2.12) | 0.02 |
| rs11053646 | |||||||
| GG | 177 (85.5%) | 191 (92.2%) | Reference | ||||
| GC | 30 (14.5%) | 16 (7.8%) | 4.7 | 2.02 (1.06–3.83) | 0.02 | 2.01 (1.03–3.8) | 0.01 |
| CC | 00 (00) | 00 (00) | 0.001 | 1.07 (0.02–54.6) | 0.96* | - | - |
| GC + CC Vs GG | 30 (14.5%) | 16 (7.8%) | 4.7 | 1.99 (1.05–3.75) | 0.03* | - | - |
| GC Vs GG + CC | 30 (14.5%) | 16 (7.8%) | - | - | - | - | - |
| CC VS GG + CC | 00 (00) | 00 (00) | - | - | - | - | - |
| G | 384 (0.93) | 398 (0.96) | Reference | ||||
| C | 30 (0.07) | 16 (0.04) | 4.5 | 1.94 (1.04–3.62) | 0.03 | 2.1 (1.09–3.82) | 0.04 |
Odds ratio (95% CI) adjusted with Age, Gender and BMI.
The other variant rs11053646 was carried out with similar subjects and frequencies of GG and GC genotypes were 85.5% and 14.5% in cases and 92.2% and 7.8% in controls. CC genotype was completely absent in all the subjects. In this statistical analysis, we find significant association in GC and C allele [(GC Vs GG; OR = 2.02, 95%CI = 1.06–3.83; p = 0.02) and (C vs G; OR = 1.94, 95%CI = 1.04–3.62; p = 0.03)]. After adjusting the Yates' correction, the dominant model was found to be associated (GC + CC vs GG; OR = 1.99, 95%CI = 1.05–3.75; p = 0.03). In this variant also positive association was observed when adjusted with the genotype data correlated with age, gender and BMI, i.e., with GC genotypes and C allele [(GC vs GG; OR = 2.01, 95%CI = 1.03–3.80; p = 0.01) and (C vs G; OR = 2.10, 95%CI = 1.09–3.82; p = 0.04)].
3.3. Association of rs464218 and rs11053646 variants with combined parameters
ANOVA analysis was carried out with the study effects of rs464218 and rs11053646 variants on the different parameters. We analysed the distribution of these variables about the various genotypes in the selected polymorphisms. BMI could not show the association with any of the genotypes in rs464218 and rs11053646 variants. Although the association was found with TG in both rs464218 (p = 0.003) and rs11053646 (p = 0.002) variants and rs11053646 variant, HDL-C was significantly associated (p = 0.04). The complete data of ANOVA analysis was documented in Table 4.
Table 4.
Correlation between BMI, lipid profile and genotypes involved in this study.
| Genotypes |
SORT1(rs464218) |
OLR1(rs11053646) |
||||||
|---|---|---|---|---|---|---|---|---|
| AA | AG | GG | P value | GG | GC | CC | P value | |
| N | 63 (30.4%) | 106 (51.2%) | 38 (18.4%) | – | 177 (85.5%) | 30 (14.5%) | 00 (00) | – |
| BMI(kg/m2) | 29.72 ± 4.44 | 29.15 ± 4.42 | 30.16 ± 4.17 | 0.89 | 29.82 ± 4.63 | 29.68 ± 4.33 | 0.0 ± 0.0 | 0.64 |
| TC (mmol/L) | 5.22 ± 1.05 | 5.09 ± 1.08 | 4.94 ± 0.85 | 0.23 | 5.13 ± 0.90 | 5.10 ± 1.06 | 0.0 ± 0.0 | 0.23 |
| TG (mmol/L) | 1.76 ± 1.01 | 1.72 ± 1.00 | 1.37 ± 0.62 | 0.003 | 1.52 ± 0.59 | 1.69 ± 1.00 | 0.0 ± 0.0 | 0.002 |
| HDL-C (mmol/L) | 0.65 ± 0.24 | 0.66 ± 0.25 | 0.69 ± 0.21 | 0.45 | 0.70 ± 0.19 | 0.66 ± 0.25 | 0.0 ± 0.0 | 0.04 |
| LDL-C (mmol/L) | 3.77 ± 0.88 | 3.64 ± 0.93 | 3.63 ± 0.80 | 0.54 | 3.73 ± 0.86 | 3.67 ± 0.90 | 0.0 ± 0.0 | 0.74 |
Data represented by Mean±standard deviation.
3.4. Gender-based association within the genotypes
Gender-based genotype association was tabulated in Table 5 as male and female subjects. We could not find any statistical association when compared in rs464218 polymorphism with male and female subjects in cases vs controls (p > 0.05). However, with rs11053646 polymorphism, a positive association was observed only in the comparison between male cases vs male controls with GC genotype and C allele [(GC vs GG; OR = 5.60, 95%CI = 1.53–20.49; p = 0.04) and (C vs G; OR = 5.08, 95%CI = 1.42–18.10; p = 0.005)]. The dominant model also seems to be positive association after the Yates correction in male controls vs male cases [GG + CC vs GG; OR = 4.96, 95%CI = 1.46–16.82; p = 0.005].
Table 5.
Association of SORT1 and OLR1 genotypes in Stroke male and female subjects.
| Genotype | Case (n = 69) | Control (n = 69) | OR (95% CI) | P value | Case (n = 138) | Control (n = 138) | OR (95% CI) | P value |
|---|---|---|---|---|---|---|---|---|
| Male Subjects | Male Subjects | Female Subjects | Female Subjects | |||||
| rs464218 | ||||||||
| AA | 18 (26.1%) | 27 (39.1%) | Reference | 45 (32.6%) | 59 (42.7%) | Reference | ||
| AG | 37 (53.6%) | 32 (46.4%) | 1.73 (0.81–3.71) | 0.15 | 69 (50.0%) | 60 (43.5%) | 1.50 (0.89–2.53) | 0.12 |
| GG | 14 (20.3%) | 10 (14.5%) | 2.1 (0.76–5.74) | 0.14 | 24 (17.4%) | 19 (13.8%) | 1.65 (0.80–3.38) | 0.16 |
| AG + GG Vs AA | 51 (73.9%) | 42 (60.9%) | 1.82 (0.88–3.75) | 0.1 | 93 (67.4%) | 79 (57.3%) | 1.54 (0.94–2.52) | 0.08 |
| A | 73 (0.53) | 86 (0.62) | Reference | 159 (0.58) | 178 (0.64) | Reference | ||
| G | 65 (0.47) | 52 (0.38) | 1.47 (0.91–2.37) | 0.11 | 117 (0.42) | 98 (0.36) | 1.33 (0.94–1.88) | 0.09 |
| rs11053646 | ||||||||
| GG | 55 (79.7%) | 66 (95.6%) | Reference | 122 (88.4%) | 125 (90.6%) | Reference | ||
| GC | 14 (20.3%) | 3 (4.4%) | 5.6 (1.53–20.49) | 0.04 | 16 (11.6%) | 13 (9.4%) | 1.26 (0.58–2.73) | 0.55 |
| CC | 00 (00) | 00 (00) | 1.19 (0.02–61.35) | 0.92a | 00 (00) | 00 (00) | 1.02 (0.02–52.03) | 0.99a |
| GC + CC Vs GG | 14 (20.3%) | 3 (4.4%) | 4.96 (1.46–16.82) | 0.005a | 16 (11.6%) | 13 (9.4%) | 1.25 (0.58–2.68) | 0.56a |
| G | 124 (0.90) | 135 (0.98) | Reference | 260(0.94) | 263 (0.95) | Reference | ||
| C | 14 (0.10) | 3 (0.02) | 5.08 (1.42–18.10) | 0.005 | 16 (0.06) | 13 (0.05) | 1.24 (0.58–2.64) | 0.56 |
Indicates Yates correction.
4. Discussion
The aim of the present study was to investigate the confirmed variants through GWAS and meta-analysis studies with stroke in the Saudi population. As per the literature, this will be the initial study performed in the Saudi population. The results of currents study confirm positive association in rs464218 (SORT1) and rs11053646 (OLR1) variants with allele, genotype and dominant models (p < 0.05). In rs11053646 polymorphisms with male subjects compared between cases and controls found to be associated with dominant model heterozygote genotypes (p < 0.05). In both the polymorphisms, genotypes were associated with triglycerides when we correlated (p < 0.05). The prevalence of stroke is increased and confirmed as the second cause of disability as well as the fourth cause of death in the global population. However, burden with high proportion is divided in the developing countries because of larger populations. Stroke as a complex disorder is quite difficult to rule out the involvement of genetic and environmental components. The prior studies have reported the identified genetic associations with stroke and other diseases (example: coronary heart disease and atrial fibrillation). There are limited number of genetic studies have been carried out in Saudi Arabia with stroke. Although, Saudi Arabia, is known to be obese country as it is existent in the Arab region. The prevalence of obesity has been drastically increasing due to western lifestyle and free diet. The women are more affected with obesity compared with the males. This study could help the Saudi origin to design the disease marker by implementing the genetic and molecular studies by enrolling non-obese subjects. A large number of genes have been identified to be associated with the risk of genetic diseases, and among them, the genes involved in the synthesis and metabolism of serum lipids have been subject of intense research due to their clinical significance and their ability to be quantified from the serum. The prognosis of stroke mainly depends on interaction amongst the baseline characteristics (Bustamante et al., 2016, Shahid et al., 2017). Currently, GWAS have identified more than thousands of SNPs connected with human diseases and complex traits which have been a success in elucidating pathophysiological mechanisms underlying diseases with the genetic influence (Traylor et al., 2016, Zheng et al., 2017). Familial studies have provided the relation between heritability and multifactorial diseases. Stroke is influenced by environmental factors and genetics is considered as a fourth leading cause of death in the world (Terni et al., 2015). Genome-wide linkage, meta-analysis, candidate gene, GWAS, twin and family association studies suggested that genetic variances are responsible for the varied risk of ischemic stroke (Zhang et al., 2016).
Several studies have documented the relation between stroke and atherosclerosis (Hollander et al., 2003, Scannapieco et al., 2003). As per the World Health Organization, stroke is defined as the clinical syndrome typified by rapidly developing signs of focal or global disturbance of cerebral functions, lasting more than 24 h or leading to death, with no apparent causes other than of vascular origin (Xiuju et al., 2016). Almost all, ∼50% of the strokes are caused by atherosclerosis, defined as a type of disease in which plaques build up in the arteries. The presence of unstable atherosclerotic plaques in the carotid artery is a known etiological factor causing cerebrovascular events, such as an ischemic stroke, transient ischemic attack, or amaurosis fugax. Thus, carotid atherosclerotic plaque represents an important disease burden, as stroke is one of the leading causes of death worldwide and a major cause of long-term disability (Gorgui et al., 2017). An earlier study by Musunuru et al. (2010) confirmed the genetic relation association through LDLR levels act the binding of a key transcription factor and shifting the expression of the SORT1 gene involved in intracellular protein transport. The rs464218 polymorphism in SORT1 gene has been identified by GWAS in Europe, East Asian, Southern Asian, Middle Eastern Asian and Africa Americans populations (Buyske et al., 2012, Consortium CADG, 2011, Gudbjartsson et al., 2009, Kathiresan et al., 2009, Ozaki et al., 2002, Saade et al., 2011, Wang et al., 2011). Our current study results with rs464218 polymorphism are in association with Zhou et al. (2015) and Jeemon et al. (2011) studies but differ in the genotype frequencies, and this could be due to the ethnicity. In our study, the rs11053646 polymorphism in OLR1 gene was associated, and our results confirm the earlier meta-analysis studies, concluding as may contribute to the risk of disease susceptibility (Au et al., 2015). In Saudi Arabia, there are limited studies have been documented with stroke, and among them, Alhazzani et al. (2017) performed the molecular study with CYPC219 genetic polymorphisms and found to be associated. Al-Shenqiti et al. (2017) performed the detailed study of stroke who were effected for the first time. There are many other studies have been carried out in stroke as hospital-based and cross-sectional studies in the kingdom (Al Rajeh and Awada, 2002, Al Tahan et al., 1997, Zafar et al., 2016). The current study has couple of limitations and among them (i) selection of single SNPs from each gene and (ii) missing the MRI scan. The advantage of this study was selection of pure Saudi patients. In conclusion, our results confirm the SORT1 and OLR1 variants were associated in the Saudi population. The current results were in the association with the prior study results documented through GWAS and meta-analysis association. However, global ethnic population studies should be performed to rule out in the human hereditary diseases in all ethnicities.
Acknowledgement
This research was funded by Sheikh Abdullah Bin Abdul Mohsen Al Tuwaijri Chair for Applied Research in Stroke Majmaah University, Saudi Arabia. The authors would like to express their gratitude towards Sheikh Abdullah Al Tuwaijri Rector Dr. Khalid Bin Saad Al Muqrin, and Chair Supervisor Dr. Raid S. Al Badarie for providing the necessary support and assistance for completing this piece of work.
Conflict of interest
All the authors declare there is no conflict of Interest towards this manuscript.
Footnotes
Peer review under responsibility of King Saud University.
References
- Al Rajeh S., Awada A. Stroke in Saudi Arabia. Cerebrovasc. Dis. 2002;13:3–8. doi: 10.1159/000047738. [DOI] [PubMed] [Google Scholar]
- Al Tahan A., Buchur J., El Khwsky F., Ogunniyi A., Al-Rajeh S., Larbi E. Risk factors of stroke at high and low altitude areas in Saudi Arabia. Arch. Med. Res. 1997;29:173–177. [PubMed] [Google Scholar]
- Alharbi K.K., Richardson T.G., Khan I.A., Syed R., Mohammed A.K., Boustred C.R. Influence of adiposity-related genetic markers in a population of saudi arabians where other variables influencing obesity may be reduced. Dis. Mark. 2014;2014:758232. doi: 10.1155/2014/758232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alharbi K.K., Ali Khan I., Syed R., Alharbi F.K., Mohammed A.K., Vinodson B. Association of JAZF1 and TSPAN8/LGR5 variants in relation to type 2 diabetes mellitus in a Saudi population. Diabetol. Metab. Syndr. 2015;7:92. doi: 10.1186/s13098-015-0091-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhazzani A.A., Munisamy M., Karunakaran G. Pharmacogenetics of CYP2C19 genetic polymorphism on clopidogrel response in patients with ischemic stroke from Saudi Arabia. Neurosci. (Riyadh, Saudi Arabia) 2017;22:31–37. doi: 10.17712/nsj.2017.1.20160303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shenqiti A.M., Ibrahim S.R., Khaled O.A., Ali A.R., Ahmed M.S. Incidence of first time stroke: a Saudi experience. Eur. Neurol. 2017;77:147–151. doi: 10.1159/000455094. [DOI] [PubMed] [Google Scholar]
- Andersen G., Vestergaard K., Ingeman-Nielsen M., Jensen T.S. Incidence of central post-stroke pain. Pain. 1995;61:187–193. doi: 10.1016/0304-3959(94)00144-4. [DOI] [PubMed] [Google Scholar]
- Arvind P., Nair J., Jambunathan S., Kakkar V.V., Shanker J. CELSR2-PSRC1-SORT1 gene expression and association with coronary artery disease and plasma lipid levels in an Asian Indian cohort. J. Cardiol. 2014;64:339–346. doi: 10.1016/j.jjcc.2014.02.012. [DOI] [PubMed] [Google Scholar]
- Au A., Griffiths L.R., Cheng K.K., Wee Kooi C., Irene L., Keat Wei L. The influence of OLR1 and PCSK9 gene polymorphisms on ischemic stroke: evidence from a meta-analysis. Scient. Rep. 2015;5:18224. doi: 10.1038/srep18224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benamer H.T., Grosset D. Stroke in Arab countries: a systematic literature review. J. Neurol. Sci. 2009;284:18–23. doi: 10.1016/j.jns.2009.04.029. [DOI] [PubMed] [Google Scholar]
- Bustamante A., García-Berrocoso T., Rodriguez N., Llombart V., Ribó M., Molina C. Ischemic stroke outcome: a review of the influence of post-stroke complications within the different scenarios of stroke care. Eur. J. Inter. Med. 2016;29:9–21. doi: 10.1016/j.ejim.2015.11.030. [DOI] [PubMed] [Google Scholar]
- Buyske S., Wu Y., Carty C.L., Cheng I., Assimes T.L., Dumitrescu L. Evaluation of the metabochip genotyping array in African Americans and implications for fine mapping of GWAS-identified loci: the PAGE study. PLoS One. 2012;7:e35651. doi: 10.1371/journal.pone.0035651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Consortium CADG A genome-wide association study in Europeans and South Asians identifies five new loci for coronary artery disease. Nat. Genet. 2011;43:339–344. doi: 10.1038/ng.782. [DOI] [PubMed] [Google Scholar]
- Das S., Roy S., Munshi A. Association between PDE4D gene and ischemic stroke: recent advancements. Int. J. Neurosci. 2016;126:577–583. doi: 10.3109/00207454.2015.1051621. [DOI] [PubMed] [Google Scholar]
- Gorgui J., Gasbarrino K., Georgakis M.K., Karalexi M.A., Nauche B., Petridou E.T. Circulating adiponectin levels in relation to carotid atherosclerotic plaque presence, ischemic stroke risk, and mortality: A systematic review and meta-analyses. Metab.: Clin. Exp. 2017;69:51–66. doi: 10.1016/j.metabol.2017.01.002. [DOI] [PubMed] [Google Scholar]
- Gudbjartsson D.F., Bjornsdottir U.S., Halapi E., Helgadottir A., Sulem P., Jonsdottir G.M. Sequence variants affecting eosinophil numbers associate with asthma and myocardial infarction. Nat. Genet. 2009;41:342–347. doi: 10.1038/ng.323. [DOI] [PubMed] [Google Scholar]
- Hassan A., Markus H.S. Genetics and ischaemic stroke. Brain. 2000;123(Pt 9):1784–1812. doi: 10.1093/brain/123.9.1784. [DOI] [PubMed] [Google Scholar]
- Hattori H., Sonoda A., Sato H., Ito D., Tanahashi N., Murata M. G501C polymorphism of oxidized LDL receptor gene (OLR1) and ischemic stroke. Brain Res. 2006;1121:246–249. doi: 10.1016/j.brainres.2006.08.091. [DOI] [PubMed] [Google Scholar]
- He, T., Wang, J., Wang, X.L., Deng, W.S., Sun, P., 2017. Association between the Matrix Metalloproteinase-9 rs3918242 polymorphism and ischemic stroke susceptibility: a meta-analysis. J. Stroke Cerebrovasc. Dis.: Off. J. Natl. Stroke Assoc. [DOI] [PubMed]
- Hollander M., Hak A.E., Koudstaal P.J., Bots M.L., Grobbee D.E., Hofman A. Comparison between measures of atherosclerosis and risk of stroke: the Rotterdam Study. Stroke. 2003;34:2367–2372. doi: 10.1161/01.STR.0000091393.32060.0E. [DOI] [PubMed] [Google Scholar]
- Hubacek J.A. Genetic determination of dyslipidemia - What tell us the results of genome-wide association studies? Vnitrni lekarstvi. 2016;62:868–876. [PubMed] [Google Scholar]
- Jeemon P., Pettigrew K., Sainsbury C., Prabhakaran D., Padmanabhan S. Implications of discoveries from genome-wide association studies in current cardiovascular practice. World J. Cardiol. 2011;3:230–247. doi: 10.4330/wjc.v3.i7.230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin X., Wang D., Zhou Y., Xiong H. Association between the interleukin-6-174 G/C polymorphism and risk of ischemic stroke: a meta-analysis. Genet. Mol Res. 2015;14:13076–13083. doi: 10.4238/2015.October.26.3. [DOI] [PubMed] [Google Scholar]
- Kathiresan S., Voight B.F., Purcell S., Musunuru K., Ardissino D., Mannucci P.M. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009;41:334–341. doi: 10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.H. Meta-analysis of genetic association studies. Ann. Lab. Med. 2015;35:283–287. doi: 10.3343/alm.2015.35.3.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Li, X., Lin, H., Fu, X., Lin, W., Li, M., et al., 2017. Metabolic syndrome and stroke: a meta-analysis of prospective cohort studies. J. Clin. Neurosci.: Off. J. Neurosurg. Soc. Australasia. [DOI] [PubMed]
- Linsel-Nitschke P., Heeren J., Aherrahrou Z., Bruse P., Gieger C., Illig T. Genetic variation at chromosome 1p13.3 affects sortilin mRNA expression, cellular LDL-uptake and serum LDL levels which translates to the risk of coronary artery disease. Atherosclerosis. 2010;208:183–189. doi: 10.1016/j.atherosclerosis.2009.06.034. [DOI] [PubMed] [Google Scholar]
- Loci associated with ischaemic stroke and its subtypes (SiGN): a genome-wide association study, 2016. Lancet Neurol., vol. 15, pp. 174–184. [DOI] [PMC free article] [PubMed]
- Luo C., Fan L.H., Zhang H., Zhao J., Li L., Zhang L. Association between C807T(C/T) polymorphism of platelet glycoprotein gene and sensitivity to ischemic stroke: a meta-analysis. Genet Mol Res. 2017;16 doi: 10.4238/gmr16019416. [DOI] [PubMed] [Google Scholar]
- Munshi A., Das S., Kaul S. Genetic determinants in ischaemic stroke subtypes: seven year findings and a review. Gene. 2015;555:250–259. doi: 10.1016/j.gene.2014.11.015. [DOI] [PubMed] [Google Scholar]
- Musunuru K., Strong A., Frank-Kamenetsky M., Lee N.E., Ahfeldt T., Sachs K.V. From noncoding variant to phenotype via SORT1 at the 1p13 cholesterol locus. Nature. 2010;466:714–719. doi: 10.1038/nature09266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalls M.A., Pankratz N., Lill C.M., Do C.B., Hernandez D.G., Saad M. Large-scale meta-analysis of genome-wide association data identifies six new risk loci for Parkinson's disease. Nat. Genet. 2014;46:989–993. doi: 10.1038/ng.3043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell M.J., Xavier D., Liu L., Zhang H., Chin S.L., Rao-Melacini P. Risk factors for ischaemic and intracerebral haemorrhagic stroke in 22 countries (the INTERSTROKE study): a case-control study. The Lancet. 2010;376:112–123. doi: 10.1016/S0140-6736(10)60834-3. [DOI] [PubMed] [Google Scholar]
- Ozaki K., Ohnishi Y., Iida A., Sekine A., Yamada R., Tsunoda T. Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nat. Genet. 2002;32:650–654. doi: 10.1038/ng1047. [DOI] [PubMed] [Google Scholar]
- Qin B., Zheng Y., Zhang W., Wang C., Wang J., Cai Z. Lack of associations between rs2910164 and rs11614913 polymorphisms and the risk of ischemic stroke. Int. J. Clin. Exp. Med. 2015;8:18359. [PMC free article] [PubMed] [Google Scholar]
- Robert A.A., Zamzami M.M. Stroke in Saudi Arabia: a review of the recent literature. Pan Afr. Med. J. 2014;17:14. doi: 10.11604/pamj.2014.17.14.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saade S., Cazier J.-B., Ghassibe-Sabbagh M., Youhanna S., Badro D.A., Kamatani Y. Large scale association analysis identifies three susceptibility loci for coronary artery disease. PloS One. 2011;6:e29427. doi: 10.1371/journal.pone.0029427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannapieco F.A., Bush R.B., Paju S. Associations between periodontal disease and risk for atherosclerosis, cardiovascular disease, and stroke. A systematic review. Ann. Periodontol. 2003;8:38–53. doi: 10.1902/annals.2003.8.1.38. [DOI] [PubMed] [Google Scholar]
- Shahid S.U., Shabana N., Cooper J.A., Rehman A., Humphries S.E. Common variants in the genes of triglyceride and HDL-C metabolism lack association with coronary artery disease in the Pakistani subjects. Lipids Health Dis. 2017;16:24. doi: 10.1186/s12944-017-0419-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M.S., Jiang T., Tan L., Yu J.T. Genome-wide association studies in neurology. Ann. Transl. Med. 2014;2:124. doi: 10.3978/j.issn.2305-5839.2014.11.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terni E., Giannini N., Brondi M., Montano V., Bonuccelli U., Mancuso M. Genetics of ischaemic stroke in young adults. BBA Clin. 2015;3:96–106. doi: 10.1016/j.bbacli.2014.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traylor M., Malik R., Nalls M.A., Cotlarciuc I., Radmanesh F., Thorleifsson G. Genetic Variation at 16q24. 2 is associated with small vessel stroke. Ann. Neurol. 2016 doi: 10.1002/ana.24840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang F., Xu C.-Q., He Q., Cai J.-P., Li X.-C., Wang D. Genome-wide association identifies a susceptibility locus for coronary artery disease in the Chinese Han population. Nat. Genet. 2011;43:345–349. doi: 10.1038/ng.783. [DOI] [PubMed] [Google Scholar]
- Wang L., Yanuck D., Beecham A., Gardener H., Slifer S., Blanton S.H. A candidate gene study revealed sex-specific association between the OLR1 gene and carotid plaque. Stroke. 2011;42:588–592. doi: 10.1161/STROKEAHA.110.596841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichmann H.E. Genetic epidemiology in Germany–from biobanking to genetic statistics. Methods Inform. Med. 2005;44:584–589. [PubMed] [Google Scholar]
- Xia W., Hu Z., Song Z. [Meta-analysis for the relationship between lipoprotein-associated phospholipase A2 and ischemic stroke] Zhong nan da xue xue bao Yi xue ban = J. Central South Univ. Med. Sci. 2017;42:208–214. doi: 10.11817/j.issn.1672-7347.2017.02.015. [DOI] [PubMed] [Google Scholar]
- Xiuju C., Zhen W., Yanchao S. A meta-analysis of adiponectin gene rs22411766 T>G polymorphism and ischemic stroke susceptibility. Open Med. (Warsaw, Poland). 2016;11:115–120. doi: 10.1515/med-2016-0022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanamala C.M., Bundhun P.K., Ahmed A. Comparing mortality between fibrinolysis and primary percutaneous coronary intervention in patients with acute myocardial infarction: a systematic review and meta-analysis of 27 randomized-controlled trials including 11 429 patients. Coron. Art. Dis. 2017 doi: 10.1097/MCA.0000000000000489. [DOI] [PubMed] [Google Scholar]
- Yang H., Guo W., Li J., Cao S., Zhang J., Pan J. Leptin concentration and risk of coronary heart disease and stroke: a systematic review and meta-analysis. PLoS One. 2017;12:e0166360. doi: 10.1371/journal.pone.0166360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zafar A., Al-Khamis F.A., Al-Bakr A.I., Alsulaiman A.A., Msmar A.H. Risk factors and subtypes of acute ischemic stroke. A study at King Fahd Hospital of the University. Neurosciences (Riyadh, Saudi Arabia) 2016;21:246–251. doi: 10.17712/nsj.2016.3.20150731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H., Zhai Q., Zhang Z., Cai B., Cai H., Zhou S. Association of GWAS-Supported Variants rs556621 on Chromosome 6p21. 1 with large artery atherosclerotic stroke in a Southern Chinese Han Population. NeuroMol. Med. 2016:1–7. doi: 10.1007/s12017-016-8433-7. [DOI] [PubMed] [Google Scholar]
- Zheng J., Rodriguez S., Laurin C., Baird D., Trela-Larsen L., Erzurumluoglu M.A. HAPRAP: a haplotype-based iterative method for statistical fine mapping using GWAS summary statistics. Bioinformatics. 2017;33:79–86. doi: 10.1093/bioinformatics/btw565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong C., Zhong X., Xu T., Xu T., Zhang Y. Sex-specific relationship between serum uric acid and risk of stroke: a dose-response meta-analysis of prospective studies. J. Am. Heart Assoc. 2017;6 doi: 10.1161/JAHA.116.005042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y.-J., Hong S.-C., Yang Q., Yin R.-X., Cao X.-L., Chen W.-X. Association of variants in CELSR2-PSRC1-SORT1 with risk of serum lipid traits, coronary artery disease and ischemic stroke. Int. J. Clin. Exp. Pathol. 2015;8:9543. [PMC free article] [PubMed] [Google Scholar]