Abstract
High cancer drug development attrition rates have provoked considerable debate about whether the two-dimensional tumor growth inhibition high-throughput screening assays used in pre-clinical lead discovery adequately reflect solid tumor complexity. We used automated high-content screening image acquisition and analysis methods to compare fluorescent drug uptake, accumulation, and distribution in Cal33 and FaDu head and neck cancer (HNC) monolayer and multicellular tumor spheroid (MCTS) models. Ellipticine, idarubicin, daunorubicin, and doxorubicin were studied because of their fluorescent properties and broad anti-tumor activities. HNC MCTSs were generated in 384-well ultra-low attachment plates where compound exposure, image acquisition, and analysis could be performed in situ. Fluorescent drug accumulation in Cal33 monolayer and MCTS cultures was linear with respect to concentration, and appeared to achieve steady-state levels within 10–15 min of drug exposure, which were maintained through 30–45 min. Drug accumulation in monolayers was independent of cell number and/or density, and every cell achieved uniform drug concentrations. In MCTSs, however, drug accumulation increased as the number of cells and sizes of the MCTSs became bigger. Drugs exhibited restricted penetration and distribution gradients, accumulating preferentially in cells in the outer layers of MCTSs relative to those in the inner cores. Cal33 monolayers were 6-, 20-, 10-, and 16-fold more sensitive than MCTSs to growth inhibition by ellipticine, idarubicin, daunorubicin, and doxorubicin, respectively. In Cal33 MCTSs exposed to ellipticine or doxorubicin for 24 h, MCTSs were smaller and although they still exhibited drug penetration and distribution gradients, the fluorescent intensity difference between outer and inner cells was reduced. After a 24 h exposure, both drugs had penetrated throughout FaDu MCTSs, consistent with drug-induced death of peripheral cell layers enhancing drug penetration. The increased resistance of MCTS cultures and their ability to recapitulate drug penetration and distribution gradients argues strongly for the deployment of these more physiological models in cancer lead discovery. MCTSs have the potential to enhance the correlation between in vitro potencies and in vivo efficacy, and ultimately may lead to improved cancer drug approval rates.
Keywords: : multicellular tumor spheroids, drug accumulation penetration, drug distribution, 2D versus 3D culture models
Introduction
New cancer drug leads are typically identified in high-throughput growth inhibition screening campaigns conducted in human tumor cell lines that are both maintained and assayed in two-dimensional (2D) culture conditions.1–6 Two-dimensional growth inhibition assays provide the throughput and capacity required to screen large compound (>100 K) libraries, and molecules that potently inhibit in vitro tumor cell line growth ultimately progress to in vivo tumor efficacy studies in mice.4,7,8 Preclinical mouse tumor models include murine tumors transplanted into syngeneic mice, genetically engineered mouse models, or, most commonly, human xenograft tumor cells implanted into mice with compromised immune systems.4 Mechanism of action studies are then initiated for leads that demonstrate in vivo tumor efficacy.4,7,8 However, despite considerable investments in cancer research, drug discovery and development in general, new cancer drug approval rates are ≤5%, greater than twofold lower than in other therapeutic areas.4,7–9 The higher attrition rates for cancer drug development have provoked considerable discussion about whether the prevailing 2D tumor growth inhibition high-throughput screening (HTS) assays used for pre-clinical lead discovery adequately recapitulate the complexities of solid tumors. For example, tumor cell lines adapted to growth in 2D proliferate quicker than cells isolated from primary tumors, do not exhibit similar drug resistance profiles, and favor the discovery of anti-proliferative agents while neglecting the self- and population-renewing tumor stem cells that contribute to recurrence and metastasis.10–14 Genomic comparisons between head and neck cancer (HNC) cell lines and primary tumors revealed subsets of unique mutations in cell lines that favor immortalization and continuous maintenance in tissue culture.15 To mitigate such concerns, patient-derived xenograft (PDX) models that have never been adapted to tissue culture but are serially passaged in nude mice have been implemented to better predict clinical trial drug responses.16–18 However, mouse PDX models have neither the throughput nor the capacity required to support cancer lead discovery from large compound libraries.
In HTS growth inhibition assays, 2D monolayer tumor cell lines are exposed to uniform drug concentrations in a homogenous environment where cell-extra cellular matrix (ECM) and cell-cell contacts either do not occur or have been minimized because low cell seeding densities were selected to ensure exponential growth throughout the compound exposure period.10–14,19–26 However, cells in solid tumors interact extensively with the ECM, form numerous cell-cell focal adhesion junctions, and experience a variety of hypoxic, acidic, or nutrient microenvironments that support differential proliferation rates in distinct regions of the tumor.27–29 Some anti-cancer agents may exhibit reduced activity in hypoxic, acidic, or nutrient-deprived microenvironments.27–29 Dormant or quiescent tumor cells that have stopped replicating, or that proliferate slowly due to reduced nutrient and/or oxygen levels in their microenvironment will be resistant to molecules targeting cell proliferation mechanisms.27–29 Permeability barriers may restrict drug penetration and distribution in solid tumors such that cells that are distal to blood vessels experience lower drug concentrations.27–29 Drug effectiveness will be compromised when agents fail to achieve therapeutic concentrations in every tumor cell, and the survival of even a few cancer cells could be the beginning of recurrence.
One strategy to improve cancer drug development success rates would be to utilize more physiologically relevant in vitro cellular models for early lead discovery. These models should recapitulate the morphologies and microenvironments experienced by cells in solid tumors in either preclinical mouse models or patients.4,30–34 For solid tumors such as HNC, this would dictate the implementation of three-dimensional (3D) in vitro tumor models that develop hypoxic, acidic, or nutrient-deprived microenvironments, exhibit drug permeability barriers, and recapitulate tumor architecture, morphology, and pathophysiology.30–34 There is a broad consensus that 3D tumor culture models will improve the correlation between in vitro cancer drug potencies and efficacy in solid tumor animal models or human clinical trials.10–14,19–26,34
In this article, we describe the use of automated image acquisition and analysis methods on a high-content imaging platform to visualize, quantify, and investigate cancer drug accumulation and distribution in 2D and 3D HNC cell culture models. Multicellular tumor spheroids (MCTSs) are self-assembled 3D tumor cell aggregates that are formed under conditions where cell-cell interactions predominate over cell-substrate interactions.11,12,22,24,34–36 Human tumor cell lines cultured in U-bottomed ultra-low attachment microtiter plates (ULA plates) treated with a hydrophilic neutrally charged coating to prevent cell adhesion to the plate surface have been shown to self-assemble into tight spheroids, compact aggregates, or loose aggregates.11,34,36–38 We have shown that several HNC cell lines cultured in 384-well ULA plates form uniform MCTSs, and that the process is compatible with automation, requires relatively few cells per well, takes days rather than weeks to occur, and compound exposure and assay detection can be performed in situ.34 We present data comparing the uptake, accumulation, and distribution of four broadly used fluorescent cancer drugs into 2D monolayer and MCTS HNC cell culture models: the dimethyl-pyrido-carbazole plant alkaloid ellipticine, and the anthracycline chemotherapeutics idarubicin, daunorubicin, and doxorubicin. In 2D HNC monolayers, drug uptake proceeded in a time- and concentration-dependent manner until cells achieved steady-state uniform drug accumulation levels that were independent of cell density. In MCTSs, however, the time- and concentration-dependent accumulation of drugs increased as the number of cells and sizes of the MCTSs became larger, with cells in the peripheral outer layers of the MCTSs accumulating considerably higher drug levels than cells in the inner core. The apparent drug penetration barriers and distribution gradients in HNC MCTSs coincided with a 6- to 20-fold enhanced resistance to growth inhibition by these agents when directly compared with 2D HNC monolayers.
Materials and Methods
Reagents
Thirty-seven percent formaldehyde was purchased from Sigma-Aldrich (St. Louis, MO). Hoechst 33342 was purchased from Life Technologies (Thermo Fisher Scientific, Waltham, MA). Dimethyl sulfoxide (DMSO) (99.9% high-performance liquid chromatography [HPLC] grade) was obtained from Alfa Aesar (Ward Hill, MA). Dulbecco's Mg2+- and Ca2+-free phosphate-buffered saline (PBS) was purchased from Corning (Tewksbury, MA). Fetal bovine serum (FBS), l-glutamine, penicillin, and streptomycin (P/S) were purchased from Thermo Fisher Scientific. CellTiter-Glo® (CTG) luminescent cell viability assay reagent was purchased from Promega Corporation (Madison, WI). Ellipticine was purchased from Tocris (Bio-Techne, Avonmouth, Bristol, United Kingdom), idarubicin was purchased from Sigma-Aldrich, and daunorubicin and doxorubicin were provided by the National Cancer Institute (NCI).
Cells and Tissue Culture
Cal33 and FaDu HNC cell lines were obtained from Dr. Jennifer Grandis in the HNC Spore at the University of Pittsburgh Cancer Institute, and they were maintained in a humidified incubator at 37°C, 5% CO2, and 95% humidity. Cal33 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS, 1% l-glutamine, and 1% P/S. FaDu cells were cultured in DMEM supplemented with 10% FBS, 1% non-essential amino acids, 1% l-glutamine, and 1% P/S. Isolated cell suspensions were prepared from tissue culture flasks by dissociating cells with trypsin and centrifugation at 1,200 rpm for 5 min at room temperature, and re-suspension in growth media. The number of trypan blue excluding viable cells in the cell suspension was counted by using a hemocytometer.
Preparation of 2D Monolayer and MCTS HNC Cultures
The production of 2D HNC monolayers in 384-well microtiter plates has been previously described.39,40 Briefly, 45 μL of the desired seeding densities of Cal33 or FaDu single-cell suspensions was transferred into the wells of a 384-well tissue culture-treated microplate (No. 781091; Greiner BioOne) by using a Matrix electronic multichannel pipette (Thermo Fisher Scientific); cells were allowed to settle by gravity for 10 min, followed by centrifugation at 100 rpm for 1 min; and plates were then placed in an incubator at 37°C, 5% CO2, and 95% humidity for 24 h.
The generation of HNC MCTSs in 384-well ULA plates has been previously described.34 Briefly, 384-well ULA plates (Cat. No. 4516; Corning) were rehydrated by the addition of 50 μL of serum-free DMEM to each well and incubation in a humidified incubator for 15 min. Media were removed from the wells of the ULA plates, and 45 μL of a Cal33 or FaDu single-cell suspension at different seeding densities (625, 1,250, 2,500, 5,000, 10,000, or 20,000 cells/well) was transferred into each well by using a Matrix electronic multichannel pipette; ULA plates were centrifuged at 100 rpm for 1 min, and then placed in an incubator at 37°C, 5% CO2, and 95% humidity for 24 h.
Two-Dimensional Monolayer and MCTS HNC Cell Line Growth Inhibition Assays
Three hundred eighty-four-well 2D HNC monolayer growth inhibition assays have been previously described.39,40 Briefly, Cal33 cells were seeded in a volume of 45 μL of growth medium at 1,000 cells/well in uncoated, white, opaque 384-well assay plates (No. 781080; Greiner BioOne) and the plates were then incubated at 37°C, 5% CO2, and 95% humidity for 24 h. After 24 h, 5 μL of compounds serially diluted in serum-free media to produce the indicated concentrations were transferred into the test wells of assay plates by using a Janus MDT Mini (PerkinElmer, Waltham, MA) automated liquid handler platform equipped with a 384-well transfer head. At that time, 5 μL of 5% DMSO (0.5% DMSO final) and 5 μL of 2 mM doxorubicin (200 μM final) were transferred into the maximum plate control (Max.) and minimum plate control (Min.) wells (n = 32 each), respectively. Compound-treated assay plates were then returned to an incubator at 37°C, 5% CO2, and 95% humidity for 72 h. After 72 h, 25 μL of the CTG (Promega Corporation) cell viability detection reagent was dispensed into the wells of assay plates, and the plates were incubated for 15 min at ambient temperature. The relative light units (RLUs) luminescence signals were then captured on a SpectraMax M5e (Molecular Devices, LLC, Sunnyvale, CA) micro-titer plate reader platform.
Three hundred eighty-four-well ULA-plate HNC MCTS growth inhibition has been previously described.34 Briefly, Cal33 cells were seeded in a volume of 45 μL of growth medium at 5,000 cells/well in 384-well ULA plates (Corning) and the plates were then incubated at 37°C, 5% CO2, and 95% humidity for 24 h. After 24 h, 5 μL of compounds serially diluted in serum-free media to produce the indicated concentrations was transferred into the replicate (n = 3) test wells of assay plates by using a Janus MDT Mini automated liquid handler platform equipped with a 384-well transfer head. Five microliters of 5% DMSO and 5 μL of 2 mM doxorubicin were transferred into the maximum and minimum plate control wells (n = 32 each), respectively. Compound-treated plates were then returned to an incubator at 37°C, 5% CO2, and 95% humidity for 72 h. After 72 h, 25 μL of the CTG (Promega Corporation) cell viability detection reagent was dispensed into the wells of assay plates, and incubated for 15 min at ambient temperature. The RLUs luminescence signals were then captured on a SpectraMax M5e (Molecular Devices, LLC) micro-titer plate reader platform.
Drug Accumulation Assays: Concentration Response, Time-Course, and Cell Seeding Density
In drug accumulation assays, 5 μL of serially diluted compounds was added to replicate (n = 3) wells of HNC monolayer and MCTS assay plates (Table 1) by using a Janus MDT Mini automated liquid handler platform equipped with a 384-well transfer head. The final concentrations of test compounds were 25, 12.5, 6.25, 3.12, 1.56, and 0.781 μM in a final volume of 50 μL. After a 15 min incubation at 37°C, 5% CO2, and 95% humidity, monolayer and MCTS assay plates were fixed by the addition of 50 μL of pre-warmed (37°C) 7.4% formaldehyde and 4 μg/mL Hoechst 33342 in PBS by using a Matrix electronic multichannel pipette and incubated at room temperature for 30 min, as previously described.39–41 The well contents (compounds, media and fixative) were aspirated by using a Janus MDT automated liquid handler equipped with a 384-well transfer head and exchanged with 50 μL of PBS three times for HNC monolayers or four times for HNC MCTSs. During MCTS washing, care was taken to avoid damaging or aspirating MCTSs by setting the aspiration height on the Janus MDT 2 mm higher than for washing cell monolayers. An additional PBS exchange was included to assure complete washing of MCTSs. Fifty microliters of PBS was then added to each well, and the plates were sealed with adhesive aluminum plate seals. Assay plates were imaged and analyzed on the ImageXpress® Micro (IXM; Molecular Devices, LLC) automated high-content screening (HCS) platform (see sections “Image Acquisition on the IXM HCS Platform” and “Multi-Wavelength Cell Scoring Image Analysis”).
Table 1.
Fluorescent Drug Uptake and Accumulation Assay Protocol in Multicellular Tumor Spheroids Formed from Head and Neck Cancer Tumor Cell Lines
| Step | Parameter | Value | Description |
|---|---|---|---|
| 1 | Rehydrate 384-well ULA plates | 50 μL of serum-free DMEM, 15 min | Rehydrate ULA-plate wells by the addition of 50 μL of serum-free DMEM to each well and incubation in a humidified incubator for 15 min. After incubation, the media are removed and 15 μL of culture medium is added to each well. |
| 2 | Harvest and centrifuge Cal33 or FaDu cells | 5 min, 500 × g | Aspirate medium, wash with PBS, trypsinize cells, add DMEM medium +10% FBS, and centrifuge |
| 3 | Viable Cal33 or FaDu cell count | Viable cell count | Re-suspend cells in culture medium and count the number of trypan blue excluding viable cells in a hemocytometer |
| 4 | Adjust Cal33 or FaDu cells to the required density and seed into 384-well ULA assay plates | Add 45 μL of cells/well and centrifuge at 100 rpm for 1 min | Seed wells at 625, 1,250, 2,500, 5,000, 10,000, or 20,000 cells/well in rehydrated 384-well ULA plates, centrifuge, and incubate overnight at 37°C, 5% CO2, and 95% humidity |
| 5 | Transfer fluorescent test compounds/DMSO to control wells | 5 μL | 0.781–25 μM final concentration in well, 0.25% DMSO |
| 6 | Incubate assay plates | 2.5–45 min | At 37°C, 5% CO2, and 95% humidity |
| 7 | Fix cells | 50 μL | 7.4% Formaldehyde containing 4 μg/mL Hoechst 33342 in Ca2+- and Mg2+-free PBS pre-warmed to 37°C |
| 8 | Incubate assay plates | 30 min | Ambient temperature |
| 9 | Aspirate fixative and wash 2 × with PBS | 50 μL | Aspirate fixative and wash twice with 50 μL Ca2+- and Mg2+-free PBS, 50 μL PBS in well |
| 10 | Seal plates | 1 × | Seal with adhesive aluminum or clear plate seals |
| 11 | Acquire images | 4 × , 0.2NA objective | Transmitted light and fluorescent images of the DAPI (Ch1), FITC (Ch2), and TRITC (Ch3) channels were sequentially acquired on the ImageXpress® Micro platform by using a 300 W Xenon lamp broad spectrum white light source, ZPS filter sets, and a 1.4 megapixel 2/3" chip Cooled CCD Camera. |
| 12 | Image analysis assay readout | Mean integrated fluorescent intensity values | Images were analyzed by using the Multi-Wavelength Cell Scoring image analysis module and by using the mean integrated fluorescent intensity values to quantify uptake and accumulation. |
Step Notes
1. The treated surface of the 384-well ULA plates was rehydrated by incubation with 50 μL of serum-free medium for 15 min—this step reduced the number of failed wells. After incubation, removing the serum-free medium and replacing with 15 μL culture medium prevents the ULA surface from drying out while cell suspensions are prepared.
2. Cal33 or FaDu cells were maintained in DMEM medium with 2 mM l-glutamine supplemented with 10% FBS, and 100 U/mL penicillin and streptomycin in a humidified incubator at 37°C, 5% CO2, and 95% humidity. Cell monolayers (<70% confluent) were washed 1 × with PBS, and they were then exposed to trypsin-EDTA until they detached from the surface of the tissue culture flasks. Cells were pelleted at 500 × g for 5 min in Sorvall ST 16 Centrifuge with a TX-400 Rotor.
3. Aspirate medium, re-suspend pelleted cells in tissue culture medium+FBS, and count the number of trypan blue excluding viable cells in a hemocytometer.
4. Cal33 or FaDu head and neck cancer cell lines were seeded into 384-well ULA plates (Corning, Tewksbury, MA) at the indicated seeding densities per well and incubated for 24 h at 37°C, 5% CO2, and 95% humidity in DMEM with 2 mM l-glutamine supplemented with 10% FBS, and 100 U/mL penicillin and streptomycin.
5. 0.781–25 μM of compounds were added to wells in columns 3–22 by using a Janus MDT Mini automated liquid handler platform equipped with a 384-well transfer head.
6. Incubate treated Cal33 or FaDu cells for 2.5–45 min at 37°C, 5% CO2, and 95% humidity.
7 and 8. Fix MCTSs for 30 min at ambient temperature by adding 50 μL of 7.4% formaldehyde containing 2 μg/mL Hoechst 33342 in Ca2+- and Mg2+-free PBS pre-warmed to 37°C.
9. Aspiration of media and fixative, and PBS washes were performed by using a Janus MDT Mini automated liquid handler platform equipped with a 384-well transfer head.
10. Plates were sealed with adhesive aluminum or clear plate seals.
11. Plates were loaded into the ImageXpress Micro HCS platform (Molecular Devices, LLC, Sunnyvale, CA) for scanning.
12. Images were analyzed by using the Multi-Wavelength Cell Scoring image analysis module of MetaXpress (Molecular Devices, LLC).
Ch1, fluorescent channel 1; Ch2, fluorescent channel 2; Ch3, fluorescent channel 3; DAPI, 4,6-diamidino-2-phenylindole; DMEM, Dulbecco's modified Eagle's medium; DMSO, dimethyl sulfoxide; FBS, fetal bovine serum; FITC, fluorescein isothiocyanate; HCS, high-content screening; MCTSs, multicellular tumor spheroids; PBS, phosphate-buffered saline; TRITC, tetramethylrhodamine; ULA plates, ultra-low attachment microtiter plates; ZPS, Zero Pixel Shift.
In time-course assays, 5 μL of test compounds (10 μM final) was transferred into replicate (n = 3) wells of HNC monolayer and MCTS assay plates (Table 1) by using a Janus MDT Mini automated liquid handler platform equipped with a 384-well transfer head, and the plates were incubated at 37°C, 5% CO2, and 95% humidity for the indicated time periods: 0, 2.5, 5, 10, 15, 30, and 45 min. Assay plates were fixed, stained with Hoechst, and well contents were exchanged for PBS as described earlier. Sealed assay plates were then imaged and analyzed on the IXM HCS platform (see sections “Image Acquisition on the IXM HCS Platform” and “Multi-Wavelength Cell Scoring Image Analysis”).
In cell seeding density assays, Cal33 and FaDu cell lines were seeded into 384-well monolayer and MCTS assay plates (Table 1) at seeding densities of 625, 1,250, 2,500, 5,000, 10,000, and 20,000 cells per well. After 24 h in culture, 5 μL of test compounds (10 μM final) was transferred into replicate (n = 3) wells of HNC monolayer and MCTS assay plates by using a Janus MDT Mini automated liquid handler platform equipped with a 384-well transfer head, and plates were then incubated at 37°C, 5% CO2, and 95% humidity for 15 min. Assay plates were fixed, stained with Hoechst, and well contents were exchanged for PBS as described earlier. Sealed assay plates were then imaged and analyzed on the IXM HCS platform (see section “Image Acquisition on the IXM HCS Platform”).
Image Acquisition on the IXM HCS Platform
The IXM is an automated wide-field high-content imaging platform integrated with the MetaXpress Imaging and Analysis software (Molecular Devices, LLC). The IXM optical drive includes a 300 W Xenon lamp broad spectrum white light source and a 1.4 megapixel 2/3" chip Cooled CCD Camera and optical train for standard fluorescence imaging and a transmitted light module with phase contrast. The IXM has the following Zero Pixel Shift (ZPS) filter sets: 4,6-diamidino-2-phenylindole (DAPI), fluorescein isothiocyanate (FITC)/ALEXA 488, CY3/tetramethylrhodamine (TRITC), CY5, and Texas Red. The four-position objective turret can be loaded with the following objectives: a 4 × Plan Apo 0.20 NA objective, a 10 × Plan Fluor 0.3 NA objective, a 20 × Ph1 Plan Fluor ELWD DM objective, a 20 × S Plan Fluor ELWD 0.45 NA objective, and a 40 × S Plan Fluor ELWD 0.60 NA objective.
Images of two fields of view of Cal33 monolayers were acquired sequentially by using a 10 × objective in each of three fluorescent channels: DAPI, FITC, and TRITC. The IXM infrared laser autofocus was used to detect the bottom of the plate and well, and then an image was acquired in each of the three fluorescent channels by using a fixed Z-offset from the autofocus point.
Single images of Cal33 MCTS were sequentially acquired by using either a 4 × or 10 × objective in both the transmitted light and fluorescent acquisition modes: DAPI, FITC, and TRITC. To create a single maximum 2D projection image, the IXM infrared laser autofocus was used to detect the bottom of the plate and well, and then a series of 10–20 Z-stack images were acquired, each separated by a step size of 20 μm in a range equally distributed above and below a set Z-position. A journal was then used to collapse the images in the Z-stack to generate a single maximum projection image. To acquire best focus images of MCTSs, we used the IXM automated image-based focus algorithm to acquire a coarse focus (large μm steps) set of images of Hoechst-stained objects in the DAPI channel for the first spheroid to be imaged, followed by a fine (small μm steps) set of images to select the best focus image. For all subsequent wells and channels to be imaged, only a fine focus set of images were acquired to select the best focus Z-plane.
Multi-Wavelength Cell Scoring Image Analysis
We used the multi-wavelength cell scoring (MWCS) image analysis module (Molecular Devices, LLC) to quantify the integrated fluorescent intensities of the test compounds from digital images acquired on the IXM as described earlier.
For HNC monolayers, the MWCS module image segmentation identified and classified Hoechst 33342 stained fluorescent objects in fluorescent channel 1 (Ch1; DAPI) that exhibited appropriate fluorescent intensities above background and morphology (size, width, length, and area) characteristics of Cal33 or FaDu nuclei and used these objects to create nuclear masks for each cell. For Cal33 monolayers, we defined the approximate minimum width of Hoechst-stained nuclei to be 8 μm and the approximate maximum width to be 30 μm, and the threshold intensity above local background to be 5. After applying user-defined background average intensity thresholds, typically 200 in both fluorescent channel 2 (Ch2; FITC) and fluorescent channel 3 (Ch3; TRITC), the MWCS module image segmentation then creates total cell masks for each cell. The nuclear mask from Ch1 was then used to quantify the mean integrated fluorescence intensity of Hoechst within the nuclear regions of Cal33 or FaDu cells, and to count the number of cells per image. The derived Cal33 or FaDu total cell masks in Ch2 and Ch3 were then used to quantify the mean integrated fluorescence intensity of the signals in the FITC and TRITC channels, respectively. The MWCS image analysis module outputs quantitative data, including: the mean integrated fluorescent intensities of the Hoechst-stained objects (compartments) in Ch1; the number of compartments or total cell count in Ch1; and the mean integrated fluorescent intensities of the Ch2 and Ch3 signals within the whole cell masks. The mean integrated intensity is the total pixel fluorescent intensity in Ch1, Ch2, or Ch3 within the stained area, nuclear (Ch1) or total cell mask (Ch2 and Ch3), of positively stained cells above the pre-set thresholds, divided by the number of positively stained cells.
For Cal33 and FaDu MCTSs, the approximate minimum width of Hoechst-stained nuclei (Ch1) to identify the whole spheroid was set to be 150 μm, the approximate maximum width of the spheroid was set to be 550 μm, and the threshold intensity above local background was set to be 70. After applying user-defined background average intensity thresholds, typically 50–70 in both Ch2 (FITC) and Ch3 (TRITC), the MWCS module image segmentation then created total MCTS masks in both fluorescent channels. The total MCTS mask from Ch1 was used to quantify the mean integrated fluorescence intensity of Hoechst within the Cal33 or FaDu spheroids and to count the number of MCTSs per image (usually one). The derived Cal33 or FaDu total MCTS masks from Ch2 and Ch3 were then used to quantify the mean integrated fluorescence intensity of the signals in the FITC and TRITC channels, respectively. The MWCS image analysis module outputs quantitative data, including: the mean integrated fluorescent intensities of the Hoechst-stained MCTSs in Ch1; the number of MCTSs in Ch1 (usually 1); and the mean integrated fluorescent intensities of the Ch2 and Ch3 signals within the MCTS masks. The mean integrated intensity is the total pixel fluorescent intensity in Ch1, Ch2, or Ch3 within the total MCTS masks of positively stained MCTSs above the pre-set thresholds, divided by the number of positively stained MCTSs.
Imaging Data Visualizations
Pseudo-color fluorescence intensity data visualizations were used to illustrate fluorescent drug uptake and accumulation in 2D-monolayer and MCTS cultures. The relative fluorescent intensities of the pixels in the image were represented as distinct colors, with the “hotter” and “brighter” colors (low to high, yellow, red, white) representing higher intensity signals and cooler colors (low to high, purple, cyan, green) representing lower intensity signals.
Line scan fluorescence intensity plots were created by using the line scanning tool of the ImageXpress image analysis software to draw a line across the image and plot the fluorescent intensity values versus distance in μm across the image to provide an intensity profile graph.
Data Processing, Analysis, and Curve Fitting
For the monolayer and MCTS HNC growth inhibition (50% growth inhibitory concentration [GI50]) assays, the DMSO control wells (Max. n = 32) and 200 μM doxorubicin control wells (Min., n = 32) were used to represent uninhibited growth and 100% cytotoxicity, respectively. The mean maximum and minimum plate control RLUs were used to normalize the RLU data from the compound-treated wells as percent of controls. Also, the DMSO and 200 μM doxorubicin control wells were used to calculate signal-to-background (S:B) ratios and Z′-factor coefficient assay performance statistics for the growth inhibition assay signal windows. The GI50 data were fit to a non-linear sigmoidal log (inhibitor) versus normalized response variable slope model by using the equation: Y = 100/(1 + 10^[(LogIC50 − X) × Hillslope]), where Y was the percent inhibition and X was the corresponding log10 of the compound concentration. The GI50 is the concentration of compound that gives a 50% response, half way between 0% and 100%. The Hillslope describes the steepness of the curve. The mean integrated fluorescent intensity data from monolayer and MCTS HNC drug accumulation concentration response assays were fit to a linear regression model. The mean integrated fluorescent intensity data from monolayer and MCTS HNC drug accumulation time-course assays were fit to a non-linear one-site binding model. The mean integrated fluorescent intensity data from monolayer HNC drug accumulation cell seeding density assays were fit to a linear regression model, and the data from the MCTS HNC drug accumulation cell seeding density assays were fit to a non-linear one-site binding model. All curve fitting, linear regression analysis, and graphs were created by using the GraphPad Prism 6 software.
Results
Fluorescent Anti-Cancer Drug Growth Inhibition in 2D Monolayer and MCTS HNC Cultures
We selected three broadly used anthracycline cancer chemotherapeutics, idarubicin, daunorubicin, and doxorubicin, and the dimethyl-pyrido-carbazole alkaloid ellipticine for our HCS drug accumulation and distribution experiments because of their fluorescent properties (Supplementary Fig. S1; Supplementary Data are available online at www.liebertpub.com/adt). Supplementary Figure 1A shows the chemical structures of the four anti-cancer drugs and the DNA stain Hoechst 33342. To determine appropriate excitation (Ex) and emission (Em) wavelengths for the four drugs, we dissolved them in PBS at 25 μM and performed 10 nm wavelength scans to measure their absorption and emission spectra (Supplementary Fig. S1B, C). The three structurally related anthracyclines had very similar absorption spectra with a broad absorption peak between 400 and 550 nm (Supplementary Fig. S1B). At a 480 nm Ex wavelength, the anthracyclines produced very similar emission spectra with a broad emission peak between 520 and 650 nm (Supplementary Fig. S1C). The relative fluorescent intensity units (RFUs) of idarubicin (Ex 480, Em 570 nm), daunorubicin (Ex 480, Em 590 nm), and doxorubicin (Ex 480, Em 590 nm) in solution increased linearly with respect to compound concentration (Supplementary Fig. S1D), providing a basis to quantify the relative concentrations of the anthracyclines within cells. Although ellipticine did not exhibit an obvious absorption peak or a fluorescent emission spectrum when excited at 480 nm, we have previously observed strong fluorescence signals in images acquired in the FITC channel of U-2 OS osteosarcoma cells exposed to ellipticine in the 1.56–25 μM range.42,43 Idarubicin also exhibited strong fluorescent signals in both the FITC and Texas Red channels.42,43 We included ellipticine because it is a fluorescent anti-cancer drug that is structurally distinct from the anthracyclines (Supplementary Fig. S1A).
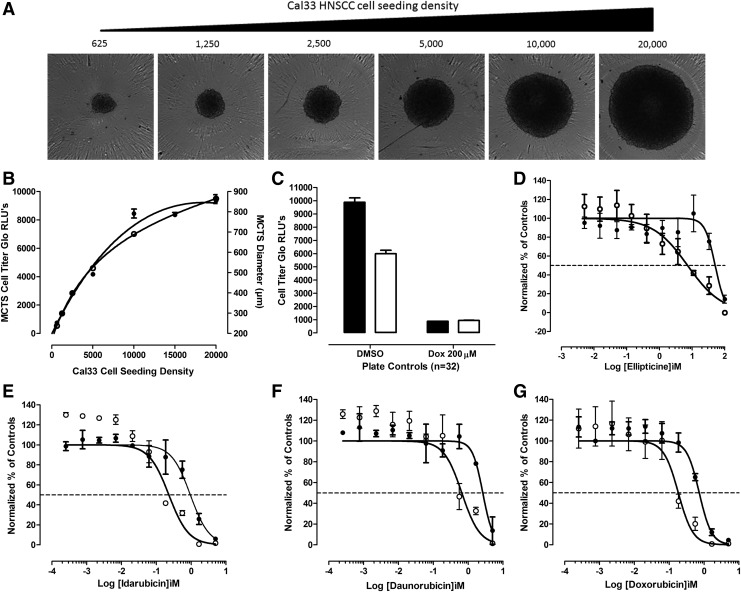
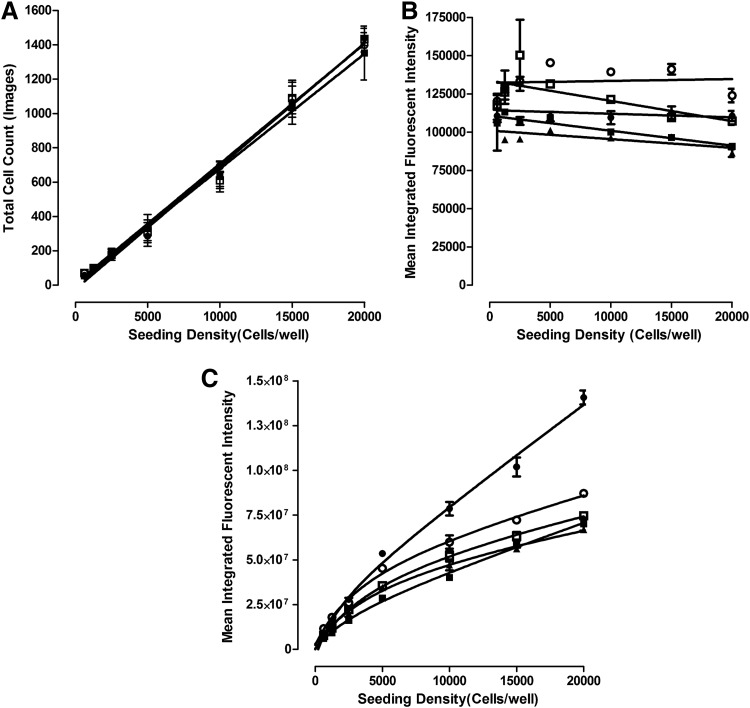
The CTG (Promega Corporation) reagent measures the luciferase luminescent signal (RLUs) produced by cellular adenosine triphosphate (ATP) levels and is frequently used as an indicator of cell number, viability and for proliferation/growth inhibition assays. We have previously developed 384-well CTG growth inhibition assays in monolayer HNC cultures to determine compound cytotoxicity,39,40 and we and others have used it to measure cell viability, cell number, and cancer drug effects in MCTSs generated in 96- or 384-well ULA plates.11,34,36–38 To demonstrate the correlation between the RLU signal and the number of viable HNC cells in MCTSs, we seeded the indicated densities of Cal33 cells into 384-well ULA plates and cultured them for 24 h to allow spheroids to form. After 24 h, we acquired 4 × transmitted light images of the MCTSs on the IXM (Fig. 1A), measured the diameters of the MCTSs by using the line scan tool of the MetaXpress image analysis software (Fig. 1B), and captured the RLUs produced after CTG was added to the wells (Fig. 1B). In the 625–5,000 cells per well range, the MCTS diameters and CTG RLU signals increased linearly with respect to the number of Cal33 cells seeded per well, and although the MCTS diameters and RLUs continued to increase thereafter as more cells were seeded into ULA-plate wells, the increase was no longer linear with respect to cell number (Fig. 1B). The Cal33 cell seeding density MCTS diameter and CTG RLU curves were almost superimposable, indicating that they are closely correlated (Fig. 1B). To define the dynamic range of 72 h compound exposure in MCTS and monolayer HNC growth inhibition assays, we used 0.5% DMSO control wells to represent uninhibited growth (Max. n = 32), and 200 μM doxorubicin +0.5% DMSO control wells to represent 100% of tumor cell cytotoxicity (Min., n = 32), respectively (Fig. 1C). In Cal33 MCTS and monolayer growth inhibitions assays, the Max.:Min. S:B ratios were 22- and 224-fold and the corresponding Z′-factor coefficients were 0.45 and 0.51, respectively (Fig. 1C), indicating that the assays performed well and provided robust and reproducible assay signal windows in which to measure the cytotoxic effects of compounds (Fig. 1D–G and Table 2). Cal33 MCTSs were substantially more resistant than the corresponding 2D monolayers to growth inhibition by all four drugs (Fig. 1D–G and Table 2): Cal33 HNC MCTS:2D monolayer GI50 ratios for ellipticine, idarubicin, daunorubicin, and doxorubicin were 6-, 20-, 10-, and 16-fold, respectively.
Fig. 1.
HNC MCTSs and comparison of cancer drug growth inhibition in 2D monolayer and 3D MCTS HNC culture models. (A) Transmitted light images of Cal33 MCTSs formed in 384-well ultra-low attachment plates seeded with different cell densities after 24 h in culture. Cal33 HNC cells were seeded into 384-well ULA plates at densities ranging from 625 to 20,000 cells per well and after 24 h in culture transmitted light images of the MCTSs were acquired on the IXM automated imaging platform by using a 4 × objective. Representative images from one of several independent experiments are shown. (B) Impact of Cal33 cell seeding density on MCTS CTG signals and diameters. The mean ± SD (n = 12) CTG RLUs (●) produced by MCTS formed in 384-well ULA plates at the indicated Cal33 cell seeding densities are plotted on the left Y-axis, and the mean ± SD (n = 2) diameters (μm) (○) of the MCTSs extracted from the transmitted light images by the single line tool of the MetaXpress image analysis software are plotted on the right Y-axis. Representative experimental data from one of several independent experiments are shown. (C) Cal33 2D monolayer and MCTS growth inhibition assay plate controls. To define the dynamic ranges of 72 h MCTS (□) and 2D monolayer (■) HNC growth inhibition assays, we used 0.5% DMSO control wells to represent uninhibited growth (Max., n = 32), and 200 μM doxorubicin +0.5% DMSO control wells to represent 100% of tumor cell cytotoxicity (Min., n = 32), respectively. The mean ± SD (n = 32) CTG RLUs from one of three to four independent experiments are shown. GI50 curves for Cal33 2D monolayer and MCTS cultures exposed to ellipticine (D), idarubicin (E), daunorubicin (F), or doxorubicin (G) for 72 h. Cal33 cells were seeded into 384-well monolayer assay plates at 1,000 cells per well (○), and into 384-well ULA plates at 5,000 cells per well (●). After 24 h in culture, the indicated concentrations of compounds were transferred into the test wells of assay plates that were then cultured for an additional 72 h before CTG detection reagent was added to the wells and the RLUs were captured on the SpectraMax M5e microtiter plate reader. The normalized mean ± SD (n = 3) growth inhibition data from triplicate wells for each compound concentration are presented. Representative experimental data from one of three to four independent experiments are shown. 3D, three-dimensional; CTG, CellTiter-Glo®; DMSO, dimethyl sulfoxide; GI50, 50% growth inhibitory concentration; HNC, head and neck cancer; IXM, ImageXpress® Micro; Max., maximum plate controls; Min., minimum plate controls; MCTS, multicellular tumor spheroid; RLUs, relative light units; SD, standard deviation; ULA plates, ultra-low attachment microtiter plates.
Table 2.
Comparison of Fluorescent Drug Potencies in Two-Dimensional Monolayer and Multicellular Tumor Spheroid Cultures of the Cal33 Head and Neck Cancer Cell Line
| 2D monolayer GI50s (μM) | MCTS GI50s (μM) | GI50 ratioa | |||||
|---|---|---|---|---|---|---|---|
| Drug | Meanb | SD | nc | Mean | SD | n | MCTS:2D |
| Ellipticine | 4.80 | 2.56 | 4 | 28.1 | 21.8 | 3 | 5.9-Fold |
| Idarubicin | 0.11 | 0.12 | 4 | 2.08 | 1.55 | 4 | 19.7-Fold |
| Daunorubicin | 0.23 | 0.29 | 4 | 2.24 | 1.17 | 3 | 9.9-Fold |
| Doxorubicin | 0.07 | 0.08 | 4 | 1.08 | 0.82 | 4 | 16.3-Fold |
GI50 ratio = the mean MCTS GI50 (μM) divided by the mean 2D monolayer GI50 (μM).
Mean = average of replicate independent experiments.
n = Number of independent experiments.
2D, two-dimensional; GI50, 50% growth inhibitory concentration; SD, standard deviation.
Fluorescent Anti-Cancer Drug Uptake and Accumulation in 2D Monolayer HNC Cultures
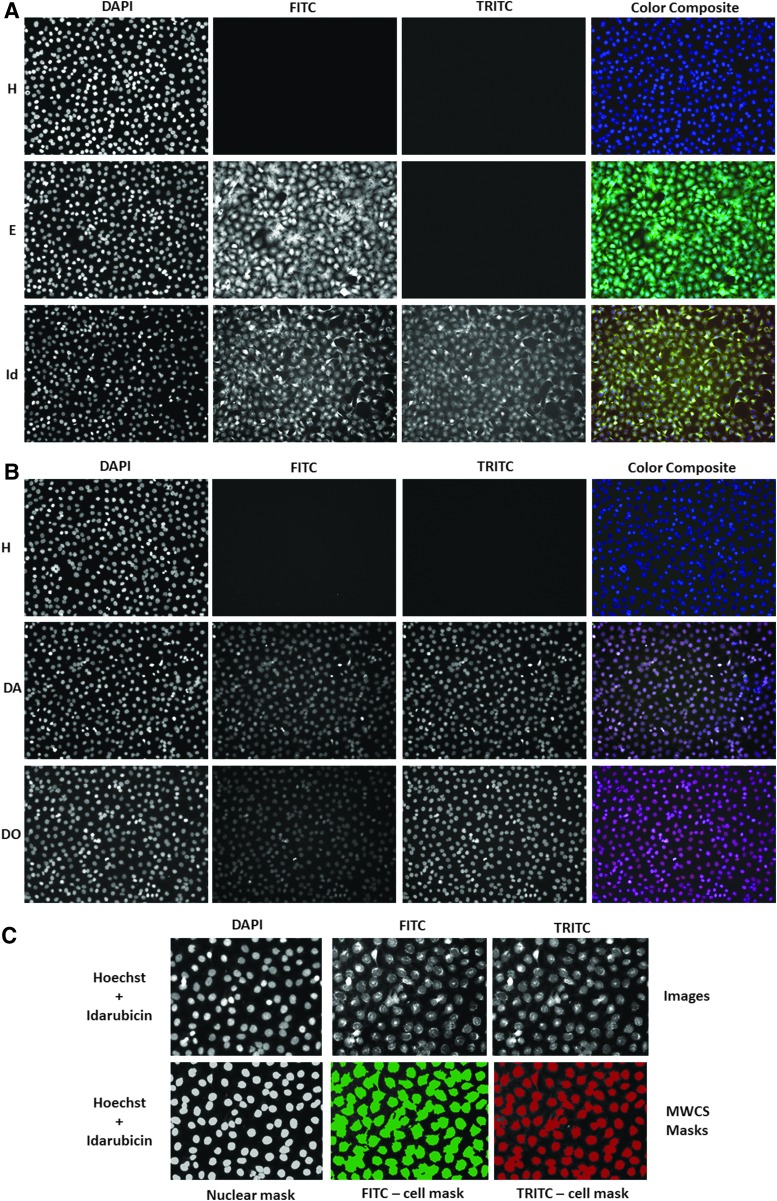
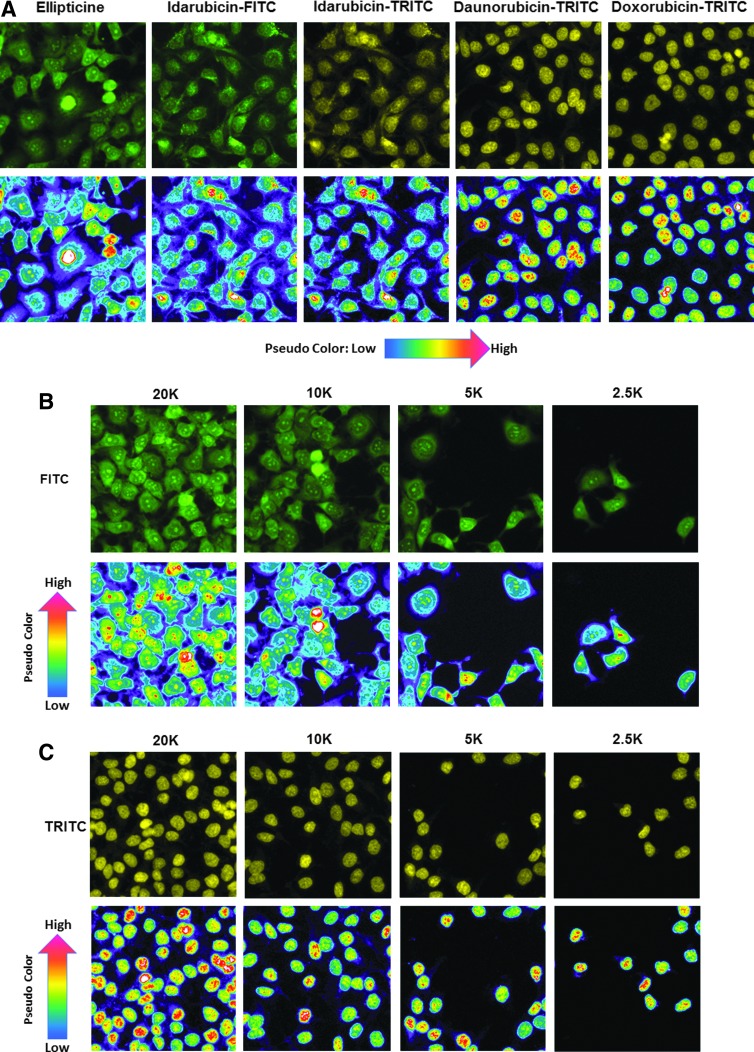
Figure 2 shows representative grayscale 10 × images of the DAPI, FITC, and TRITC channels and corresponding fluorescent color composite images of Cal33 2D monolayers exposed to 25 μM ellipticine, idarubicin, daunorubicin, or doxorubicin for 15 min before fixation in paraformaldehyde containing Hoechst for 45 min. In Cal33 monolayers not exposed to any fluorescent drugs, only Hoechst-stained nuclei were apparent in DAPI channel images, and the corresponding color composite images of the three channels were blue (Fig. 2A, B). In Cal33 monolayers exposed to ellipticine, Hoechst-stained nuclei were observed in the DAPI channel, the cytoplasmic and nuclear regions of cells were fluorescent in the FITC channel, and the resulting color composite images were green-blue (Fig. 2A). In Cal33 monolayers exposed to idarubicin, Hoechst-stained nuclei were detected in the DAPI channel, and both the cytoplasmic and nuclear regions of cells were fluorescent in the FITC and TRITC channels, at roughly equivalent intensities, resulting in yellow-blue composite images (Fig. 2A). In Cal33 monolayers exposed to daunorubicin or doxorubicin, Hoechst-stained nuclei were apparent in the DAPI channel, and cell nuclei were also fluorescent in the FITC and TRITC channels, but because the TRITC intensity was higher, the composite images are red-blue (Fig. 2B). We utilized the MWCS image analysis module of the MetaXpress software on the IXM platform to extract and analyze quantitative data from the images of Cal33 monolayers exposed to Hoechst and the four fluorescent compounds (Fig. 2C). The MWCS image segmentation identified the Hoechst-stained objects as nuclei and used these to create a nuclear mask and count cells (Fig. 2C). Using the nuclear mask and pre-set intensity thresholds in the FITC (Ch2, green) and TRITC (Ch3, red) channels, the algorithm created whole cell masks (cytoplasm and nuclear regions) in both channels (Fig. 2C), and we used the mean integrated fluorescent intensity output parameter to measure drug accumulation (Fig. 3).
Fig. 2.
Images and image analysis of fluorescent drug accumulation in HNC monolayers. (A, B) Images of Cal33 monolayers stained with Hoechst alone, or Hoechst plus ellipticine, idarubicin, daunorubicin, or doxorubicin. Cal33 cells were seeded into 384-well monolayer assay plates at 1,000 cells per well and cultured for 24 h before 10 μM of the indicated compounds was added to wells that were incubated for an additional 15 min before fixation in formaldehyde containing Hoechst for 30 min. After washing with PBS, images of two fields of view of 2D Cal33 monolayers were acquired sequentially on the IXM by using a 10 × objective in each of three fluorescent channels: DAPI, FITC, and TRITC. Representative grayscale images from numerous independent experiments of each of the fluorescent channels are presented along with the corresponding color composite images. (C) MWCS image analysis module nuclear and whole cell masks. The MWCS module image segmentation identified the Hoechst 33342 stained Cal33 nuclei and created nuclear masks for each cell. After applying user-defined background average intensity thresholds, typically 200 in both Ch2 (FITC) and Ch3 (TRITC), the MWCS module image segmentation created total cell masks for each cell. The nuclear mask was used to quantify the mean integrated fluorescence intensity of Hoechst within Cal33 nuclei, and to count the number of cells. The derived Cal33 whole cell masks in Ch2 and Ch3 were then used to quantify the mean integrated fluorescence intensity of the drugs in the FITC and TRITC channels, respectively. Ch2, fluorescent channel 2; Ch3, fluorescent channel 3; DAPI, 4,6-diamidino-2-phenylindole; FITC, fluorescein isothiocyanate; MWCS, multi-wavelength cell scoring; PBS, phosphate-buffered saline; TRITC, tetramethylrhodamine.
Fig. 3.
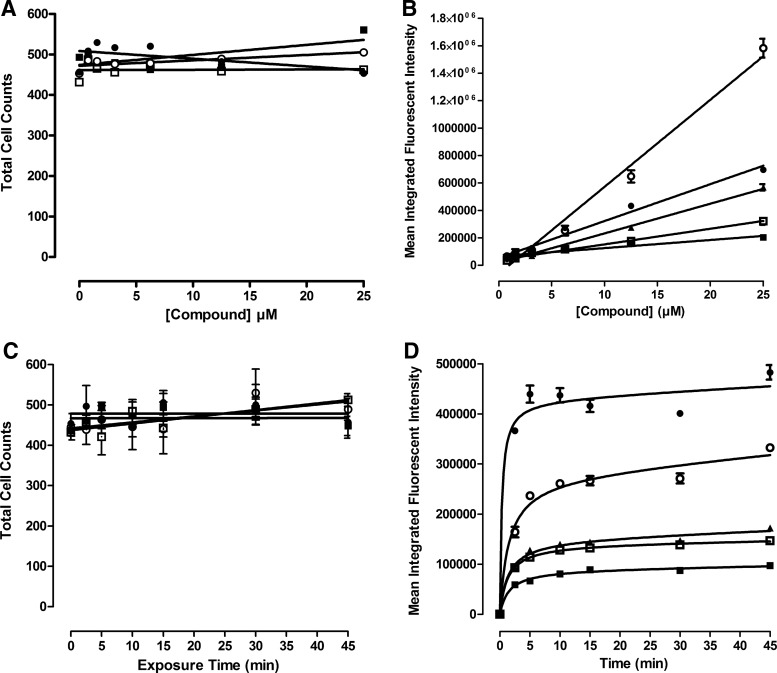
Fluorescent drug accumulation in Cal33 HNC monolayers. (A) Cell counts and (B) mean integrated fluorescent intensity versus drug exposure concentrations. Cal33 cells were seeded into 384-well monolayer assay plates at 1,000 cells per well and were incubated for 24 h before the addition of test drugs at final concentrations of 25, 12.5, 6.25, 3.12, 1.56, or 0.781 μM and plates were incubated for an additional 15 min before being fixed in formaldehyde containing Hoechst for 30 min. After washing with PBS, images of two fields of view of 2D Cal33 monolayers were acquired sequentially on the IXM by using a 10 × objective in the DAPI, FITC, and TRITC channels, and they were then analyzed by using the MMCS image analysis module. (C) Cell counts and (D) mean integrated fluorescent intensity versus drug exposure times. In time-course assays, test drugs were added to wells at 10 μM final concentrations and the assay plates were incubated at 37°C, 5% CO2, and 95% humidity for 2.5, 5, 10, 15, 30, and 45 min. Assay plates that were fixed and stained with Hoechst were washed with PBS and then imaged and analyzed on the IXM HCS platform by using the MWCS image analysis module. The mean ± SD (n = 3) total cell counts from triplicate wells for each compound concentration and timepoint are presented in (A, C), ellipticine (●), idarubicin (○), doxorubicin (■), and daunorubicin (□). The mean ± SD (n = 3) mean integrated fluorescent intensities from triplicate wells for each compound concentration and timepoint are presented in (B, D), ellipticine-FITC (●), idarubicin-FITC (○), idarubicin-TRITC (▲), doxorubicin-TRITC (■), and daunorubicin-TRITC (□). Representative data from one of three independent experiments are shown. HCS, high-content screening.
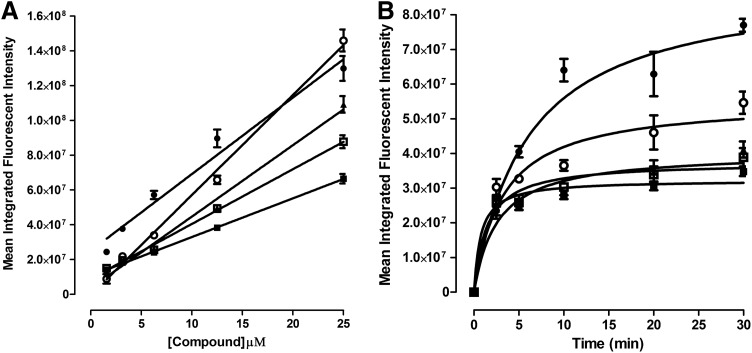
Figure 3 shows the concentration- and time-dependent uptake and accumulation of fluorescent compounds in Cal33 monolayer cultures. The number of Hoechst-stained objects detected in Ch1 demonstrated that the number of cells analyzed in the acquired images of Cal33 monolayer cultures exposed to fluorescent compounds at the indicated concentrations (up to 25 μM), and for the periods designated (up to 45 min), were consistent for all conditions (Fig. 3A, C). The mean integrated fluorescent intensity values observed in Cal33 monolayers exposed to different concentrations of ellipticine, idarubicin, daunorubicin, and doxorubicin for 15 min revealed that drug uptake and accumulation increased linearly with respect to compound concentration (Fig. 3B), consistent with the linear relationship between RFUs and compound concentrations observed in solution (Supplementary Fig. S1D). To evaluate the time-dependent uptake and accumulation of the drugs into Cal33 monolayers, we exposed cells to 10 μM of the compounds for the indicated periods (Fig. 3D). The time-course of drug uptake and accumulation in Cal33 monolayers followed a very similar profile for all four compounds. There was a rapid initial drug uptake phase from 0 to 5 min that appeared linear, between 5 and 10 min drug uptake slowed and accumulation approached a maximum plateau, and thereafter accumulation reached steady-state equilibrium levels that were maintained through 45 min (Fig. 3D).
Fluorescent Anti-Cancer Drug Uptake and Accumulation in MCTS HNC Cultures
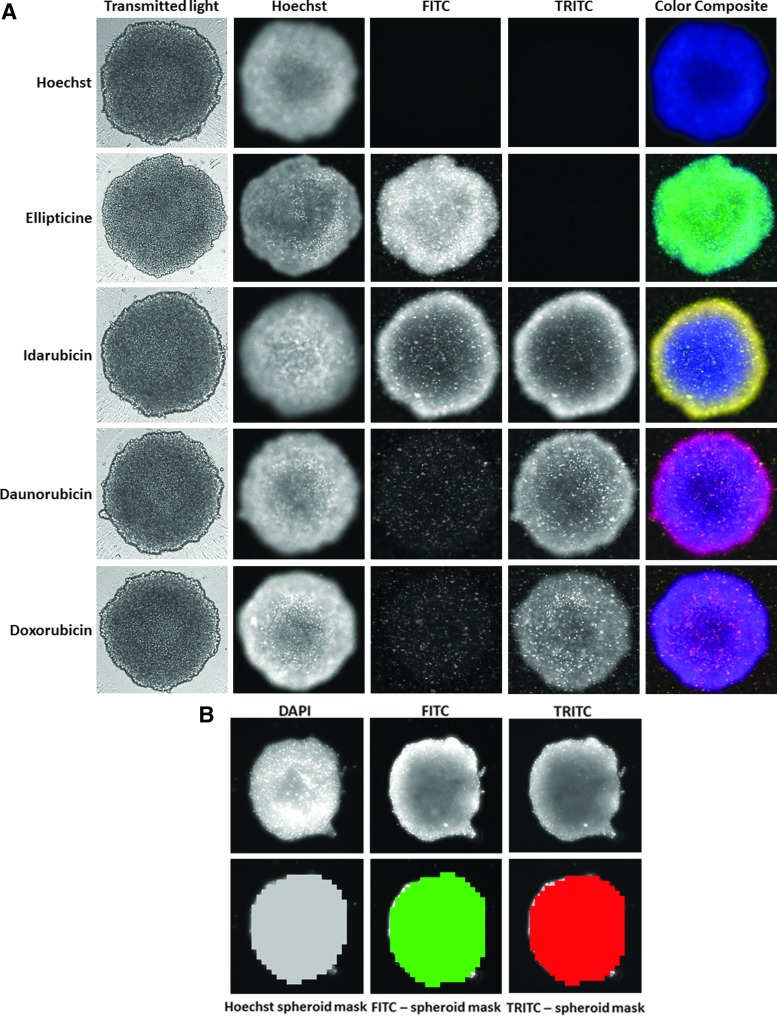
Figure 4A shows representative grayscale 4 × images of the transmitted light, DAPI, FITC, and TRITC channels, and corresponding florescent color composite images of 5K Cal33 MCTSs exposed to 10 μM ellipticine, idarubicin, daunorubicin, or doxorubicin for 15 min before fixation in paraformaldehyde containing Hoechst for 45 min. The transmitted light images of MCTS exposed to drugs under these conditions revealed no apparent changes in size or morphology. Cal33 MCTSs exposed to ellipticine were fluorescent in the DAPI (Hoechst-stained nuclei) and FITC channels, and the resulting color composite images were green-blue (Fig. 4A). Cal33 MCTSs exposed to idarubicin were fluorescent in the DAPI, FITC, and TRITC channels, and the color composite images were yellow-blue because the FITC and TRITC intensities were roughly equivalent (Fig. 4A). Cal33 MCTSs exposed to daunorubicin or doxorubicin were also fluorescent in the DAPI, FITC, and TRITC channels, but the color composite images were red-blue because the TRITC intensity was higher (Fig. 4A). The fluorescent images of Cal33 MCTSs exposed to Hoechst and/or the four fluorescent compounds all exhibited an apparent donut-like staining pattern, where cells in the outer edges of the MCTSs had higher intensities relative to cells in the inner cores (Fig. 4A). The MWCS image segmentation created a total MCTS mask in Ch1 for Hoechst-stained objects that were >150 μm but <550 μm and a threshold intensity ≥70 gray levels over local background (Fig. 4B). The algorithm then used the total MCTS mask in Ch1 and user preset intensity thresholds, typically 50–70 gray levels over local background, in the FITC (Ch2, green) and TRITC (Ch3, red) channels to create MCTS masks in both channels (Fig. 4B), and we used the mean integrated fluorescent intensity output to measure drug accumulation (Fig. 5).
Fig. 4.
Images and image analysis of fluorescent drug accumulation in HNC multicellular tumor spheroids. (A) Images of Cal33 MCTSs stained with Hoechst alone, or Hoechst plus ellipticine, idarubicin, daunorubicin, or doxorubicin. Cal33 cells were seeded into 384-well ULA plates at 5,000 cells per well and were incubated for 24 h before 10 μM of the indicated drugs was added to wells and incubated for an additional 15 min before fixation in formaldehyde containing Hoechst and washing with PBS. Images of Cal33 MCTSs were acquired sequentially on the IXM by using a 4 × objective in the transmitted light, DAPI, FITC, and TRITC channels. Representative grayscale images from numerous independent experiments of the transmitted light and fluorescent channels are presented along with a corresponding color composite image of the fluorescent channels. (B) MWCS image analysis module whole spheroid masks. Hoechst-stained objects in Ch1 (DAPI) that were >150 μm but <550 μm wide with threshold intensities ≥70 above local background were used to create whole MCTS masks. After applying user-defined background average intensity thresholds, typically 50–70 in both Ch2 (FITC) and Ch3 (TRITC), the MWCS module image segmentation created total MCTS masks in all three fluorescent channels. The derived total MCTS masks from Ch2 and Ch3 were then used to quantify the mean integrated fluorescence intensity of the drugs in the FITC and TRITC channels, respectively. Ch1, fluorescent channel 1.
Fig. 5.
Fluorescent drug accumulation in Cal33 HNC MCTSs. (A) Concentration dependence. Test drugs were added to wells containing Cal33 MCTSs, formed overnight from seeding 5,000 cells per well in ULA plates, at final concentrations of 25, 12.5, 6.25, 3.12, 1.56, or 0.781 μM and plates were incubated for 15 min before fixation in formaldehyde containing Hoechst for 30 min. After washing with PBS, DAPI, FITC, and TRITC images of Cal33 MCTSs were acquired on the IXM by using a 4 × objective and analyzed by using the MMCS image analysis module. (B) Time-dependence test compounds were added to wells containing MCTSs at a final concentration of 10 μM and incubated for 2.5, 5, 10, 20, and 30 min. Assay plates were then fixed and stained with Hoechst for 30 min, and after washing with PBS were imaged and analyzed on the IXM HCS platform by using the MWCS image analysis module. The mean ± SD (n = 3) of the mean integrated fluorescent intensity values from triplicate wells for each compound concentration (A) and timepoint (B) are presented: ellipticine-FITC (●), idarubicin-FITC (○), idarubicin-TRITC (▲), doxorubicin-TRITC (■), and daunorubicin-TRITC (□). Representative data from one of three independent experiments are shown.
Figure 5 shows the concentration- and time-dependent uptake and accumulation of fluorescent compounds in 5K Cal33 MCTSs. A single Hoechst-stained object was detected in Ch1 images and was classified by the MWCS segmentation as an MCTS. The mean integrated fluorescent intensity values observed in Cal33 MCTSs exposed to different concentrations of ellipticine, idarubicin, daunorubicin, and doxorubicin for 15 min demonstrated that drug accumulation increased linearly with respect to compound concentration (Fig. 5A). The time course of drug uptake and accumulation of all four compounds followed a very similar profile in Cal33 MCTSs. The initial uptake phase appeared linear through 5 min, began to slow and drug accumulation approached a maximum plateau between 5 and 10 min, and thereafter steady-state equilibrium levels were maintained through 30 min (Fig. 5B). Although fluorescent drug uptake and accumulation behaved similarly with respect to compound concentration and time dependence in HNC monolayers seeded at 1,000 cells/well, and MCTS cultures seeded at 5,000 cells/well (Figs. 3 and 5), the mean integrated intensity values were ∼100-fold higher in MCTSs. A direct comparison of the total integrated intensity values between monolayers and MCTS cultures would perhaps be a more appropriate parameter to evaluate; however, the MWCS image analysis module does not provide this output.
The MCTS drug accumulation studies described earlier used an imaging method that involved acquisition of 10–20 Z-stack images each separated by 20 μm that were collapsed into a single 2D maximum projection image to increase the number of cells and focal planes contributing to the data. We were concerned that the maximum projection image might be significantly elevating and/or exaggerating the mean integrated intensity values of MCTSs relative to 2D monolayers where only a single focal plane image was acquired. We, therefore, compared the MCTS maximum projection image acquisition and analysis method with an image-based focus method where only a single best focus image of each MCTS was acquired per channel (Supplementary Fig. S2). Supplementary Figure S2A shows a comparison of the transmitted light and TRITC maximum projection and best optical focus images for 5K Cal33 MCTSs that had been exposed to either 10 μM of idarubicin or doxorubicin for 15 min before fixation in paraformaldehyde containing Hoechst for 30 min. Supplementary Figure S2B shows the mean integrated fluorescent intensity values output by the MWCS module from maximum projection and best optical focus images of 5K Cal33 MCTSs exposed to Hoechst (2 μg/mL) and/or 10 μM of ellipticine, idarubicin, daunorubicin, or doxorubicin. Although the MCTS mean integrated fluorescent intensity values for the compounds were slightly higher for the maximum projection image acquisition and analysis method, the values were not significantly elevated relative to the data obtained with the single image best focus method. Based on the images and quantitative data (Supplementary Fig. S2), we concluded that the single best focus image acquisition method produces comparable data with minimal loss of information, and requires much less time to acquire a 384-well plate.
Cell Seeding Density Effects on Fluorescent Anti-Cancer Drug Uptake and Accumulation in 2D Monolayer and MCTS HNC Cultures
We next evaluated the effects of cell seeding density on drug accumulation in monolayer and MCTS HNC cultures (Figs. 6 and 7). As anticipated, increasing the cell seeding density of the Cal33 monolayers produced a linear increase in the number of cells acquired and analyzed in the two 10 × images captured in each well (Fig. 6A). The average total Cal33 cell counts in the two images ranged between 50 and 1,400 cells in monolayer wells seeded at 625 and 20,000 cells per well, respectively. In monolayers, the mean integrated fluorescent intensity values for all four fluorescent drugs were largely equivalent at all cell seeding densities (Fig. 6B), indicating that drug accumulation was independent of the number of cells in the cultures. Figure 7 shows representative FITC and TRITC fluorescent images with their corresponding pseudo-color fluorescence pixel intensity visualizations for Cal33 monolayers exposed to 10 μM of each of the four compounds for 15 min. Figure 7A shows representative FITC and TRITC fluorescent images and pseudo-color visualizations for all four compounds from Cal33 cells seeded at 10,000 cells per well, and Figure 7B and C show the corresponding images and data visualizations for Cal33 cells seeded at different densities and exposed to either 10 μM ellipticine or doxorubicin for 15 min. Consistent with the quantitative data in Figure 6, the FITC and TRITC images and pseudo-color data visualizations for each of the four compounds were remarkably consistent at all cell seeding densities, and they confirmed that drug accumulation was independent of the number of Cal33 cells in the monolayers (Fig. 7).
Fig. 6.
Effects of Cal33 HNC cell seeding density on fluorescent drug accumulation in 2D monolayers (A, B) and MCTSs (C). Cal33 cells were seeded into 384-well monolayer and MCTS assay plates at seeding densities of 625, 1,250, 2,500, 5,000, 10,000, and 20,000 cells per well. After 24 h in culture, test drugs were added at final concentrations of 10 μM and plates were then incubated for 15 min before fixation in formaldehyde containing Hoechst and washing with PBS. DAPI, FITC, and TRITC images of Cal33 monolayers and MCTSs were acquired on the IXM and analyzed by using the MWCS image analysis module as described earlier. (A) Cal33 monolayer cell counts versus seeding density. The mean ± SD (n = 3) total cell counts from triplicate wells for each monolayer cell seeding density are presented: ellipticine (●), idarubicin (○), doxorubicin (■), and daunorubicin (□). (B) Cal33 monolayer fluorescent drug accumulation versus cell seeding density. The mean ± SD (n = 3) of the mean integrated fluorescent intensities from triplicate wells for each monolayer cell seeding density are presented: ellipticine-FITC (●), idarubicin-FITC (○), idarubicin-TRITC (▲), doxorubicin-TRITC (■), and daunorubicin-TRITC (□). (C) Cal33 MCTS fluorescent drug accumulation versus cell seeding density. The mean ± SD (n = 3) of the mean integrated fluorescent intensities from triplicate wells for each MCTS cell seeding density are presented: ellipticine-FITC (●), idarubicin-FITC (○), idarubicin-TRITC (▲), doxorubicin-TRITC (■), and daunorubicin-TRITC (□). Representative data from one of three independent experiments are shown.
Fig. 7.
Effects of Cal33 cell number on fluorescent drug distribution in 2D monolayers: images and pseudo-color pixel intensity visualizations for (A) test compounds at a seeding density of 10,000 cells/well, (B) ellipticine, and (C) doxorubicin at different seeding densities. Cal33 cells were seeded into 384-well monolayer assay plates at seeding densities of 625, 1,250, 2,500, 5,000, 10,000, and 20,000 cells per well, and after 24 h in culture, 10 μM of test compounds was added to wells and incubated for 15 min before being fixed in formaldehyde and washed with PBS. FITC and TRITC images of Cal33 monolayers were acquired on the IXM. Representative images colored by the image acquisition channel, green for FITC and yellow for TRITC, are presented along with the corresponding pseudo-color pixel intensity visualizations, where the relative fluorescent intensities of the pixels in the images are indicated as distinct colors. The “hotter” and “brighter” colors (low to high, yellow, red, white) represent higher intensity signals, and cooler colors (low to high, purple, cyan, green) represent lower intensity signals. Representative data from one of three independent experiments are shown.
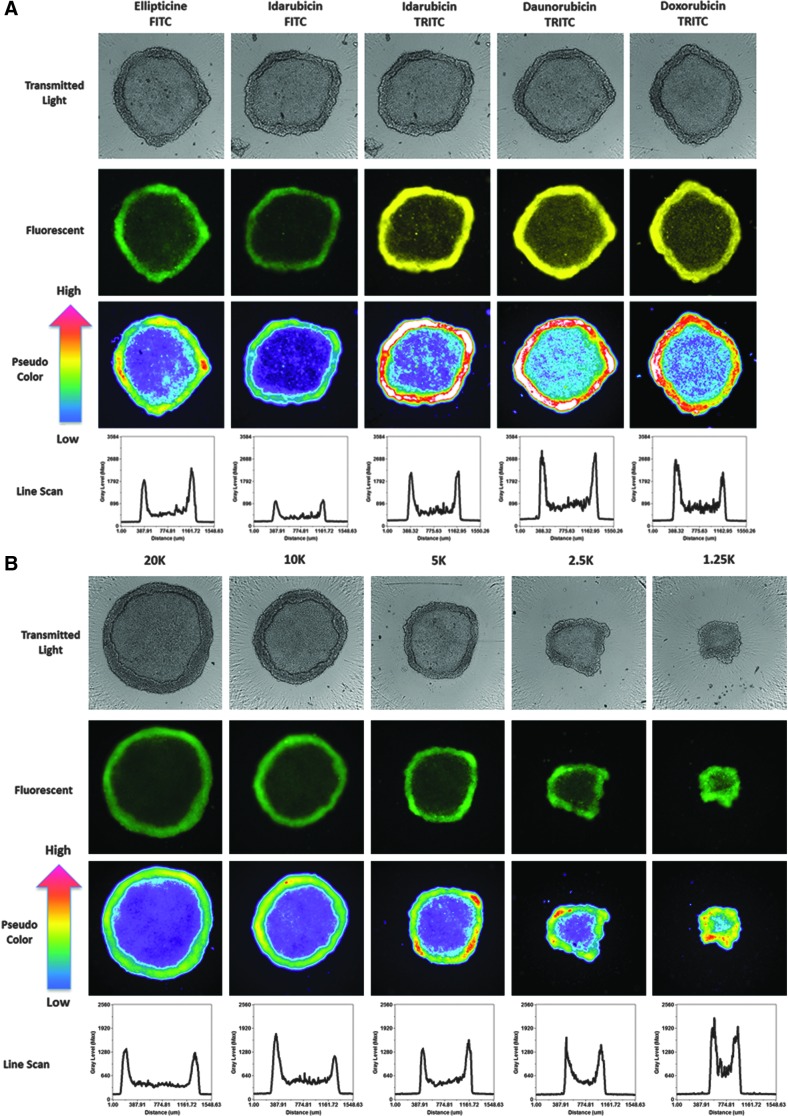
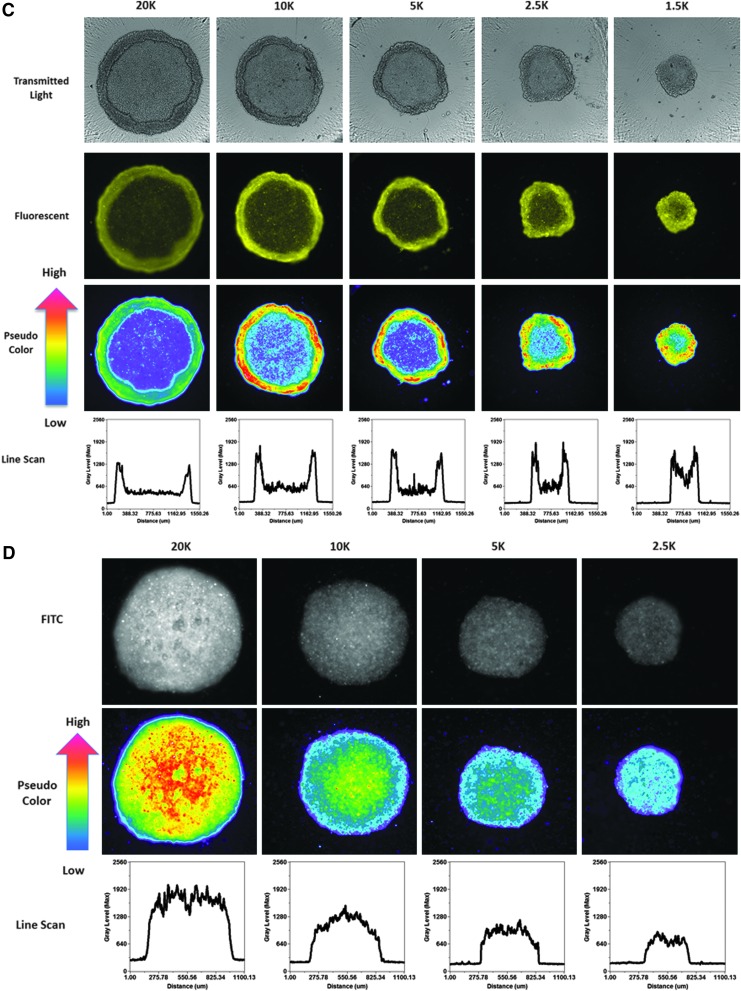
In MCTSs, however, the mean integrated fluorescent intensity values for the four fluorescent drugs increased as the cell seeding density increased (Fig. 6C), indicating that drug accumulation was strongly influenced by the number of cells and sizes of the MCTSs formed. Figure 8A–C shows representative transmitted light, FITC and TRITC images together with the corresponding pseudo-color visualizations and line scan fluorescence intensity plots for Cal33 MCTSs exposed to 10 μM of each of the four compounds for 15 min. Transmitted light images of Cal33 MCTSs show two distinct spheroid regions or phenotypes, an inner core surrounded by an outer layer that is several layers of cells thick (Fig. 8A–C). Although the outer layers of cells were more apparent in the transmitted light images of Cal33 MCTSs presented in Figure 8, scrutiny of the images in Figures 1A and 4A also supports two distinct spheroid regions or cellular phenotypes. Transmitted light images of Cal33 MCTSs exposed to drugs at 10 μM for 15 min revealed no apparent changes in size or morphology. Figure 8A shows representative transmitted light, FITC and TRITC images with pseudo-color visualizations, and line scan plots for the 4 compounds in Cal33 MCTSs formed by cells seeded at 5,000 cells per well, and Figure 8B and C shows the corresponding images and data visualizations/plots for Cal33 MCTSs formed from cells seeded at different densities and exposed to either 10 μM ellipticine or doxorubicin for 15 min. The fluorescent images, pseudo-color visualizations, and line scan plots indicated that there was a significant drug penetration gradient in Cal33 HNC MCTS cultures (Fig. 8A–C). Although the mean drug integrated fluorescent intensities increased as the numbers of cells and spheroid sizes increased (Fig. 6C), substantially lower fluorescent drug intensities were detected in cells located in the MCTS inner cores relative to cells present in the outer layers, indicating that drugs were preferentially taken up by and accumulated in cells in the outer layers of the MCTSs (Fig. 8A–C). Although the fluorescent drug intensity levels in cells in the MCTS inner cores were higher than background levels outside the MCTS, inner core intensity values decreased as the number of cells and sizes of the MCTSs increased (Fig. 8B, C). When we prelabeled Cal33 cells with CM-FDA cell tracker green before seeding them into 384-well ULA plates and culturing them for 24 h to form MCTSs (Fig. 8D), the MCTS FITC images, pseudo-color visualizations, and line scan plots indicated that the CM-FDA fluorescence was distributed uniformly throughout the MCTSs, and if anything, the fluorescent intensities were slightly higher in the inner cores of MCTSs relative to the outer layers (Fig. 8D). Similar fluorescence distribution patterns were observed in MCTSs formed by Cal33 cells prelabeled with the cell tracker orange dye, and in HNC MCTSs formed by FaDu cells prelabeled with cell tracker dyes (data not shown). The uniform distribution of fluorescence observed in MCTSs formed from HNC cell lines prelabeled with fluorescent dyes before spheroid formation indicates that the differential distribution of fluorescent drugs in MCTSs is the result of preferential uptake and accumulation in cells in the outer layers of the MCTSs, and not due to an imaging artifact caused by inefficient excitation light penetration and/or fluorescent emission light detection in MCTSs.
Fig. 8.
Effects of Cal33 cell number on fluorescent drug distribution in MCTSs: images, pseudo-color pixel intensity visualizations, and line scan fluorescent intensity profiles for (A) test drugs in MCTSs formed in ULA plates at a seeding density of 5,000 cells/well, (B) ellipticine, (C) doxorubicin, and (D) Cell Tracker Green in MCTSs formed at different seeding densities. Cal33 cells were seeded into 384-well ULA plates at seeding densities of 625, 1,250, 2,500, 5,000, 10,000, and 20,000 cells per well, and after 24 h in culture, 10 μM of test compounds was added to the MCTSs and incubated for 15 min before fixation in formaldehyde and washing with PBS. Transmitted light, FITC and TRITC images of Cal33 MCTSs were then acquired on the IXM. Transmitted light images and fluorescent images colored by the image acquisition channel, green for FITC and yellow for TRITC, are presented along with the corresponding pseudo-color pixel intensity visualizations, and line scan fluorescent intensity profiles. Representative data from one of three independent experiments are shown.
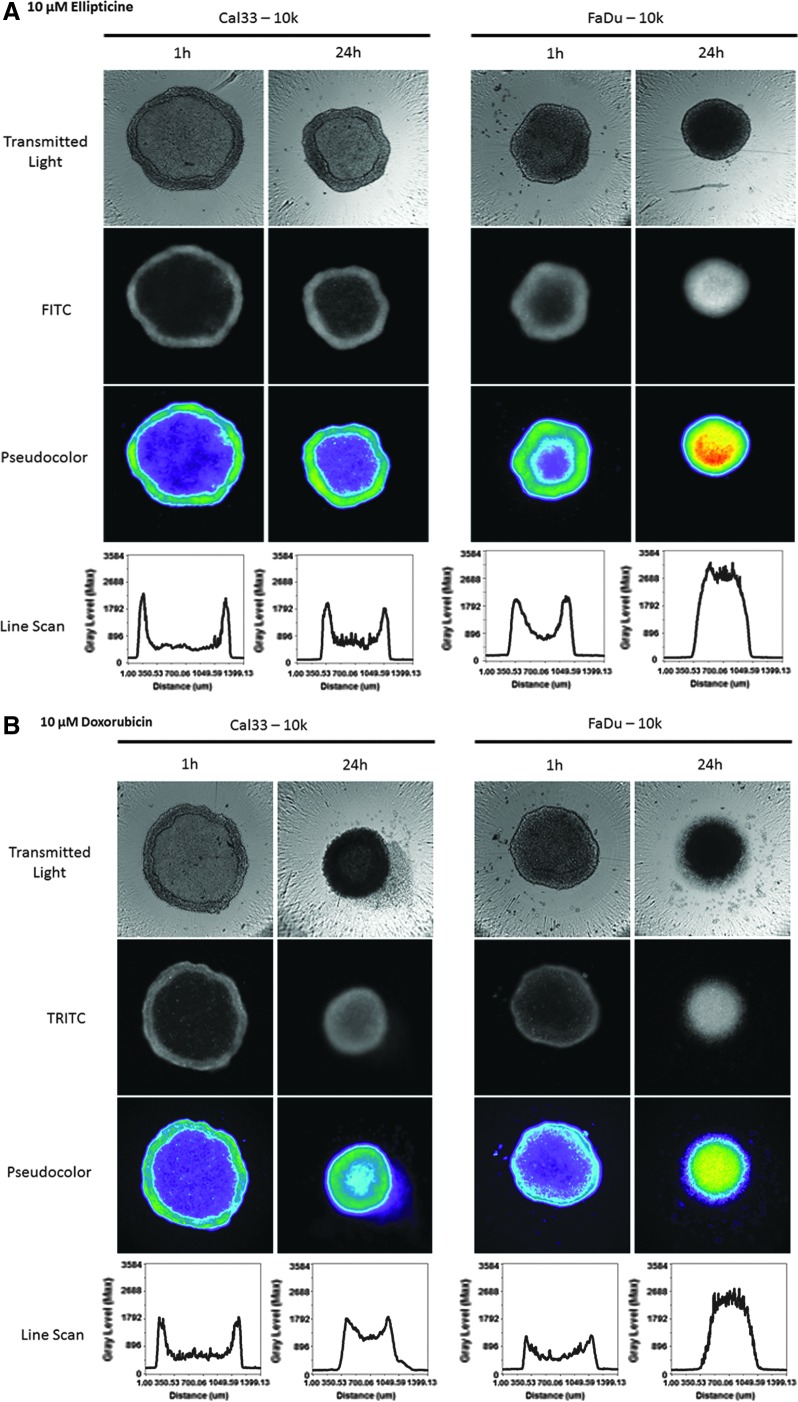
Although Cal33 MCTSs were more resistant than 2D monolayers to growth inhibition by the four drugs, 72 h exposure to higher drug concentrations effectively killed MCTSs (Fig. 1D–G). We, therefore, wanted to examine whether the drug distribution gradient observed after 1 h of drug exposure (Figs. 6C and 8A–C) would be maintained at later time points. Figure 9 shows representative transmitted light, FITC or TRITC fluorescent images with the corresponding pseudo-color visualizations, and line scan plots for 10K Cal33 and FaDu MCTSs exposed to 10 μM ellipticine or doxorubicin for 1 and 24 h. Consistent with the data presented in Figure 8A–C, after 1 h of drug exposure, ellipticine and doxorubicin preferentially accumulated in cells in the outer layers of Cal33 and FaDu MCTSs (Fig. 9A, B). Transmitted light images of the Cal33 and FaDu MCTSs exposed to 10 μM ellipticine or doxorubicin for 1 and 24 h revealed changes in both the sizes and morphologies of the MCTSs at 24 h (Fig. 9A, B). Twenty-four hours of drug exposure substantially decreased the sizes of the MCTSs relative to 1 h exposure, and in three of the four 24 h conditions MCTS outer cell layers were either missing, smaller, or disintegrating, and MCTS inner core regions appeared much darker. Although Cal33 MCTSs exposed to ellipticine for 24 h were smaller than MCTSs that were exposed for 1 h, the MCTSs still exhibited two distinct regions or phenotypes and a drug penetration intensity gradient with preferential accumulation in the cells of the outer layers relative to cells in the inner core (Fig. 9A). The outer layers of Cal33 MCTSs exposed to doxorubicin for 24 h appeared to be disintegrating or sloughing off the inner core, and although the smaller MCTSs exhibited a drug penetration gradient with preferential accumulation in cells of the outer layers, the intensity differential between cells in the outer layers and inner core was substantially reduced (Fig. 9B). FaDu MCTSs exposed to ellipticine or doxorubicin for 24 h were smaller than MCTSs that were exposed for only 1 h, the outer MCTS cell layers were either missing, smaller, or disintegrating, and higher fluorescent intensities were observed in the inner cores of the MCTSs, indicating that the drugs had penetrated and were distributed throughout the MCTSs (Fig. 9A, B).
Fig. 9.
Effects of drug exposure times on fluorescent drug distribution in MCTSs: images, pseudo-color pixel intensity visualizations, and line scan fluorescent intensity profiles for 1 and 24 h exposure of Cal33 and FaDu MCTSs to (A) ellipticine and (B) doxorubicin. Cal33 and FaDu cells were seeded into 384-well ULA plates at 10,000 cells per well, and after 24 h in culture, 10 μM of ellipticine or doxorubicin was added to the MCTSs and incubated for either 1 or 24 h before being fixed in formaldehyde and washed with PBS. Transmitted light, FITC and TRITC images of MCTSs were then acquired on the IXM. Transmitted light images and grayscale fluorescent images are presented along with the corresponding pseudo-color pixel intensity visualizations, and line scan fluorescent intensity profiles. Representative data from one of three independent experiments are shown.
Discussion
MCTSs and multilayer cell cultures (MCCs) are the in vitro 3D tumor culture models most commonly used for the investigation of drug uptake, accumulation, and distribution.27–29,44–54 MCCs are tumor cells cultured on permeable membranes to form 200–250 μm thick disks of tissue that separate donor and acceptor media reservoirs.28,29,48,49,51–53 Drugs are added to one reservoir, and passage through the culture can be measured by sampling both compartments as a function of time. Similar to MCTSs, MCCs have been shown to replicate some of the properties of solid tumors with respect to ECM formation, cell-cell adhesion, nutrient and proliferation gradients, and the development of hypoxic and/or necrotic regions.28,29,48,49,51–53 HPLC and mass spectrometry (MS) analytical methods are used to quantify drug concentrations in MCC donor and acceptor reservoirs.28,29,48,49,51–53 Drug accumulation and distribution in both MCTSs and MCCs has been studied by using radiolabeled drugs that are characterized by scintillation counting and/or autoradiography, or with fluorescent molecules.27–29,44–54 Fluorescent and radiolabeled drug distributions can also be visualized directly in MCTSs or MCCs that have been sectioned, or by using HPLC and MS to measure drug concentrations in cells isolated after sequential exposure to trypsin.27–29,44–54 Images acquired on stand-alone custom fluorescent microscopy platforms have been used to visualize the distribution of doxorubicin and other fluorescent anthracyclines in cell monolayers, MCTSs, MCCs, and tumor tissues.27–29,44–49,51,53,54 In a few of these studies, custom image analysis algorithms were developed to quantify drug distribution and accumulation.44,47,51,54 We report here the use of automated image acquisition and analysis methods on an HCS platform to quantify and compare fluorescent drug accumulation and distribution in HNC monolayers and MCTS models. We used 384-well ULA plates to make HNC MCTSs because the process is compatible with automation, requires relatively few cells to generate MCTSs within 1–3 days, and compound exposure, image acquisition, and analysis can be performed in situ.11,34,36–38
We selected the anthracycline chemotherapeutics idarubicin, daunorubicin, and doxorubicin, and the plant alkaloid ellipticine, for our studies because these molecules are fluorescent and have broad anti-tumor activities (Supplementary Fig. S1 and Figs. 2 and 4).55,56 Anthracyclines inhibit topoisomerase II activity by intercalation between adjacent DNA base pairs, are active against a wide variety of hematological and solid tumor malignancies, and are approved components of the standard of care therapy for several cancers.55 Doxorubicin (Adriamycin) and other fluorescent anthracyclines have also been used to study drug uptake and distribution in monolayers, MCTSs, MCCs, and tumor tissues.27,28,44–48,51,53,54 Ellipticine is structurally distinct from the anthracyclines (Supplementary Fig. S1A), but it also inhibits topoisomerase II activity by DNA intercalation, and exhibits antiproliferative activity against many human cancer lines, and toward murine and human leukemias in vivo.56 Cells exposed to ellipticine exhibit fluorescence in the FITC channel, but we believe that this is the first time that it has been used as a probe to investigate drug uptake, accumulation, and distribution in monolayer and MCTS cultures.42,43,56 Cal33 HNC monolayers were significantly more sensitive than MCTSs to growth inhibition by ellipticine, idarubicin, daunorubicin, and doxorubicin, by 6-, 20-, 10-, and 16-fold, respectively (Fig. 1 and Table 2). The enhanced resistance of HNC MCTSs to doxorubicin is consistent with MCTSs formed by cell lines from other tumor tissues. MCF-7 breast cancer monolayers were 50-fold more sensitive to doxorubicin than MCF-7 MCTSs.47 MGH-U1 colon cancer monolayers were 5-fold more sensitive to doxorubicin than MGH-U1 MCTSs.46 HCT-8 colon carcinoma monolayers were substantially more sensitive to doxorubicin than HCT-8 MCC cultures.48
The uptake and accumulation of all four fluorescent drugs in Cal33 monolayers was linear with respect to concentration, and after an initial (<5 min) rapid uptake phase, accumulation appeared to reach steady-state levels within 10 min that were maintained through 45 min (Fig. 3). Cells in Cal33 monolayers that were seeded at different densities and exposed to the same compound concentrations for the same exposure time accumulated equivalent and uniform drug levels in every cell (Figs. 6A, B, and 7). Drug accumulation in HNC monolayers was independent of cell number and/or density, and every cell experienced the same drug concentration. Fluorescent drug accumulation in Cal33 MCTSs was also linear with respect to concentration, and it appeared to achieve steady-state levels within 10–15 min of drug exposure that were maintained through 30 min (Fig. 5). However, in Cal33 MCTS cultures, drug accumulation increased as the number of cells and sizes of the MCTSs became bigger (Fig. 6C). MCTSs also exhibited very distinct drug penetration and distribution gradients, with drugs preferentially accumulating in cells in the outer layers of MCTSs compared with the substantially lower fluorescent drug intensities detected in cells located in the inner cores (Fig. 8A–C). Ellipticine and the anthracyclines exhibited similar drug penetration and distribution gradients in MCTSs formed by Cal33 or FaDu HNC cell lines (Figs. 8 and 9). The pseudo-color pixel intensity representations and line scan plots further highlight the fluorescent drug penetration and distribution gradient between the periphery and inner cores of HNC MCTSs (Figs. 8 and 9). Transmitted light images of HNC MCTSs revealed two distinct spheroid regions or phenotypes, an inner core surrounded by an outer layer that was several layers of cells thick, and the fluorescent images indicated that the drugs accumulated preferentially in the peripheral cell layers (Figs. 8 and 9). The increased drug accumulation levels observed as MCTS cell numbers and sizes became bigger (Fig. 6C) are consistent with drugs accumulating preferentially in the outer cell layers of MCTSs and the increase in surface area of a sphere (4πr2) (Figs. 4, 8, and 9). As the MCTS radius and surface area increases, more cells can occupy the outer cell layers. The uniform distribution of fluorescence intensities observed in HNC MCTSs formed from cells prelabeled with fluorescent cell tracker dyes indicates that the differential drug penetration and distribution gradient of fluorescent drugs in MCTSs was not due to an imaging artifact produced by either inefficient excitation light penetration and/or the detection of fluorescent light emission from MCTSs (Fig. 8D). Doxorubicin, daunorubicin, epirubicin, and mitoxantrone have previously been shown to exhibit uneven drug penetration and distribution gradients in MCTSs, MCCs, and tumor tissues.27–29,44–48,51,53,54 These anthracyclines were also shown to exert reduced activity against the centrally located cells of MCTSs, or cells on the far side of MCCs away from the donor reservoir.27–29,44–48,51,53,54 In vitro studies in MCTSs or MCCs formed by tumor cell lines of different tissue origins indicate that many chemotherapeutics exhibit limited penetration and uneven distribution patterns: vinblastine, paclitaxel, docetaxel, 5-fluorouracil, methotrexate, and cisplatin.29,45–47,49,51–53 Ellipticine joins the list of anti-cancer agents that exhibit reduced drug penetration and uneven accumulation and distribution patterns that are primarily confined to the peripheral cell layers of MCTSs.
Tumor cell adhesion, both cell-ECM and cell-cell, and higher tumor cell packing densities contribute to the poor penetration and drug resistance observed in MCTS or MCC cultures exposed to paclitaxel, doxorubicin, methotrexate, or 5-fluorouracil.28,44,48–50,54 However, the penetration of these agents increased when drug concentrations were high enough and/or exposures were long enough to induce apoptosis and reduce tumor cell packing densities in these cultures.28,44,48–50,54 After 24 h of drug exposure to 10 μM ellipticine or doxorubicin, Cal33 and FaDu MCTSs were smaller and displayed changes in morphology; the outer cell layers were either missing, smaller, or disintegrating, and the inner core regions appeared much darker (Fig. 9). Although Cal33 MCTSs exposed to ellipticine or doxorubicin for 24 h still exhibited a drug penetration gradient with preferential accumulation in peripheral cells relative to the inner core, the fluorescent intensity differences between outer and inner cells were smaller than after only 1 h of exposure (Fig. 9). FaDu MCTSs exposed to ellipticine or doxorubicin for 24 h exhibited higher fluorescent intensities in the inner cores of the MCTSs, indicating that the drugs had penetrated throughout the MCTSs (Fig. 9). Our observations in HNC MCTSs are consistent with the hypothesis that drug-induced cell death of peripheral cell layers is a major determinant of solid tumor drug penetration.28,44,48–50,54 It has been proposed that basic drugs such as doxorubicin and related anthracyclines may become sequestered in the peripheral layers of MCTSs, MCCs, and solid tumors because they accumulate and are trapped in acidic compartments such as endosomes or lysosomes, or because they bind so avidly to DNA.28,29,51 Similarly, paclitaxel binding to tubulin may lead to its sequestration in the peripheral layers of MCTSs and human solid tumor histocultures.50
Fluorescent images of patient tumors and mouse xenografts demonstrated that systemically administered doxorubicin only penetrates the periphery of prostate cancer tumors.44,54 Similarly, fluorescent imaging of mouse mammary and human breast cancer tumors after systemic administration of doxorubicin showed that drug penetration and accumulation was concentrated in only a few cell layers surrounding tumor blood vessels.28 Radiolabeled paclitaxel penetration in patient-derived HNC tumors and FaDu xenografts was also confined to the periphery for up to 24 h, until drug-induced apoptosis reduced epithelial cell packing density and enhanced penetration.50 Most cells in human tissues reside within a few cell diameters of blood vessels, but as solid tumors expand and grow, populations of tumor cells develop that are distant (>100 μm) from blood vessels.27–29,53 As the distances between tumor cells and functioning blood vessels increase, nutrient and oxygen concentrations decrease, thereby creating gradients of declining tumor cell proliferation and the development of hypoxic regions.27–29,53 Similarly, acidic metabolic end-product (e.g., lactic and carbonic acid) concentrations rise and extracellular pH drops as distances from blood vessels increase.27–29,53 The effects of increased separation distances from blood vessels on cellular microenvironments will be exacerbated in tumors with poor vascular organization, irregular blood flow, or when tumor cells compress blood and/or lymphatic vessels.27–29,53 Drug penetration and distribution in tumor tissues is a balance between the flow of fluid out of blood vessels and resorption by the lymphatics. These processes are influenced by the permeability of the wall of the blood vessel delivering the agent, and by both diffusion and convection within the interstitial fluid of the tumor tissue.27–29,44,53 Drug physiochemical properties (size, shape, pKa, logP, solubility, etc.) affect the rates of diffusion and convection in interstitial fluid, and the composition and structure of the ECM, cell-ECM contacts, and focal adhesion junctions between adjacent cells regulate the movement and distribution of molecules within tumor tissues.27–29,44,48,49,53 Water-soluble drugs diffuse around and between cells and are often found associated with the ECM, whereas lipophilic drugs penetrate the lipid membranes of cells.27–29,44,48,49,53 Tumor cell adhesion and the higher cell packing densities of solid tumors reduce drug penetration and contribute to drug resistance.27,28,48,49 Drug distribution in solid tumors is also affected by consumptive processes, including xenobiotic metabolism, non-specific and specific (target-mediated) binding to tissue components, and by the uptake and retention of agents by tumor and/or stromal cell populations.27–29,44,48,49,53
Drug resistance continues to be a major obstacle to the realization of long-term clinical benefits for cancer therapies. Intrinsic drug resistance is exemplified by a de novo lack of response to treatment, and acquired resistance refers to the disease progression that occurs after an initial response to therapy.57,58 Primary or intrinsic resistance factors pre-exist at treatment onset, limiting the effectiveness of many therapies.57,58 Intrinsic drug resistance falls into two main categories. Tumor intrinsic factors include coexistent genetic alterations to drug targets, mutations in other signaling pathway genes, and inactivation of pro-apoptotic pathways.57,58 Patient- or drug-specific pharmacokinetic features include absorption, distribution, metabolism and excretion (ADME) properties, drug efflux transporter expression, drug metabolism, and drug-drug interactions that also confer intrinsic drug resistance.57,58 Acquired drug resistance is a complex and diverse process that evolves while the patient is receiving therapy, implying that the tumor develops “escape” mechanisms to evade drug action.57,58 Acquired resistance may involve target modifications, bypass or compensatory signaling, and histologic transformation, among other mechanisms.57,58 However, cells in solid tumors that are distal to blood vessels may be resistant to anti-cancer agents for other reasons: cells that are growing slowly or that have stopped replicating due to reduced nutrient and/or oxygen levels will be resistant to molecules targeting cell proliferation mechanisms; some agents may be less effective under hypoxic, acidic, or nutrient-deprived conditions; and cells that are distal to blood vessels may experience lower drug concentrations due to reduced drug access.27–29,44,48,49,53,57 Cell contacts with the ECM, adhesion junctions formed between adjacent cells, and high tumor cell packing densities constitute drug permeability barriers that limit drug penetration, distribution, and efficacy.27,28,48,49 Reduced drug penetration contributes to the resistance of solid tumors to several chemotherapeutic agents, including; doxorubicin, epirubicin, daunorubicin, mitoxantrone, methotrexate, 5-fluorouracil, vinblastine, paclitaxel, and docetaxel.27–29,44–54 MCTSs develop microenvironments similar to those of avascular tumor nodules, micro-metastases, or the intervascular regions of large solid tumors with respect to volume growth kinetics, morphology, and the formation and upkeep of gradients of nutrient distribution, oxygen concentration, and cell proliferation.11,12,22,24,34–36 Cells in MCTSs also experience similar adhesive, topographical, and mechanical forces to cells in solid tumors, and how these cellular cues alter biological responses.10–14,19–26 We show here that HNC MCTSs exhibited permeability barriers that resulted in uneven drug distribution and exposure gradients that coincided with enhanced resistance relative to monolayer cultures. In contrast, cells in 2D HNC monolayers experienced uniform drug concentrations in a consistent environment and all cells accumulated the same steady-state drug concentrations. The enhanced resistance of MCTS cultures and their ability to recapitulate drug penetration and distribution gradients strongly argues for the deployment of these models in cancer lead discovery. The implementation of more physiologically relevant 3D tumor models has the potential to improve the correlation between in vitro potencies and in vivo efficacy, and ultimately may lead to increased cancer drug approval rates.
Supplementary Material
Abbreviations Used
- 2D
two-dimensional
- 3D
three-dimensional
- Ch1
fluorescent channel 1
- Ch2
fluorescent channel 2
- Ch3
fluorescent channel 3
- CTG
CellTiter-Glo®
- DAPI
4,6-diamidino-2-phenylindole
- DMEM
Dulbecco's modified Eagle's medium
- DMSO
dimethyl sulfoxide
- ECM
extra cellular matrix
- Em
emission
- Ex
excitation
- FBS
fetal bovine serum
- FITC
fluorescein isothiocyanate
- GI50
50% growth inhibitory concentration
- HCS
high-content screening
- HNC
head and neck cancer
- HPLC
high-performance liquid chromatography
- HTS
high-throughput screening
- IXM
ImageXpress® Micro
- Max.
maximum plate controls
- MCCs
multilayer cell cultures
- MCTSs
multicellular tumor spheroids
- Min.
minimum plate controls
- MS
mass spectrometry
- MWCS
multi-wavelength cell scoring
- PBS
phosphate-buffered saline
- PDX
patient-derived xenograft
- P/S
penicillin and streptomycin
- RFUs
relative fluorescent intensity units
- RLUs
relative light units
- S:B
signal-to-background
- TRITC
tetramethylrhodamine
- ULA plates
ultra-low attachment microtiter plates
Acknowledgments
The studies were supported in part by a Development Research Project award (Johnston, Principal Investigator) from the HNC Spore P50 (Ferris and Grandis, CA097190) of the University of Pittsburgh Cancer Institute.
Disclosure Statement
No competing financial interests exist.
References
- 1.Al-Lazikani B, Banerji U, Workman P: Combinatorial drug therapy for cancer in the post-genomic era. Nat Biotechnol 2012;30:679–692 [DOI] [PubMed] [Google Scholar]
- 2.Barretina J, Caponigro G, Stransky N, et al. : The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012;483:603–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim N, He N, Yoon S: Cell line modeling for systems medicine in cancers. Int J Oncol 2014;44:371–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ocana A, Pandiella A, Siu LL, Tannock IF: Preclinical development of molecular-targeted agents for cancer. Nat Rev Clin Oncol 2011;8:200–209 [DOI] [PubMed] [Google Scholar]
- 5.Ocaña A, Pandiella A: Personalized therapies in the cancer “omics” era. Mol Cancer 2010;9:202–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shoemaker R: The NCI60 human tumour cell line anticancer drug screen. Nat Rev Cancer 2006;6:813–823 [DOI] [PubMed] [Google Scholar]
- 7.Hait W: Anticancer drug development: the grand challenges. Nat Rev Drug Discov 2010;9:253–254 [DOI] [PubMed] [Google Scholar]
- 8.Hutchinson L, Kirk R: High drug attrition rates—where are we going wrong? Nat Rev Clin Oncol 2011;8:189–190 [DOI] [PubMed] [Google Scholar]
- 9.Kamb A, Wee S, Lengauer C: Why is cancer drug discovery so difficult? Nat Rev Drug Discov 2007;6:115–120 [DOI] [PubMed] [Google Scholar]
- 10.Baker B, Chen CS: Deconstructing the third dimension: how 3D culture microenvironments alter cellular cues. J Cell Sci 2012;125:3015–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ekert J, Johnson K, Strake B, Pardinas J, Jarantow S, Perkinson R, Colter DC: Three-dimensional lung tumor microenvironment modulates therapeutic compound responsiveness in vitro—implication for drug development. PLoS One 2014;125:3015–3024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich J, Seidel C, Ebner R, Kunz-Schughart LA: Spheroid-based drug screen: considerations and practical approach. Nat Protoc 2009;4:309–324 [DOI] [PubMed] [Google Scholar]
- 13.Härmä V, Virtanen J, Mäkelä R, et al. : A comprehensive panel of three-dimensional models for studies of prostate cancer growth, invasion and drug responses. PLoS One 2010;5:e10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hongisto V, Jernström S, Fey V, et al. : High-throughput 3D screening reveals differences in drug sensitivities between culture models of JIMT1 breast cancer cells. PLoS One 2013;8:e77232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li H, Wawrose JS, Gooding WE, et al. : Genomic analysis of head and neck squamous cell carcinoma cell lines and human tumors: a rational approach to preclinical model selection. Mol Cancer Res 2014;12:571–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao HKJ, Ferretti S, Monahan JE, et al. : Sellers high-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat Med 2015;21:1318–1325 [DOI] [PubMed] [Google Scholar]
- 17.Hidalgo M, Amant F, Biankin AV, et al. : Patient-derived xenograft models: an emerging platform for translational cancer research. Cancer Discov 2014;4:998–1013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Izumchenko E, Meir J, Bedi A, Wysocki PT, Hoque MO, Sidransky D: Patient-derived xenografts as tools in pharmaceutical development. Clin Pharmacol Ther 2016;99:612–621 [DOI] [PubMed] [Google Scholar]
- 19.Fischbach C, Chen R, Matsumoto T, et al. : Engineering tumors with 3D scaffolds. Nat Methods 2007;4:855–860 [DOI] [PubMed] [Google Scholar]
- 20.Horman S, To J, Orth AP: An HTS-compatible 3D colony formation assay to identify tumor specific chemotherapeutics. J Biomol Screen 2013;18:1298–1308 [DOI] [PubMed] [Google Scholar]
- 21.Lovitt C, Shelper TB, Avery VM: Miniaturized three-dimensional cancer model for drug evaluation. Assay Drug Dev Technol 2013;11:435–448 [DOI] [PubMed] [Google Scholar]
- 22.Lovitt C, Shelper TB, Avery VM: Advanced cell culture techniques for cancer drug discovery. Biology 2014;3:345–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shin C, Kwak B, Han B, Park K: Development of an in vitro 3D tumor model to study therapeutic efficiency of an anticancer drug. Mol Pharm 2013;10:2167–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang C, Tang Z, Zhao Y, Yao R, Li L, Sun W: Three-dimensional in vitro cancer models: a short review. Biofabrication 2014;6:22001. [DOI] [PubMed] [Google Scholar]
- 25.Wenzel C, Riefke B, Gründemann S, et al. : 3D high-content screening for the identification of compounds that target cells in dormant tumor spheroid regions. Exp Cell Res 2014;323:131–143 [DOI] [PubMed] [Google Scholar]
- 26.Yip D, Cho CH: A multicellular 3D heterospheroid model of liver tumor and stromal cells in collagen gel for anti-cancer drug testing. Biochem Biophys Res Commun 2013;433:327–332 [DOI] [PubMed] [Google Scholar]
- 27.Kerr D, Kaye SB: Aspects of cytotoxic drug penetration, with particular reference to anthracyclines. Cancer Chemother Pharmacol 1987;19:1–5 [DOI] [PubMed] [Google Scholar]
- 28.Minchinton A, Tannock IF: Drug penetration in solid tumours. Nat Rev Cancer 2006;6:583–592 [DOI] [PubMed] [Google Scholar]
- 29.Tannock I, Lee CM, Tunggal JK, Cowan DS, Egorin MJ: Limited penetration of anticancer drugs through tumor tissue: a potential cause of resistance of solid tumors to chemotherapy. Clin Cancer Res 2002;8:878–884 [PubMed] [Google Scholar]
- 30.Abbot A: Biology's new dimension. Nature 2003;424:870–872 [DOI] [PubMed] [Google Scholar]
- 31.Pampaloni F, Reynaud EG, Stelzer EH: The third dimension bridges the gap between cell culture and live tissue. Nat Rev Mol Cell Biol 2007;8:839–845 [DOI] [PubMed] [Google Scholar]
- 32.Ryan S, Baird AM, Vaz G, et al. : Drug discovery approaches utilizing three-dimensional cell culture. Assay Drug Dev Technol 2016;14:19–28 [DOI] [PubMed] [Google Scholar]
- 33.Zips D, Thames HD, Baumann M: New anticancer agents: in vitro and in vivo evaluation. In Vivo 2005;19:1–7 [PubMed] [Google Scholar]
- 34.Sant S, Johnston PA: The production of 3D tumor spheroids for cancer drug discovery. Drug Discov Today Technol 2017;23:27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Foty R. A simple hanging drop cell culture protocol for generation of 3D spheroids. J Vis Exp 2011;51:pii: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vinci M, Gowan S, Boxall F, et al. : Advances in establishment and analysis of three-dimensional tumor spheroid-based functional assays for target validation and drug evaluation. BMC Biol 2012;10:29–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Howes A, Richardson RD, Finlay D, Vuori K: 3-Dimensional culture systems for anti-cancer compound profiling and high-throughput screening reveal increases in EGFR inhibitor-mediated cytotoxicity compared to monolayer culture systems. PLoS One 2014;9:e108283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rotem A, Janzer A, Izar B, Ji Z, Doench JG, Garraway LA, Struhl K: Alternative to the soft-agar assay that permits high-throughput drug and genetic screens for cellular transformation. Proc Natl Acad Sci U S A 2015;112:5708–5713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnston P, Sen M, Hua Y, et al. : High-content pSTAT3/1 imaging assays to screen for selective inhibitors of STAT3 pathway activation in head and neck cancer cell lines. Assay Drug Dev Technol 2014;12:55–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Johnston P, Sen M, Hua Y, et al. : HCS campaign to identify selective inhibitors of IL-6-induced STAT3 pathway activation in head and neck cancer cell lines. Assay Drug Dev Technol 2015;13:356–376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hua Y, Shun TY, Strock CJ, Johnston PA: High-content positional biosensor screening assay for compounds to prevent or disrupt androgen receptor and transcriptional intermediary factor 2 protein-protein interactions. Assay Drug Dev Technol 2014;12:395–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dudgeon D, Shinde SN, Shun TY, et al. : Characterization and optimization of a novel protein-protein interaction biosensor HCS assay to identify disruptors of the interactions between p53 and hDM2. Assay Drug Dev Technol 2010;8:437–458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Johnston PA, Shinde SN, Hua Y, Shun TY, Lazo JS, Day BW: Development and validation of a high-content screening assay to identify inhibitors of cytoplasmic Dynein-mediated transport of glucocorticoid receptor to the nucleus. Assay Drug Dev Technol 2012;10:432–456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Au J, Jang SH, Zheng J, et al. : Determinants of drug delivery and transport to solid tumors. J Control Release 2001;74:31–46 [DOI] [PubMed] [Google Scholar]
- 45.Durand R: Flow Cytometry Studies of intracellular Adriamycin in multicell spheroids in vitro. Cancer Res 1981;41:3495–3498 [PubMed] [Google Scholar]
- 46.Erlichman C, Vidgen D: Cytotoxicity of Adriamycin in MGH-U1 cells grown as monolayer cultures, spheroids, and xenografts in immune-deprived mice. Cancer Res 1984;44:5369–5375 [PubMed] [Google Scholar]
- 47.Gong X, Lin C, Cheng J, et al. : Generation of multicellular tumor spheroids with microwell-based agarose scaffolds for drug testing. Plos One 2015;10:e0130348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Grantab R, Sivananthan S, Tannock IF: The penetration of anticancer drugs through tumor tissue as a function of cellular adhesion and packing density of tumor cells. Cancer Res 2006;66:1033–1039 [DOI] [PubMed] [Google Scholar]
- 49.Grantab R, Tannock IF: Penetration of anticancer drugs through tumour tissue as a function of cellular packing density and interstitial fluid pressure and its modification by bortezomib. BMC Cancer 2012;12:214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kuh H, Jang SH, Wientjes MG, Weaver JR, Au JL: Determinants of paclitaxel penetration and accumulation in human solid tumor. J Pharmacol Exp Ther 1999;290:871–880 [PubMed] [Google Scholar]
- 51.Kyle A, Huxham LA, Chiam AS, Sim DH, Minchinton AI: Direct assessment of drug penetration into tissue using a novel application of three-dimensional cell culture. Cancer Res 2004;64:6304–6309 [DOI] [PubMed] [Google Scholar]
- 52.Kyle A, Huxham LA, Yeoman DM, Minchinton AI: Limited tissue penetration of taxanes: a mechanism for resistance in solid tumors. Clin Cancer Res 2007;13:2804–2810 [DOI] [PubMed] [Google Scholar]
- 53.Phillips R, Loadman PM, Cronin BP: Evaluation of a novel in vitro assay for assessing drug penetration into avascular regions of tumours. Br J Cancer 1998;77:2112–2119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zheng J, Chen CT, Au JL, Wientjes MG: Time- and concentration-dependent penetration of doxorubicin in prostate tumors. AAPS PharmSci 2001;3:E15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Rabbani A, Finn RM, Ausio J: The anthracylcine antibiotics: antitumor drugs that alter chromatin structure. BioEssays 2004;27:50–56 [DOI] [PubMed] [Google Scholar]
- 56.Sailer B, Valdez JG, Steinkamp JA, Darzynkiewicz Z, Crissman HA: Monitoring uptake of ellipticine and its fluorescence lifetime in relation to the cell cycle phase by flow cytometry. Exp Cell Res 1997;236:259–267 [DOI] [PubMed] [Google Scholar]
- 57.Holohan C, Van Schaeybroeck S, Longley DB, Johnston PG: Cancer drug resistance: an evolving paradigm. Nat Rev Cancer 2013;13:714–726 [DOI] [PubMed] [Google Scholar]
- 58.Lovly C, Shaw AT: Molecular pathways: resistance to kinase inhibitors and implications for therapeutic strategies. Clin Cancer Res 2014;20:2249–2256 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.