Abstract
The dual specificity phosphatase DUSP1 was the first mitogen activated protein kinase phosphatase (MKP) to be identified. It dephosphorylates conserved tyrosine and threonine residues in the activation loops of mitogen activated protein kinases ERK2, JNK1 and p38‐alpha. Here, we report the crystal structure of the human DUSP1 catalytic domain at 2.49 Å resolution. Uniquely, the protein was crystallized as an MBP fusion protein in complex with a monobody that binds to MBP. Sulfate ions occupy the phosphotyrosine and putative phosphothreonine binding sites in the DUSP1 catalytic domain.
Keywords: crystallization chaperone, dual specificity phosphatase, DUSP, maltose‐binding protein, monobody, sulfate ions
Short abstract
PDB Code(s): 6APX
Introduction
Mitogen‐activated protein kinases (MAPKs), a group of highly conserved tyrosine and serine/threonine protein kinases in eukaryotes,1 play vital roles in a myriad of cellular processes.2, 3, 4 MAPKs are inactive until they become phosphorylated by map kinase kinases (MAPKKs), usually at two sites within their activation loops, which contain a characteristic TxY motif that needs to be phosphorylated on both the threonine and tyrosine residues in order to lock the enzyme into its active conformation. Dephosphorylation, mediated by protein tyrosine phosphatases (PTPs) or dual‐specificity phosphatases (DUSPs), returns the MAPKs to their inactive state. Tight control and balance between phosphorylation and deophosphorylation of MAPKs is pivotal for the proper execution of downstream signaling pathways. DUSPs can dephosphorylate both phosphotyrosine and phosphothreonine in the TxY motif of MAPKs.4 The 26 unique DUSPs encoded by the human genome are classified either as MAPK phosphatases (MKPs) or atypical DUSPs.5 The MKPs are composed of an N‐terminal non‐catalytic MAPK‐binding domain (MKBD) and a highly conserved C‐terminal catalytic domain (CD). Atypical DUSPs have only the CD and show high sequence homology to the Vaccinia virus H1 gene product.6, 7, 8 As key regulators in many MAPK signaling pathways, DUSPs are emerging as potential therapeutic targets in human diseases such as cancer.9
DUSP1, the first MKP demonstrated to be specific for MAPKs, targets ERK2, JNK1 and p38‐alpha.10 It is an inducible nuclear protein11, 12 that is 367 amino acids in length and broadly expressed in a variety of human tissues.13 DUSP1 is an important regulator of both adaptive14 and innate immune systems.15 Mice lacking DUSP1 are resistant to diet‐induced obesity, implicating DUSP1 in metabolic control as well.16 Elevated or diminished expression of DUSP1 has also been observed in various cancers. Elevated DUSP1 levels are associated with melanoma,17 pancreatic,18 prostate,19 glial20 and gastric21 cancer cell lines, whereas decreased DUSP1 expressions are associated with ovarian22 and testicular23 cancers. Moreover, while DUSP1 is overexpressed in early stages of colon, prostate and bladder cancer, its expression is diminished as the tumor becomes more aggressive.24, 25 Overexpression of DUSP1 has also been linked to central nervous system diseases such as multiple sclerosis26 and major depressive disorder.27 In short, the perturbation of DUSP1 expression has been linked to a wide variety of maladies, making it a potential molecular target for therapeutic intervention.
As the archetypal member of the MKP family, DUSP1 has been extensively studied. Even so, the structures of neither the full‐length enzyme nor its CD are known. The lack of structural information may explain why very few inhibitors have been reported to target this enzyme.28 Here, we report the crystal structure of a catalytically inactive DUSP1‐CD (Cys258Ser mutant) at 2.49 Å resolution. We expect that the availability of this structure will facilitate efforts to develop DUSP1 inhibitors and elucidate structure/function relationships for this important enzyme.
Results and Discussion
Construct design and overall structure of the DUSP1‐CD
A truncated DUSP1 protein lacking 53 C‐terminal residues had significantly higher basal phosphatase activity than the full‐length protein.29 Therefore, the CD employed in the present study encompasses residues 172–314 (of 367). Attempts to obtain diffraction quality crystals of the wild‐type DUSP1‐CD or the DUSP1‐CD(Cys258Ser) mutant, either alone or fused to the C‐terminus of E. coli maltose binding protein (MBP), failed. The MBP used in these experiments contained several surface entropy reduction mutations that were designed to facilitate the crystallization of MBP fusion proteins.30 Finally, the MBP‐DUSP1‐CD(Cys258Ser) fusion protein was purified in complex with a “monobody” (YSX1) that specifically binds to the maltose‐free form of MBP.31 This time diffraction quality crystals were obtained and the structure was solved at 2.49 Å resolution with one heterodimer per asymmetric unit [Fig. 1(A)]. Data collection and refinement statistics are summarized in Table 1.
Figure 1.
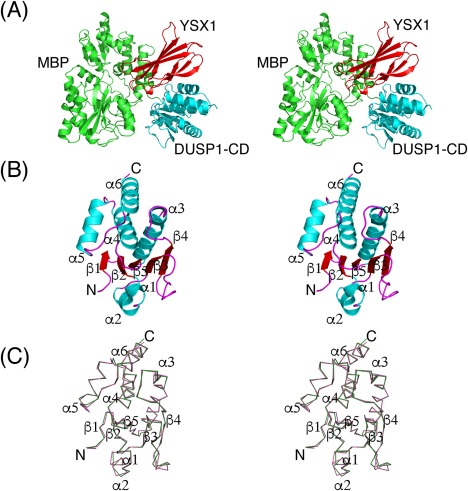
(A) Stereoview illustrating the crystal structure of the YSX1 monobody (red) bound to the MBP (green)‐DUSP1‐CD (cyan) fusion protein. (B) Cartoon diagram representing the overall structure of human DUSP1‐CD in stereoview. Secondary structural elements are labeled. (C) Stereoview showing the Cα trace of DUSP1‐CD (green) superimposed with that of DUSP4‐CD (pink). Secondary structure elements are labeled. The point of view is the same as that in (B).
Table 1.
Crystallographic statistics
| Data Collection | |
| X‐ray source | MicroMax‐007 HF |
| Wavelength (Å) | 1.5418 |
| Resolution (Å)a | 50.00–2.49 (2.53–2.49) |
| Space group | P43212 |
| Unit‐cell parameters | |
| a, b (Å) = | 105.8 |
| c (Å) = | 181.4 |
| α, β, γ (degree) = | 90 |
| Total reflections | 646916 |
| Unique reflectionsa | 36681 (1728) |
| Completeness (%)a | 99.7 (95.6) |
| Multiplicitya | 17.6 (9.0) |
| Mean I/σ (I)a | 40.1 (2.0) |
|
Rmerge
a
Rpim CC1/2 (highest resolution shell) |
0.099 (0.830) 0.023 (0.251) 0.881 |
| Refinement | |
| Rwork | 0.20 |
| Rfree | 0.23 |
| No. of atoms/B‐factors (Å2) | |
| Protein | |
| MBP | 2727/66.2 |
| DUSP1‐CD | 1119/53.7 |
| Monobody | 710/50.7 |
| Ligands | |
| Sulfates | 15/57.1 |
| Glycerol | 6/76.4 |
| Water | 81/49.0 |
| RMSD from ideal geometry | |
| Bond lengths (Å) | 0.008 |
| Bond angles (degree) | 0.867 |
| MolProbity validationb | |
| MolProbity clash score | 5.25 (99th percentile) |
| MolProbity protein geometry score | 1.60 (99th percentile) |
| Ramachandran plot | |
| Favored regions (%) | 89.4 |
| Allowed regions (%) | 10.1 |
| Generously allowed region (%) | 0.5 |
| Outliers (%) | 0.0 |
| PDB code | 6APX |
Values in parentheses are for the highest‐resolution shell.
MolProbity was used to analyze the structure.
The fold of the DUSP1‐CD closely resembles those of other MKPs with a central twisted five‐stranded β‐sheet surrounded by six α‐helices [Fig. 1(B)]. One side of the β‐sheet is covered by two α‐helices while four α‐helices straddle the other. In the crystal lattice, the DUSP1‐CD makes crystal contacts with MBP and the monobody but not with any of the neighboring symmetry‐related DUSP1‐CD molecules.
The coordinates of the DUSP1‐CD were submitted to the PDBeFold server32 to identify the three‐dimensional protein structures in the PDB that are most similar to DUSP1. DUSP4,33 DUSP5,34 and DUSP1535 were the most closely related structures with root mean square deviation (RMSD) values of 0.48, 0.52, and 0.57 Å, respectively. When the structure of the DUSP1‐CD is aligned with that of the DUSP4‐CD (with the highest Z‐score of 14.7), 133 Cα atoms can be superimposed with a RMSD of 0.48 Å out of a total of 143 and 144 residues in the DUSP1 and DUSP4 CDs, respectively [Fig. 1(C)]. Thus, the overall fold of DUSP1‐CD is highly similar to that of the DUSP4‐CD.
The active site
The active site of the DUSP1‐CD is composed of His257‐Cys258Ser‐Gln259‐Ala260‐Gly261‐Ile262‐Ser263‐Arg264 in the phosphotyrosine binding loop (PTP‐loop) and Asp227, which acts as a general acid/base during catalysis. In the crystal structure of the DUSP1‐CD, a sulfate ion from the crystallization solution is bound in the PTP‐loop (sulfate1), mimicking the phosphate moiety of phosphotyrosine (Fig. 2). It is held in the active site by an extensive network of hydrogen bonds between O atoms of the sulfate and the backbone amide NH atoms of Ala260 (3.1 Å), Gly261 (3.0 Å), Ile262 (3.2 Å), Ser263 (3.1 Å), and Arg264 (2.9 Å). The guanidinium nitrogens of the highly conserved Arg264 form two hydrogen bonds (3.0 Å, 2.8 Å) with two oxygen atoms of the bound sulfate. The side chain oxygen atom of Ser258 is located 2.6 Å away from the oxygen atom of the sulfate; a cysteine in this position serves as the catalytic nucleophile in the wild‐type enzyme.36 The side chain carbonyl oxygen of Gln259 forms a hydrogen bond (3.0 Å) with the side chain amide nitrogen of Gln72 in MBP.
Figure 2.
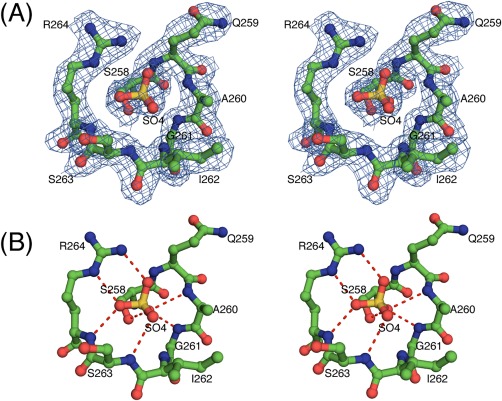
(A) Stereoview of the fit of the residues of the PTP‐loop of DUSP1‐CD to the final 2mFo – DFc electron density map (blue, 2.49 Å resolution, contoured at the 1σ level) with the bound sulfate ion. C atoms are colored green, O atoms red, N atoms blue, and the S atom orange. (B) Stereoview illustrating the interactions of the bound sulfate ion in the PTP‐loop of DUSP1‐CD in ball and stick representation. Hydrogen bonds are indicated by red dashes. The point of view is the same as that in (A).
Bound sulfate ions
The DUSP1‐CD was crystallized in 75 mM MES pH 5.9 and 2.4 M ammonium sulfate. In the current structure, three sulfate ions are bound to the DUSP1‐CD. As detailed above, sulfate1 is bound in the PTP‐loop. To identify the putative phosphothreonine‐binding site in DUSP1, the structure of the DUSP1‐CD was aligned with that of the phosphorylated p38 peptide bound to VHR [PDB code: 1J4X].37 Sulfate2 of the DUSP1‐CD is situated close to the region where the phosphate of the peptide phosphothreonine binds to VHR. Therefore, it seems feasible that sulfate2 mimics the binding of the phosphothreonine phosphate in the TxY motif of MAPKs to the DUSP1‐CD. However, whereas the p38 phosphothreonine binds weakly in the VHR/peptide complex, sulfate2 in the DUSP1‐CD structure exhibits strong H‐bonding interactions with the side chain of Arg292 (2.9 Å, 2.8 Å), the main chain amide of Gly172 (3.0 Å) and a water molecule (2.9 Å). A third sulfate (Sulfate3) was found in a pocket adjacent to the PTP‐loop. This sulfate makes H‐bonds with the main chain of Asn209 (3.0 Å) and the side chains of Arg264 (2.7 Å, 3.2 Å) and Ser207 (2.4 Å) of DUSP1, as well as with Gln72 (2.7 Å) of MBP. Arg264 is a conserved residue of the PTP‐loop and its guanidinium side chain is pivotal for the binding of the phosphate in the active site.
Conclusions
In summary, we have determined the crystal structure of the DUSP1‐CD at 2.49 Å resolution. To obtain this structure, it was necessary to employ a monobody that binds to MBP as a co‐crystallization chaperone. In chaperone‐assisted crystallography, an oft‐raised question is whether the chaperone has induced any changes in the structure of the target protein. The fact that the structure of DUSP1‐CD aligned quite well with that of the DUSP4‐CD, which was crystallized in the absence of chaperones, implies that no structural changes in the DUSP1‐CD were caused by the monobody. Although inhibitors of several DUSPs have been described, the overall progress in this field has been limited by the conserved PTP‐loop architecture. In this regard, the structure of the DUSP1‐CD may be useful for the development of specific inhibitors against the DUSP family.
Materials and Methods
Cloning, expression, and purification
The DUSP1‐CD, harboring a Cys258Ser mutation and including a C‐terminal His6‐tag was cloned into the pMALX(E) vector (a gift from Dr. Lars Pedersen, NIEHS) at the NotI and BamHI restriction sites.30 The open reading frame of monobody YSX1 was synthesized by Biomatik and supplied in a pUC57 vector. It was digested with NdeI and BamHI and ligated between the same two restriction sites in pET‐11a. The MBP‐DUSP1‐CD(Cys258Ser)‐His6 and YSX1 proteins were expressed separately in E. coli strain BL21‐CodonPlus(DE3)‐RIL (Agilent Technologies). Cultures were grown to mid‐log phase (OD600 = 0.4–0.6) at 37°C in Luria‐Bertani medium supplemented with 100 μg mL−1 ampicillin, 30 μg mL−1 chloramphenicol and 0.2% glucose. Proteins were induced by isopropyl β‐D‐1‐thiogalactopyranoside (IPTG) at a concentration of 1 mM. The MBP‐DUSP1‐CD(Cys258Ser)‐His6 fusion protein was induced at 37°C for 4 h. To produce the YSX1 monobody, first the culture was cooled to 18°C for 30 min and then induced overnight. The cells were harvested by centrifugation and stored at −80°C until further use.
All purification steps were performed at 4°C using a Bio Rad NGC Chromatography System with pre‐packed chromatography columns (GE Healthcare). Co‐lysis was employed to prepare the monobody/MBP‐DUSP1‐CD(Cys258Ser)‐His6 complex. Cells were suspended in buffer A (50 mM Tris‐HCl pH 7.5, 150 mM NaCl, 10% (v/v) glycerol and 25 mM imidazole) supplemented with complete EDTA‐free protease inhibitor cocktail tablets (Roche Molecular Biochemicals), lysed using an APV‐1000 homogenizer (Invensys APV Products) and centrifuged at 30,000 g for 30 min. The supernatant was filtered through a 0.22 μm polyethersulfone membrane (Thermo Scientific) and applied onto a 5 ml His Trap FF column pre‐equilibrated in buffer A. The column was washed with 10 column volumes of buffer A and eluted with a linear gradient of imidazole in buffer A to 500 mM. Fractions containing the fusion protein/monobody complex were pooled, concentrated by ultrafiltration (30 kDa cutoff, Millipore) and diluted with 25 mM Tris‐HCl pH 7.5 (buffer B) to reduce the NaCl concentration to about 25 mM. The diluted sample was applied onto a 5 ml Hi Trap Q HP anion exchange column equilibrated with buffer B. The column was washed with 10 column volumes of buffer B containing 25 mM NaCl and the bound protein was eluted with a linear gradient of NaCl to 1 M. The eluted fractions of the complex were pooled, concentrated and applied onto a HiPrep 26/60 Sephacryl S‐200 HR column equilibrated in 25 mM Tris pH 7.5, 150 mM NaCl, 2 mM Tris (2‐carboxyethyl) phosphine (TCEP), 10% (v/v) glycerol (buffer C). The peak fractions containing the complex were pooled, concentrated to 33 mg mL−1, centrifuged at 15,700 g for 10 min and filtered through a 0.22 μm Ultra free centrifugal filter unit (Millipore). Aliquots were flash frozen in liquid nitrogen and stored at −80°C.
Crystallization and data collection
The purified complex (33 mg mL−1) was screened for crystals using a Gryphon crystallization robot (Art Robbins Instruments) against commercially available crystal screens at 19°C by the sitting drop vapor diffusion method. Crystals suitable for data collection were optimized in a 2 μL drop (1 μL protein + 1 μL crystallization solution) against 500 μL of reservoir solution by the hanging drop method in 75 mM MES pH 5.9, 2.4 M ammonium sulfate. After a week, a single crystal was retrieved, cryo‐protected with a short sweep in the mother liquor supplemented with 10% (v/v) glycerol and immediately flash cooled in liquid nitrogen. X‐ray diffraction data were collected using a MAR345 detector mounted on Rigaku MicroMax‐007 HF high intensity microfocus generator equipped with VariMAX HF optics (Rigaku Corporation) and operated at 40 kV and 30 mA. Crystals were held at 93K. The data were integrated and scaled with HKL‐3000.38
Structure solution and refinement
The structure of the MBP fusion protein/monobody complex was solved by molecular replacement using PHASER39 from the CCP4 program suite. A search model containing three ensembles comprised of the coordinates of MBP (PDB code 3H4Z),40 the monobody YSX1 (PDB code 3CSB),31 and DUSP4 (PDB code 3EZZ)33 was employed as implemented in PHASER. The model was rebuilt manually in COOT41 and refined with phenix.refine in the PHENIX software suite.42 Model validation was performed with MolProbity.43 The Ramachandran plots were prepared with PROCHECK44 and they revealed that 88.4% of the residues fall within the most favored region, 10.1% are in the allowed region, and 0.5% are in the generously allowed region. There are no residues in the disallowed regions. Figures were prepared with PyMOL (The PyMOL Molecular Graphics System, Version 1.8 Schrödinger, LLC).
Conflict of Interest Statement
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commerical products or organizations imply endorsement by the US Government.
Data Deposition
Coordinates and structure factors were deposited in the Protein Data Bank under accession code 6APX.
References
- 1. Su B, Karin M (1996) Mitogen‐activated protein kinase cascades and regulation of gene expression. Curr Opin Immunol 8:402–411. [DOI] [PubMed] [Google Scholar]
- 2. Duan W, Wong WS (2006) Targeting mitogen‐activated protein kinases for asthma. Curr Drug Targets 7:691–698. [DOI] [PubMed] [Google Scholar]
- 3. Chang L, Karin M (2001) Mammalian MAP kinase signalling cascades. Nature 410:37–40. [DOI] [PubMed] [Google Scholar]
- 4. Boutros T, Chevet E, Metrakos P (2008) Mitogen‐activated protein (MAP) kinase/MAP kinase phosphatase regulation: roles in cell growth, death, and cancer. Pharmacol Rev 60:261–310. [DOI] [PubMed] [Google Scholar]
- 5. Jeong DG, Wei CH, Ku B, Jeon TJ, Chien PN, Kim JK, Park SY, Hwang HS, Ryu SY, Park H, Kim DS, Kim SJ, Ryu SE (2014) The family‐wide structure and function of human dual‐specificity protein phosphatases. Acta Cryst 70:421–435. [DOI] [PubMed] [Google Scholar]
- 6. Alonso A, Burkhalter S, Sasin J, Tautz L, Bogetz J, Huynh H, Bremer MC, Holsinger LJ, Godzik A, Mustelin T (2004) The minimal essential core of a cysteine‐based protein‐tyrosine phosphatase revealed by a novel 16‐kDa VH1‐like phosphatase, VHZ. J Biol Chem 279:35768–35774. [DOI] [PubMed] [Google Scholar]
- 7. Guan KL, Broyles SS, Dixon JE (1991) A Tyr Ser protein phosphatase encoded by Vaccinia virus. Nature 350:359–362. [DOI] [PubMed] [Google Scholar]
- 8. Koksal AC, Nardozzi JD, Cingolani G (2009) Dimeric quaternary structure of the prototypical dual specificity phosphatase VH1. J Biol Chem 284:10129–10137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Rios‐Fernandez R, Callejas‐Rubio JL, Caba‐Molina M, Rios‐Peregrina R, Ortego‐Centeno N (2014) Mesenteric inflammatory venoocclusive disease in a patient with Sjogren's Syndrome. Case Rep Med 2014:423420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Slack DN, Seternes OM, Gabrielsen M, Keyse SM (2001) Distinct binding determinants for ERK2/p38alpha and JNK map kinases mediate catalytic activation and substrate selectivity of map kinase phosphatase‐1. J Biol Chem 276:16491–16500. [DOI] [PubMed] [Google Scholar]
- 11. Hutter D, Chen P, Barnes J, Liu Y (2000) Catalytic activation of mitogen‐activated protein (MAP) kinase phosphatase‐1 by binding to p38 MAP kinase: critical role of the p38 C‐terminal domain in its negative regulation. Biochem J 352:155–163. [PMC free article] [PubMed] [Google Scholar]
- 12. Misra‐Press A, Rim CS, Yao H, Roberson MS, Stork PJ (1995) A novel mitogen‐activated protein kinase phosphatase. Structure, expression, and regulation. J Biol Chem 270:14587–14596. [DOI] [PubMed] [Google Scholar]
- 13. Wancket LM, Frazier WJ, Liu YS (2012) Mitogen‐activated protein kinase phosphatase (MKP)‐1 in immunology, physiology, and disease. Life Sci 90:237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Salojin KV, Owusu IB, Millerchip KA, Potter M, Platt KA, Oravecz T (2006) Essential role of MAPK phosphatase‐1 in the negative control of innate immune responses. J Immunol 176:1899–1907. [DOI] [PubMed] [Google Scholar]
- 15. Liu Y, Shepherd EG, Nelin LD (2007) MAPK phosphatases–regulating the immune response. Nat Rev Immunol 7:202–212. [DOI] [PubMed] [Google Scholar]
- 16. Wu JJ, Roth RJ, Anderson EJ, Hong EG, Lee MK, Choi CS, Neufer PD, Shulman GI, Kim JK, Bennett AM (2006) Mice lacking MAP kinase phosphatase‐1 have enhanced MAP kinase activity and resistance to diet‐induced obesity. Cell Metab 4:61–73. [DOI] [PubMed] [Google Scholar]
- 17. Kundu S, Fan K, Cao M, Lindner DJ, Tuthill R, Liu L, Gerson S, Borden E, Yi T (2010) Tyrosine phosphatase inhibitor‐3 sensitizes melanoma and colon cancer to biotherapeutics and chemotherapeutics. Mol Cancer Ther 9:2287–2296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liao Q, Guo J, Kleeff J, Zimmermann A, Buchler MW, Korc M, Friess H (2003) Down‐regulation of the dual‐specificity phosphatase MKP‐1 suppresses tumorigenicity of pancreatic cancer cells. Gastroenterology 124:1830–1845. [DOI] [PubMed] [Google Scholar]
- 19. Srikanth S, Franklin CC, Duke RC, Kraft RS (1999) Human DU145 prostate cancer cells overexpressing mitogen‐activated protein kinase phosphatase‐1 are resistant to Fas ligand‐induced mitochondrial perturbations and cellular apoptosis. Mol Cell Biochem 199:169–178. [DOI] [PubMed] [Google Scholar]
- 20. Weber RG, Rieger J, Naumann U, Lichter P, Weller M (2001) Chromosomal imbalances associated with response to chemotherapy and cytotoxic cytokines in human malignant glioma cell lines. Int J Cancer 91:213–218. [DOI] [PubMed] [Google Scholar]
- 21. Bang YJ, Kwon JH, Kang SH, Kim JW, Yang YC (1998) Increased MAPK activity and MKP‐1 overexpression in human gastric adenocarcinoma. Biochem Biophys Res Commun 250:43–47. [DOI] [PubMed] [Google Scholar]
- 22. Murty VV, Reuter VE, Bosl GJ, Chaganti RS (1996) Deletion mapping identifies loss of heterozygosity at 5p15.1‐15.2, 5q11 and 5q34‐35 in human male germ cell tumors. Oncogene 12:2719–2723. [PubMed] [Google Scholar]
- 23. Peng HQ, Liu L, Goss PE, Bailey D, Hogg D (1999) Chromosomal deletions occur in restricted regions of 5q in testicular germ cell cancer. Oncogene 18:3277–3283. [DOI] [PubMed] [Google Scholar]
- 24. Zhang L, Zhou W, Velculescu VE, Kern SE, Hruban RH, Hamilton SR, Vogelstein B, Kinzler KW (1997) Gene expression profiles in normal and cancer cells. Science 276:1268–1272. [DOI] [PubMed] [Google Scholar]
- 25. Loda M, Capodieci P, Mishra R, Yao H, Corless C, Grigioni W, Wang YB, MagiGalluzzi C, Stork PJS (1996) Expression of mitogen‐activated protein kinase phosphatase‐1 in the early phases of human epithelial carcinogenesis. Am J Pathol 149:1553–1564. [PMC free article] [PubMed] [Google Scholar]
- 26. Eljaschewitsch E, Witting A, Mawrin C, Lee T, Schmidt PM, Wolf S, Hoertnagl H, Raine CS, Schneider‐Stock R, Nitsch R, Ullrich O (2006) The endocannabinoid anandamide protects neurons during CNS inflammation by induction of MKP‐1 in microglial cells. Neuron 49:67–79. [DOI] [PubMed] [Google Scholar]
- 27. Duric V, Banasr M, Licznerski P, Schmidt HD, Stockmeier CA, Simen AA, Newton SS, Duman RS (2010) A negative regulator of MAP kinase causes depressive behavior. Nat Med 16:1328–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bakan A, Lazo JS, Wipf P, Brummond KM, Bahar I (2008) Toward a molecular understanding of the interaction of dual specificity phosphatases with substrates: insights from structure‐based modeling and high throughput screening. Curr Med Chem 15:2536–2544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hutter D, Chen PL, Barnes J, Liu YS (2002) The carboxyl‐terminal domains of MKP‐1 and MKP‐2 have inhibitory effects on their phosphatase activity. Mol Cell Biochem 233:107–117. [DOI] [PubMed] [Google Scholar]
- 30. Moon AF, Mueller GA, Zhong X, Pedersen LC (2010) A synergistic approach to protein crystallization: combination of a fixed‐arm carrier with surface entropy reduction. Protein Sci 19:901–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gilbreth RN, Esaki K, Koide A, Sidhu SS, Koide S (2008) A dominant conformational role for amino acid diversity in minimalist protein–protein interfaces. J Mol Biol 381:407–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Krissinel E, Henrick K (2004) Secondary‐structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Cryst 60:2256–2268. [DOI] [PubMed] [Google Scholar]
- 33. Jeong DG, Jung SK, Yoon TS, Woo EJ, Kim JH, Park BC, Ryu SE, Kim SJ (2009) Crystal structure of the catalytic domain of human MKP‐2 reveals a 24‐mer assembly. Proteins 76:763–767. [DOI] [PubMed] [Google Scholar]
- 34. Jeong DG, Cho YH, Yoon TS, Kim JH, Ryu SE, Kim SJ (2007) Crystal structure of the catalytic domain of human DUSP5, a dual specificity MAP kinase protein phosphatase. Proteins 66:253–258. [DOI] [PubMed] [Google Scholar]
- 35. Yoon TS, Jeong DG, Kim JH, Cho YH, Son JH, Lee JW, Ryu SE, Kim SJ (2005) Crystal structure of the catalytic domain of human VHY, a dual‐specificity protein phosphatase. Proteins 61:694–697. [DOI] [PubMed] [Google Scholar]
- 36. Zhou G, Denu JM, Wu L, Dixon JE (1994) The catalytic role of Cys124 in the dual specificity phosphatase VHR. J Biol Chem 269:28084–28090. [PubMed] [Google Scholar]
- 37. Schumacher MA, Todd JL, Rice AE, Tanner KG, Denu JM (2002) Structural basis for the recognition of a bisphosphorylated MAP kinase peptide by human VHR protein Phosphatase. Biochemistry 41:3009–3017. [DOI] [PubMed] [Google Scholar]
- 38. Minor W, Cymborowski M, Otwinowski Z, Chruszcz M (2006) HKL‐3000: the integration of data reduction and structure solution–from diffraction images to an initial model in minutes. Acta Cryst 62:859–866. [DOI] [PubMed] [Google Scholar]
- 39. McCoy AJ, Grosse‐Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ (2007) Phaser crystallographic software. J Appl Cryst 40:658–674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mueller GA, Edwards LL, Aloor JJ, Fessler MB, Glesner J, Pomes A, Chapman MD, London RE, Pedersen LC (2010) The structure of the dust mite allergen Der p 7 reveals similarities to innate immune proteins. J Allergy Clin Immunol 125:909–917. e904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Emsley P, Cowtan K (2004) Coot: model‐building tools for molecular graphics. Acta Cryst 60:2126–2132. [DOI] [PubMed] [Google Scholar]
- 42. Afonine PV, Grosse‐Kunstleve RW, Echols N, Headd JJ, Moriarty NW, Mustyakimov M, Terwilliger TC, Urzhumtsev A, Zwart PH, Adams PD (2012) Towards automated crystallographic structure refinement with phenix.refine. Acta Cryst 68:352–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen VB, Arendall WB, 3rd , Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC (2010) MolProbity: all‐atom structure validation for macromolecular crystallography. Acta Cryst 66:12–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Laskowski RA, Macarthur MW, Moss DS, Thornton JM (1993) Procheck ‐ a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291. [Google Scholar]


