Abstract
Phosphorylase kinase (PhK), a 1.3 MDa regulatory enzyme complex in the glycogenolysis cascade, has four copies each of four subunits, (αβγδ)4, and 325 kDa of unique sequence (the mass of an αβγδ protomer). The α, β and δ subunits are regulatory, and contain allosteric activation sites that stimulate the activity of the catalytic γ subunit in response to diverse signaling molecules. Due to its size and complexity, no high resolution structures have been solved for the intact complex or its regulatory α and β subunits. Of PhK's four subunits, the least is known about the structure and function of its largest subunit, α. Here, we have modeled the full‐length α subunit, compared that structure against previously predicted domains within this subunit, and performed hydrogen‐deuterium exchange on the intact subunit within the PhK complex. Our modeling results show α to comprise two major domains: an N‐terminal glycoside hydrolase domain and a large C‐terminal importin α/β‐like domain. This structure is similar to our previously published model for the homologous β subunit, although clear structural differences are present. The overall highly helical structure with several intervening hinge regions is consistent with our hydrogen‐deuterium exchange results obtained for this subunit as part of the (αβγδ)4 PhK complex. Several low exchanging regions predicted to lack ordered secondary structure are consistent with inter‐subunit contact sites for α in the quaternary structure of PhK; of particular interest is a low‐exchanging region in the C‐terminus of α that is known to bind the regulatory domain of the catalytic γ subunit.
Keywords: hydrogen‐deuterium exchange, mass spectrometry, calmodulin, oligomeric proteins, phosphorylase kinase, glycoside hydrolase, molecular modeling
Abbreviations
- PhK
phosphorylase kinase
- BKNR
β‐karyopherin nuclear transport receptor family
- CaM
calmodulin
- CBL
calcineurin B‐like
- GH
glycoside hydrolase
- GHL
glycoside hydrolase‐like
- HDX
hydrogen‐deuterium exchange
- IBL
importin α/β‐like
- MS
mass spectrometer, ‐try, ‐tric
- PDB
Protein Data Bank
- TFA
trifluoroacetic acid
Introduction
In the cascade activation of glycogen utilization leading to energy production in mammalian skeletal muscle, phosphorylase kinase (PhK*) phosphorylates and activates glycogen phosphorylase, which then degrades glycogen.1 The (αβγδ)4 PhK complex is among the largest and most complex enzymes known, with 90% of its 1.3 MDa mass involved in its regulation, namely the integration of activating signals. The activity of PhK, catalyzed by its γ subunit, is markedly enhanced by various metabolic, hormonal and neural stimuli, which PhK integrates through allosteric sites on its three regulatory subunits, α, β and δ. The δ subunit is an endogenous molecule of tight‐binding calmodulin (CaM),2 which accounts for PhK's activation by Ca2+ ions. The enzyme is also activated by phosphorylation of its α and β subunits,3, 4 although the β subunit predominates in this form of activation, because it is the first subunit to be phosphorylated and its phosphorylation parallels activation of γ.4 Much less is known about the structure and function of α, the largest of PhK's subunits. Apart from its secondary role in activation by phosphorylation,4 the α subunit has been shown to be part of a Ca2+‐dependent subunit communication network in PhK that also involves the γ and δ subunits.5
Crystal structures for the δ subunit (CaM) and the catalytic domain of γ are available,6, 7 but not for the large α (138.4 kDa) or β (125.2 kDa) subunits. These last two subunits are homologous, and are likely products of an early gene duplication event.8 From sequence analysis, Pallen9 first reported that both the α and β subunits possess N‐terminal glycoside hydrolase‐like (GHL) domains. These domains were later modeled using threading and additional informatics approaches,10 using as templates related glycoside hydrolase‐15 (GH‐15) family members, which are predominately starch degrading enzymes that contain (α/α)6 barrel catalytic domains.11 The actual presence of such a domain in PhK was later indicated by its tight binding of the glucoamylase inhibitor acarbose, which perturbs PhK's structure and stimulates its kinase activity.12 In addition to the N‐terminal GHL domains in α and β, Callebaut and coworkers13 predicted that both subunits contain two C‐terminal calcineurin B‐like (CBL) domains, suggesting a similar overall domain structure for both regulatory subunits; however, structural predictions for remaining domains in these proteins remained elusive. Nadeau et al.14 first predicted a structure for the full‐length β subunit using an integrated structural approach, combining multiple threading, chemical crosslinking, mass spectrometry (MS), CD spectroscopy, cryoelectron microscopy and bioinformatics to analyze the structure of this subunit and localize it in the quaternary structure of the PhK complex. In that report, multiple threading of the complete subunit indicated a two‐domain structure, containing an N‐terminal GHL domain and a C‐terminal HEAT‐repeat, protein phosphatase 2A subunit PR65/A‐like domain. Here we report the first atomic model of the full‐length α subunit and examine its structure in the PhK complex using hydrogen‐deuterium exchange (HDX), coupled with high‐resolution MS.
In HDX, amide backbone exchange of protons with deuterons provides information on a protein's secondary and tertiary structure, as well as dynamics, by measuring the number of hydrogens that exchange for deuterons on the amide backbone of the protein when incubated in D2O buffer. Exchange of these hydrogens depends on solvent exposure, dynamics, and secondary structure, as hydrogen‐bonds and burial of residues inhibit exchange.15, 16, 17 We have measured HDX for the α subunit as part of the non‐activated (αβγδ)4 PhK complex to gain insight into its structure. For organizational convenience, we will discuss our HDX results in the context of previously predicted structural domains within regions of the α subunit,13 as well as a full‐length structural model presented in this current work. It must be remembered, however, that HDX results cannot corroborate any particular predicted model. At best, they could be consistent with it, or conversely, inconsistent with it. This caveat is particularly pertinent for a highly helical structure, which seems to be the case for α and the entire PhK complex.14 An added caveat is that our HDX results on α pertain to it as part of a hexadecameric complex, not as an isolated protein. PhK's extensive quaternary structure would likely distort the structure of any free‐standing isolated subunit, regardless of whether that structure was generated from modeling or from crystallography. Moreover, the quaternary structure will definitely shield intersubunit contact areas from exchange. In fact, mapping of regions protected from exchange is one of the major reasons for performing HDX on a complex, as it identifies potential regions of subunit interactions, such as in the current study in which a putative interaction site for γ is identified on the α subunit.
Results and Discussion
Full‐length model of the α subunit
Because a structure for the full‐length α subunit was not available, we generated a 3‐D model using the I‐TASSER hierarchical structural modeling approach.18 Multiple threading alignments of the α sequence were carried out to identify template structures from the Protein Data Bank (PDB) library,19 followed by structural assembly and refinement steps, with subsequent reconstruction of atomic models.20 In the best fit structure (Fig. 1), two major domains comprising residues 1–436 (GHL) and 437–1237 (IBL: importin α/β‐like) were predicted and modeled, using as templates members of the GH‐15 family (blue‐grey trace) and IBL nuclear transport receptors of the β‐karyopherin (BKNR) family of proteins (yellow trace).21, 22 Although the thread content and coverage for the GHL domain in our model are in agreement with previous results,10 our threading templates differ significantly for the remainder of the subunit. The tertiary structure calculated for the IBL domain in our model is, however, consistent with the overall domain structure previously predicted for this region of the subunit.13 Thus, our model will be directly compared to that of Carrière et al.13 using the regions of α that were initially delineated in their model (Table 1). This not only allows a ready comparison of the two predicted structures, but a convenient organizational tool for discussing the HDX results, rather than simply dividing the α subunit into arbitrary segments.
Figure 1.
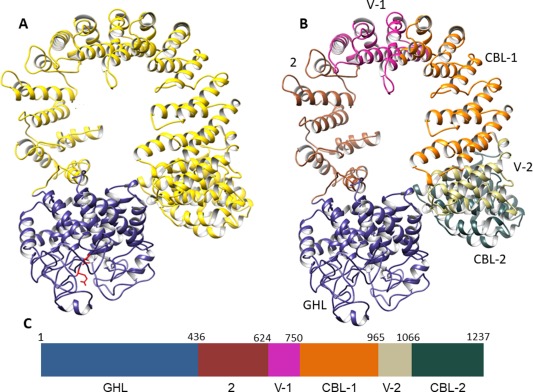
Theoretical 3‐D structure of the PhK α subunit. (A) Hierarchical protein structural modeling of the α subunit carried out using I‐TASSER.20 X‐ray crystal structures of glucodextranase (PDB ID: 1ULV) from Anthrobacter globiformis bound to acarbose (not shown) and importin β (IBL)(PDB ID: 4C0O) from human were used to thread, respectively, residues 1–436 (blue‐gray ribbon trace) and 437–1237 (gold trace) of the multi‐domain α sequence. Putative catalytic Glu residues 185 and 371 are shown in red. (B) Subdomains of the large IBL domain labeled and color coded to match the schematic of the linear structure of α shown in (C). Further modeling details of the GHL domain and IBL subdomains are listed in Table I.
Table 1.
GHL Domain and IBL Subdomains of the α Subunit as Described Herein and Shown in Figure 2
| Template/threading Structures | ||||
|---|---|---|---|---|
| Domains | Subdomains | Residues | Previous Modelinga | Our Model |
| GHL | 1–436 | Glycoside hydrolase | Glycoside hydrolase | |
| IBL | 2 | 437–624 | No previous model | BKNR |
| IBL | Variable‐1 | 625–750 | No previous model | BKNR |
| IBL | CBL‐1 | 751–965 | Calcineurin B‐like proteins | BKNR |
| IBL | Variable‐2 | 966–1066 | No previous model | BKNR |
| IBL | CBL‐2 | 1067–1237 | Calcineurin B‐like proteins | BKNR |
Previous modeling templates from Carrière et al.13 are listed for comparison to our modeling templates.
Structural comparisons of the homologous α and β subunits
In a previous report we modeled the full‐length β subunit (1092 residues) and demonstrated that two major domains of that subunit were good topographical fits with GH and HEAT‐repeat proteins from the PDB.14 Our current results for the homologous α subunit demonstrate, however, that the extent of GH thread coverage of its N‐terminus differs significantly from that of β. For example, thread coverage of the β‐GHL spanned the first 629 residues, which include the (α/α)6 GH‐15 catalytic domain and auxiliary B and C domains found in bacterial and archaeal GH‐15 members; in contrast, GH‐15 templates for α primary structure extending beyond the GH‐15 catalytic domain, ending at residue 436, produced models with low TM scores (data not shown), suggesting structural differences between α and β immediately C‐terminal to the conserved (α/α)6 subdomain. As discussed previously, residues (437–624) following the (α/α)6 domain of the α subunit are best modeled by nuclear receptors with β‐karyopherin folds as templates, and thus have a greater percentage of helical composition than their counterpart region in the β subunit.14 The highest ranking templates and their extent coverage of the large C‐terminal domains of α and β also differed, with the best templates corresponding, respectively, to the BKNR family member transportin 3 and the protein phosphatase 2A subunit PR65/A.14 The differences in the modeled structures for α and β likely reflect not only their different functions,4, 5, 23 but also regions that are unique to α, namely two large inserts that comprise both its Variable subdomains.8 These subdomains, which have been modeled successfully for the first time in this report, demonstrate the presence of a hinge region and several helix‐loop‐helix structures that were not previously observed in the full14 and partial10, 13 models of the β subunit.
HDX of nonactivated PhK
Although theoretically there are no size limitations for proteins when using HDX‐MS, large complexes, such as PhK with 325 kDa of unique sequence (the αβγδ protomer), are more challenging to study. To do so requires using a protocol that reduces potential proton back‐exchange through the use of short gradients, but without negatively affecting peptide coverage due to co‐eluting peptides from overly‐short gradients. We have successfully analyzed PhK with good HDX‐MS coverage and minimal back‐exchange (controls demonstrate <18% back exchange for the gradient used; data not shown). Of the 325 kDa of unique sequence in the αβγδ protomer, 138.4 kDa is contributed by the regulatory α subunit. Our initial peptide mapping of non‐activated PhK identified 192 unique peptides that corresponded to the α subunit, providing overlapping coverage for 88% of its sequence. To analyze the structure of this subunit in the context of the intact complex, HDX was carried out on non‐activated PhK for different time intervals up to 6 h. Peptides with either overlapping envelopes or low‐intensity peaks were then excluded, the latter because the intensity and quality of the spectrum peaks decreased further with the incorporation of deuterium. A total of 69 peptides with well‐defined, non‐overlapping envelopes were selected, resulting in accurate mass measurements of HDX for 74% of the α subunit (Supplementary Information, Fig. S1). The time‐courses of exchange for 64 of these peptides are presented in Supplementary Information, Figure S1 with the remaining 5, representing a range of exchange extents, shown in the text (Fig. 2).
Figure 2.
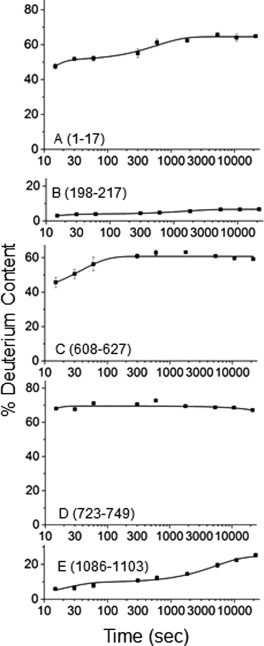
Representative HDX time‐course plots. Peptides correspond to residues 1–17 (A), 198–217 (B), 608–627 (C), 723–749 (D), and 1086–1103 (E). Error bars are present on all data points, though some are smaller than the symbols and thus not readily visible, and are the average deviation of independent exchange experiments as described under Materials and Methods. The time‐courses of exchange for all remaining peptides are shown in Supplementary Information, Figure S1.
HDX was used to examine the structural and functional regions of the α subunit, either predicted9, 10, 13 or determined experimentally.5, 8, 24, 25, 26, 27, 28, 29 Analyses of the deuterium content for peptides that overlapped specific regions in the primary structure allowed us to further resolve the extent incorporation occurring at multiple or even single amino acids at discrete sites within these regions. To simplify comparison of the amount of exchange in different regions of α, the extent of incorporation in each region was converted to percent of the total theoretical possible exchange, i.e., the number of residues in the respective peptide that can theoretically exchange (all residues in the peptide excluding the first two residues, which lose deuterium in proteolysis/MS and any prolines). The relative amount of exchange for a given region was then arbitrarily classified as low (≤ 30%), medium (31–60%) or high (> 60%). The relative exchange rates in the various regions were then compared.
GHL domain (1–436)
As discussed above, predictions of the structures of the α and β subunits suggested both subunits to have multiple domains, with the first being an N‐terminal GHL domain.9, 10 Our current multi‐domain model derived from threading is consistent with those previous reports. The GH‐15 thread coverage of α shown in Figure 1 (blue‐gray trace) displays the canonical (α/α)6‐barrel fold for the catalytic domains found in bacterial and archaeal glucoamylases and glucodextranases.30 Thirteen α‐helices are proximal to the active sites in these enzymes, with two groups of six internally and externally packed α‐helices forming the (α/α)6‐barrel.30 GH‐15 enzymes commonly contain a general acid‐base pair of catalytic Glu residues spaced approximately 200 residues apart;30 for the α subunit, but not β, Carrière et al.10 noted that two such Glu residues were appropriately situated in the active site funnel to effect catalysis. We found that the protein structurally closest to the best model for this region of α is Anthrobacter globiformus glucodextranase (GDase; PDB # 1UG9),30 with a TM score of 0.77 and a RMSD of 2.04 Å, indicating a good topographical match.31 The best of the top 10 templates for the GHL domain from the PDB was a crystal structure of glucodextranase in complex with the pseudo‐tetrasaccharide acarbose, a potent transition state inhibitor of glucoamylases.32 Correspondingly, we have found that PhK also binds acarbose, stabilizing a conformation of the complex that stimulates its kinase activity, indicating communication between the acarbose binding site(s) and the kinase catalytic site.12
As would be expected for a highly helical structure, the majority of the predicted GHL domain is protected from exchange (Table 2). At 15 s, over 80% of the domain undergoes low exchange, with only the N‐terminal residues 3–17 and a region from 362–372 exhibiting medium exchange and residue 220 high exchange (Table 2). The extent incorporation measured for residue 220 (resolved using overlapping peptides covering residues 218–242) is consistent with its location in a short loop in our model that was previously suggested by partial proteolysis.28 This predicted short loop connects two helices that correspond to helices 6 and 7 in GH‐15 structures. Helix 6 (199–218) is packed internally (Fig. 3), and undergoes very low exchange, even after 6 h (Table 2, residues 200–217). Helix 7 (224–241) is external in the GHL (α/α)6 barrel fold (Fig. 3), and correspondingly, shows higher exchange than the internally located helix 6: residues 221–233 and 235–242 reach medium levels of exchange at later time points, and residue 234 exhibits high exchange within 10 min (Table 2). These data are consistent with helix 7 being partially melted in the PhK complex. These exchange levels parallel results from a recent report showing that thermal denaturation of the GH‐15 catalytic domain from A. awamori undergoes a predicted pattern of unfolding, in which helices 6 and 7 are among the last and first, respectively, of the 12 barrel helices to unfold.33
Table 2.
Extent Deuterium Incorporation for the α‐subunit
| Data Consolidated Regionsa | 15 sb (%D) | 10 minb (%D) | 90 minb (%D) | 6 hb (%D) | ||||
|---|---|---|---|---|---|---|---|---|
| 3–17 | 48 | Medium | 61 | High | 66 | High | 65 | High |
| 26–43 | 19 | Low | 28 | Low | 33 | Medium | 36 | Medium |
| 44–49 | 2 | Low | 0 | Low | 5 | Low | 5 | Low |
| 50–52 | 0 | Low | 0 | Low | 0 | Low | 0 | Low |
| 55–60 | 2 | Low | 2 | Low | 2 | Low | 2 | Low |
| 61–81 | 9 | Low | 11 | Low | 16 | Low | 17 | Low |
| 82–91 | 0 | Low | 0 | Low | 0 | Low | 0 | Low |
| 94–112 | 11 | Low | 19 | Low | 26 | Low | 31 | Medium |
| 113–122 | 0 | Low | 7 | Low | 23 | Low | 24 | Low |
| 125–135 | 24 | Low | 39 | Medium | 42 | Medium | 45 | Medium |
| 136–144 | 0 | Low | 0 | Low | 4 | Low | 18 | Low |
| 173–181 | 1.5 | Low | – | Lowc | 8 | Low | 11 | Low |
| 184–199 | 12 | Low | 42 | Medium | 51 | Medium | 52 | Medium |
| 200–217 | 3 | Low | 5 | Low | 7 | Low | 7 | Low |
| 220 | 100 | High | 100 | High | 100 | High | – | Highc |
| 221–233 | 15 | Low | 20 | Low | 48 | Medium | – | Mediumc |
| 234 | 40 | Medium | 80 | High | 90 | High | 100 | High |
| 235–242 | 10 | Low | 15 | Low | 23 | Low | 45 | Medium |
| 245–259 | 17 | Low | 32 | Medium | 40 | Medium | 40 | Medium |
| 262–264 | 0 | Low | 0 | Low | 0 | Low | 0 | Low |
| 265–275 | 23 | Low | 29 | Low | 36 | Medium | 77 | High |
| 278–294 | 3 | Low | 6 | Low | 7 | Low | 11 | Low |
| 295–296 | 5 | Low | 10 | Low | 20 | Low | 15 | Low |
| 299–311 | 13 | Low | 30 | Low | 38 | Medium | 41 | Medium |
| 312–325 | 9 | Low | 43 | Medium | 51 | Medium | 58 | Medium |
| 362–372 | 35 | Medium | 39 | Medium | 37 | Medium | 37 | Medium |
| 375–384 | 13 | Low | 18 | Low | 29 | Low | 42 | Medium |
| 385 | 0 | Low | 20 | Low | 80 | High | 90 | High |
| 386–406 | 13 | Low | 21 | Low | 32 | Medium | 37 | Medium |
| 408–412 | 0.7 | Low | 0.9 | Low | 1.4 | Low | 2 | Low |
| 415–445 | 8 | Low | 21 | Low | 30 | Low | 37 | Medium |
| 448–461 | 16 | Low | 28 | Low | 42 | Medium | 50 | Medium |
| 462–466 | 100 | High | – | Highc | 100 | High | 88 | High |
| 513–531 | 21 | Low | 36 | Medium | 42 | Medium | 52 | Medium |
| 545–571 | 12 | Low | 27 | Low | 36 | Medium | 44 | Medium |
| 573–585 | 32 | Medium | 38 | Medium | 39 | Medium | 46 | Medium |
| 586–607 | 10 | Low | 24 | Low | 32 | Medium | 39 | Medium |
| 610–614 | 30 | Low | 68 | High | 74 | High | 68 | High |
| 615–627 | 39 | Medium | 50 | Medium | 48 | Medium | 47 | Medium |
| 634–639 | 49 | Medium | 60 | Medium | 55 | Medium | 52 | Medium |
| 662–699 | 24 | Low | 28 | Low | – | 33 | Medium | |
| 702–704 | 80 | High | 80 | High | 83 | High | 77 | High |
| 705–722 | 65 | High | 64 | High | 62 | High | 60 | Medium |
| 725–732 | 71 | High | 71 | High | 68 | High | 66 | High |
| 733–749 | 55 | Medium | 61 | High | 57 | Medium | 56 | Medium |
| 750–763 | 49 | Medium | 51 | Medium | 51 | Medium | 50 | Medium |
| 773–798 | 18 | Low | 30 | Low | 38 | Medium | 45 | Medium |
| 799–825 | 4 | Low | 8 | Low | 8 | Low | 9 | Low |
| 850–876 | 14 | Low | 24 | Low | 27 | Low | 32 | Medium |
| 877–879 | 20 | Low | 67 | High | 73 | High | 70 | High |
| 930–966 | 7 | Low | 11 | Low | 15 | Low | 20 | Low |
| 1037–1078 | 41 | Medium | 47 | Medium | 47 | Medium | 46 | Medium |
| 1081–1087 | 21 | Low | 39 | Medium | 50 | Medium | 54 | Medium |
| 1088–1103 | 6 | Low | 13 | Low | 19 | Low | 26 | Low |
| 1106–1121 | 46 | Medium | 68 | High | 65 | High | 64 | High |
| 1124–1131 | 4 | Low | 6 | Low | 28 | Low | 35 | Medium |
| 1132–1143 | 1.4 | Low | 15 | Low | 16 | Low | 18 | Low |
| 1167–1176 | 1.3 | Low | 3 | Low | 6 | Low | 7 | Low |
| 1178–1189 | 19 | Low | 38 | Medium | 46 | Medium | 51 | Medium |
| 1204–1215 | 4 | Low | 7 | Low | 15 | Low | 16 | Low |
Regions showing the deuterated coverage of the entire α‐subunit, with the percent deuterium incorporated and the resulting classification of that region listed for each time point. Data consolidation () has been applied to all regions, and the number of deuterons incorporated has been converted to percentages. Overlapping peptides used for further resolution are not included. Redundant peptides (same peptide, with different charge) are also excluded; entire time courses for every peptide can be found in the supplementary material.
Four time points, out of the nine, are shown here to represent early, middle and late exchange.
If no data were available for a specific time point, the classification for the peptide that could be reasonably extrapolated from the prior and subsequent time point is listed for that point.
Figure 3.
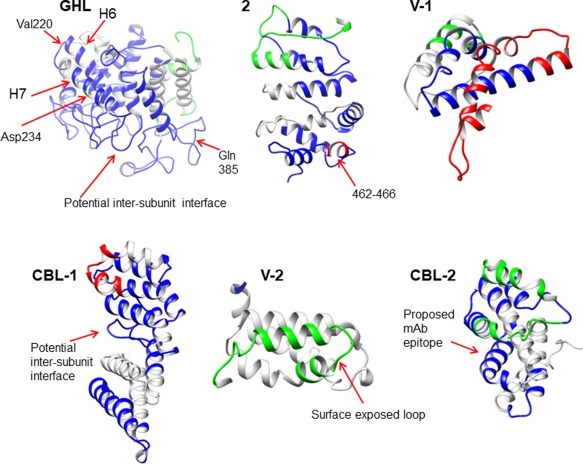
HDX after 30 s in the α subunit GHL domain and IBL subdomains. The locations of the domain/subdomains are outlined in Figure 1 and details of modeling are in Table I. Specific regions of interest discussed in the text for the domain/subdomains are labeled. The lowest deuterium levels are denoted by blue, medium levels by green, and the highest deuterium incorporation by red.
One other residue, 385, also progresses from low to high levels of exchange by 30 min; however, the residues surrounding it (375–384 and 386–400) are part of a predicted large extended loop that undergoes considerably slower and lower levels of exchange (Table 2). The slow exchange observed for this long loop structure [and several others along one face of the GHL domain (Fig. 3)], is consistent with its occurring at an inter‐subunit interface in the PhK complex, which would be consistent with crosslinking12, 34, 35 and other known interactions of the α subunit within the complex.24
Even after 5 min, greater than 80% of the predicted GHL domain undergoes only low or medium levels of exchange (Supplementary Information, Fig. S2), again consistent with a largely helical structure. One of the medium exchanging regions, 362–372 (Table 2), rapidly reaches that extent, followed by regions 125–135, 245–259 and 299–325, with all plateauing at medium levels of exchange. Peptides covering the majority of the regions 26–43, 265–275 and 375–445 also slowly reach medium levels of exchange. These measurements are consistent with a simulation demonstrating that the catalytic domain of a related GH‐15 member glucoamylase unexpectedly underwent thermal denaturation at moderate temperatures, suggesting that regions of the (α/α)6 barrel may be moderately dynamic,33 which could apply to both glycoside hydrolase (GH) monomers and the GHL domain of α within the PhK complex. All other regions of the α‐GHL failed to reach more than low levels of exchange, even after 6 h (Fig. 4).
Figure 4.
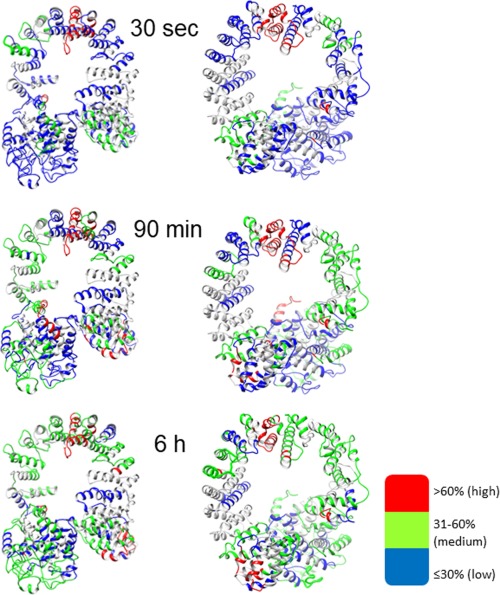
The deuterium level of the α subunit from non‐activated PhK mapped onto the model of α at three different time points. The left row shows the front orientation and the right row the back, with the lowest deuterium levels in blue, medium levels in green, and the highest deuterium incorporation in red, as indicated by the inset.
IBL domain (437–1237)
The predicted large IBL structure for the C‐terminal 65% of the α model was a good topographical match (TM score = 0.922; RMSD = 2.01Å) with an importin‐β complex in the protein data bank (PDB 3EA5),36 and its predominately helical HEAT repeat structure within the β‐karyopherin fold corresponded well with the high helical content detemined previously for the α subunit by CD spectroscopy.14 In addition to the structural similarities between the α model and importin‐β, both proteins share additional functional and structural features, including self‐association,22, 24 alternative splicing in different tissues to form isoforms with large internal deletions,21, 25 and multiple protein interaction sites.1, 5, 11, 24, 26, 34 The IBL domain has several subdomains with varying secondary structure that appear to overlap with multiple domains predicted for this region of α by Carrière et al.13
To facilitate comparisons, throughout the remainder of this paper, our predicted large IBL structure for α will be discussed in terms of the smaller, individual domains (termed subdomains herein) previously predicted to lie within it,13 which are denoted in Table 1. Thus, we begin working toward the C‐terminus of the IBL structure by first examining the sequence from 437–624, which was referred to by Carrière et al.13 as Domain B; but for consistency in terminology within our discussion, we refer to as subdomain 2.
Subdomain 2 (437–624)
Our model predicts the region of the IBL corresponding to subdomain 2 to be primarily an anti‐parallel α‐helical structure, which is consistent with the low H/D exchange rates generally observed at short times for this region (Fig. 3, Table 2). In fact, the majority of the domain undergoes low levels of exchange until later time points, and still has only medium levels of exchange by 6 h (Table 2), indicating that it is relatively well protected. Only three exceptions to the observed low levels of exchange were found, residues 462–466 and 610–614 with high exchange, and 615–627 with medium (Table 2). These peptides overlap with two loop structures in our predicted model (458–463 and 607–622).
Variable subdomain 1 (625–750)
At the beginning of Variable subdomain 1 (Fig. 3), a small loop (residues 627–628) is predicted to connect two small 1‐ and 5‐turn helices. Peptides adjacent to this predicted loop rapidly undergo medium levels of exchange (Table 2), in agreement with minimal levels of secondary structure. The α subunit of PhK is mostly homologous with β, but has several regions that are unique, one of them being 676–766.8 Residues 654–712 from within this region are deleted in the splice variant isoform α′ found in cardiac PhK,25 and as a whole this α‐unique region has been reported to be highly variable among different species.13 We observed high or medium levels of exchange for the majority of a large predicted stretch (residues 702–763, Table 2) of Variable subdomain 1 merging into CBL‐1, consistent with the model's large loop structures connecting several small helices. Similarly, Variable subdomain 1 was hypothesized to be a hinge region by Carrière et al.13 Our model, with its highly ordered helical structure of subdomain 2 contrasting significantly with the more random helix‐(large loop)‐helix structure of Variable subdomain 1, agrees well with the previous prediction13 that these two regions have distinctly different structures. Certainly, the generally low level of deuterium incorporation observed for subdomain 2 compared to the high levels for Variable subdomain 1 indicates these two subdomains are structurally distinct.
Regarding residues 654–712, which are deleted in the α′ splice variant, our model predicts that they are predominantly helical. For example, the stretch from 662–699 is largely helical, and correspondingly only slowly deuterated, exhibiting low levels of exchange even after 3 h (Supplementary Information, Fig. S1). In contrast, the remainder (702–712) of the alternatively spliced region is contained within peptides that undergo rapid and high exchange by 15 s (Table 2); our model predicts these residues to occupy the last two turns of a helix and extend into a long loop. The remainder of Variable subdomain 1 (713–750) also exhibits high or high medium levels of exchange, in agreement with its predicted structure of a series of small helices connected by large loops.
Based on its high content of hydrophilic amino acids, the region from 676–766 was hypothesized to be predominantly on the surface of the PhK complex, where it could participate in interactions of the kinase with its environment.8 This hypothesis of surface exposure is supported by our high exchange data [e.g., peptide 723–749, Fig. 1(D)], as well as partial proteolysis of the complex,28 which exposed a loop (699–748) overlapping this region. This loop was the most rapidly proteolyzed region of α in the PhK complex, and was targeted by a variety of proteases having different specificities.
CBL‐1 subomain (751–965)
The first 13 residues of this region, proximal to the hinge region of Variable subdomain 1, rapidly exchange (15 s) at a high medium level (Table 2), and in our model compose the initial turns of two helices and a small connecting loop between them. The CBL‐1 subdomain was predicted by Carrière et al.13 to contain two EF‐hand motifs, which are helix‐loop‐helix structures. Although our model does not predict a calcineurin‐like structure for this region of α, it does predict a high overall helical content (Fig. 3), with the exception of a relatively large loop (851–869). This predicted loop, located on the inner face of the molecule, showed low levels of exchange even after 90 min (Table 2), which is consistent with its being at an interfacial contact site between subunits (Fig. 3). It might be noted that this loop is proximal to a leucine zipper (833–854) that was previously implicated in the self‐association of α subunits.24 In fact, zero‐length crosslinking of PhK by transglutaminase results in the formation of an α–α dimer,34 although the specific identities of the crosslinked residues have yet to be determined.
Variable subdomain 2 (966–1066)
This 101‐residue domain is highly helical in our IBL threading model, with distorted helix‐loop structures occurring at its predicted N‐ and C‐termini (Fig. 3). All but 3 residues at the termini of Variable subdomain 2 constitute a region (967–1064) that is unique to α (i.e., not present in β) and which contains the regulatory phosphorylatable region (970–1030).37 Adjacent to the N‐terminus of this domain, residue 967 has been identified as part of an exposed loop by partial proteolysis,28 which is consistent with our model. The C‐terminus of this domain, residues 1035–1066, occurs within a region (1037–1078) that rapidly (15 s) undergoes exchange to a medium level, which is maintained through 6 h (Table 2); our model predicts this stretch to be a loop‐short helix‐loop structure. The previous identification of an exposed loop end by partial proteolysis at residue 104128 occurs at a distorted helix‐loop (1037–1041) in the model (Fig. 3).
CBL‐2 subdomain (1067–1237)
This second of the two CBL subdomains predicted by Carrière et al.13 represents the remainder of the α subunit and contains three known functional regions: 1) a region that binds to the γ subunit's apparent master allosteric activation switch for the PhK complex;23 2) a site that binds exogenous CaM;26 and 3) an epitope for a monoclonal antibody against the α subunit.29 The first of these regions, the α‐γ interaction site, which was determined by 2‐hybrid screening and near‐zero‐length chemical crosslinking with formaldehyde,5 would likely be protected against exchange by PhK's quaternary structure. The last two sites, which are targeted by exogenous CaM and the anti‐α antibody, are by definition surface accessible, and therefore susceptible to exchange at levels dictated by the secondary structure of each binding site. These inferences were applied to our model and used in conjunction with the HDX data to evaluate and refine our predictions regarding the location and structure of the three functional regions within the CBL‐2 subdomain.
The γ binding site, known to lie somewhere between residues 1060 and 1237,5 occurs within a predicted structure [ellipsed and colored cyan in Fig. 5(A)] that projects perpendicularly from the plane bisecting the large toroid formed by the remainder of the subunit. This cyan structure comprises the last seven residues of the Variable 2 and entire CBL‐2 subdomains of the IBL domain, the remainder of which is colored yellow, as in Figure 2(A). A blow‐up of the ellipsed cyan region, but now with color‐coded HDX levels depicted, reveals multiple exchange surfaces that are oriented primarily along either the inner [Fig. 5(C)] or outer [Fig. 5(B)] faces of the toroid. Several low exchanging regions (1088–1103 and 1124–1143; Table 2) along the inner face with both helical (1135–1143) and loop [1131–1134; blue arrow in Fig. 5(C)] structures, are consistent with the presence of an inter‐subunit contact site, and agree with our previous mapping of the γ interaction site to CBL‐2.5
Figure 5.
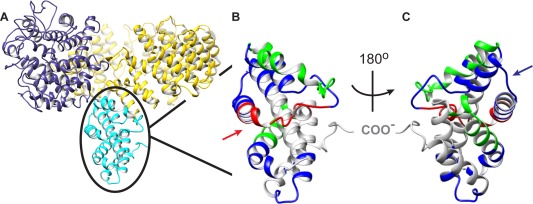
The proposed γ subunit binding site of the regulatory α subunit. (A) The atomic model of the α subunit shown in Figure 1(A) is rotated 90°, placing the toroid perpendicular to the plane of the page. The GHL and IBL domains are shown in the same color scheme as in Figure 1(A) (blue and yellow, respectively), with the C‐terminal 178 residues (1060–1237, now in cyan instead of yellow) of the latter domain projecting from the toroid. Opposite sides of this projection, enclosed by an ellipse, are shown blown up with their 90‐min HDX heat maps in Panels B and C. The low exchanging surface of Panel C (blue arrow) reveals the putative binding area for the γ subunit, whereas the opposite high exchanging surface (Panel B, red arrow) overlaps the known binding site for exogenous calmodulin.
Medium exchange at later time points was also observed in CBL‐2 for residues within a predicted primarily helical structure along the outer face [1067–1087, Table 2, Fig. 5(B)], and high exchange was observed for a peptide containing residues 1106–1121 [Table 2, Fig. 5(B), red arrow] on this same outer face, suggesting exposure of this region in the complex. This exposed region overlaps with a CaM‐crosslinked α peptide (1080–1114) that was isolated from a digest of PhK after chemical crosslinking in the presence of exogenous Ca2+/CaM.26 That the region between 1088 and 1103 only slowly exchanges over a 6‐h period (Table 2) argues that the residue crosslinked to CaM lies within one of two stretches, 1080–1085 or 1104–1114.
Previously, the epitope for a monoclonal antibody against the α subunit was mapped to within the last 106 residues (1132–1237)29 of CBL‐2. HDX results identified four peptides covering three regions (Table 2) within this broad epitope, the majority of which undergo only low exchange. Residues 1178–1189, however, undergo medium levels of exchange by 5 min, and are proximal to a previously identified exposed loop (1156–1170),28 suggesting that the actual epitope is centrally localized within the 1132–1237 stretch.
Conclusions
A model of the full‐length α subunit of PhK was generated that predicted it to be a two‐domain structure comprising GHL and β‐karyopherin folds. Previous predictions for the structures of individual regions of α13 correspond reasonably well with the distinct subdomains observed in the two major threading domains of our model, as do known structural and functional sites on the subunit. The HDX results for the α subunit as a component of the (αβγδ)4 complex provide useful reference information for predicting potential intersubunit contact sites. For example, considering the modeled structure in light of the HDX results provides for the first time a structural rationale for previous chemical crosslinking and 2‐hybrid screening results involving the α and γ subunits,5 localizing an intramolecular γ‐binding site to the CBL‐2 subdomain of α. This same subdomain has additional potential interfaces for interactions with exogenous Ca2+/CaM and a subunit specific anti‐α monoclonal antibody.26, 29
Materials and Methods
Enzymes and reagents
PhK (non‐activated) was purified from New Zealand White rabbit psoas muscle as previously described38 and stored at −80°C. Deuterium oxide, pepsin, and trifluoroacetic acid (TFA) were from Sigma‐Aldrich Corp. (St. Louis, MO), ammonium phosphate from Thermo Fisher Scientific (Pittsburgh, PA), and MS grade acetonitrile from J.T. Baker Chemical (Center Valley, PA).
Hdx
PhK stock (3.5 mg/mL) was diluted to 0.4 mg/mL in 90% D2O Hepes buffer (10 mM Hepes, 10% sucrose, 0.2 mM EDTA, pD corrected for equivalent pH of 6.8) and incubated at 30°C. At various times of incubation between 15 s and 6 h, 10‐μL aliquots were removed, and exchange was quenched by the addition of 10 μL of cold 0.12 M ammonium phosphate buffer (pH 2.0) containing 0.4 mg/mL pepsin, for a final pH of 2.3. Each sample was immediately injected onto the HPLC loop and kept at −2°C for a 3‐min digestion using a custom‐built cooling chamber to reduce back exchange,39 after which the resulting peptides were analyzed by on‐line reversed‐phase HPLC‐MS as described below.
The amount of back‐exchange in the custom‐built cooling chamber for HDX‐MS is 17%, independently of the peptide chemistry (sequence) or retention time.19 This is very similar to the amount of back exchange described with other equipment,40, 41, 42, 43 which is about 10–25% of the total theoretical labile amide linkages.
Ms
Initial peptide identification was performed as above, but in the absence of D2O. Following an equilibration of 2.5 min at 30°C, the PhK was diluted with an equal volume of the cold ammonium phosphate/pepsin solution and immediately injected onto the HPLC loop and digested as above. The resultant peptides were then loaded onto an on‐line reversed‐phase C18 column (Zorbax C8SB Wide Pore Guard Column, Micro‐Tech Scientific, 1 cm × 0.32 mm) and desalted for 4 min at a rate of 75 μL/min with solvent A (0.1% TFA). The peptides were then eluted at a flow rate of 25 μL/min with a gradient of 20–70% solvent B (0.1% TFA in acetonitrile) over 30 min. The ESI source was at 1.8 kV and the MS (LTQ FT, Thermo Fisher Scientific) was equipped with a 7T magnet. Survey MS spectra (m/z range of 400–2000) were acquired in the FT‐ICR cell with a resolution of 50,000 in profile mode, and the 6 most intense ions in each FT scan were selected for MS/MS in the linear ion trap in centroid mode using a 120‐ min exclusion list window. Peptide identification was performed by Proteome Discoverer (Version 2.0, Thermo Fisher Scientific), using the Sequest HT algorithm to search a database containing the sequences of all four PhK subunits and common contaminants. The following constraints were used for high quality control of mass spectra: peptide mass range of 500–8000; threshold of 1000; Xcorr of 1.5, 2 and 2.5 for ions with a charge of 1, 2 and 3, respectively, a delta correlation score greater than 0.08, and minimum ion count of 9. All mass spectra were examined manually to ensure that only high quality assignments were used for correct peptide identification. Peptide map protein coverage was 88% for the α subunit, 94% for β, 90% for γ and 91% for δ.
Deuterated samples were analyzed in a similar manner, with the omission of the tandem mass analysis; only ICR FT scans were used. HDX data were analyzed using HDExaminer (Sierra Analytics), and manual analyses were performed using Qual Browser (Thermo‐Finnigan). All peptides could not be analyzed in the deuterated samples, and the final coverage of the α subunit decreased from 88% in the non‐deuterated sample to 74%; however, many of these peptides were overlapping, providing good data for amino acid level resolution in many regions (see Supplementary Information, Fig. S2 for subunit coverage map). HDX kinetics were fit using OriginPro version 9.2 to a two‐parameter exponential
where is the deuterium content at time , and are the number of fast and slow exchanging hydrogens, respectively, and and are the respective rate constants. The data points for the exchange time courses of the individual peptides (Fig. 1, Supplementary Information, Fig. S1) represent a minimum of two, but up to four, individual exchange experiments.
Threading and structure modeling
The theoretical atomic model of the α subunit was constructed using I‐TASSER.18, 44 With the rabbit muscle α subunit sequence (Accession # = P18688) as query, multiple sequence‐template alignments were initially generated by the meta‐server LOMETS.19 Domain boundaries were delimited by ThreaDom,45 and the α subunit sequence was divided into two domains, based on the threading alignments and the template structures. The first, corresponding to a GHL domain, matched well with several high scoring templates, with a TM score of 0.63 and RMSD of 8.5 Å for this model. The second, corresponding to an IBL domain, had lower scoring templates, and a TM score and RMSD of 0.34 and 17.1 Å, respectively. The domains were constructed by I‐TASSER, with spatial restraints (Cα distance map and side‐chain contacts) extracted from the templates being used to guide replica‐exchange Monte Carlo assembly simulation, with sparse distance and contact restraints from short template fragments used as additional restraints to assist the simulations.18 Decoys generated during the structural assembly simulations were clustered by SPICKER,46 and the cluster centroid models were further refined by REMO to build the full‐atomic model.20 The entire α subunit model was constructed by connecting both domain models together with the domain orientation repacked based on the I‐TASSER energy potential.
Supporting information
Supporting Information
Statement: Phosphorylase kinase (PhK) is among the largest and most complex protein kinases, with a mass of 1.3 MDa comprising sixteen subunits (αβγδ)4. The least structural information is available for its largest (138.4 kDa) subunit, α. This article describes the first 3D model for the α subunit, as well as the first hydrogen‐deuterium exchange studies carried out on PhK, identifying among other things exposed regions of the α subunit within the PhK complex.
References
- 1. Brushia RJ, Walsh DA (1999) Phosphorylase kinase: the complexity of its regulation is reflected in the complexity of its structure. Front Biosci 4:D618–D641. [DOI] [PubMed] [Google Scholar]
- 2. Cohen P, Burchell A, Foulkes JG, Cohen PT, Vanaman TC, Nairn C (1978) Identification of the Ca2+‐dependent modulator protein as the fourth subunit of rabbit skeletal muscle phosphorylase kinase. FEBS Lett 92:287–293. [DOI] [PubMed] [Google Scholar]
- 3. Cohen P, Watson DC, Dixon GH (1975) The hormonal control of activity of skeletal muscle phosphorylase kinase. Amino‐acid sequences at the two sites of action of adenosine‐3':5'‐monophosphate‐dependent protein kinase. Eur J Biochem 51:79–92. [DOI] [PubMed] [Google Scholar]
- 4. Ramachandran C, Goris J, Waelkens E, Merlevede W, Walsh DA (1987) The interrelationship between cAMP‐dependent alpha and beta subunit phosphorylation in the regulation of phosphorylase kinase activity. Studies using subunit specific phosphatases. J Biol Chem 262:3210–3218. [PubMed] [Google Scholar]
- 5. Rice NA, Nadeau OW, Yang Q, Carlson GM (2002) The calmodulin‐binding domain of the catalytic gamma subunit of phosphorylase kinase interacts with its inhibitory alpha subunit: evidence for a Ca2+ sensitive network of quaternary interactions. J Biol Chem 277:14681–14687. [DOI] [PubMed] [Google Scholar]
- 6. Babu YS, Bugg CE, Cook WJ (1988) Structure of calmodulin refined at 2.2 A resolution. J Mol Biol 204:191–204. [DOI] [PubMed] [Google Scholar]
- 7. Owen DJ, Noble ME, Garman EF, Papageorgiou AC, Johnson LN (1995) Two structures of the catalytic domain of phosphorylase kinase: an active protein kinase complexed with substrate analogue and product. Structure 3:467–482. [DOI] [PubMed] [Google Scholar]
- 8. Kilimann MW, Zander NF, Kuhn CC, Crabb JW, Meyer HE, Heilmeyer LM Jr (1988) The alpha and beta subunits of phosphorylase kinase are homologous: cDNA cloning and primary structure of the beta subunit. Proc Natl Acad Sci USA 85:9381–9385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pallen MJ (2003) Glucoamylase‐like domains in the alpha‐ and beta‐subunits of phosphorylase kinase. Protein Sci 12:1804–1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Carriere C, Jonic S, Mornon JP, Callebaut I (2008) 3D mapping of glycogenosis‐causing mutations in the large regulatory alpha subunit of phosphorylase kinase. Biochim Biophys Acta 1782:664–670. [DOI] [PubMed] [Google Scholar]
- 11. Bott R, Saldajeno M, Cuevas W, Ward D, Scheffers M, Aehle W, Karkehabadi S, Sandgren M, Hansson H (2008) Three‐dimensional structure of an intact glycoside hydrolase family 15 glucoamylase from Hypocrea jecorina. Biochemistry 47:5746–5754. [DOI] [PubMed] [Google Scholar]
- 12. Nadeau OW, Liu W, Boulatnikov IG, Sage JM, Peters JL, Carlson GM (2010) The glucoamylase inhibitor acarbose is a direct activator of phosphorylase kinase. Biochemistry 49:6505–6507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Carriere C, Mornon JP, Venien‐Bryan C, Boisset N, Callebaut I (2008) Calcineurin B‐like domains in the large regulatory alpha/beta subunits of phosphorylase kinase. Proteins 71:1597–1606. [DOI] [PubMed] [Google Scholar]
- 14. Nadeau OW, Lane LA, Xu D, Sage J, Priddy TS, Artigues A, Villar MT, Yang Q, Robinson CV, Zhang Y, Carlson GM (2012) Structure and location of the regulatory beta subunits in the (alphabetagammadelta)4 phosphorylase kinase complex. J Biol Chem 287:36651–36661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Artigues A, Nadeau OW, Rimmer MA, Villar MT, Du X, Fenton AW, Carlson GM (2016) Protein structural analysis via mass spectrometry‐based proteomics. Exp Med Biol 919:397–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hoofnagle AN, Resing KA, Ahn NG (2003) Protein analysis by hydrogen exchange mass spectrometry. Annu Rev Biophys Biomol Struct 32:1–25. [DOI] [PubMed] [Google Scholar]
- 17. Truhlar SM, Croy CH, Torpey JW, Koeppe JR, Komives EA (2006) Solvent accessibility of protein surfaces by amide H/2H exchange MALDI‐TOF mass spectrometry. J Am Soc Mass Spectrom 17:1490–1497. [DOI] [PubMed] [Google Scholar]
- 18. Zhang Y (2008) I‐TASSER server for protein 3D structure prediction. BMC Bioinform 9:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wu S, Zhang Y (2007) LOMETS: a local meta‐threading‐server for protein structure prediction. Nucleic Acids Res 35:3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Y, Zhang Y (2009) REMO: A new protocol to refine full atomic protein models from C‐alpha traces by optimizing hydrogen‐bonding networks. Proteins 76:665–676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Maertens GN, Cook NJ, Wang W, Hare S, Gupta SS, Oztop I, Lee K, Pye VE, Cosnefroy O, Snijders AP, KewalRamani VN, Fassati A, Engelman A, Cherepanov P (2014) Structural basis for nuclear import of splicing factors by human Transportin 3. Proc Natl Acad Sci USA 111:2728–2733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Strom AC, Weis K (2001) Importin‐beta‐like nuclear transport receptors. Genome Biol 2:3008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Nadeau OW, Anderson DW, Yang Q, Artigues A, Paschall JE, Wyckoff GJ, McClintock JL, Carlson GM (2007) Evidence for the location of the allosteric activation switch in the multisubunit phosphorylase kinase complex from mass spectrometric identification of chemically crosslinked peptides. J Mol Biol 365:1429–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ayers NA, Wilkinson DA, Fitzgerald TJ, Carlson GM (1999) Self‐association of the alpha subunit of phosphorylase kinase as determined by two‐hybrid screening. J Biol Chem 274:35583–35590. [DOI] [PubMed] [Google Scholar]
- 25. Harmann B, Zander NF, Kilimann MW (1991) Isoform diversity of phosphorylase kinase alpha and beta subunits generated by alternative RNA splicing. J Biol Chem 266:15631–15637. [PubMed] [Google Scholar]
- 26. James P, Cohen P, Carafoli E (1991) Identification and primary structure of calmodulin binding domains in the phosphorylase kinase holoenzyme. J Biol Chem 266:7087–7091. [PubMed] [Google Scholar]
- 27. Newsholme P, Angelos KL, Walsh DA (1992) High and intermediate affinity calmodulin binding domains of the alpha and beta subunits of phosphorylase kinase and their potential role in phosphorylation‐dependent activation of the holoenzyme. J Biol Chem 267:810–818. [PubMed] [Google Scholar]
- 28. Rimmer MA, Artigues A, Nadeau OW, Villar MT, Vasquez‐Montes V, Carlson GM (2015) Mass spectrometric analysis of surface‐exposed regions in the hexadecameric phosphorylase kinase complex. Biochemistry 54:6887–6895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wilkinson DA, Marion TN, Tillman DM, Norcum MT, Hainfeld JF, Seyer JM, Carlson GM (1994) An epitope proximal to the carboxyl terminus of the alpha‐subunit is located near the lobe tips of the phosphorylase kinase hexadecamer. J Mol Biol 235:974–982. [DOI] [PubMed] [Google Scholar]
- 30. Mizuno M, Tonozuka T, Suzuki S, Uotsu‐Tomita R, Kamitori S, Nishikawa A, Sakano Y (2004) Structural insights into substrate specificity and function of glucodextranase. J Biol Chem 279:10575–10583. [DOI] [PubMed] [Google Scholar]
- 31. Xu J, Zhang Y (2010) How significant is a protein structure similarity with TM‐score = 0.5?. Bioinformatics 26:889–895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Svensson B, Sierks MR (1992) Roles of the aromatic side chains in the binding of substrates, inhibitors, and cyclomalto‐oligosaccharides to the glucoamylase from Aspergillus niger probed by perturbation difference spectroscopy, chemical modification, and mutagenesis. Carbohydr Res 227:29–44. [DOI] [PubMed] [Google Scholar]
- 33. Liu HL, Wang WC (2003) Predicted unfolding order of the 13 alpha‐helices in the catalytic domain of glucoamylase from Aspergillus awamori var. X100 by molecular dynamics simulations. Biotechnol Prog 19:1583–1590. [DOI] [PubMed] [Google Scholar]
- 34. Nadeau OW, Carlson GM (1994) Zero length conformation‐dependent cross‐linking of phosphorylase kinase subunits by transglutaminase. J Biol Chem 269:29670–29676. [PubMed] [Google Scholar]
- 35. Nadeau OW, Sacks DB, Carlson GM (1997) Differential affinity cross‐linking of phosphorylase kinase conformers by the geometric isomers of phenylenedimaleimide. J Biol Chem 272:26196–26201. [DOI] [PubMed] [Google Scholar]
- 36. Forwood JK, Lonhienne TG, Marfori M, Robin G, Meng W, Guncar G, Liu SM, Stewart M, Carroll BJ, Kobe B (2008) Kap95p binding induces the switch loops of RanGDP to adopt the GTP‐bound conformation: implications for nuclear import complex assembly dynamics. J Mol Biol 383:772–782. [DOI] [PubMed] [Google Scholar]
- 37. Zander NF, Meyer HE, Hoffmann‐Posorske E, Crabb JW, Heilmeyer LM, Jr , Kilimann MW (1988) cDNA cloning and complete primary structure of skeletal muscle phosphorylase kinase (alpha subunit). Proc Natl Acad Sci USA 85:2929–2933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. King MM, Carlson GM (1981) Synergistic activation by Ca2+ and Mg2+ as the primary cause for hysteresis in the phosphorylase kinase reactions. J Biol Chem 256:11058–11064. [PubMed] [Google Scholar]
- 39. Villar MT, Miller DE, Fenton AW, Artigues A (2010) SAIDE: A Semi‐Automated Interface for Hydrogen/Deuterium Exchange mass spectrometry. Proteomica 6:63–69. [PMC free article] [PubMed] [Google Scholar]
- 40. Smith DL, Deng Y, Zhang Z (1997) Probing the non‐covalent structure of proteins by amide hydrogen exchange and mass spectrometry. J Mass Spectrom 32:135–146. [DOI] [PubMed] [Google Scholar]
- 41. Wang L, Smith DL (2003) Downsizing improves sensitivity 100‐fold for hydrogen exchange‐mass spectrometry. Anal Biochem 314:46–53. [DOI] [PubMed] [Google Scholar]
- 42. Chalmers MJ, Busby SA, Pascal BD, He Y, Hendrickson CL, Marshall AG, Griffin PR (2006) Probing protein ligand interactions by automated hydrogen/deuterium exchange mass spectrometry. Anal Chem 78:1005–1014. [DOI] [PubMed] [Google Scholar]
- 43. Iacob RE, Bou‐Assaf GM, Makowski L, Engen JR, Berkowitz SA, Houde D (2013) Investigating monoclonal antibody aggregation using a combination of H/DX‐MS and other biophysical measurements. J Pharm Sci 102:4315–4329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Yang J, Yan R, Roy A, Xu D, Poisson J, Zhang Y (2015) The I‐TASSER Suite: protein structure and function prediction. Nat Meth 12:7–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Xue Z, Xu D, Wang Y, Zhang Y (2013) ThreaDom: extracting protein domain boundary information from multiple threading alignments. Bioinformatics 29:i247–i256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y, Skolnick J (2004) SPICKER: a clustering approach to identify near‐native protein folds. J Comput Chem 25:865–871. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information


