Abstract
Self-cleaving ribozymes were discovered 30 years ago and have been found throughout nature, from bacteria to animals, but little is known about their biological functions and regulation, particularly how cofactors and metabolites alter their activity. A hepatitis delta virus-like self-cleaving ribozyme maps upstream of a phosphoglucosamine mutase (glmM) open reading frame in the genome of the human gut bacterium Faecalibacterium prausnitzii. The presence of a ribozyme in the untranslated region of glmM suggests a regulation mechanism of gene expression. In the bacterial hexosamine biosynthesis pathway, the enzyme glmM catalyzes the isomerization of glucosamine 6-phosphate into glucosamine 1-phosphate. In this study, we investigated the effect of these metabolites on the co-transcriptional self-cleavage rate of the ribozyme. Our results suggest that glucosamine 6-phosphate, but not glucosamine 1-phosphate, is an allosteric ligand that increases the self-cleavage rate of drz-Fpra-1, providing the first known example of allosteric modulation of a self-cleaving ribozyme by the substrate of the adjacent gene product. Given that the ribozyme is activated by the glmM substrate, but not the product, this allosteric modulation may represent a potential feed-forward mechanism of gene expression regulation in bacteria.
Graphical Abstract
Ribozymes are RNA molecules that can catalyze a chemical transformation in the absence of a protein cofactor.1–3 Self-cleaving ribozymes comprise a group of RNA molecules that promote a site-specific self-scission reaction. In all known self-cleaving ribozymes, the cleavage reaction is a trans-esterification that involves a nucleophilic attack by a 2′-oxygen on the adjacent phosphodiester bond, producing a 2′–3′cyclic phosphate and a 5′-hydroxyl product.4–14 To date, nine self-cleaving ribozyme families have been discovered, comprising the hairpin,10 hammerhead,8,9 hepatitis delta virus (HDV),6,7 glucosamine-6-phosphate synthase (glmS),11 Neurospora Varkud satellite (VS),12 twister,13 twister sister (TS), pistol, and hatchet motifs.14 First characterized 30 years ago,6,7,15 the HDV self-cleaving ribozyme has been extensively studied, with elucidated crystal structures and a mechanism of self-scission. 16–21 HDV-like self-cleaving ribozymes exhibit great sequence diversity but fold into a conserved secondary structure that includes a nested double pseudoknot.22 HDV-like ribozymes have been found in many eukaryotes, including humans, as well as in Chilo iridescent virus, in several bacteria, and most recently in several microbial metagenomic data sets.14,22–27 These self-cleaving RNAs likely play a number of distinct roles in biology,22–27 but little is known about their regulation, particularly with regard to the role ligands or metabolites may have in modulating HDV-like ribozyme self-cleavage as either allosteric effectors or cofactors.
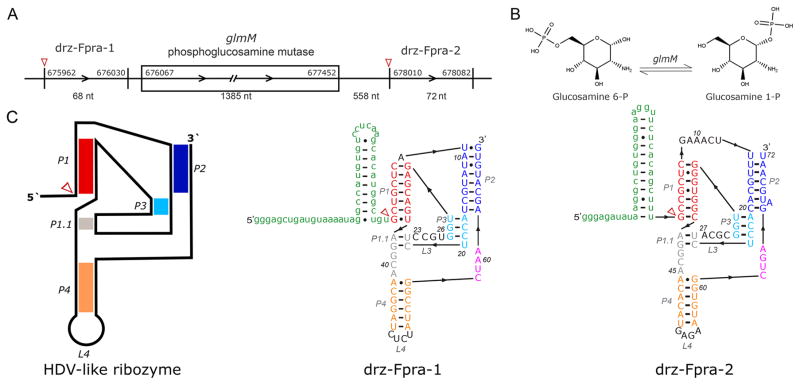
One particular case of interest is the drz-Fpra-1 HDV-like ribozyme, found in the human gut bacterium Faecalibacterium prausnitzii.22 F. prausnitzii is a Gram-positive Firmicute that represents >5% of the total bacterial population in the fecal microbiota of a healthy human and is suggested to be negatively correlated with certain pathologies, such as Crohn’s disease and ulcerative colitis.28–30 The ribozyme cleavage site maps 106 nucleotides upstream of the phosphoglucosamine mutase (glmM) open reading frame (ORF)22 (Figure 1A). The enzyme glmM is a component of the hexosamine biosynthesis pathway, catalyzing the transformation of glucosamine 6-phosphate (GlcN6P) into glucosamine 1-phosphate (GlcN1P)31 (Figure 1B). The final product of the hexosamine biosynthesis pathway is uridine diphosphate N-acetyl-glucosamine (UDP-GlcNAc), a key substrate used for cell wall biosynthesis.32 The secondary structure of the ribozyme is shown in Figure 1C.
Figure 1.
HDV-like ribozymes in F. prausnitzii. (A) Genome locus of F. prausnitzii showing the glmM ORF, the upstream drz-Fpra-1 ribozyme, and the newly discovered drz-Fpra-2 ribozyme. (B) Isomeration reaction promoted by the glmM enzyme. (C) Schematic representation of the secondary structure of HDV-like ribozymes (left) and predicted secondary structures of drz-Fpra-1 and drz-Fpra-2 ribozymes, including hairpins in the 5′-leader sequence. Red arrowheads mark the cleavage sites.
In bacteria, a common mode of regulation of gene expression involves sensing a metabolite related to the adjacent gene product through an RNA regulatory element called a riboswitch.33 In the case of glmS, a GlcN6P-sensing riboswitch,34 the RNA is also a self-cleaving ribozyme that resides at the 5′-untranslated region (UTR) of the transcript encoding glutamine fructose-6-phosphate amidotransferase, which catalyzes the transformation of fructose 6-phosphate into GlcN6P.11 The metabolite GlcN6P acts as a cofactor of the glmS ribozyme, accelerating its self-scission nearly 1 million-fold11,35–39 and promoting the degradation of the adjacent ORF through an RNase J-dependent mechanism.40 Thus, this feedback system senses the amount of GlcN6P in the cell and represses gene expression through the ribozyme-dependent activity. This metabolite-responsive regulation system has been well-characterized, by structural, biochemical, and mechanistic studies.11,35–39,41–44 Other metabolites, such as glucose 6-phosphate (Glc6P), may compete with GlcN6P and thus upregulate gene expression by inhibiting the self-cleavage of the RNA.39,41 Surprisingly, only three mutations in the active site are necessary to convert the ribozyme into a coenzyme-independent self-cleaving ribozyme, revealed by an in vitro selection of a GlcN6P-insensitive glmS ribozyme that used a divalent cation for catalysis without changing the overall fold of the ribozyme.42
Because of the proximity of the HDV-like ribozyme and the glmM gene, we hypothesized that the drz-Fpra-1 ribozyme and the metabolites involved in the hexosamine biosynthesis pathway may contribute to gene expression regulation through modulation of the ribozyme activity. During the course of this work, we found another HDV-like ribozyme, named drz-Fpra-2 (Figure 1C), downstream of the glmM gene, and we chose to study the two ribozymes in parallel. To explore the mechanism of this putative regulation, we studied the in vitro ribozyme self-cleavage kinetics in the presence of several metabolites. The kinetic and structural probing of drz-Fpra-1 in the presence of the metabolites shows an allosteric modulation of the ribozyme by the substrate, but not the product, of the adjacent gene product. The effect on drz-Fpra-2 was largely the opposite.
MATERIALS AND METHODS
In Vitro RNA Transcription
RNA was transcribed at 37 °C for 1 h in a 20 μL volume containing 10 mM dithiothreitol (DTT), 2 mM spermidine, rNTPs (1.25 mM each), 7.5 mM MgCl2, 1 unit of T7 RNA polymerase, and 0.5 pmol of DNA template. The transcripts were purified by 10% polyacrylamide gel electrophoresis (PAGE) under denaturing conditions (7 M urea). RNA was eluted from the gel into 300 μL of 300 mM KCl and precipitated by adding 700 μL of 100% ethanol at −20 °C.
In Vitro Co-Transcriptional Cleavage Kinetics
In vitro transcription was performed in a manner similar to that used for the RNA transcription assay described above with the following modifications: 4.5 mM MgCl2; GTP, UTP, and CTP (1.25 mM each); 250 μM ATP; 4.5 μCi of [α-32P]ATP (PerkinElmer); and 40 mM HEPES (pH 7.4). A 10 μL transcription reaction was initiated by the addition of DNA and the mixture incubated at 24 °C for 10 min. A 1.0 μL aliquot of the reaction mixture was withdrawn, and its transcription and self-scission were terminated by the addition of urea loading buffer. The remaining 4.0 μL volume was diluted 25-fold (final volume of 100 μL) into a physiological-like buffer [50 mM HEPES (pH 7.4), 10 mM NaCl, 140 mM KCl, and the desired concentrations of MgCl2 and the metabolite]. Control experiments showed that this dilution efficiently prevented any new RNA synthesis; therefore, our kinetic analysis did not need to account for the kinetics of transcription, contrasting with the previously described analysis of co-transcriptional cleavage by Long and Uhlenbeck.45 For conditions requiring a consistent ionic strength, the buffer and metabolite stocks were pH-adjusted by the addition of KOH and the contribution of K+ and Na+ from the metabolite stocks was tracked. The concentration of K+ was adjusted by the addition of KCl for a final reaction concentration of 140 mM. Aliquots (5 μL) were collected at the indicated times following the dilution of the transcription reaction mixture into the physiological-like buffer at 37 °C, and self-scission was terminated by adding 5 μL of stop buffer containing 20 mM EDTA, 5 mM Tris (pH 7.4), and 8 M urea, with xylene cyanol and bromophenol blue loading dyes. The denaturing PAGE gel of cleavage products was exposed to phosphorimage screens and analyzed using a Typhoon phosphorimager and ImageQuant software (GE Healthcare). The band intensities were analyzed by creating line profiles of each lane using ImageQuant, exporting the data to Microsoft Excel. Self-cleavage data were fit to a monoexponential decay function (eq 1)
| (1) |
where A and C represent the relative fractions of the ribozyme population cleaving with a rate constant k and remaining uncleaved, respectively. The model was fit to the data using a linear least-squares analysis and the Solver module of Microsoft Excel.
Labeling of the 3′-Terminus of RNA
RNA was transcribed in vitro and purified via PAGE. The appropriate RNA species was excised, precipitated, and resuspended in water. RNA was then ligated at 37 °C for 3 h in a volume of 10 μL, in RNA ligase buffer containing 50 mM Tris-HCl, 10 mM MgCl2, 1 mM DTT (pH 7.5) (NEB), 2 μCi of [5′-32P]cytidine 3′,5′-bisphosphate (PerkinElmer), and 1 unit of T4 RNA ligase (NEB), and again purified via PAGE.
In-Line Probing
The 3′-end-labeled RNA was incubated with varying amounts of ligand for up to 2 days at 37 °C in a buffer containing 140 mM KCl, 10 mM NaCl, 20 mM Tris chloride (pH 8.5), 1 mM MgCl2, and 1 mM spermidine, on the basis of the in-line probing technique of Soukup and Breaker.46 The partially hydrolyzed RNAs were resolved using denaturing PAGE, exposed to phosphorimage screens (Molecular Dynamics/GE Healthcare), and scanned with a GE Typhoon phosphorimager. The sequences in the degradation pattern were assigned by running α-phosphorothioate nucleotide-modified RNA cleaved by treatment with iodoethanol in parallel lanes.47,48 The band intensities were analyzed by creating line profiles of each lane using ImageQuant and exporting the data to Microsoft Excel. The areas of the fitted curves were used to measure changes in intensity related to the binding, divided by intensities of a control band. The resulting ratios were plotted in Excel as a function of ligand concentration and modeled with a dissociation constant equation for a single ligand:
| (2) |
The model was fit to the data using a linear least-squares analysis and the Solver module of Microsoft Excel.
Metabolites
All the metabolites used in this study were purchased from Sigma-Aldrich. To measure the concentration of free phosphate, which may affect the ribozyme kinetics by forming an insoluble complex with the Mg2+ ions, we used a fluorescent phosphate sensor based on the bacterial phosphate-sensing protein (Thermo Fisher). The molar ratios of free phosphate in GlcN1P, GlcN6P, and GlcP were found to be 0.0001, 0.002, and 0.0004, respectively.
RESULTS
Discovery of a Second HDV-like Ribozyme in the Same Locus of the F. prausnitzii Genome
We decided to analyze the F. prausnitzii genome to verify the mapping of the drz-Fpra-1 ribozyme near the glmM gene. To our surprise, we found another HDV-like ribozyme, drz-Fpra-2, with a sequence highly similar to that of drz-Fpra-1 in the same locus. This second ribozyme was found 558 nucleotides downstream of the glmM gene (Figure 1A). Although this ribozyme is similar in structure and sequence, some differences near the active site were observed in the proposed secondary structures (Figure 1C), as discussed below. One unique feature found in both drz-Fpra ribozymes is an A–U base pair at the top of the P1.1 region, where a G-C base pair is usually found in other HDV-like ribozymes.49
Self-Scission Kinetics of drz-Fpra Ribozymes in the Presence of Metabolites
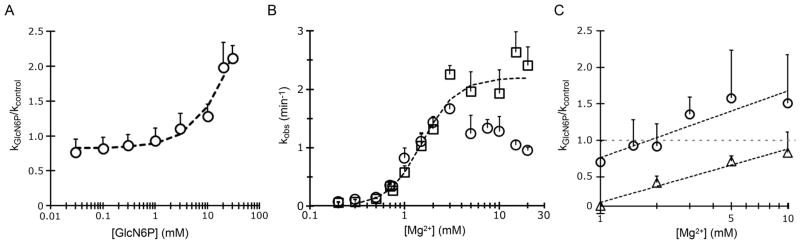
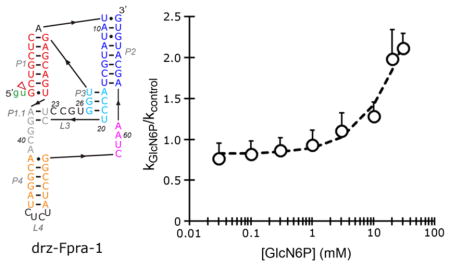
To study the effect of the metabolite on the rate of cleavage of the ribozymes, we started with an in vitro co-transcriptional cleavage kinetics performed in the presence or absence of 20 mM GlcN6P at 5 mM Mg2+. Surprisingly, the effect of the metabolite on each ribozyme was different. GlcN6P accelerates the self-scission of the drz-Fpra-1 ribozyme, whereas it decreases the rate of self-cleavage of the drz-Fpra-2 ribozyme (Figure 2). The low amplitude of the effect of the metabolite on the two ribozymes indicated that GlcN6P is an allosteric effector (modulator) and not a cofactor or coenzyme that directly participates in catalysis. To further investigate the effect of the ligand on the activity of drz-Fpra-1, we performed a titration of GlcN6P, keeping the ionic strength of the solution constant and the Mg2+ concentration fixed at 5 mM. Self-cleavage kinetics under conditions approximating those of co-transcriptional self-scission showed a dose response between the cleavage rate constant and the metabolite, increasing the kGlcN6P >2-fold at higher concentrations of GlcN6P, when compared to the no-metabolite kcontrol (Figure 3A). At low concentrations of GlcN6P, the cleavage rate was similar to that of the no-metabolite control, indicating that the metabolite may not be affecting the catalytic mechanism of the ribozyme.
Figure 2.
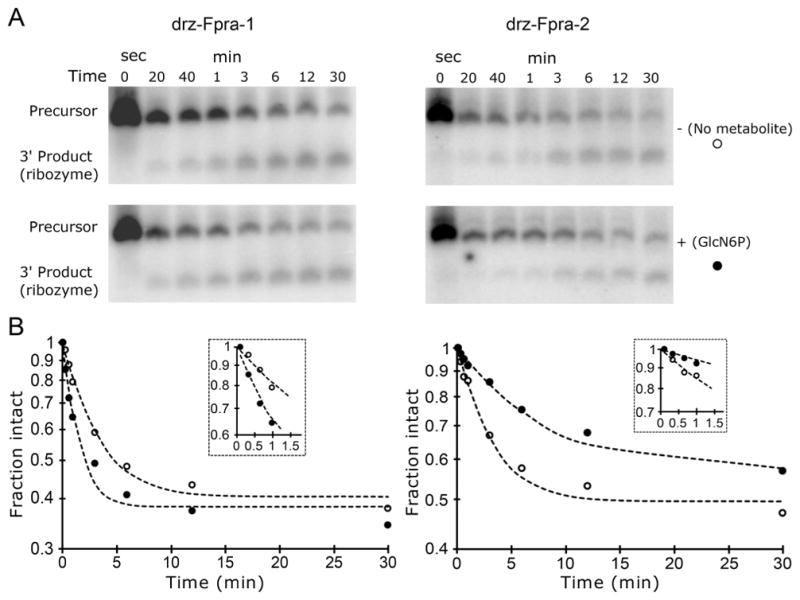
Influence of GlcN6P on the co-transcriptional self-scission of drz-Fpra-1 and drz-Fpra-2. To study the effect of the metabolite on the cleavage rate of the ribozymes, in vitro co-transcriptional cleavage kinetics were performed in presence and absence of GlcN6P. (A) PAGE analysis of the co-transcriptional self-scission of the two ribozymes in the absence (top) and presence (bottom) of 20 mM GlcN6P. (B) Log-linear graphs of ribozyme self-scission. GlcN6P accelerates the self-cleavage of drz-Fpra-1 [left; no GlcN6P (○) and 20 mM GlcN6P (●)] but inhibits that of drz-Fpra-2 (right). Early time points are shown in the insets.
Figure 3.
Effect of GlcN6P and Mg2+ on the drz-Fpra ribozymes. (A) drz-Fpra-1 self-scission dose response to GlcN6P at a constant Mg2+ concentration (5 mM), normalized to the no-metabolite control. (B) Mg2+ dependence of drz-Fpra-1 self-scission in the presence of 20 mM GlcN6P (□) or no metabolite (○). (C) Mg2+ dependence of drz-Fpra-1 (○) and drz-Fpra-2 (△) self-scission in 20 mM GlcN6P normalized to a no-metabolite control.
To investigate the origin of this dose response, we varied the concentration of Mg2+, the divalent metal ion used by the ribozyme for catalysis,17,18,50–52 in the presence of 20 mM GlcN6P. At low concentrations of Mg2+, both kGlcN6P and kcontrol behave similarly, with a strong overlap between both data sets (Figure 3B). This trend changes at approximately physiological Mg2+, where the cleavage rate increases in the presence of GlcN6P. At higher concentrations of the divalent metal ion, kcontrol decreases, possibly due to misfolding. In contrast, this effect is not seen for kGlcN6P, which continues to gradually increase. This result is consistent with a model in which the metabolite interacts with the ribozyme but does not act directly in the catalytic step of the reaction (Figure 3B). We also performed the same titration of Mg2+ in the presence of 20 mM GlcN6P for drz-Fpra-2. As expected, an opposite effect was observed, where a decrease in the cleavage rate was found under the same conditions when compared to a no-metabolite control (Figure 3C).
To investigate the specificity of the ribozyme-metabolite interaction, we decided to test compounds related to GlcN6P. The ribozyme self-cleavage kinetics were tested in the presence of the following molecules: GlcN6P, the precursor of the reaction promoted by the glmM enzyme; GlcN1P, the isomer of GlcN6P and the product of the reaction; glucose (Glc); Glc6P; and glucosamine (GlcN). The last two were chosen because of their chemical similarity to the metabolites, because each carries a different chemical group present in both metabolites of the enzymatic reaction (the amino and the phosphate groups). The kinetic rate constants were normalized to no-metabolite cleavage kinetics under the same conditions and showed that GlcN1P, the product of the reaction catalyzed by glmM, activated only the Fpra-2 ribozyme, albeit with low statistical significance, whereas Glc slightly increased the activity of both ribozymes (Figure S1). GlcN and Glc6P increased the rate of cleavage of both drz-Fpra-1 and drz-Fpra-2 ribozymes to a similar extent (Figure S1). This is an unexpected result for drz-Fpra-2 because it was not inhibited by GlcN and Glc6P, as it was in the presence of GlcN6P. These results suggest that both the amino and the phosphate groups are important in modulating the self-scission rate of the ribozymes, and their relative positioning around the sugar ring leads to the differential effect on the two ribozymes. Finally, we decided to probe the ribozymes in the presence of UDP-GlcNAc, the final product of the hexosamine biosynthesis pathway, but no significant effect was observed (Figure S2).
Another way to compare the effects of GlcN6P and GlcN1P is to calculate the ratio of the self-scission rate constants at the same concentrations of the metabolite, while varying the Mg2+ concentration. At a metabolite concentration of 20 mM, the kGlcN6P/kGlcN1P ratios for drz-Fpra-1 were 1.4 ± 0.1, 2.2 ± 0.4, and 2.5 ± 0.5 at 1, 5, and 10 mM Mg2+, respectively. Interestingly, when both GlcN1P and GlcN6P were included in the drz-Fpra-1 reaction mixture, the effect of GlcN6P was enhanced (Figure S3).
The Structure of the drz-Fpra-1 Ribozyme Is Stabilized by GlcN6P
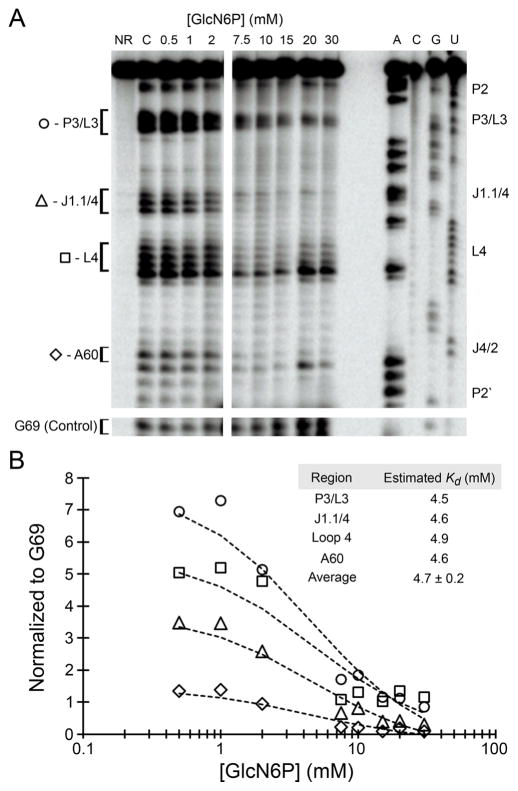
To investigate the effect of the metabolite on the ribozyme structure, we performed an in-line probing experiment in the presence of different concentrations of GlcN6P. This technique profiles the natural degradation of an RNA molecule under different conditions, providing information about the relative stability of individual phosphodiesters. 46 More flexible, solvent-exposed regions (such as single-strand regions) and specific conformations are more likely to promote a 2′-OH attack on the scissile phosphate, cleaving the molecule. The pattern of cleavage under different conditions can indicate structural changes, suggesting differences in the flexibility and protection promoted by a ligand. Our results showed that GlcN6P promotes an apparent stabilization of the ribozyme at specific sites, suggesting that the metabolite directly interacts with the RNA (Figure 4A). The regions showing the most prominent change in degradation at higher GlcN6P concentrations are P3/L3, J1.1/4, L4, and A60 (J4/2). Three of those regions (P3/L3, J1.1/4, and A60) surround the active site of the ribozyme, with A60 only two nucleotides from the catalytic cytosine residue (C58). No region with an increased rate of degradation was observed in this experiment.
Figure 4.
Structural probing of GlcN6P binding of the drz-Fpra-1 ribozyme. (A) In-line probing of the 3′-labeled drz-Fpra-1 ribozyme in the presence of GlcN6P. The band intensities that change with GlcN6P concentration are indicated in the figure by the nucleotide identity. The sequence of the RNA was determined using iodoethanol cleavage of ribozymes with a phosphorothioate-modified backbone at positions indicated above each lane. The intensities of the control band (G69) from the same experiment are shown below the rest of the gel. (B) Graph of band intensities for the regions P3/L3 (○), J1.1/4 (△), L4 (□), and A60 (◇), normalized to a control band at G69. The data were fit to a model based on eq 2. The average dissociation constant is estimated to be 4.7 ± 0.2 mM. The positions where the band intensities respond to metabolite concentration are also indicated to the left of the gel in panel A.
The data obtained for each concentration of metabolite, including the no-metabolite control, were normalized to G69 near the 3′-terminus of the ribozyme, used as a control position with ligand-independent degradation. The data were modeled by a dissociation constant equation (eq 2) for a single ligand, and a Kd was calculated independently for each affected region (Figure 4B). The estimations of Kd for all four regions were consistent, resulting in an average Kd of 4.7 ± 0.2 mM. Performing the same experiment with the metabolite GlcN1P, the isomer of GlcN6P, and the product of the enzymatic reaction that does not affect the rate of cleavage of the ribozyme, we did not observe any region with significant changes in its degradation pattern (Figure S4A,B). We measured the degradation for the regions P3/L3, L4, and A60 (J4/2) and used three different control bands for normalization. No apparent Kd was revealed for any of the regions, demonstrating that GlcN1P does not affect the degradation pattern of the ribozyme.
The Metabolite Effect Is Ribozyme- and Position-Specific
To verify that the effects promoted by GlcN6P in both drz-Fpra-1 and drz-Fpra-2 were specific, we decided to test the influence of the ligand on different ribozymes of the same family, starting with two inactive drz-Fpra-1 mutants, C58A and C58U. On the basis of previous mutagenesis of HDV ribozymes,16,17,19,22,23,51,53 we expected a very slow self-cleavage of C58A and an abolishment of catalysis for C58U. In 2 h experiments, neither mutant of drz-Fpra-1 showed self-scission in the presence of GlcN6P (data not shown). These results demonstrate that GlcN6P does not rescue the activity of the ribozyme, further supporting a role as an allosteric modulator and not a catalytic cofactor for the ligand. To further investigate the specificity of GlcN6P, we probed two control ribozymes with the metabolite, the genomic HDV (gHDV) and the antigenomic HDV (aHDV), the two distinct HDV ribozymes from the hepatitis delta virus.6,7 No significant change in the cleavage rate was observed for these ribozymes (Figure 5), suggesting that the kinetic modulation by the metabolite is specific to the drz-Fpra ribozymes.
Figure 5.
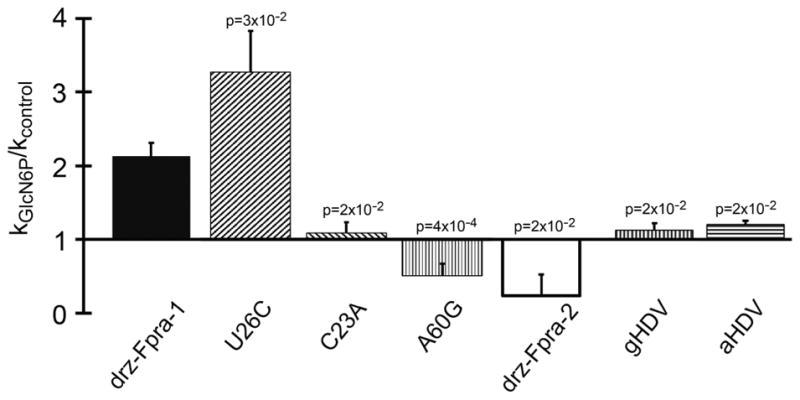
Effect of GlcN6P on self-scission of drz-Fpra-1/drz-Fpra-2 hybrids and HDV ribozymes. Comparison of modulation of the ribozyme activity by 20 mM GlcN6P normalized to the no-metabolite control at 5 mM Mg2+. U26C, C23A, and A60G are drz-Fpra-1/drz-Fpra-2 hybrids referenced to the drz-Fpra-1 positions. gHDV and aHDV are the genomic and antigenomic HDV ribozymes, respectively. All p values were calculated with respect to the wild-type Fpra-1 ribozyme and show the statistical significance of the results.
To narrow the range of causes of the opposite effect promoted by GlcN6P on drz-Fpra-1 and drz-Fpra-2, we decided to study the discrepant nucleotides near the active sites of the two otherwise similar ribozymes. The catalytic cores of the two ribozymes differ by only three nucleotides. We tested three hybrids, C23A and U26C in the L3 region and A60G in the J4/2 region of the drz-Fpra-1 sequence. We found that U26C retains and even slightly increases the effect of GlcN6P on the cleavage rate constant; C23A seems to abrogate the metabolite effect, and A60G reverts the metabolite effect, decreasing the rate of cleavage of the ribozyme when compared to the kWT (Figure 5), to a level similar to that of drz-Fpra-2. These results indicate that nucleotides C23 and A60 are involved in the allosteric modulation of drz-Fpra-1.
DISCUSSION
One of the nine natural families of self-cleaving ribozymes, the HDV-like ribozymes are found in several bacteria, many eukaryotes (including humans), Chilo iridescent virus, and microbial metagenomic data sets.14,22–27 Little is known about their biological functions and regulation, particularly any roles that cofactors and metabolites have in altering their activity. drz-Fpra-122 and drz-Fpra-2 HDV-like ribozymes were found in the human gut bacterium F. prausnitzii genome, surrounding the phosphoglucosamine mutase (glmM) open reading frame (ORF).49 The enzyme glmM catalyzes the transformation of glucosamine 6-phosphate (GlcN6P) into glucosamine 1-phosphate (GlcN1P) in the hexosamine biosynthesis pathway. 31 We have studied the effect of the metabolites from this pathway on the self-cleavage rate of the drz-Fpra ribozymes. Whereas GlcN6P increases the rate of cleavage of the drz-Fpra-1 ribozyme and appears to stabilize its structure, GlcN1P, the product of the glmM enzymatic reaction, does not significantly affect the ribozyme.
The effect of GlcN6P on drz-Fpra-1 showed that the ligand modestly increases the rate of self-cleavage of the ribozyme when compared to the no-metabolite control. Titration of Mg2+ at a constant GlcN6P concentration or titration of a ligand at a constant Mg2+ concentration showed similar results, increasing the rate of self-cleavage of the ribozyme. When we examined the influence of GlcN6P on the Mg2+ dependence of the ribozymes, we observed that the ligand effect is not seen at low concentrations of the divalent cation, behaving like the no-metabolite control. Mg2+ coordinated with a hydroxide anion likely acts as the general base in the self-cleavage reaction of HDV ribozymes, deprotonating the 2′-OH of the base that promotes the attack in the scissile phosphate,17 while the essential cytosine in J4/2 (C58 in drz-Fpra-1) acts as a general acid, mediating a proton transfer to the 5′-oxygen of G1, the leaving group of the self-scission reaction.17 If the effect promoted by the metabolite directly influenced the role of Mg2+ in the mechanism of the reaction, the kGlcN6P would be expected to increase the cleavage rate in all instances throughout the ligand titration, including at low concentrations of Mg2+. This result suggests that the effect caused by the ligand does not directly influence the catalytic role of the metal ion in the self-cleavage reaction.
In the case of titration of GlcN6P, the ribozyme responds to the ligand, cleaving faster than the no-metabolite control does. Again, this effect is not seen at low concentrations of the metabolite, indicating that the effect is not directly related to the catalysis in the active site; otherwise, we would expect to see an appreciably higher kGlcN6P even at lower concentrations of GlcN6P. Moreover, testing the effect of the metabolite in the two inactive mutants of drz-Fpra-1, C58A and C58U, we found that the ligand does not rescue their activity, indicating that GlcN6P does not act as the general acid. This finding contrasts with the role of GlcN6P in the glmS ribozyme, where the cofactor actively participates in the active site, mediating the protonation of the 5′-oxyanion leaving group.36–39,42–44,54 The glmS cofactor has been implicated in other roles in catalysis, including helping to align the active site, stabilizing the developing charge during the self-cleavage reaction, and participating in a set of competing hydrogen bonds to ensure potent activation and regulation of the catalysis.54,55 In contrast, our results suggest that the effect produced by GlcN6P on drz-Fpra-1 is not directly in the catalytic step of the self-cleavage reaction, suggesting an allosteric modulation of the ribozyme activity.
The structural probing of drz-Fpra-1 in the presence of GlcN6P and GlcN1P showed the decreased conformational flexibility of the RNA solely in the presence of GlcN6P (Figure 4 and Figure S4). The active site of HDV ribozymes is formed by the P3/L3, P1.1, and J4/2 elements,16,18,19 and our probing data show that the ligand decreases the rate of degradation of P3/L3, J1.1/4, L4, and A60 in J4/2. Even though GlcN6P clearly affects the active site stability and promotes faster self-scission, the in-line probing experiment does not pinpoint the site of interaction between the metabolite and the ribozyme, because the changes in the degradation pattern may result from a conformational change promoted by the intermolecular interaction, and additional studies would be needed to map the exact position of interaction.
For all regions with changes in the intensity of the in-line probing bands, the obtained estimated Kd was similar, with an average value of 4.7 ± 0.2 mM. The intracellular concentration of GlcN6P in F. prausnitzii is not known, but the reported steady-state concentration in Escherichia coli cells is 1.2 mM.56 Although it is ~4 times lower than the estimated Kd for the drz-Fpra-1 ribozyme, subcellular localization and spikes in metabolic activity may result in a concentration of the metabolite that is higher than the cell culture average. Thus, the metabolite concentration may reach intracellular levels that significantly alter the activity of the ribozyme.
The drz-Fpra-2 ribozyme, located 558 nucleotides down-stream of the glmM gene, was found by sequence similarity when searching for drz-Fpra-1. Interestingly, GlcN6P promoted an opposite effect on drz-Fpra-2 and on drz-Fpra-1, decreasing the rate of self-cleavage compared to the kcontrol. This effect was unexpected and intriguing, bringing to our attention those nucleotides of the active site where the ribozymes differ. We tested the effect of GlcN6P on the activity of the hybrids of drz-Fpra-1 and drz-Fpra-2, with the following mutations made to drz-Fpra-1: C23A and U26C in L3 and A60G in J4/2. The results demonstrate that U26C retains the effect of GlcN6P on the cleavage rate, maintaining the elevated kGlcN6P and suggesting that the metabolite does not interact with the ribozyme at this nucleotide. On the other hand, the C23A mutation abrogates the metabolite effect, bringing the rate of self-cleavage to the same level as the no-metabolite control. The third mutation, A60G, reverts the metabolite effect, decreasing the rate of cleavage of the ribozyme below the kcontrol, a case similar to what is observed in drz-Fpra-2. Thus, our data suggest that C23A and A60G are the discrepant nucleotides from the catalytic core that play a role in the opposite effect caused by GlcN6P in the two ribozymes. C23 is part of L3 and is important for maintaining the active site structure, stacking on C24 and possibly hydrogen bonding with neighboring residues.16,18,19 C23 may also be important for the stacking with U-1, a role similar to the role of U23 in the HDV ribozyme.16,18,19 A mutation at C23 may promote a conformational change that prevents the interaction of GlcN6P with the ribozyme. A60 is part of the trefoil turn of the ribozyme structure and makes an A-minor interaction with P3, participating in a network of hydrogen bonds between P3/L3 and J4/2 regions.16,18,19 Thus, the A60G mutation would sterically hinder the interaction of the nucleobase, causing the J4/2 region to move away from the P3 region and potentially disrupting the interactions of the A60 ribose, as well. This change may be sufficient to abrogate the already weak interaction between the ribozyme and the metabolite.
Testing ribozyme kinetics in the presence of other metabolites elucidated the functional moieties critical in GlcN6P sensitivity in drz-Fpra-1. The aforementioned GlcN1P, the product of the enzymatic reaction of glmM, showed no significant effect on drz-Fpra-1 and a small effect on drz-Fpra-2. Similarly, Glc and UDP-GlcNAc had little to no effect on the ribozymes. On the other hand, GlcN and Glc6P increased the rates of cleavage for both ribozymes, leading us to speculate that both the amino and phosphate groups are relevant for the effect of the ligand in the ribozyme, as long as they are not at adjacent carbons of the sugar ring, as seen in GlcN1P. The concentration of Glc6P in the cytosol is likely very low, and a previous report about the concentration of metabolites in E. coli with a detection limit of 130 nM did detect Glc6P.56 Thus, despite an in vitro effect of Glc6P on the ribozymes, it is unlikely that it affects their activity in vivo.
The cleavage rate constants obtained for gHDV and aHDV ribozymes in the presence of GlcN6P were similar to those of the no-metabolite control. These results suggest that the interaction between the metabolite and drz-Fpra-1 is specific. With regard to the structure of the ribozymes, to the best of our knowledge, the first P1.1 base pair is found to be A–U only in bacteria,22,24,25,27,49 and drz-Fpra-1 was the only case of a bacterial HDV-like ribozyme upstream of a neighboring gene. However, a recent discovery of several ribozymes using comparative genomic analysis14 also revealed HDV-like ribozymes in environmental samples, of which four showed the glmM gene 34–42 nucleotides downstream of the ribozymes, as in drz-Fpra-1 in F. prausnitzii (37 nucleotides). Seven other HDV-like ribozymes showed the glmM gene upstream of the ribozymes. All of these ribozymes have an A–U base pair in P1.1, suggesting that they originate from closely related bacteria. Alignment of the drz-Fpra-1, drz-Fpra-2, and other microbial HDV-like ribozymes with the secondary structure is shown in Figure S5. The alignment, combined with the results presented here, suggests that the env-26 HDV-like ribozyme would respond to GlcN6P, because it is the only sequence with a cytosine in the same position of L3 as in drz-Fpra-1 (C23), whereas all other ribozymes bear an adenosine at this position, like in drz-Fpra-2, where we found no effect of the metabolite. With regard to the J4/2 strand, only drz-Fpra-2 contains a guanosine residue at the position equivalent to A60 in drz-Fpra-1, here reported to be responsible for the inhibitory effect of GlcN6P on the activity of the ribozyme.
We show that the metabolite GlcN6P interacts with the drz-Fpra-1 ribozyme and increases its self-cleavage activity; however, we do not know the biological significance of these findings, and future studies will include biological assays that address this question. In contrast to the glmS riboswitch ribozyme, which is part of a feedback regulatory loop utilizing the product of the downstream metabolic step to induce ribozyme self-scission and mRNA degradation, the activity of drz-Fpra-1 increases with the concentration of the substrate, providing an example of a putative feed-forward mechanism. The downstream RNA product, which starts with the cleaved drz-Fpra-1 ribozyme and is followed by the GlmM open reading frame, is terminated with a 5′-OH that is sequestered by the structure of the cleaved ribozyme.16,18,19 This structure may protect the 5′-terminus against endonucleases, thus increasing the mRNA stability. Moreover, RNAs with 5′-OH termini have an extended half-life when compared with those of 5′-phosphorylated mRNAs, because 5′-hydroxyls are inferior substrates for the endonucleolytic degradation by RNase E.57,58 The extended half-life of mRNAs with 5′-OH termini over phosphorylated 5′-termini has been used in bacterial metabolic engineering to design a number of aptazymes that increased the stability of the downstream transcripts upon ligand-dependent self-scission,59 and a similar mechanism may be acting in the case of the F. prausnitzii ribozyme-terminated glmM mRNA. To the best of our knowledge, drz-Fpra-1 activation by GlcN6P is the first example of an allosteric modulation of a natural self-cleaving ribozyme by a metabolite. We believe that this is not a unique case and that other examples of natural allosterically regulated self-cleaving ribozymes exist, providing another example of gene expression regulation at the RNA level.
Supplementary Material
Acknowledgments
Funding
This work was supported by Science Without Boarders Program-CAPES Foundation, Ministry of Education of Brazil [Process 99999.013571/2013-03 (L.F.M.P.)], National Institutes of Health Grant 5F31GM103241-02 (R.M.J.), and National Science Foundation Grant MCB 1330606 and a grant from the John Templeton Foundation (to A.L.).
We thank all members of the Lupták laboratory for helpful discussions.
ABBREVIATIONS
- HDV
hepatitis delta virus
- glmS
glucosamine-6-phosphate synthase
- glmM
phosphoglucosamine mutase
- ORF
open reading frame
- GlcN6P
glucosamine 6-phosphate
- GlcN1P
glucosamine 1-phosphate
- UDP-GlcNAc
uridine diphosphate N-acetyl-glucosamine
- Glc
glucose
- Glc6P
glucose 6-phosphate
- GlcN
glucosamine
- PAGE
polyacrylamide gel electrophoresis
Footnotes
Author Contributions
L.F.M.P., R.M.J., and A.L. designed the study. L.F.M.P., R.M.J., and J.Y.F. performed the experiments. All authors analyzed the data. L.F.M.P., R.M.J., and A.L. wrote the manuscript. L.F.M.P. and R.M.J. contributed equally to this work.
Notes
The authors declare no competing financial interest.
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.biochem. 7b00879.
Effect of multiple ligands on drz-Fpra-1 and drz-Fpra-2 (Figure S1), effect of UDP-GlcNAc on both ribozymes (Figure S2), activity of drz-Fpra-1 in mixtures of GlcN6P and GlcN1P (Figure S3), in-line probing of drz-Fpra-1 in the presence of GlcN1P (Figure S4), secondary structure alignment of ribozymes (Figure S5), and sequences of constructs used in this study (Table S1) (PDF)
References
- 1.Doudna JA, Cech TR. The chemical repertoire of natural ribozymes. Nature. 2002;418:222–228. doi: 10.1038/418222a. [DOI] [PubMed] [Google Scholar]
- 2.Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- 3.Guerrier-Takada C, Gardiner K, Marsh T, Pace N, Altman S. The RNA moiety of ribonuclease P is the catalytic subunit of the enzyme. Cell. 1983;35:849–857. doi: 10.1016/0092-8674(83)90117-4. [DOI] [PubMed] [Google Scholar]
- 4.Jimenez RM, Polanco JA, Luptak A. Chemistry and biology of self-cleaving ribozymes. Trends Biochem Sci. 2015;40:648–661. doi: 10.1016/j.tibs.2015.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fedor MJ. Comparative enzymology and structural biology of RNA self-cleavage. Annu Rev Biophys. 2009;38:271–299. doi: 10.1146/annurev.biophys.050708.133710. [DOI] [PubMed] [Google Scholar]
- 6.Wu HN, Lin YJ, Lin FP, Makino S, Chang MF, Lai MM. Human hepatitis delta virus RNA subfragments contain an autocleavage activity. Proc Natl Acad Sci U S A. 1989;86:1831–1835. doi: 10.1073/pnas.86.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharmeen L, Kuo MY, Dinter-Gottlieb G, Taylor J. Antigenomic RNA of human hepatitis delta virus can undergo self-cleavage. J Virol. 1988;62:2674–2679. doi: 10.1128/jvi.62.8.2674-2679.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Prody GA, Bakos JT, Buzayan JM, Schneider IR, Bruening G. Autolytic processing of dimeric plant virus satellite RNA. Science. 1986;231:1577–1580. doi: 10.1126/science.231.4745.1577. [DOI] [PubMed] [Google Scholar]
- 9.Hutchins CJ, Rathjen PD, Forster AC, Symons RH. Self-cleavage of plus and minus RNA transcripts of avocado sunblotch viroid. Nucleic Acids Res. 1986;14:3627–3640. doi: 10.1093/nar/14.9.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Buzayan JM, Gerlach WL, Bruening G. Non-enzymatic cleavage and ligation of RNAs complementary to a plant virus satellite RNA. Nature. 1986;323:349–353. [Google Scholar]
- 11.Winkler WC, Nahvi A, Roth A, Collins JA, Breaker RR. Control of gene expression by a natural metabolite-responsive ribozyme. Nature. 2004;428:281–286. doi: 10.1038/nature02362. [DOI] [PubMed] [Google Scholar]
- 12.Saville BJ, Collins RA. A site-specific self-cleavage reaction performed by a novel RNA in Neurospora mitochondria. Cell. 1990;61:685–696. doi: 10.1016/0092-8674(90)90480-3. [DOI] [PubMed] [Google Scholar]
- 13.Roth A, Weinberg Z, Chen AGY, Kim PB, Ames TD, Breaker RR. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat Chem Biol. 2014;10:56–60. doi: 10.1038/nchembio.1386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weinberg Z, Kim PB, Chen TH, Li S, Harris KA, Lunse CE, Breaker RR. New classes of self-cleaving ribozymes revealed by comparative genomics analysis. Nat Chem Biol. 2015;11:606–610. doi: 10.1038/nchembio.1846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuo MY, Sharmeen L, Dinter-Gottlieb G, Taylor J. Characterization of self-cleaving RNA sequences on the genome and antigenome of human hepatitis delta virus. J Virol. 1988;62:4439–4444. doi: 10.1128/jvi.62.12.4439-4444.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferre-D’Amare AR, Zhou K, Doudna JA. Crystal structure of a hepatitis delta virus ribozyme. Nature. 1998;395:567–574. doi: 10.1038/26912. [DOI] [PubMed] [Google Scholar]
- 17.Das SR, Piccirilli JA. General acid catalysis by the hepatitis delta virus ribozyme. Nat Chem Biol. 2005;1:45–52. doi: 10.1038/nchembio703. [DOI] [PubMed] [Google Scholar]
- 18.Chen JH, Yajima R, Chadalavada DM, Chase E, Bevilacqua PC, Golden BL. A 1.9 A crystal structure of the HDV ribozyme precleavage suggests both Lewis acid and general acid mechanisms contribute to phosphodiester cleavage. Biochemistry. 2010;49:6508–6518. doi: 10.1021/bi100670p. [DOI] [PubMed] [Google Scholar]
- 19.Ke A, Zhou K, Ding F, Cate JH, Doudna JA. A conformational switch controls hepatitis delta virus ribozyme catalysis. Nature. 2004;429:201–205. doi: 10.1038/nature02522. [DOI] [PubMed] [Google Scholar]
- 20.Koo SC, Lu J, Li NS, Leung E, Das SR, Harris ME, Piccirilli JA. Transition state features in the hepatitis delta virus ribozyme reaction revealed by atomic perturbations. J Am Chem Soc. 2015;137:8973–8982. doi: 10.1021/jacs.5b01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Golden BL, Hammes-Schiffer S, Carey PR, Bevilacqua PC. An integrated picture of HDV ribozyme catalysis. In: Russell R, editor. Biophysics of RNA Folding. Springer; New York: 2013. pp. 135–167. [Google Scholar]
- 22.Webb CH, Riccitelli NJ, Ruminski DJ, Lupták A. Widespread occurrence of self-cleaving ribozymes. Science. 2009;326:953. doi: 10.1126/science.1178084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Salehi-Ashtiani K, Lupták A, Litovchick A, Szostak JW. A genomewide search for ribozymes reveals an HDV-like sequence in the human CPEB3 gene. Science. 2006;313:1788–1792. doi: 10.1126/science.1129308. [DOI] [PubMed] [Google Scholar]
- 24.Ruminski DJ, Webb CH, Riccitelli NJ, Lupták A. Processing and translation initiation of non-long terminal repeat retrotransposons by hepatitis delta virus (HDV)-like self-cleaving ribozymes. J Biol Chem. 2011;286:41286–41295. doi: 10.1074/jbc.M111.297283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Eickbush DG, Eickbush TH. R2 retrotransposons encode a self-cleaving ribozyme for processing from an rRNA cotranscript. Mol Cell Biol. 2010;30:3142–3150. doi: 10.1128/MCB.00300-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Riccitelli NJ, Delwart E, Luptak A. Identification of minimal HDV-like ribozymes with unique divalent metal ion dependence in the human microbiome. Biochemistry. 2014;53:1616–1626. doi: 10.1021/bi401717w. [DOI] [PubMed] [Google Scholar]
- 27.Sanchez-Luque FJ, Lopez MC, Macias F, Alonso C, Thomas MC. Identification of an hepatitis delta virus-like ribozyme at the mRNA 5′-end of the L1Tc retrotransposon from Trypanosoma cruzi. Nucleic Acids Res. 2011;39:8065–8077. doi: 10.1093/nar/gkr478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miquel S, Martin R, Rossi O, Bermudez-Humaran LG, Chatel JM, Sokol H, Thomas M, Wells JM, Langella P. Faecalibacterium prausnitzii and human intestinal health. Curr Opin Microbiol. 2013;16:255–261. doi: 10.1016/j.mib.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 29.Heinken A, Khan MT, Paglia G, Rodionov DA, Harmsen HJ, Thiele I. Functional metabolic map of Faecalibacterium prausnitzii, a beneficial human gut microbe. J Bacteriol. 2014;196:3289–3302. doi: 10.1128/JB.01780-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sokol H, Pigneur B, Watterlot L, Lakhdari O, Bermudez-Humaran LG, Gratadoux JJ, Blugeon S, Bridonneau C, Furet JP, Corthier G, Grangette C, Vasquez N, Pochart P, Trugnan G, Thomas G, Blottiere HM, Dore J, Marteau P, Seksik P, Langella P. Faecalibacterium prausnitzii is an anti-inflammatory commensal bacterium identified by gut microbiota analysis of Crohn disease patients. Proc Natl Acad Sci U S A. 2008;105:16731–16736. doi: 10.1073/pnas.0804812105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mengin-Lecreulx D, van Heijenoort J. Characterization of the essential gene glmM encoding phosphoglucosamine mutase in Escherichia coli. J Biol Chem. 1996;271:32–39. doi: 10.1074/jbc.271.1.32. [DOI] [PubMed] [Google Scholar]
- 32.Raetz CR. Molecular genetics of membrane phospholipid synthesis. Annu Rev Genet. 1986;20:253–295. doi: 10.1146/annurev.ge.20.120186.001345. [DOI] [PubMed] [Google Scholar]
- 33.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat Rev Mol Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 34.McCown PJ, Corbino KA, Stav S, Sherlock ME, Breaker RR. Riboswitch diversity and distribution. RNA. 2017;23:995–1011. doi: 10.1261/rna.061234.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Klein DJ, Ferre-D’Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 36.Cochrane JC, Lipchock SV, Strobel SA. Structural investigation of the GlmS ribozyme bound to Its catalytic cofactor. Chem Biol. 2007;14:97–105. doi: 10.1016/j.chembiol.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Viladoms J, Fedor MJ. The glmS ribozyme cofactor is a general acid-base catalyst. J Am Chem Soc. 2012;134:19043–19049. doi: 10.1021/ja307021f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cochrane JC, Lipchock SV, Smith KD, Strobel SA. Structural and chemical basis for glucosamine 6-phosphate binding and activation of the glmS ribozyme. Biochemistry. 2009;48:3239–3246. doi: 10.1021/bi802069p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.McCarthy TJ, Plog MA, Floy SA, Jansen JA, Soukup JK, Soukup GA. Ligand requirements for glmS ribozyme self-cleavage. Chem Biol. 2005;12:1221–1226. doi: 10.1016/j.chembiol.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 40.Collins JA, Irnov I, Baker S, Winkler WC. Mechanism of mRNA destabilization by the glmS ribozyme. Genes Dev. 2007;21:3356–3368. doi: 10.1101/gad.1605307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Watson PY, Fedor MJ. The glmS riboswitch integrates signals from activating and inhibitory metabolites in vivo. Nat Struct Mol Biol. 2011;18:359–363. doi: 10.1038/nsmb.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lau MW, Ferre-D’Amare AR. An in vitro evolved glmS ribozyme has the wild-type fold but loses coenzyme dependence. Nat Chem Biol. 2013;9:805–810. doi: 10.1038/nchembio.1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Soukup GA. Core requirements for glmS ribozyme self-cleavage reveal a putative pseudoknot structure. Nucleic Acids Res. 2006;34:968–975. doi: 10.1093/nar/gkj497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jansen JA, McCarthy TJ, Soukup GA, Soukup JK. Backbone and nucleobase contacts to glucosamine-6-phosphate in the glmS ribozyme. Nat Struct Mol Biol. 2006;13:517–523. doi: 10.1038/nsmb1094. [DOI] [PubMed] [Google Scholar]
- 45.Long DM, Uhlenbeck OC. Kinetic characterization of intramolecular and intermolecular hammerhead RNAs with stem II deletions. Proc Natl Acad Sci U S A. 1994;91:6977–6981. doi: 10.1073/pnas.91.15.6977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Soukup GA, Breaker RR. Relationship between internucleotide linkage geometry and the stability of RNA. RNA. 1999;5:1308–1325. doi: 10.1017/s1355838299990891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ryder SP, Strobel SA. Nucleotide analog interference mapping. Methods. 1999;18:38–50. doi: 10.1006/meth.1999.0755. [DOI] [PubMed] [Google Scholar]
- 48.Vu MM, Jameson NE, Masuda SJ, Lin D, Larralde-Ridaura R, Luptak A. Convergent Evolution of Adenosine Aptamers Spanning Bacterial, Human, and Random Sequences Revealed by Structure-Based Bioinformatics and Genomic SELEX. Chem Biol. 2012;19:1247–1254. doi: 10.1016/j.chembiol.2012.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Webb CH, Lupták A. HDV-like self-cleaving ribozymes. RNA Biol. 2011;8:719–727. doi: 10.4161/rna.8.5.16226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Cerrone-Szakal AL, Siegfried NA, Bevilacqua PC. Mechanistic characterization of the HDV genomic ribozyme: solvent isotope effects and proton inventories in the absence of divalent metal ions support C75 as the general acid. J Am Chem Soc. 2008;130:14504–14520. doi: 10.1021/ja801816k. [DOI] [PubMed] [Google Scholar]
- 51.Nakano S, Chadalavada DM, Bevilacqua PC. General acid-base catalysis in the mechanism of a hepatitis delta virus ribozyme. Science. 2000;287:1493–1497. doi: 10.1126/science.287.5457.1493. [DOI] [PubMed] [Google Scholar]
- 52.Chen J, Ganguly A, Miswan Z, Hammes-Schiffer S, Bevilacqua PC, Golden BL. Identification of the catalytic Mg(2)(+) ion in the hepatitis delta virus ribozyme. Biochemistry. 2013;52:557–567. doi: 10.1021/bi3013092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Perrotta AT, Shih I, Been MD. Imidazole rescue of a cytosine mutation in a self-cleaving ribozyme. Science. 1999;286:123–126. doi: 10.1126/science.286.5437.123. [DOI] [PubMed] [Google Scholar]
- 54.Bingaman JL, Zhang S, Stevens DR, Yennawar NH, Hammes-Schiffer S, Bevilacqua PC. The GlcN6P cofactor plays multiple catalytic roles in the glmS ribozyme. Nat Chem Biol. 2017;13:439–445. doi: 10.1038/nchembio.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bingaman JL, Gonzalez IY, Wang B, Bevilacqua PC. Activation of the glmS ribozyme Nnucleophile via over-determined hydrogen bonding. Biochemistry. 2017;56:4313–4317. doi: 10.1021/acs.biochem.7b00662. [DOI] [PubMed] [Google Scholar]
- 56.Bennett BD, Kimball EH, Gao M, Osterhout R, Van Dien SJ, Rabinowitz JD. Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat Chem Biol. 2009;5:593–599. doi: 10.1038/nchembio.186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. [DOI] [PubMed] [Google Scholar]
- 59.Carothers JM, Goler JA, Juminaga D, Keasling JD. Model-driven engineering of RNA devices to quantitatively program gene expression. Science. 2011;334:1716–1719. doi: 10.1126/science.1212209. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.