Abstract
The two-component signal transduction system PhoP/PhoQ is an important regulator for stress responses and virulence in most Gram-negative bacteria, but characterization of PhoP/PhoQ in Shigella has not been thoroughly investigated. In the present study, we found that deletion of phoPQ (ΔphoPQ) from Shigella flexneri 2a 301 (Sf301) resulted in a significant decline (reduced by more than 15-fold) in invasion of HeLa cells and Caco-2 cells, and less inflammation (− or +) compared to Sf301 (+++) in the guinea pig Sereny test. In low Mg2+ (10 μM) medium or pH 5 medium, the ΔphoPQ strain exhibited a growth deficiency compared to Sf301. The ΔphoPQ strain was more sensitive than Sf301 to polymyxin B, an important antimicrobial agent for treating multi-resistant Gram-negative infections. By comparing the transcriptional profiles of ΔphoPQ and Sf301 using DNA microarrays, 117 differentially expressed genes (DEGs) were identified, which were involved in Mg2+ transport, lipopolysaccharide modification, acid resistance, bacterial virulence, respiratory, and energy metabolism. Based on the reported PhoP box motif [(T/G) GTTTA-5nt-(T/G) GTTTA], we screened 38 suspected PhoP target operons in S. flexneri, and 11 of them (phoPQ, mgtA, slyB, yoaE, yrbL, icsA, yhiWX, rstA, hdeAB, pagP, and shf–rfbU-virK-msbB2) were demonstrated to be PhoP-regulated genes based on electrophoretic mobility shift assays and β-galactosidase assays. One of these PhoP-regulated genes, icsA, is a well-known virulence factor in S. flexneri. In conclusion, our data suggest that the PhoP/PhoQ system modulates S. flexneri virulence (in an icsA-dependent manner) and stress responses of Mg2+, pH and antibacterial peptides.
Keywords: S. flexneri, two-component signal transduction system, PhoP/PhoQ, Mg2+, virulence, icsA
Introduction
Shigella is a facultative intracellular Gram-negative pathogen which causes shigellosis by penetrating and replicating within human colonic epithelial cells. This invasive process causes ulcerative lesions that result in a bloody and purulent diarrhea characteristic of bacillary dysentery. The bacteria are transmitted via the fecal-oral route and invade the mucosa of the colon. Based on the biotype, Shigella is divided into four subgroups and Shigella flexneri 2a is the main subgroup in China. Shigella is highly infectious and it causes shigellosis by infection with only 10 to 100 organisms (Watanabe, 1988). Due to the non-standard use of antibiotics and the spread of drug resistant genes, Shigella drug resistance in clinical settings is becoming more and more prominent, which makes it difficult for shigellosis clinical treatment (Benny et al., 2014; Chen et al., 2014; Zhang et al., 2014).
Bacterial infection of the hosts relies on the ability of bacteria to cope with the challenge of environmental pressures. The two-component system (TCS) is a multivariate regulation mechanism which widely exists in the prokaryotes and contributes to the adaptation of bacteria to environmental challenges. In these TCSs, the activation of a sensor histidine kinase leads to autophosphorylation and then transfers the phosphoryl group to the cognate response regulator, which further regulates the expression of downstream genes (Gooderham and Hancock, 2009; Singh et al., 2014). TCSs play important roles in Shigella virulence. For example, the OmpR/EnvZ TCS was reported to control the virulence of S. flexneri (Bernardini et al., 1990). It was noticed that the PhoP/PhoQ TCS is involved in virulence regulation in S. typhimurium (Miller et al., 1989; Perez et al., 2009). In 2000, Moss and coworkers showed that a phoP mutant decreased the inflammatory response and was more sensitive to polymorphonuclear leucocytes (PMNs) in S. flexneri (Moss et al., 2000), and our previous studies have shown that inhibitors of PhoQ reduced the virulence of S. flexneri (Cai et al., 2011). These results indicate that PhoP/PhoQ has the function of virulence regulation in Shigella, but the regulatory mechanism of PhoPQ in Shigella has not been confirmed.
PhoP/PhoQ is a broadly conserved TCS among many pathogenic and non-pathogenic bacteria. In most of these organisms the PhoPQ system has an original function of monitoring the extracellular Mg2+, while in pathogenic bacteria it also plays an important role in regulation of bacterial virulence phenotypes (Miller et al., 1989; Johnson et al., 2001; Grabenstein et al., 2006; Perez et al., 2009). The PhoP/PhoQ TCS consists of the histidine kinase PhoQ and the response regulator PhoP. PhoQ can respond to environmental signals by autophosphorylation. Phosphorylated PhoQ transfers the phosphate to PhoP, and activated PhoP further regulates the expression of downstream genes. Although the PhoP/PhoQ system has similar functions in regulating bacterial virulence in pathogenic bacteria such as Salmonella typhimurium, Yersinia pestis and Mycobacterium tuberculosis (Oyston et al., 2000; Cano et al., 2001; Perez et al., 2001), the regulons of PhoPQ vary in different species of bacteria. Groisman compared the PhoP-regulated genes in Salmonella and E. Coli and found that only a limited number of genes were in common between the two PhoP regulons (Groisman, 2001).
The virulence gene icsA (also named virG) is located on the virulence plasmid of Shigella and encodes the outer membrane protein IcsA, which is a key virulence factor in the Shigella pathogenesis (Bernardini et al., 1989). In the early stage of Shigella infection, the bacteria reach the intestinal lumen in which IcsA binds to the still unknown receptor to help Shigella adhere on the surface of the host cell (Brotcke Zumsteg et al., 2014). After invasion into the host cell, IcsA functions in activating the neural Wiskott-Aldrich syndrome protein (N-WASP) to mediate the intracellular actin-based motility (ABM), which is important for Shigella to survive within the host cell (Goldberg and Theriot, 1995). The Shigella icsA mutant strain shows a defect of bacterial intracellular actin assembly and cell-to-cell spread, followed by a significant decrease of virulence both in cells and animal models (Teh and Morona, 2013; Brotcke Zumsteg et al., 2014; Leupold et al., 2017).
In the present study, we have investigated the regulation functions of PhoP/PhoQ in S. flexneri. PhoPQ knocking out caused a decrease of S. flexneri virulence in HeLa cells and Caco-2 cells invasion models and guinea pig Sereny test, which was similar to that reported by Moss et al. (2000). The activity of PhoPQ allowed Shigella to tolerate low environmental Mg2+, acidic pH, and the presence of polymyxin B. Those environmental input signals promoted the expression of PhoPQ. We screened out 11 PhoP-regulated genes or operons in Shigella by using electrophoretic mobility shift assays (EMSAs) and β-galactosidase assays, in which a well-known virulence factor, icsA, was found and validated to be regulated by PhoPQ for the first time. It indicates that the PhoPQ system modulates S. flexneri virulence in an icsA-dependent manner.
Materials and methods
Ethics statement
All guinea pig infection procedures were approved by the Institutional Animal Care and Use Committee (IACUC) of School of Basic Medical Sciences, Fudan University (IACUC Animal Project Number 20140226-022-qu) according to national guidelines (Regulations for the Administration of Affairs Concerning Experimental Animals, China).
Bacterial strains, plasmids, and growth conditions
S. flexneri 2a 301 (Sf301, GenBank accession number AE005674) was kindly provided by Pr. Qi Jin (MOH Key Laboratory of Systems Biology of Pathogens, Institute of Pathogen Biology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China). The bacterial strains and plasmids used in this study are listed in Table 1. S. flexneri and E. coli were grown in Luria–Bertani medium (LB; Oxoid, Basingstoke, UK) at 37°C. Antibiotics were used at the following concentrations: ampicillin (100 μg/ml), kanamycin (50 μg/ml), tetracycline (10 μg/ml) and gentamicin (50 μg/ml) (Sigma-Aldrich, St. Louis, MO, USA).
Table 1.
Bacterial strains and plasmids used in this study.
| Name | Descriptiona | Source or references |
|---|---|---|
| BACTERIAL STRAINS | ||
| Shigella | ||
| Sf301 | Wild-type S. flexneri 2a 301 | Jin et al., 2002 |
| ΔphoPQ | phoPQ deletion of wild-type Sf301, Kan | This study |
| ΔphoPQc | ΔphoPQ complemented with pphoPQ, Amp, Kan | This study |
| ΔphoPQ(pGEMT) | ΔphoPQ introduced with plasmid pGEMT | This study |
| ΔphoPQ(picsA) | ΔphoPQ introduced with plasmid picsA | This study |
| ΔicsA | icsA deletion of wild-type Sf301, Kan | This study |
| ΔicsAc | ΔicsA complemented with picsA, Amp, Kan | This study |
| ΔicsA(pGEMT) | ΔicsA introduced with plasmid pGEMT | This study |
| Sf301(pphoP::lacZ) | Sf301 introduced with plasmid pphoP::lacZ | This study |
| Sf301(pshf::lacZ) | Sf301 introduced with plasmid pshf::lacZ | This study |
| Sf301(picsA::lacZ) | Sf301 introduced with plasmid picsA::lacZ | This study |
| Sf301(placZ) | Sf301 introduced with plasmid placZ | This study |
| ΔphoPQ(pphoP::lacZ) | ΔphoPQ introduced with plasmid pphoP::lacZ | This study |
| ΔphoPQ(pshf::lacZ) | ΔphoPQ introduced with plasmid pshf::lacZ | This study |
| ΔphoPQ(picsA::lacZ) | ΔphoPQ introduced with plasmid picsA::lacZ | This study |
| ΔphoPQ(placZ) | ΔphoPQ introduced with plasmid placZ | This study |
| E. coli | ||
| DH5α | supE44 ΔlacU169 hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| BL21(DE3) | F-ompT hsdSB(rB-mB-) gal dcm (DE3) | Invitrogen |
| PLASMIDS | ||
| pKD46 | Red recombinase expression plasmids, low copy number, Amp | Datsenko and Wanner, 2000 |
| pKD13 | oriR6K, Amp, Kan | Datsenko and Wanner, 2000 |
| pGEMT | PCR cloning vector, high copy number, Amp | Promega |
| pphoPQ | Wild-type gene phoPQ cloned into pGEMT | This study |
| picsA | Wild-type gene icsA cloned into pGEMT | This study |
| pET28a | oriR, IPTG induced, Kan | Novagen |
| pET28a-phoP | pET28a with insertion of the gene phoP, for PhoP expression | This study |
| pBAD/His/LacZ | pBAD, LacZ ORF, pBR322 ori, Amp | Invitrogen |
| pACYC184 | Medium copy number vector, p15A ori, Cm, Tc | Chang and Cohen, 1978 |
| placZ | pACYC184 inserted with the promoterless lacZ gene PCR amplified from pBAD/His/LacZ | This study |
| pphoP::lacZ | placZ inserted with promoter region of phoP | This study |
| pshf::lacZ | placZ inserted with promoter region of shf | This study |
| picsA::lacZ | placZ inserted with promoter region of icsA | This study |
Kan, kanamycin resistance; Amp, ampicillin resistance; Cm, chloramphenicol resistance; Tc, tetracycline resistance.
Construction of S. flexneri deletion mutant and complementation strains
S. flexneri phoQ/phoP deletion mutant strain was constructed by one-step inactivation of chromosomal genes using PCR products (Datsenko and Wanner, 2000). First, Sf301 was transformed with pKD46 (Table 1) to express the λ Red recombinase and was selected by ampicillin. The transformants were induced with L-arabinose (1 mM) and made electro-competent cells. A phoPQ homologous recombination fragment with kanamycin resistance gene cassette (kan, 1,394 bp) was amplified from pKD13 with primers PhoPQus-kan-F/PhoPQds-kan-R (containing upstream and downstream regions of Sf301 phoPQ; Table 2). The purified PCR products were then digested with DpnI, suspended in 10 mM Tris (pH 8.0), transferred into Sf301 by electroporation, and grown at 30°C. The phoPQ knockout mutant was screened out by kanamycin, and verified by PCR and sequencing (Sangon Biotech, Shanghai, China). The phoPQ knockout mutant was grown at 43°C to remove pKD46, plasmid removal was confirmed by PCR, and the strain was named ΔphoPQ (Figure S1).
Table 2.
Primers used in this study.
| Primera | Sequence (5′−3′) | Location (bp)b | Product length (bp) | Annotationc |
|---|---|---|---|---|
| CONSTRUCTION AND IDENTIFICATION OF ΔphoPQ | ||||
| PhoPQus-kan-F | AATCGCGTTACACTATTTTAATAATTAAGACAGGGAGAAATAAAAGTGTAGGCTGGAGCTGCTTCG | 1191632–1191676 | Underline: up and downstream | |
| PhoPQds-kan-R | GAATCAATGACTTGATGTAGTGGTAAAAGGACATATTTATTCATCATTCCGGGGATCCGTCGACC | 1189464–1189508 | 1,394 | regions of phoPQ |
| InterPQ-F | CTGGTTGTTGAAGACAATGCG | 1191602–1191622 | 2,087 | |
| InterPQ-R | AATCACCTCCATCCGCGCACC | 1189536–1189556 | ||
| OuterPQ-F | AATCAGTGCCGGATGGCGATG | 1191773–1191793 | 2,413 | |
| OuterPQ-R | TTCATACAGTGCACCGAACGG | 1189381–1189401 | ||
| pKD46-F | GCAGAACACATCCGGTACATG | 1592–1612 | 641 | |
| pKD46-R | CTGACGTTCTGCAGTGTATGC | 2212–2232 | ||
| CONSTRUCTION OF ΔphoPQc | ||||
| ΔphoPQc-F | GCCTCAAATCAGTGCCGGATG | 1191779–1191799 | 2,414 | |
| ΔphoPQc-R | ACAGTGCACCGAACGGTGTAG | 1189386–1189406 | ||
| CONSTRUCTION OF pET28a-phoP | ||||
| pET28a-phoP-F | CGCGGATCCATGCGCGTACTGGTTGTTGAA | 1191611–1191631 | 669 | Underline: BamHI |
| pET28a-phoP-R | CCGCTCGAGGCGCAATTCGAACAGGTAGCC | 1190963–1190983 | Underline: XhoI | |
| AMPLIFICATION OF GENE PROMOTER REGIONS | ||||
| PphoP-F | GCCTCAAATCAGTGCCGGATG | 1191779–1191799 | 176 | |
| PphoP-R | ACGCGCATTTTTATTTCTCCC | 1191624–1191644 | ||
| PmgtA-F | CTGTTGTCCCATAACGTGTTG | 4419849–4419869 | 187 | |
| PmgtA-R | CCATATAACCTCCGGTAAGTG | 4419683–4419703 | ||
| PslyB-F | CGTGAATACCATGCGGAATGA | 1697757–1697777 | 186 | |
| PslyB-R | AGCATCCCTCATGGTCAAAGT | 1697922–1697942 | ||
| PyoaE-F | GATCCGTAATTTAACTTTCGA | 1448408–1448428 | 214 | |
| PyoaE-R | AGAAAGGCAGGCGTTAAAAGG | 1448601–1448621 | ||
| PrstA-F | GTGGAATCAGCCCGGCGATAT | 1656568–1656588 | 220 | |
| PrstA-R | CGGTAGATATAAAAACGTCAC | 1656767–1656787 | ||
| Pshf-F | GAGTACCTGTGTTGTTCTGAG | 191446–191466 | 224 | |
| Pshf-R | AACCCAATAAAGCTGGTGCAT | 191649–191669 | ||
| PicsA-F | TTATCGAACATATAGCTTTCC | 149445–149465 | 189 | |
| PicsA-R | ATCAGTAAGTGGTTGATAAAC | 149613–149633 | ||
| PhdeA-F | ATCCCCTGCTATCAATCTATG | 3634002–3634022 | 209 | |
| PhdeA-R | TAAAGTGAAAGAGCCGTCACG | 3633814–3633834 | ||
| PyrbL-F | AATCACGTACTGAAATCGTTC | 3340458–3340478 | 168 | |
| PyrbL-R | GAATCATGCCATCTCCTGGAA | 3340605–3340625 | ||
| PyhiW-F | GGAAACTTTGTGCTCTCAGTA | 3698224–3698244 | 244 | |
| PyhiW-R | CTGCGATTATTTCAATTTCAG | 3698447–3698467 | ||
| PpagP-F | AGATGATTGTTGTATCTCGTA | 694218–694238 | 246 | |
| PpagP-R | TCTACTACTAGCATAGCAAAG | 693993–694013 | ||
| PipaH7.8-F | CCTCTGGAGCTTTATCCAGTC | 61790–61810 | 210 | |
| PipaH7.8-R | AGGAAATGTAAGCCGAGTAAG | 61979–61999 | ||
| PvirA-F | TTCTGTACGCTTGCCCAAAGT | 149370–149390 | 203 | |
| PvirA-R | ATGGAATGTTATTCTTCTCTT | 149188–149208 | ||
| CONSTRUCTION OF THE LacZ FUSION | ||||
| lacZ-F | AAAAGTACTGACGATGACGATAAGGATCCA | 3,102 | Underline: ScaI | |
| lacZ-R | CATGCCATGGCATCCGCCAAAACAGCCAAGC | Underline: NcoI | ||
| PphoP-lacZ-F | TCCCTCGGGGCCTCAAATCAGTGCCGGATG | 1191779–1191799 | 176 | Underline: AvaI |
| PphoP-lacZ-R | AAAAGTACTACGCGCATTTTTATTTCTCCC | 1191624–1191644 | Underline: ScaI | |
| Pshf-lacZ-F | TCCCTCGGGTTATCGAACATATAGCTTTCC | 191446–191466 | 224 | Underline: AvaI |
| Pshf-lacZ-R | AAAAGTACTATCAGTAAGTGGTTGATAAAC | 191649–191669 | Underline: ScaI | |
| PicsA-lacZ-F | TCCCTCGGGGAGTACCTGTGTTGTTCTGAG | 149445–149465 | 189 | Underline: AvaI |
| PicsA-lacZ-R | AAAAGTACTAACCCAATAAAGCTGGTGCAT | 149613–149633 | Underline: ScaI | |
Primers were designed according to the genomic sequence of S. flexneri 2a 301 (GenBank accession number AE005674). F, forward primer; R, reverse primer.
Location is the locus of the primer in the genomic sequence of S. flexneri 2a 301.
Underlined sequences represent the upstream and downstream regions of phoPQ or restriction enzyme sites.
For construction of complementation of phoPQ for ΔphoPQ, the phoPQ operon with its promoter region (2,414 bp) was amplified by PCR with primers ΔphoPQc-F/ΔphoPQc-R (Table 2), which were designed based on the genome of Sf301. The PCR products were ligated with pGEMT (Table 1), and the bacteria with the insertion were selected by ampicillin. After verification by sequencing, the phoPQ complementary plasmid (pphoPQ) was transformed into ΔphoPQ, and the transformants were selected by kanamycin and ampicillin, verified by PCR and sequencing, and named ΔphoPQc. pGEMT was introduced into ΔphoPQ as a vector control, and was named ΔphoPQ (pGEMT).
The construction of S. flexneri icsA, yoaE, yrbL, or rstA deletion mutant strains and their complementation strains used the same method as that of phoPQ with primers listed in Table S4. For construction of the icsA complementary plasmid picsA, the icsA gene with its promoter region (3,642 bp) was amplified from wild-type Sf301 with primers ΔicsAc-F/ΔicsAc-R, and ligated into the pGEMT vector. After verification by sequencing, picsA was then transformed into ΔphoPQ to construct the icsA expression strain ΔphoPQ(picsA), verified by PCR and sequencing.
Invasion assay with S. flexneri
The invasion ability of strains of S. flexneri was determined by gentamicin protection assay on HeLa cells and Caco-2 cells (Hale and Formal, 1981; Mounier et al., 1992). Cells were grown in 24-well plates at 5% CO2 [HeLa cells in Dulbecco's Modified Eagle Medium (DMEM) with 10% fetal bovine serum (FBS) and Caco-2 cells in DMEM medium with 20% FBS]. Bacterial strains were inoculated into LB containing 0.3 M NaCl until the OD600 reached 1.0. Bacteria were added to semi-confluent HeLa cells or Caco-2 cells at a multiplicity of infection (MOI) of 10. The plates were then centrifuged at 900 g for 10 min. After incubating at 37°C for 5, 15, or 30 min, gentamicin was added to the medium with a final concentration of 100 μg/ml for 60 min at 37°C. The cells were then lysed with 1 ml 0.1% Triton X-100 in PBS for 10 min. The lysates were diluted 1:10 in PBS and plated onto LB agar plates in triplicate, and colonies were counted. The invasion rate was calculated as the number of bacteria recovered from gentamicin-treated cells divided by the total number of inoculated bacteria.
For immunofluorescence assay, HeLa cells grown in 24-well tissue culture plates with coverslips were infected with strains of S. flexneri for 15 min, fixed with 3.7% formaldehyde in PBS for 15 min, and then permeabilized with 0.2% Triton X-100 for 5 min. Bacteria were stained with anti-Shigella O-Ag serum (BD Biosciences, New York, USA) and then with IgG Alexa 488 conjugate (Life Technologies, New York, USA), actin was stained with Texas Red phalloidin (Life Technologies), and nuclei were stained with 4′,6-Diamidino-2-Phenylindole (DAPI, Life Technologies) for 30 min. The coverslips were mounted and observed under a confocal laser scanning microscope (CLSM; Leica TCSSP5, Mannheim, Germany) at × 100 magnification (Chang et al., 2005).
S. flexneri sereny test and pathological examination
The virulence of S. flexneri was determined by guinea pig keratoconjunctivitis Sereny test (Sereny, 1957). Female guinea pigs (age 6 weeks, about 300 g) were inoculated with 109 Colony-Forming Units (CFU) of bacteria per eye (six guinea pig eyes in each group), and observed at 24, 48, and 72 h. Inoculation with LB served as a negative control. The keratoconjunctivitis induced by the bacteria was scored as follows: −, no disease or mild irritation; +, mild conjunctivitis or late development and/or rapid clearing of symptoms; ++, keratoconjunctivitis without purulence; and +++, fully developed keratoconjunctivitis with purulence. At 72 h post-inoculation, guinea pigs were euthanized with pentobarbital (40 mg/kg) and the eyes were removed and fixed in 4% formalin in PBS (pH 7.2). After hematoxylin and eosin (H&E) staining, the eye sections were examined under a microscope.
Bacterial growth curves under low Mg2+, acidic pH and the presence of polymyxin B conditions
Growth curves of the strains were determined by measuring the OD600 with an Eppendorf spectrophotometer at 60 min intervals over a period of 14 h. For the low Mg2+ growth assay, N medium was used containing 0.1 M Tris-HCl (pH 7.4), 38 mM glycerol, 0.1% (wt/vol) Casamino Acids, 0.37 g/l KCl, 0.087 g/l K2SO4, 0.99 g/l (NH4)2SO4 and 0.14 g/l KH2PO4 (Barchiesi et al., 2012). Overnight cultures of bacterial strains were inoculated into N medium supplemented with 10 μM or 10 mM MgCl2 (at 1:50 dilution) and incubated at 37°C with shaking. To assay acid resistance of bacteria, E glucose broth was used containing 0.2 g/l MgSO4•7H2O, 2 g/l citric acid, 13.1 g/l K2HPO4•3H2O, 3.5 g/l Na(NH4) HPO4•4H2O, and 0.4% glucose. Overnight cultures were inoculated into E glucose broth at pH 7 or pH 5 (at 1:50 dilution) and incubated at 37°C with shaking (Barchiesi et al., 2012). For the polymyxin B resistance assay, overnight cultures were inoculated into LB, grown with shaking until the OD600 reached 0.6, then bacteria were diluted in sterile 0.85% saline to about 5 × 103 cells per ml and exposed into different concentrations of polymyxin B (5, 10, 20, and 40 μg/ml) for 1 h at 37°C. Surviving bacteria were determined by plating on LB agar plates in triplicate. The survival rate was calculated as the number of bacteria treated with polymyxin B divided by that of the untreated control. All experiments were repeated at least three times.
Microarray analysis and qRT-PCR
For microarray analysis, Sf301 and ΔphoPQ were inoculated into LB medium and grown to mid-log phase (6 h) or early-stationary phase (10 h), with three biological replicates. Cells were harvested by centrifugation at 10,000 g for 1 min, and total RNAs were extracted using the RNeasyH Mini Kit (QIAGEN, Hilden, Germany) following the manufacturer's instructions. Agilent custom-specific design GeneChip of Sf301 genomic DNA were used. Each microarray (4*44k) contained spots with 4168 specific 60-mer oligonucleotides representing the 4168 ORFs of Sf301 in triplicate, carried out by Shanghai Biotechnology Co. Ltd. (Shanghai, China) according to standard protocols provided by Agilent Technologies (Palo Alto, USA). Briefly, the quality and quantity of RNA samples were determined and checked by Agilent 2100 bioanalyzer (Agilent Technologies). The RNA samples were then reverse transcribed to cDNA by MMLV reverse transcriptase (Invitrogen, Carlsbad, USA), followed by transcription with T7 RNA polymerase (New England BioLabs, Beverly, UK) to generate aminoacyl-UTP-labeled cRNA. Amino allyl modified cRNAs were purified and labeled with Cy3 (Cy3 NHS ester, GE Healthcare, Piscataway, NJ). Labeled cRNAs were then fragmented in fragmentation buffer (Agilent Technologies) and mixed with the Gene Expression Hybridization Kit (Agilent Technologies) at 65°C for 17 h with a constant rotation rate of 10 rpm for hybridization. The arrays were scanned by an Agilent DNA Microarray scanner. Microarray data were normalized in the Agilent Feature Extraction software. The ratio of gene expression (ΔphoPQ vs. Sf301) was calculated from the normalized signal intensities. A false discovery rate of 5% (P-value cutoff; 0.05) was used for variance analysis of three biological replicates and an arbitrary threshold of 2.0-fold or 0.5-fold was used for defining significant differences in expression ratios.
For validating the differential expression genes of microarray, qRT-PCR was carried out. Total RNA of bacteria was extracted using the RNeasyH Mini Kit. The extracted RNA was reverse transcribed into cDNA using iScript reverse transcriptase (Bio-Rad, Hercules, CA, USA) with incubation for 5 min at 25°C, followed by 30 min at 42°C and 5 min at 85°C. Subsequently, qRT-PCRs were performed using SYBR green PCR reagents (Premix EX TaqTM, Takara Biotechnology, Dalian, China) in the Mastercycler realplex system (Eppendorf AG, Hamburg, Germany) with amplification conditions of 95°C for 30 s, 40 cycles of 95°C for 5 s and 60°C for 34 s, followed by melting curve analysis. The 16S rRNA methyltransferase coding gene rsmC was used to normalize the transcriptional levels of genes in the qRT-PCRs. All qRT-PCRs were carried out in triplicate with at least three independent RNA samples. The primers (Table S1) were designed based on the genome of Sf301 using Beacon designer software (Premier Biosoft International Ltd., Palo Alto, CA, USA).
EMSA
For analyzing the interaction of the recombinant PhoP and the promoter regions of putative target genes, EMSA were performed using the DIG Gel Shift Kit (Roche, Basel, Switzerland) according to the manufacturer's instructions. The phoP gene was amplified with primers pET28a-phoP-F/pET28a-phoP-R (Table 2) from the genomic DNA of Sf301 and inserted into the vector pET-28a (+) to obtain the recombinant plasmid pET28a-phoP. The recombinant plasmid was then transformed into Escherichia coli BL21 (DE3). Bacteria were grown to an OD600 of 0.6 at 37°C and 0.8 mM IPTG was then added to induce PhoP protein expression for 6 h at 30°C. The expressed His-tagged PhoP protein was purified using the ProBondTM Purification System (Invitrogen, Carlsbad, CA, USA) according to the manufacturer's instructions. PhoP was phosphorylated prior to gel shift reaction by incubating PhoP with 50 mM acetylphosphate for 1 h. The predicted promoter regions of putative target genes were amplified with primers in Table 2 and labeled with digoxigenin using terminal transferase. Each gel shift assay included the probe labeled with digoxigenin plus increasing concentrations of the phosphorylated PhoP (PhoP-P, ranging from 0.16 to 1.6 μM). The coding sequence of virA in Sf301 without PhoP box sequence was designated as a negative control. All samples were incubated at 25°C for 20 min, separated by electrophoresis on 6% non-denaturing polyacrylamide gel, blotted onto a positively charged nylon membrane (Millipore), and detected by an enzyme immunoassay following the manufacturer's instructions.
DNase I footprinting assay
DNase I footprinting assays were performed according to Wang et al. (2012). The promoter regions of yoaE, mgtA and shf were amplified with primers listed in Table 2, and separately cloned into the pUC18BT vector (Shanghai Biotechnology Corporation, China), which was further used as the template for preparation of fluorescent FAM labeled probes. The probes were purified by the Wizard® SV Gel and PCR Clean-Up System (Promega) and quantified with a NanoDrop 2000C (Thermo Fisher Scientific Waltham, MA, USA). For each assay, 500 ng probes were incubated with different amounts of PhoP in 40 μl binding buffer at 30°C for 30 min. Then 10 μl DNase I (0.01 unit) (Promega) and 100 nmol CaCl2 were added, incubated at 25°C for 1 min, and the reactions were stopped by adding 140 μl DNase I stopping solution (200 mM unbuffered sodium acetate, 30 mM EDTA and 0.15% SDS). The DNA samples were extracted with phenol/chloroform, dissolved in 30 μl MilliQ water and loaded for carrying out capillary electrophoresis. The data were collected using the GeneScan-500 LIZ dye Size Standard (Applied Biosystems, Foster City, CA, USA).
LacZ fusion and β-galactosidase assay
A promoterless lacZ reporter gene was amplified by PCR with primers lacZ-F/lacZ-R (Table 2) using the pBAD/His/LacZ vector (Invitrogen, Carlsbad, CA, USA) as a template. Purified PCR products were digested with ScaI and NcoI endonucleases (MBI Fermentas, Vilnius, Lithuania) and inserted into the medium-copy-number plasmid pACYC184 (Chang and Cohen, 1978). The bacteria with the insertion were selected using tetracycline, verified by PCR and sequencing, and designated placZ. The promoter-proximal DNA region of phoP, shf, and icsA were amplified by PCR with Pfu DNA polymerase (Takara Biotechnology, Dalian, China) using the primers listed in Table 2. PCR products were digested with AvaI and ScaI endonucleases, and inserted into plasmid placZ. The bacteria with the insertion were selected by tetracycline, verified by PCR and sequencing, and named pphoP::lacZ, pshf::lacZ, and picsA::lacZ. These recombinant plasmids were introduced into Sf301 and the ΔphoPQ strain, respectively. The promoterless placZ was transferred into the bacterial strains as negative control. The reporter bacterial strains were separately grown in LB, or N medium with 10 μM/10 mM MgCl2, or E glucose broth at pH 7/5.5 or LB with 25 μg/ml polymyxin B. The bacteria were harvested, lysed with 400 μl lysozyme (1 mg/ml) at 37°C for 30 min, and then the β-galactosidase activity in the cellular extracts was measured by the β-Galactosidase Enzyme Assay System (Promega) following the manufacturer's instructions. All experiments were repeated at least three times independently.
Statistical analysis
Experiments were performed in triplicate and repeated at least three times. The data were analyzed with Student's t-test or one-way factorial analysis of variance in SPSS version 14.0 (Chicago, IL). Differences in means with a P < 0.05 were considered significant.
Microarray accession number
The complete microarray data set is uploaded in the Gene Expression Omnibus database (http://www.ncbi.nlm.nih.gov/geo/) under accession numbers GPL24308 for the platform design and GSE107365 for the original data set.
Results
Deletion of phoPQ diminished S. flexneri virulence
To analyze the regulatory mechanism of PhoPQ in Shigella, a S. flexneri phoPQ deletion mutant strain was constructed by using homologous recombination (Datsenko and Wanner, 2000). After transformation of the λ Red recombinase expression plasmid pKD46, the Sf301 was transformed with a fragment of a kanamycin resistance cassette with long flanking regions homologous of phoPQ. The phoPQ knockout mutant was then screened out by kanamycin, verified by PCR and sequencing, pKD46 was removed by growth at 43°C, and the strain named as ΔphoPQ (Figure S1). For construction of the phoPQ complemented strain, the phoPQ operon with its promoter region was amplified from wild-type Sf301, and ligated into the pGEMT vector. The phoPQ expression plasmid was then transformed into ΔphoPQ, selected by growth in the presence of kanamycin and ampicillin, verified by PCR as well as sequencing, and named as ΔphoPQc.
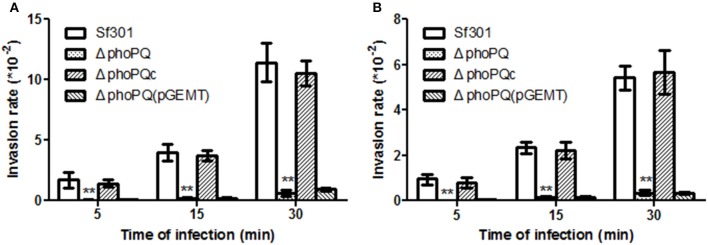
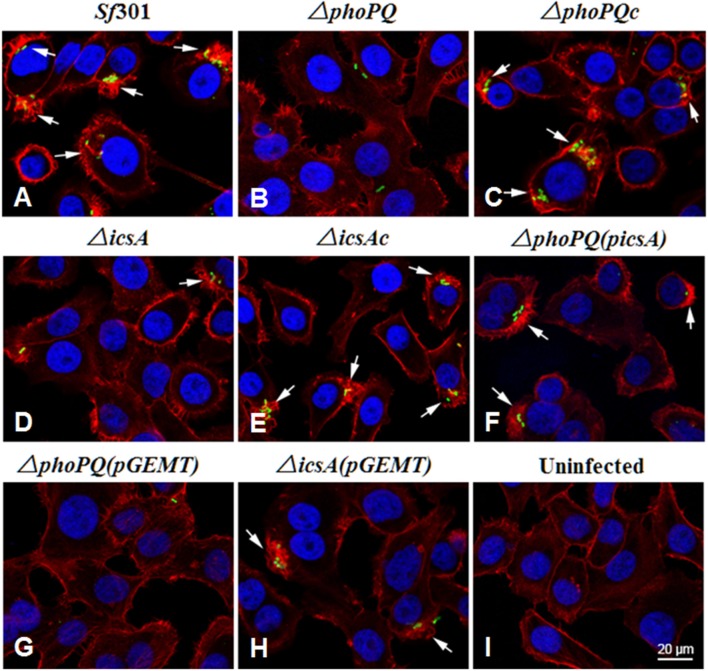
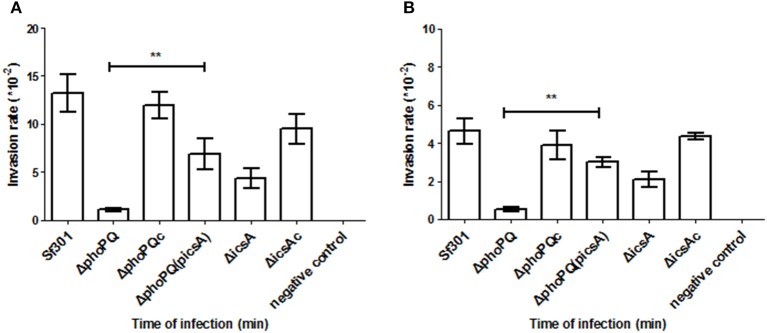
The invasion ability of Sf301, ΔphoPQ and ΔphoPQc were evaluated by the gentamicin protection assay on HeLa cells and Caco-2 cells. Bacterial strains were separately inoculated into HeLa cells or Caco-2 cells at a MOI of 10, incubated for 5, 15, or 30 min, and then gentamicin was added to kill extracellular bacteria. In HeLa cells, invasion rates of ΔphoPQ at 5, 15, or 30 min post-inoculation were reduced by 56-fold, 22-fold, and 15-fold, respectively, compared with those of Sf301. The invasion ability of ΔphoPQc (complementary phoPQ) was recovered to levels of Sf301 (Figure 1A). In Caco-2 cells, invasion rates of ΔphoPQ were similar to those in HeLa cells (Figure 1B). Next, Shigella-infected HeLa cells were observed by confocal immunofluorescence microscopy. HeLa cells grown on coverslips in 24-well tissue culture plates were infected with the strains of S. flexneri at a MOI of 10 and incubated for 15 min. The infected cells were stained with rabbit polyclonal anti-shigella anti-serum, Texas Red-labeled phalloidin, and DAPI (Figure 2). In Sf301 infected cells, more cells displayed membrane ruffling (more than 5 cells showing obvious membrane ruffling in a microscope field of view at ×100 magnification), which indicated actin cytoskeleton changes of HeLa cells, while in ΔphoPQ infected cells no obvious membrane ruffling observed. The membrane ruffling of ΔphoPQc infected cells could be restored to the level of Sf301 infected cells by complementation with pphoPQ (Figures 2A–C).
Figure 1.
The invasion ability of Sf301, ΔphoPQ and ΔphoPQc in HeLa cells and Caco-2 cells. The gentamicin protection assay was used as a cellular model to evaluate the effect of phoPQ deletion on virulence of Shigella. Bacteria grown to logarithmic phase were added into the cells for 5, 15, or 30 min. Then gentamicin was added into the medium to kill extracellular bacteria. Colonies of lysates on LB plates were counted. The invasion rate is calculated as the number of intracellular bacteria divided by the number of inoculated bacteria and multiplied by 10,000. The ΔphoPQ(pGEMT) was used as an empty plasmid control. (A) Bacterial ability to invade HeLa cells. (B) Bacterial ability to invade Caco-2 cells. Values are means ± standard deviations from three independent wells. **P < 0.01.
Figure 2.
Changes in the cytoskeleton of HeLa cells infected with S. flexneri strains. HeLa cells were infected with S. flexneri strains for 15 min. Actin was visualized by staining with Texas Red-labeled phalloidin (red), bacteria were stained with rabbit polyclonal anti-Shigella anti-serum (green), and nuclei of HeLa cells and bacterial DNA were stained with DAPI (blue). The coverslips were mounted and observed under a confocal laser scanning microscope at ×100 magnification (A–I). Arrows indicate locations of membrane ruffles. The ΔphoPQ(pGEMT) and ΔicsA(pGEMT) were used as empty plasmid controls.
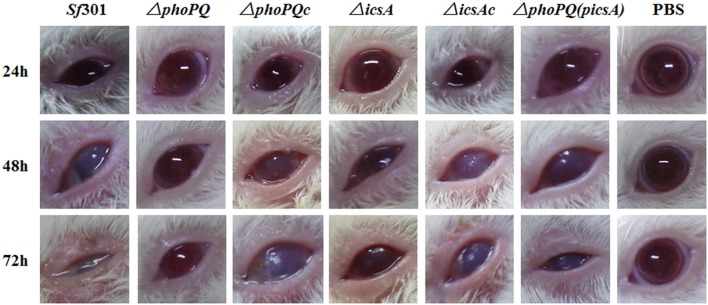
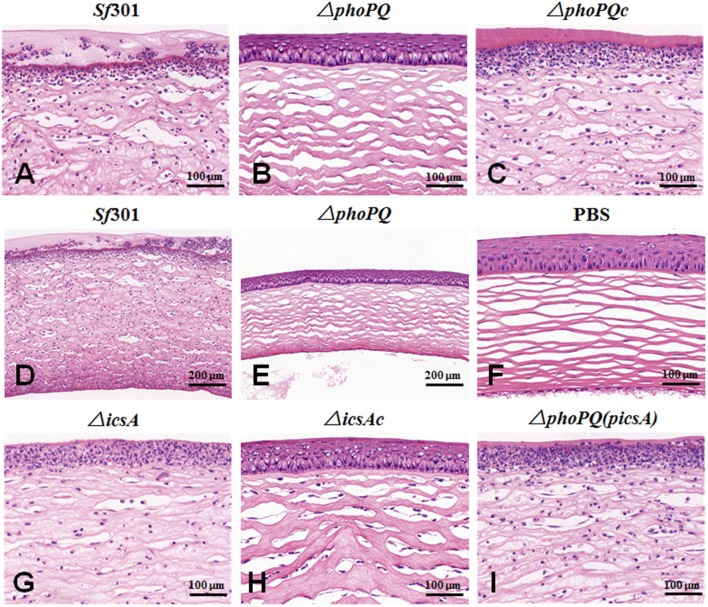
The Guinea pig Sereny test was used to determine the virulence of Sf301, ΔphoPQ, and ΔphoPQc. Guinea pigs were infected with 109 CFU per eye and observed at different time points. At 24 h post-infection, the guinea pig eyes infected with Sf301 displayed keratoconjunctivitis with or without purulence (+ or ++). At 48 and 72 h post-infection, the inflammation exacerbated with great purulence (+++). The ΔphoPQ-infected guinea pig eyes displayed no obvious inflammation (–) at 24 h post-infection, and a slight keratoconjunctivitis without purulence (+) after 48 h. In ΔphoPQc-infected guinea pig eyes, the inflammation reaction was similar to that of Sf301. Guinea pigs inoculated with PBS were used as a negative control (Figure 3, Table 3). At 72 h post-infection, all guinea pigs were euthanized with pentobarbital and the eyes were removed, stained with hematoxylin and eosin and pathological examinations were carried out. Sf301-infected eyes showed typical inflammatory reactions including luminal debris, epithelial desquamation, neutrophil infiltration and submucosal edema along with severe ulcers (Figures 4A,D). In contrast, ΔphoPQ-infected eyes showed very small numbers of neutrophils in the mucosal lamina or submucosa, and minimal edema in the submucosa (Figures 4B,E). The inflammatory lesions induced by ΔphoPQc recovered to the Sf301 level (Figure 4C).
Figure 3.
Keratoconjunctivitis in guinea pig eyes produced by S. flexneri strains. Female guinea pigs (age 6 weeks, about 300 g) were inoculated with 109 CFU of bacteria per eye (six guinea pig eyes in each group), and observed at 24, 48, and 72 h. Guinea pig eye inoculation with PBS served as a negative control.
Table 3.
Degree of keratoconjunctival inflammation in guinea pig eyes infected with S. flexneri strains.
| Bacterial strain | 24 h | 48 h | 72 h | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 1 | 2 | 3 | 4 | 5 | 6 | 1 | 2 | 3 | 4 | 5 | 6 | |
| Sf301 | ++ | + | ++ | + | ++ | + | +++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ | +++ | +++ | +++ |
| ΔphoPQ | – | – | – | – | – | – | – | – | + | – | – | – | – | + | + | – | + | – |
| ΔphoPQc | ++ | + | + | + | + | ++ | +++ | +++ | ++ | ++ | +++ | +++ | +++ | +++ | ++ | +++ | +++ | +++ |
| ΔicsA | – | + | + | – | + | – | + | + | + | – | + | + | + | ++ | + | – | + | ++ |
| ΔicsAc | ++ | ++ | + | ++ | + | + | +++ | +++ | ++ | +++ | ++ | ++ | +++ | +++ | ++ | +++ | +++ | ++ |
| ΔphoPQ(picsA) | + | ++ | + | + | + | + | ++ | ++ | ++ | ++ | ++ | + | ++ | +++ | ++ | ++ | +++ | ++ |
| PBS | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
The degree of keratoconjunctival inflammation in each of the six guinea pigs infected with S. flexneri strains and PBS at 24, 48, and 72 h (n = 6). Guinea pig keratoconjunctivitis test was rated as follows: –, no disease or mild irritation; +, mild conjunctivitis or late development and/or rapid clearing of symptoms; ++, keratoconjunctivitis without purulence; and +++, fully developed keratoconjunctivitis with purulence.
Figure 4.
Pathological examination of guinea pig eyes infected with S. flexneri strains in Sereny test. Guinea pigs of the Sereny test were euthanized at 72 h post-infection and the eyes were removed and fixed in 4% formalin in PBS (pH 7.2). After hematoxylin and eosin (H&E) staining, eye sections were examined using a light microscope at ×400 (A–I) or ×200 (D,E) magnification.
Deletion of phoPQ decreased the ability of S. flexneri to withstand the challenge of environmental stress
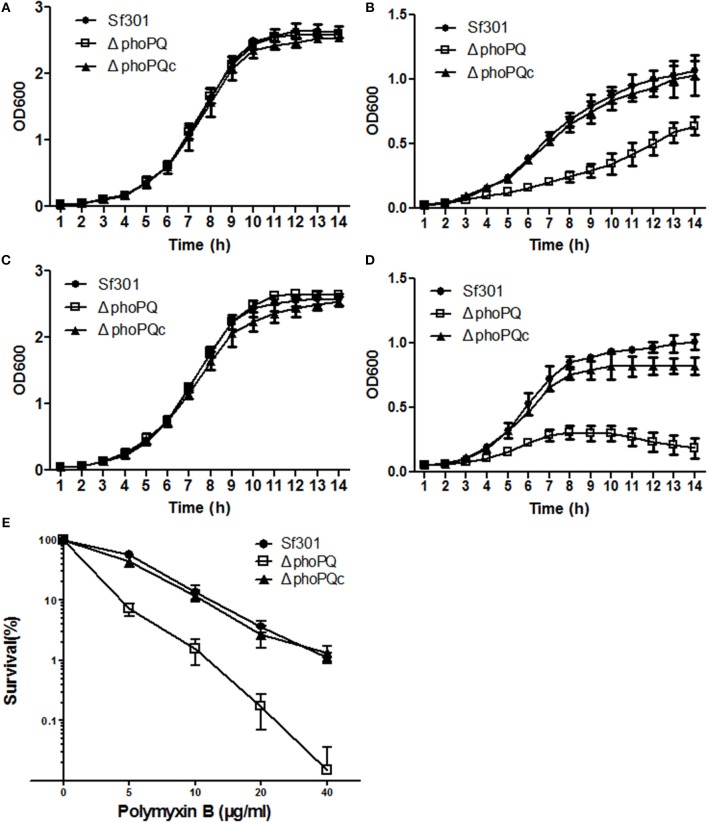
Extracellular Mg2+, pH and antimicrobial peptides have been described as input signals of the PhoP/PhoQ system (Garcia Vescovi et al., 1996; Gunn and Miller, 1996; Bearson et al., 1998; Groisman, 2001; Lejona et al., 2003; Barchiesi et al., 2012). Therefore, the role of PhoPQ in S. flexneri ability to withstand low Mg2+, acidic pH, or the presence of polymyxin B was analyzed. Sf301, ΔphoPQ and ΔphoPQc were grown in different conditions and growth curves were determined. In high Mg2+ medium (10 mM), the three strains showed no difference in growth pattern (Figure 5A), while in low Mg2+ medium (10 μM), ΔphoPQ showed limited growth compared to that of Sf301. At mid-log phase (8 h), Sf301 reached an OD600 of 0.68, but ΔphoPQ only reached an OD600 of 0.25 and took about 6 h more to reach an OD600 of 0.64. This growth deficiency was rescued by complementation with pphoPQ (Figure 5B). In neutral medium (pH 7), growth curves showed no difference among the three strains (Figure 5C). However, in the acidic medium (pH 5), Sf301 and ΔphoPQc showed only limited growth while ΔphoPQ was unable to grow (Figure 5D). At different concentrations of polymyxin B (5, 10, 20, 40 μg/ml), the survival rates of ΔphoPQ were significantly lower than that of Sf301 (P < 0.05), and the resistance defect of ΔphoPQ was restored by complementation with pphoPQ (Figure 5E).
Figure 5.
PhoPQ is required for Shigella to resist an environment of low Mg2+, acidic pH, or the presence of polymyxin B. Sf301, ΔphoPQ and ΔphoPQc were grown in N minimal medium supplemented with 10 mM MgCl2 (A) or 10 μM MgCl2 (B). The OD600 was determined every 60 min for 14 h. Sf301, ΔphoPQ and ΔphoPQc were grown in E glucose broth at pH 7 (C) or pH 5 (D). The OD600 was determined every 60 min for 14 h. Sf301, ΔphoPQ and ΔphoPQc were diluted in sterile 0.85% saline to about 5 × 103 cells per ml, and exposed for 1 h at 37°C to various concentrations of polymyxin B. Survivors were measured by plating on LB. The survival rate was calculated as the number of bacteria treated with polymyxin B divided by that of the untreated control (E). Results are mean from three independent assays performed in duplicate. Error bars correspond to standard deviations.
Identification of PhoP-regulated genes in S. flexneri
DNA microarray was used to compare the transcriptional profiles of Sf301 and ΔphoPQ at middle-log phase (6 h) or early-stationary phase (10 h) under LB growth conditions. At middle-log phase, 117 differentially expressed genes (DEGs) were identified. Among them, 32 genes were up-regulated and 85 genes were down-regulated in ΔphoPQ (Table 4). At early-stationary phase, 54 DEGs were identified, with 19 genes up-regulated and 35 genes down-regulated in ΔphoPQ (Table S5). The 117 DEGs were involved in metal ion transport (katE, narU, bfr), acid resistance (hdeABCD, gadAB, yhiWX, xasA), lipopolysaccharides (LPS) modification and antibacterial peptide tolerance (rfbU, mdoB, slyB, pagP, msbB2, pmrD), signal transduction (phoPQ, rstA, cstA), bacterial virulence (icsA, virK), respiratory and energy metabolism (hyaABCDEF, appABC) (Table 4). Among them, 44 DEGs were verified by qRT-PCR, with 38 giving a result consistent with that of the DNA microarray (Table 4).
Table 4.
Differentially expressed genes of ΔphoPQ compared to Sf301 by microarray and qRT-PCR at middle-log phase.
| Gene | Expression ratio (mutant/WT)a | Location | Description or predicted function | ||
|---|---|---|---|---|---|
| Microarrayb | P-valuesc | qRT-PCRd | |||
| AMINO ACID TRANSPORT AND METABOLISM | |||||
| gntT | 0.09 | <0.0001 | ND | Chromosome | High-affinity transport permease for gluconate |
| tdcC | 3.19 | 0.0224 | 1.01 ± 0.21 | Chromosome | Threonine/serine transporter TdcC |
| edd | 0.27 | 0.0011 | ND | Chromosome | Phosphogluconate dehydratase |
| nanA | 0.27 | 0.0129 | 0.45 ± 0.13 | Chromosome | N-Acetylneuraminate lyase |
| exuT | 0.32 | 0.0252 | ND | Chromosome | Transport protein of hexuronates |
| ybaS | 0.32 | 0.0002 | 0.15 ± 0.04 | Chromosome | Glutaminase |
| gntU | 0.33 | 0.0002 | ND | Chromosome | Low affinity gluconate transporter |
| xasA | 0.35 | 0.0227 | 0.48 ± 0.01 | Chromosome | Acid sensitivity protein, putative transporter |
| ybaT | 0.39 | 0.0006 | ND | Chromosome | Putative amino acid/amine transport protein |
| ggt | 0.41 | 0.0001 | ND | Chromosome | Gamma-glutamyltranspeptidase |
| poxB | 0.41 | 0.0011 | ND | Chromosome | Pyruvate dehydrogenase |
| sdaA | 0.44 | 0.0499 | ND | Chromosome | L-serine deaminase |
| ybdR | 0.44 | 0.0005 | 0.68 ± 0.12 | Chromosome | Putative oxidoreductase |
| nanT | 0.48 | 0.0297 | ND | Chromosome | Putative sialic acid transporter |
| CARBOHYDRATE TRANSPORT AND METABOLISM | |||||
| gntK | 0.11 | <0.0001 | 0.65 ± 0.24 | Chromosome | Gluconate kinase 1 |
| shf | 0.35 | 0.0001 | 0.25 ± 0.09 | pCP301 | Putative carbohydrate transport protein |
| ptsG | 0.32 | 0.0200 | ND | Chromosome | Glucose-specific PTS system IIBC components |
| amyA | 0.33 | 0.0003 | 0.43 ± 0.04 | Chromosome | Cytoplasmic alpha-amylase |
| gapC | 0.42 | 0.0045 | ND | Chromosome | Glyceraldehyde-3-phosphate dehydrogenase |
| yhcH | 0.43 | 0.0174 | ND | Chromosome | Hypothetical protein |
| treA | 0.44 | 0.0004 | ND | Chromosome | Trehalase |
| talA | 0.47 | 0.0001 | ND | Chromosome | Transaldolase A |
| yhcI | 0.47 | 0.0196 | ND | Chromosome | N-acetylmannosamine kinase |
| glk | 0.50 | 0.0002 | ND | Chromosome | Glucokinase |
| fucA | 2.19 | 0.0005 | ND | Chromosome | L-Fuculose phosphate aldolase |
| VIRULENCE | |||||
| icsA/virG | 0.33 | 0.0001 | 0.24 ± 0.08 | pCP301 | Intra- and intercellular spread, adhesion |
| virK | 0.43 | 0.0002 | 0.18 ± 0.05 | pCP301 | Hypothetical protein |
| CELL WALL/MEMBRANE/ENVELOPE BIOGENESIS | |||||
| rfbU | 0.32 | 0.0010 | 0.45 ± 0.12 | pCP301 | UDP-sugar hydrolase |
| mdoB | 0.34 | <0.0001 | 0.22 ± 0.08 | Chromosome | Phosphoglycerol transferase I |
| slyB | 0.41 | 0.0025 | 0.19 ± 0.06 | Chromosome | Putative outer membrane protein |
| slp | 0.32 | 0.0006 | 0.22 ± 0.08 | Chromosome | Outer membrane protein induced after carbon starvation |
| ecnB | 0.48 | 0.0007 | 0.65 ± 0.22 | Chromosome | Entericidin B membrane lipoprotein |
| pagP | 0.48 | 0.0120 | 0.15 ± 0.04 | Chromosome | Palmitoyl transferase |
| nmpC | 6.35 | 0.0007 | 3.15 ± 0.42 | Chromosome | Putative outer membrane porin protein C precursor |
| msbB2 | 0.46 | 0.0031 | 0.46 ± 0.08 | pCP301 | Lipid A biosynthesis |
| pmrD | 0.48 | 0.0166 | 0.42 ± 0.12 | Chromosome | Polymyxin resistance protein B |
| ACID RESISTANCE | |||||
| hdeD | 0.30 | 0.0078 | ND | Chromosome | Acid-resistance membrane protein |
| hdeB | 0.31 | 0.0132 | ND | Chromosome | Acid-resistance protein |
| hdeA | 0.32 | 0.0048 | 0.18 ± 0.03 | Chromosome | Acid-resistance protein |
| gadA | 0.27 | 0.0067 | 0.01 ± 0.003 | Chromosome | Glutamate decarboxylase isozyme |
| gadB | 0.31 | 0.0008 | ND | Chromosome | Glutamate decarboxylase isozyme |
| yhiW | 0.22 | <0.0001 | 0.27 ± 0.06 | Chromosome | Putative ARAC-type regulatory protein |
| yhiX | 0.24 | 0.0070 | ND | Chromosome | DNA-binding transcriptional regulator GadX |
| ENERGY PRODUCTION AND CONVERSION | |||||
| fadE | 2.31 | 0.0006 | ND | Chromosome | Acyl-CoA dehydrogenase |
| gltA | 2.66 | 0.0143 | 3.25 ± 0.72 | Chromosome | Type II citrate synthase |
| tdcD | 3.38 | 0.0427 | ND | Chromosome | Propionate/acetate kinase |
| tdcE | 3.46 | 0.0058 | ND | Chromosome | Formate acetyltransferase 3 |
| hyaC | 0.30 | 0.0014 | ND | Chromosome | Hydrogenase 1 b-type cytochrome subunit |
| hyaF | 0.30 | 0.0015 | 0.11 ± 0.03 | Chromosome | Hydrogenase-1 operon protein hyaF |
| hyaB | 0.30 | 0.0020 | 0.07 ± 0.01 | Chromosome | Hydrogenase 1 large subunit |
| appC | 0.32 | 0.0018 | ND | Chromosome | Third cytochrome oxidase, subunit I |
| hyaD | 0.32 | 0.0040 | ND | Chromosome | Hydrogenase 1 maturation protease |
| hyaA | 0.32 | 0.0004 | ND | Chromosome | Hydrogenase-1 small subunit |
| hyaE | 0.33 | 0.0057 | ND | Chromosome | Hydrogenase-1 operon protein hyaE |
| appB | 0.36 | 0.0028 | ND | Chromosome | Third cytochrome oxidase, subunit II |
| appA | 0.47 | 0.0016 | 0.6 ± 0.02 | Chromosome | Phosphoanhydride phosphorylase |
| fucO | 2.01 | 0.0025 | ND | Chromosome | L-1,2-propanediol oxidoreductase |
| sdhD | 2.07 | 0.0425 | ND | Chromosome | Succinate dehydrogenase cytochrome b556 small membrane subunit |
| sucC | 2.07 | 0.0258 | ND | Chromosome | Succinyl-CoA synthetase subunit beta |
| sdhC | 2.26 | 0.0288 | 2.94 ± 0.75 | Chromosome | Succinate dehydrogenase cytochrome b556 large membrane subunit |
| ykgF | 2.30 | 0.0241 | ND | Chromosome | Hypothetical protein |
| ykgE | 2.37 | 0.0301 | 3.21 ± 0.72 | Chromosome | Putative dehydrogenase subunit |
| INORGANIC ION TRANSPORT AND METABOLISM | |||||
| katE | 0.39 | 0.0103 | 0.32 ± 0.05 | Chromosome | Hydroperoxidase II |
| narU | 0.43 | 0.0004 | ND | Chromosome | Nitrite extrusion protein 2 |
| bfr | 0.48 | 0.0001 | ND | Chromosome | Bacterioferritin |
| LIPID TRANSPORT AND METABOLISM | |||||
| SF2149 | 0.45 | 0.0005 | ND | Chromosome | Lipid kinase |
| ybhO | 0.50 | 0.0147 | ND | Chromosome | Cardiolipin synthase 2 |
| glpF | 2.33 | 0.0129 | ND | Chromosome | Glycerol diffusion facilitator protein |
| POST-TRANSLATIONAL MODIFICATION, PROTEIN TURNOVER, CHAPERONES | |||||
| ybjX | 0.04 | <0.0001 | 0.58 ± 0.13 | Chromosome | Putative enzyme |
| cbpA | 0.31 | 0.0061 | ND | Chromosome | Curved DNA-binding protein CbpA |
| yccD | 0.44 | 0.0002 | 0.32 ± 0.09 | Chromosome | Chaperone-modulator protein CbpM |
| yeaA | 2.02 | 0.0026 | 1.75 ± 0.33 | Chromosome | Methionine sulfoxide reductase B |
| cysU | 2.08 | 0.0264 | 1.64 ± 0.52 | Chromosome | Sulfate/thiosulfate transporter subunit |
| ipgA | 2.38 | 0.0003 | ND | pCP301 | IpgA, similarities to IpgE, putative chaperone |
| SECONDARY METABOLITES BIOSYNTHESIS, TRANSPORT AND CATABOLISM | |||||
| ycaC | 0.43 | 0.0022 | ND | Chromosome | Hypothetical protein |
| SIGNAL TRANSDUCTION MECHANISMS | |||||
| phoP | 0.002 | 0.0008 | 0 | Chromosome | DNA-binding transcriptional regulator PhoP |
| phoQ | 0.002 | 0.0003 | 0 | Chromosome | Sensor protein PhoQ |
| rstA | 0.07 | 0.0003 | 0.23 ± 0.05 | Chromosome | DNA-binding transcriptional regulator RstA |
| cstA | 2.08 | 0.0256 | 1.02 + 0.32 | Chromosome | Carbon starvation protein |
| TRANSCRIPTION | |||||
| glcC | 2.47 | 0.0186 | 3.52 + 1.34 | Chromosome | DNA-binding transcriptional regulator GlcC |
| yhiE | 0.25 | 0.0002 | ND | Chromosome | Hypothetical protein |
| cbl | 0.41 | 0.0328 | ND | Chromosome | Transcriptional regulator Cbl |
| adiY | 0.44 | 0.0016 | ND | Chromosome | Putative ARAC-type regulatory protein |
| cspH | 2.06 | 0.0308 | ND | Chromosome | Cold shock-like protein |
| melR | 2.09 | 0.0364 | 2.54 ± 0.83 | Chromosome | DNA-binding transcriptional regulator MelR |
| hcaR | 2.11 | 0.0096 | ND | Chromosome | DNA-binding transcriptional regulator HcaR |
| yjjM | 3.06 | 0.0004 | ND | Chromosome | Hypothetical protein |
| TRANSLATION, RIBOSOMAL STRUCTURE AND BIOGENESIS | |||||
| SF4448 | 0.49 | 0.0355 | ND | Chromosome | tRNA |
| yhaR | 3.65 | 0.0041 | 5.07 ± 1.21 | Chromosome | Hypothetical protein |
| SF4512 | 0.50 | 0.0293 | ND | Chromosome | tRNA |
| GENERAL FUNCTION PREDICTION ONLY | |||||
| yoaE | 7.14 | <0.0001 | 3.18 ± 0.83 | Chromosome | Putative transport protein |
| yjgB | 0.41 | 0.0004 | ND | Chromosome | Putative oxidoreductase |
| yjbJ | 0.41 | 0.0007 | ND | Chromosome | Putative stress-response protein |
| SF1795 | 0.48 | <0.0001 | ND | Chromosome | Putative glyceraldehyde-3-phosphate dehydrogenase A |
| yciG | 0.49 | 0.0003 | ND | Chromosome | Hypothetical protein |
| ykgG | 2.15 | 0.0080 | ND | Chromosome | Putative transporter |
| SF3152 | 3.57 | 0.0110 | ND | Chromosome | Putative L-serine deaminase |
| FUNCTION UNKNOWN | |||||
| yrbL | 0.02 | <0.0001 | 0.07 ± 0.01 | Chromosome | Hypothetical protein |
| SF1400 | 0.04 | 0.0005 | 0.21 ± 0.04 | Chromosome | Hypothetical protein |
| ycgW | 0.09 | <0.0001 | ND | Chromosome | Hypothetical protein |
| SF2261 | 0.17 | 0.0051 | 0.04 ± 0.01 | Chromosome | Hypothetical protein |
| SF1401 | 0.34 | 0.0034 | ND | Chromosome | Hypothetical protein |
| ygaM | 0.44 | 0.0029 | ND | Chromosome | Hypothetical protein |
| yejG | 2.72 | 0.0114 | 3.88 ± 0.92 | Chromosome | Hypothetical protein |
| SF0979 | 0.30 | 0.0018 | ND | Chromosome | Hydrogenase-1 operon protein |
| SF1736 | 0.31 | 0.0013 | ND | Chromosome | Hypothetical protein |
| elaB | 0.41 | 0.0002 | ND | Chromosome | Hypothetical protein |
| yjiD | 0.41 | 0.0303 | ND | Chromosome | Hypothetical protein |
| ybfG | 0.43 | 0.0054 | ND | Chromosome | Hypothetical protein |
| SF4340 | 0.43 | 0.0016 | ND | Chromosome | Putative carnitine operon oxidoreductase |
| SF3143 | 0.45 | 0.0002 | ND | Chromosome | Hypothetical protein |
| SF2823 | 0.47 | 0.0092 | ND | Chromosome | Hypothetical protein |
| yqjE | 0.48 | 0.0065 | 0.58 ± 0.21 | Chromosome | Hypothetical protein |
| yqjD | 0.49 | 0.0031 | ND | Chromosome | Hypothetical protein |
| SF0551 | 0.50 | 0.0197 | ND | Chromosome | Putative homeobox protein |
| ipgB1 | 2.03 | 0.0017 | ND | Chromosome | IpgB1, secreted by the Mxi-Spa machinery, function unknown |
| SF1446 | 2.09 | 0.0030 | ND | Chromosome | Hypothetical protein |
| SF0572 | 2.16 | 0.0447 | ND | Chromosome | Hypothetical protein |
WT, wild type; ND, not determined.
The differentially expressed genes of microarrays were defined by change ratio> = 2, P < 0.05.
The P-values for the DEGs of microarrays.
qRT-PCR data are given as means ± standard deviations of results from three independent experiments.
To identify S. flexneri PhoP-regulated genes, the online relational databases (http://genolist.pasteur.fr) was used to search the genes with putative PhoP-binding motif in their promoter regions. A PhoP recognition motif was generated based on conserved pattern PhoP box [5′-(T/G) GTTTA-N5-(T/G) GTTTA- 3′] identified in S. typhimurium and E. coli (Kato et al., 1999; Lejona et al., 2003). The Sf301 genome was searched and 38 putative PhoP recognition motifs were detected (Table 5). Among these genes/operons, phoPQ, mgtA, slyB, rstAB, hdeAB, pagP, yrbL, yoaE, yhiWX, and shf –rfbU-virK-msbB2 were reported as members of the PhoPQ regulon in other bacteria (Kato et al., 1999; Lejona et al., 2003; Minagawa et al., 2003; Zwir et al., 2005), while icsA and ipaH7.8 have not been reported as PhoPQ-regulated genes before (Table 5). The transcriptional levels of these 12 genes or operons showed differential expression both in the microarray and qRT-PCR analyses (Table 4).
Table 5.
Prediction of PhoP-regulated genes in Sf301.
| Gene | Location | Predicted PhoP binding sites | Description or predicted function |
|---|---|---|---|
| phoP | Chromosome | tGGTTTAtttaaTGTTTAc | DNA-binding transcriptional regulator PhoP |
| yoaE | Chromosome | aTGTTTAactccCGTTTAa | Transporter |
| SF1755 | Chromosome | cCGTTTAaaattCGTTTAg | Porin |
| yrbL | Chromosome | tTGTTTAggtttTGTTTAa | Hypothetical protein |
| mgtA | Chromosome | tGGTTTAtcgttGGTTTAg | Magnesium-transporting ATPase MgtA |
| manX | Chromosome | tTAAACGggagtTAAACAt | PTS system mannose-specific transporter subunits IIAB |
| insA | Chromosome | cTAAACGaatttTAAACGg | Insertion element IS1 protein InsA |
| treR | Chromosome | cTAAACCaacgaTAAACCa | Trehalose repressor |
| mdoB | Chromosome | tTAAACGttggcTAAACGg | Phosphoglycerol transferase I |
| uspF | Chromosome | cGcTTTAggtctGGTTTAt | Stress-induced protein |
| SF1625 | Chromosome | aGGaTTAaaattGGTTTAa | Hypothetical protein |
| sbcD | Chromosome | aGaTTTAtgacaGaTTTAt | Exonuclease SbcD |
| icsA | pCP301 | tGGTTgAggcttTGTTTAa | Hypothetical protein |
| yffB | Chromosome | tGaTTTAattctGGTTaAa | Reductase |
| ygaU | Chromosome | tGaTTTAattctGGTTaAa | LysM domain/BON superfamily protein |
| ygaC | Chromosome | aGGTTcAtcgcgGcTTTAt | Hypothetical protein |
| SF2987 | Chromosome | gTGTTTAcctctGcTTTAt | Hypothetical protein |
| rpsL | Chromosome | cGtTTTAttacgTGTTTAc | 30S ribosomal protein S12 |
| malP | Chromosome | tGGTTTgcactaGcTTTAa | Maltodextrin phosphorylase |
| SF4150 | Chromosome | aGGaTTAtctgcGGTTTtt | Hypothetical protein |
| ubiC | Chromosome | aGGTTcAacagcGtTTTAc | Chorismate pyruvate lyase |
| SF1773 | Chromosome | gaGTTTAatggcGGTTaAg | Acetyltransferase |
| yjjM | Chromosome | tGtTTTAaatcgGGTTTta | Hypothetical protein |
| lpdA | Chromosome | tTGTTTAaaaatTGTTaAc | Dihydrolipoamide dehydrogenase |
| cbpA | Chromosome | cTGTTTAaaataTGTTcAg | Curved DNA-binding protein CbpA |
| yajG | Chromosome | aGGTTTcgtcctGGTTTtt | Polymerase/proteinase |
| ycbK | Chromosome | tGcTTTAcgggcGGTTaAg | Hypothetical protein |
| ycjY | Chromosome | aGGTcTAatcatGaTTTAg | Hypothetical protein |
| sdaA | Chromosome | cGGTTTttgattaGTTTAa | L-serine deaminase |
| SF1507.1 | Chromosome | tGaTTTAttagaGcTTTAt | Transmembrane anchor protein |
| slyB | Chromosome | ttGTTTAtaattGGTTgAt | Hypothetical protein |
| ybiC | Chromosome | aTGgTTAactccTGTTTAt | Hypothetical protein |
| mipA | Chromosome | tTGTTTAaggaaTGaTTAa | structural protein MipA |
| shf | pCP301 | tTGTTTAtgaatTGTTgAt | Carbohydrate transport protein |
| dppA | Chromosome | tTtTTTAatcttTGTTTgt | Dipeptide transport protein |
| hdeA | Chromosome | cTGTaTAtgtcaTGTTgAt | Acid stress chaperone HdeA |
| yhiW | Chromosome | aTGTTTgggcgaTtTTTAt | Putative ARAC-type regulatory protein |
| ipaH7.8 | pCP301 | aTGTgTAtcgttTtTTTAc | Invasion plasmid antigen |
The genes with a putative PhoP-binding motif in Sf301 were searched based on the PhoP box pattern [5′-(T/G) GTTTA-N5-(T/G) GTTTA-3′]. The putative PhoP binding sites in the promoter region were restricted to 400 bp before the start codon with at most 2 nt not matching. The bold sequences represent the PhoP box pattern in the predicted PhoP binding sites.
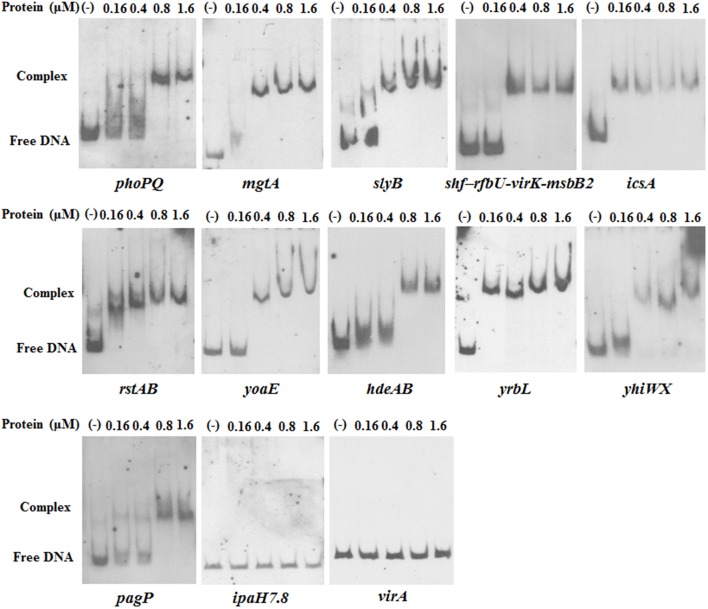
To verify those 12 predicted PhoP target genes/operons above in Shigella, EMSAs were performed. The recombinant PhoP-P resulted in a mobility shift of the fragments upstream of 11 genes/operons (phoPQ, mgtA, slyB, icsA, shf –rfbU-virK-msbB2, rstAB, yoaE, hdeAB, yrbL, yhiWX, and pagP) in a concentration-dependent manner, but did not bind to the fragment upstream of ipaH7.8 (Figure 6). As a negative control, the DNA fragment of virA coding sequence (without PhoP box motif) did not form a complex with PhoP under the same conditions (Figure 6).
Figure 6.
EMSA analysis of PhoP with the putative promoter regions. His-tagged PhoP was purified and phosphorylated (PhoP-P) by incubation with 50 mM acetylphosphate. The putative promoter regions of the genes/operons phoPQ, mgtA, slyB, shf–rfbU-virK-msbB2, icsA, rstAB, yoaE, hdeAB, yrbL, yhiWX, pagP, and ipaH7.8 were PCR-amplified. DNA probes were purified and labeled with digoxigenin. Gel shift reactions were performed by incubating the labeled probe with increasing concentrations of PhoP-P (range, 0.16–1.6 μM). Lane 1 of each blot contained a no-protein control. All samples were electrophoresed on a non-denaturing polyacrylamide gel and blotted onto a nylon membrane. After incubation with anti-digoxigenin antibody, CSPD chemiluminescent reagent was added. The DNA fragment within the virA coding region was used as a negative control.
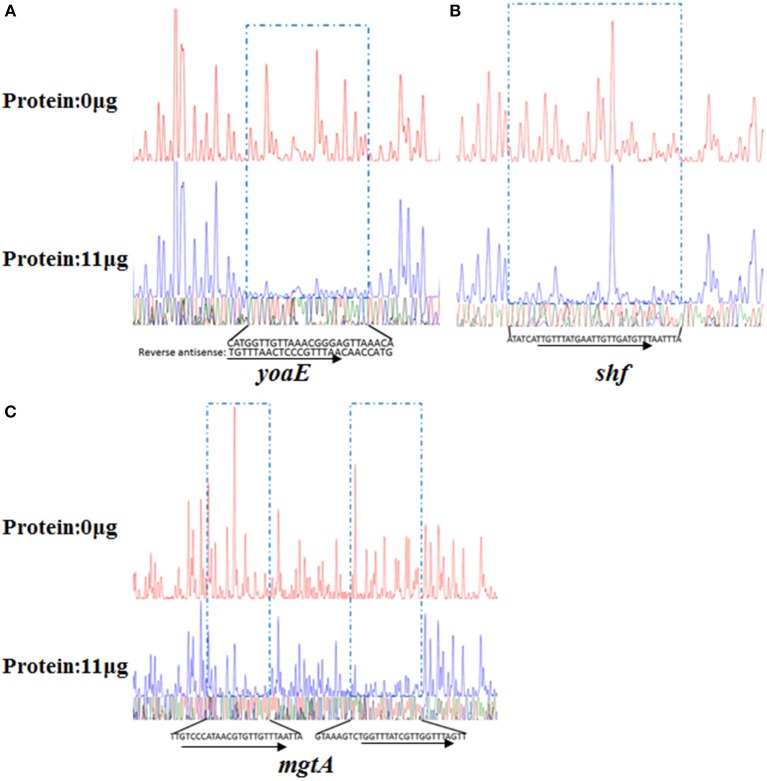
The PhoP-P binding motif sequences in the promoters of yoaE, shf and mgtA were identified by DNase I footprinting assay. A 25-nt protected sequence in the promoter region of yoaE (−107 to −83 bp) was identified (Figure 7A), and a 35-nt protected sequence was located at −115 to −81 bp in the upstream of the translational start site of shf (Figure 7B). There exist two protected sequences in the mgtA promoter (−184 to −159 bp and −152 to −124 bp) (Figure 7C). All protected sequences were in accordance with PhoP binding consensus motif (Figure 7, Table 5).
Figure 7.
Identification of the PhoP-protected cis-elements in the promoter regions of mgtA, yoaE, and shf using DNase I footprinting assay. Probes used were labeled with FAM dye and were illustrated in Material and Methods. Different amounts of PhoP protein were used for the assay. The fmol DNA Cycle Sequencing System was used for DNA sequencing reactions and the four sequencing results (G, A, T, and C) were marked with four different colors separately and then merged together. The electropherograms were aligned together with the usage of GeneScan-LIZ500. The areas protected of yoaE (A), shf (B), and mgtA (C) from DNase I by PhoP were sequenced, and inside the sequence, bases consistent with the PhoP box motif are arrowed.
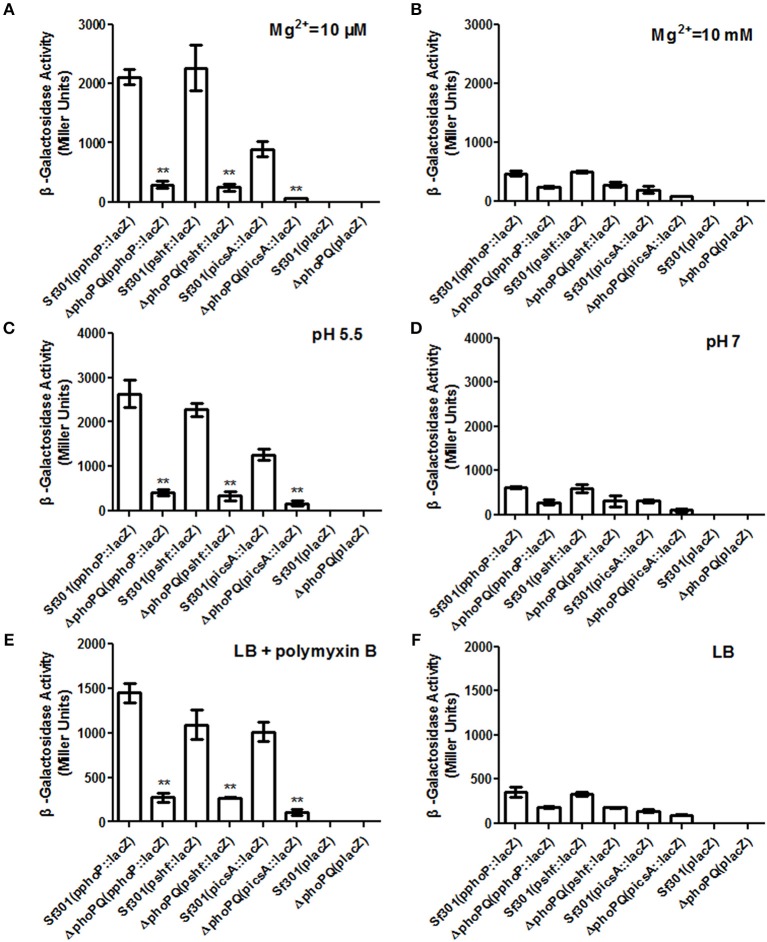
Furthermore, the transcriptional levels of genes with PhoP binding activities in low Mg2+, acidic pH, or presence of polymyxin B were detected by qRT-PCR. Among those genes, transcriptional levels of phoP, shf and icsA were up-regulated significantly in all three environmental stress conditions (Tables S6–S8). Three LacZ reporter plasmids with the promoter regions of the genes (pphoP::lacZ, pshf ::lacZ, and picsA::lacZ) were then constructed to confirm PhoPQ regulation on those genes expression. After transformation of the plasmids into ΔphoPQ or Sf301 and culture in different mediums, β-galactosidase activity was detected. In low Mg2+ (10 μM) medium, the expression of phoP, shf, and icsA in Sf301 was 7.2, 9, and 12.9-fold higher, respectively, than that in ΔphoPQ (Figure 8A), while in high Mg2+ (10 mM) medium, the expression of phoP, shf, and icsA in Sf301 was only 1.9, 1.8, and 2.2-fold higher, respectively, than that in ΔphoPQ (Figure 8B). Under acidic pH (pH 5.5) conditions, the expression of phoP, shf, and icsA in Sf301 was 6.4, 6.6, and 7.1-fold higher, respectively, than that in ΔphoPQ (Figure 8C). In contrast, at pH 7, the expression of phoP, shf, and icsA in Sf301 was only 2.2, 2, and 3-fold higher, respectively, than that in ΔphoPQ (Figure 8D). In the presence of polymyxin B (25 μg/ml) in LB medium, the expression of phoP, shf and icsA in Sf301 was 5.2, 4, and 9.2-fold higher, respectively, than that in ΔphoPQ (Figure 8E), while in LB medium only, the expression of phoP, shf and icsA in Sf301 was only 2, 1.9, and 1.5-fold higher, respectively, than that in ΔphoPQ (Figure 8F). It suggested that the expressions of phoP, shf and icsA were regulated by PhoPQ.
Figure 8.
β-Galactosidase activity of phoP, shf, and icsA LacZ reporter strains in conditions of low Mg2+, acidic pH, or presence of polymyxin B. β-Galactosidase activity from the pphoP::lacZ, pshf ::lacZ and picsA::lacZ transcriptional fusion in Sf301 and ΔphoPQ were determined. Bacteria were grown for 8 h in N medium with 10 μM MgCl2 for low Mg2+ (A) or N medium with 10 mM MgCl2 for high Mg2+ (B). Bacteria were grown for 8 h in E glucose broth at pH 5.5 (C) or pH 7 (D). Bacteria were grown for 6 h in LB then treated with 25 μg/ml polymyxin B (E) or without polymyxin B (F) for 1 h. The Sf301(placZ) and ΔphoPQ(placZ) were used as empty plasmid controls. The data correspond to mean values from three independent experiments performed in each case. Error bars correspond to standard deviations. **P < 0.01.
Validation of IcsA regulation by PhoPQ in Shigella virulence
As virulence is the key factor in Shigella pathogenesis, we focused on searching for PhoP target genes that are associated with Shigella virulence. Four PhoP-regulated genes (rstA, icsA, yrbL, and yoaE) that may be involved in Shigella virulence were deleted from Sf301, respectively. The virulence of these mutant strains was evaluated by the gentamicin protection assay on HeLa cells and only ΔicsA decreased virulence in HeLa cells compared to Sf301 (Figure S2). IcsA is one of the virulence factors required for Shigella pathogenesis (Bernardini et al., 1989; Brotcke Zumsteg et al., 2014), and its expression is regulated by PhoPQ based on results of the microarray, qRT-PCR, EMSA and β-galactosidase activity assay in our study. The transcriptional level of icsA in ΔphoPQ was significantly reduced both in the microarray (3-fold down-regulated) and qRT-PCR (4.2-fold down-regulated) compared to that of Sf301 (Table 4). A highly conserved motif is found in the promoter region of icsA and PhoP-P results in a mobility shift of the fragments upstream of icsA (Table 5, Figure 6). The β-galactosidase activities of icsA in Sf301 were significantly higher than that in ΔphoPQ (12.9, 7.1, and 9.2-fold higher, respectively) in the environments of low Mg2+, acidic pH or presence of polymyxin B (Figures 8A,C,E). As the phoPQ knockout diminished S. flexneri virulence, an icsA expression plasmid (picsA) was introduced into the ΔphoPQ strain [ΔphoPQ(picsA)] to observe whether virulence of ΔphoPQ could recover. A Shigella icsA deletion mutant strain (ΔicsA) and its complementation strain (ΔicsAc) served as controls. The invasion rate of HeLa cells or Caco-2 cells by ΔphoPQ(picsA) was 6.4 and 5.7-fold higher than that of ΔphoPQ, respectively (Figures 9A,B). ΔphoPQ(picsA) resulted in more membrane ruffles indicative of actin cytoskeleton changes in HeLa cells compared to ΔphoPQ (Figure 2F). In the guinea pig Sereny test, guinea pigs inoculated with ΔphoPQ(picsA) showed restored virulence (24 h + or ++ and 48 h ++ or +++) compared to ΔphoPQ (24 h– and 48 h– or +) (Figure 3, Table 3). Guinea pig eyes infected with ΔphoPQ(picsA) showed markedly more inflammatory reactions including epithelial desquamation and neutrophil infiltration in the pathological examination compared to ΔphoPQ-infected eyes (Figure 4I). As a control, the virulence of ΔicsA was decreased both in the gentamicin protection assay (2.7 and 2.2-fold lower, Figures 9A,B) and guinea pig Sereny test (24 h– or + and 48 h +, Figure 3, Table 3), compared to Sf301. It indicated that the virulence of ΔphoPQ(picsA) could be restored partly by complementation with picsA, but it still did not reach the level of Sf301.
Figure 9.
Invasion ability of ΔphoPQ(picsA) and ΔicsA into HeLa cells and Caco-2 cells. The gentamicin protection assay was used as a cellular model to evaluate the effect of ΔphoPQ complemented with icsA on the virulence of Shigella. Bacteria grown to logarithmic phase were added to the cells for 30 min. Gentamicin was then added to the medium to kill extracellular bacteria. Colonies of lysates on LB plates were counted. The invasion rate refers to the number of intracellular bacteria divided by the number of inoculated bacteria and multiplied by 10,000. (A) Bacterial ability to invade HeLa cells. (B) Bacterial ability to invade Caco-2 cells. Values are means ± standard deviations from three independent wells. **P < 0.01.
Discussion
The PhoPQ TCS is widely involved in the regulation of virulence in a variety of pathogenic bacteria, including Salmonella, Yersinia, Neisseria, Mycobacterium, Erwinia, Pseudomonas, and Serratia. The deletion of phoPQ in these organisms have shown a significantly decrease in virulence (Miller et al., 1989; Flego et al., 2000; Oyston et al., 2000; Johnson et al., 2001; Perez et al., 2001; Gooderham et al., 2009; Bozue et al., 2011; Barchiesi et al., 2012). Moss's previous works have shown that a phoP mutant decreased the inflammatory response and was more sensitive to PMNs in S. flexneri (Moss et al., 2000), which indicates that PhoPQ has the function of virulence regulation in Shigella. In the present study, we demonstrate that the PhoPQ system regulates the virulence of Shigella both in vivo and in vitro. In the HeLa cell and Caco-2 cell invasion models, the invasion ability of ΔphoPQ declined significantly compared with that of Sf301 (Figure 1) and no obvious membrane ruffling was observed in ΔphoPQ infection cells (Figure 2). In the guinea pig keratoconjunctivitis model, guinea pigs infected with ΔphoPQ displayed a slight conjunctival inflammation (Figure 3, Table 3) and fewer pathologic changes in the pathological examination (Figure 4).
Extracellular Mg2+, pH and antimicrobial peptides have been reported as input signals of the PhoPQ system and these signals can regulate the expression of PhoP in Salmonella and other bacteria (Garcia Vescovi et al., 1996; Gunn and Miller, 1996; Bearson et al., 1998; Lejona et al., 2003; Barchiesi et al., 2012; Shprung et al., 2012). Polymyxin B is an important antimicrobial agent extensively used clinically for the effective treatment of multi-drug resistant Gram-negative infections (Bergen et al., 2015; Brown and Dawson, 2017). As the Shigella phoPQ shares high similarity with that of Salmonella (Tables S2, S3), we predict that the Shigella PhoPQ also functions in responding the signals of extracellular Mg2+, pH and antimicrobial peptides. In the present study, we demonstrate that the PhoPQ system allows Shigella to tolerate scarce environmental Mg2+ availability, acidic pH, and high concentrations of polymyxin B. The ΔphoPQ showed growth deficiency in low Mg2+ or acidic pH conditions compared with Sf301 (Figures 5B,D). The survival rates of ΔphoPQ were significantly lower than those of Sf301 in the presence of polymyxin B (Figure 5E). We also demonstrate that the expression of PhoPQ is promoted under those three environmental stress conditions (low Mg2+, acidic pH or presence of polymyxin B) (Tables S6–S8).
Though the PhoPQ system shares similar functions in Gram-negative bacteria, the regulons of PhoPQ are diverse in different bacteria (Groisman, 2001). In the present study, we have screened the PhoPQ-regulated genes in Shigella. Firstly, DNA microarray was performed to compare the transcriptional profiles of Sf301 and ΔphoPQ, and 117 DEGs were found. The function of these genes were involved in metal ion transport (katE, narU, bfr), acid resistance (hdeABCD, gadAB, yhiWX, xasA), LPS modification and antibacterial peptide tolerance (rfbU, mdoB, slyB, pagP, msbB2, pmrD), signal transduction (phoPQ, rstA, cstA), bacterial virulence (icsA, virK), respiratory and energy metabolism (hyaABCDEF, appABC) (Table 4).
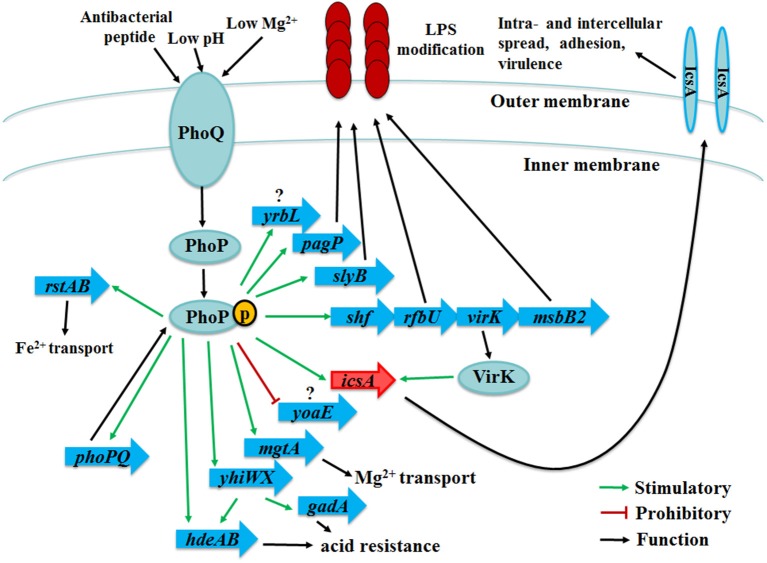
The promoter of PhoP-regulated genes contains a PhoP recognition motif [(T/G)GTTTA-5nt-(T/G)GTTTA] that has been termed the PhoP box in S. typhimurium and E. coli (Kato et al., 1999; Lejona et al., 2003). Considering the high conservation of phoPQ genes (Tables S2, S3), we screened 38 suspected PhoP target operons in Sf301 genome based on the PhoP box motif using the online relational databases (http://genolist.pasteur.fr). The putative PhoP-regulated genes were verified by EMSA (Figure 6). Eleven PhoP-regulated genes or operons were found. The phoPQ operon demonstrated autoregulation in Shigella (Figure 8). MgtA is involved in magnesium transport (Smith et al., 1998; Gall et al., 2016). PagP (Pilione et al., 2004; Bishop, 2005), SlyB (Plesa et al., 2006), RfbU (Yao and Valvano, 1994) and MsbB2 (Somerville et al., 1999; D'Hauteville et al., 2002) act on LPS modification and antibacterial peptide tolerance. HdeAB (Gajiwala and Burley, 2000) and YhiWX (Ma et al., 2002) function in acid resistance. RstAB is another two-component system that senses environmental pH and is required for the virulence of pathogenic E. coli (Cabeza et al., 2007; Jeon et al., 2008; Gao et al., 2015). IcsA is the first time discovered to be regulated by PhoPQ in our study and is involved in the cell-to-cell spreading process and bacterial virulence (Bernardini et al., 1989; Goldberg and Theriot, 1995; Brotcke Zumsteg et al., 2014). VirK is an essential virulence determinant involved in the expression of the gene icsA at the post-transcriptional level (Nakata et al., 1992; Detweiler et al., 2003). The functions of YrbL and YoaE are unknown (Figure 10). Through the DNase I footprinting assay, we demonstrated the Shigella PhoP binding sequences fit the PhoP box motif (Figure 7).
Figure 10.
Proposed regulation model of PhoPQ in Shigella. In this work, we have determined 11 genes or operons regulated by PhoPQ. Among these genes/operons, mgtA is involved in Mg2+ transport. pagP, slyB, rfbU and msbB2 act on LPS modification and antibacterial peptide tolerance. hdeAB and yhiWX function in acid resistance. icsA is the first time discovered to be regulated by PhoPQ and is involved in the cell-to-cell spreading process and bacterial virulence. virK is an essential virulence determinant involved in the expression of the icsA gene at the post-transcriptional level. rstAB is another two-component system that senses the environmental pH and regulates Fe2+ transport. phoPQ is autoregulated by PhoP. The functions of yrbL and yoaE are still unknown. Green arrows represent positive regulatory pathways. Red arrows represent negative regulatory pathways. Black arrows represent functions of PhoP-regulated genes.
To search for PhoP target genes that are associated with Shigella virulence, four genes that may be involved in virulence (rstA, icsA, yrbL, and yoaE) were deleted from Sf301, respectively. The virulence of those mutant strains was evaluated using the gentamicin protection assay on HeLa cells, and deletion of icsA decreased Shigella virulence (Figure S2). IcsA is a virulence factor involved in the cell-to-cell spreading process and required for Shigella pathogenesis (Bernardini et al., 1989; Ogawa et al., 2005). In the present study, we have demonstrated icsA is a positively PhoP-regulated gene and PhoPQ regulates S. flexneri virulence in an icsA-dependent manner. The transcriptional level of icsA in ΔphoPQ decreased significantly both in the microarray and qRT-PCR. The PhoP box motif was found in the promoter region of icsA and PhoP-P resulted in a mobility shift of the fragments upstream of icsA (Table 5, Figure 6). The promoter activities of icsA in Sf301 were significantly higher than that in ΔphoPQ in the environments of low Mg2+, acidic pH or presence of polymyxin B (Figures 8A,C,E). We introduced the icsA expression plasmid picsA into ΔphoPQ and found that the virulence of the ΔphoPQ(picsA) strain could be restored partly (Figures 2–4, 9, Table 3). Since the down-regulated level of virulence in ΔicsA is not as low as that in ΔphoPQ (Figures 2–4, 9, Table 3), we hypothesize icsA is not the only PhoP-regulated virulence factor. Besides icsA, the shf-rfbU-virK-msbB2 operon could be another virulence factor regulated by PhoP. This operon is demonstrated as being regulated by PhoP in this study and previous reports (Zwir et al., 2005). MsbB2 acts by catalyzing lipid A acylation (D'Hauteville et al., 2002; Goldman et al., 2008) and RfbU functions in the synthesis of O-antigen (Yao and Valvano, 1994). These two proteins are important in the synthesis of LPS, which is responsible for inflammation of the host. VirK is a cytoplasmic polypeptide required for the bacteria to spread into host cells by being involved in the full expression of the IcsA protein (Nakata et al., 1992; Detweiler et al., 2003).
In summary, we found that the two-component signal transduction system PhoP/PhoQ is involved in the regulation of S. flexneri virulence and ability to tolerate low environmental Mg2+, acidic pH, and antimicrobial peptide polymyxin B. We identified 117 DEGs, which were involved in Mg2+ transport, acid resistance, LPS modification, adhesion and invasion, respiratory and energy metabolism by comparing the transcriptional profiles of ΔphoPQ and Sf301. We screened out 38 potential PhoP target operons in S. flexneri by a bioinformatics search approach and 11 of them were identified to be PhoP-regulated genes/operons by EMSA assays and β-galactosidase assays. One of these genes, icsA (a well-known virulence factor), was the first time discovered to be regulated by PhoP. It indicates that the PhoPQ system modulates S. flexneri virulence in an icsA-dependent manner.
Author contributions
DQ, XC, and ZLi designed the study; ZLi, XC, MC, LY, YW, XW, ZLv, and YS completed all the experiments. ZLi performed the statistically analysis and made the figures; ZLi, DQ, and XC wrote and revised the manuscript.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Pr. Qi Jin (MOH Key Laboratory of Systems Biology of Pathogens, Institute of Pathogen Biology, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, China) for providing the S. flexneri 2a 301 strain generously.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (81571955, 81271791, 31400123), the National High-Tech and Development Plan of China (2014AA021404), the National Science and Technology Major Project of China (2012ZX10002002) and the Shanghai Municipal Committee of Science and Technology (14431900300, 15431900400).
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2017.02689/full#supplementary-material
The phoPQ gene disruption strategy. H1 and H2 refer to the homology regions of phoPQ, P1 and P2 refer to the primers of kan resistance gene.
The invasion ability of ΔicsA, ΔyoaE, ΔyrbL, and ΔrstA to HeLa cells. The bacteria that grew to logarithmic phase were added into the cells for 30 min infection. Then gentamicin was added into the medium to kill extracellular bacteria. Colonies of lysates on LB plates were counted. The invasion rate refered to the number of intracellular bacteria divided by that of inoculated bacteria and multiplied by 10,000. Values are means ± standard deviations from 3 independent wells. **P < 0.01.
Primers used for qRT-PCR verification of differentially expressed genes in ΔphoPQ.
Homological analysis of Sf301 phoP/phoQ with other homologues.
Homological analysis of Sf301 PhoP/PhoQ with other homologues.
Primers used for construction of ΔicsA, ΔyoaE, ΔyrbL, ΔrstA and their complementation strain.
Differentially expressed genes of ΔphoPQ compared to Sf301 by microarray and qRT-PCR at early-stationary phase.
The transcriptional levels of phoPQ and its regulated genes in different concentrations of Mg2+ condition.
The transcriptional levels of phoPQ and its regulated genes in different pH condition.
The transcriptional levels of phoPQ and its regulated genes with or without polymyxin B condition.
References
- Barchiesi J., Castelli M. E., Di Venanzio G., Colombo M. I., Garcia Vescovi E. (2012). The PhoP/PhoQ system and its role in Serratia marcescens pathogenesis. J. Bacteriol. 194, 2949–2961. 10.1128/JB.06820-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearson B. L., Wilson L., Foster J. W. (1998). A low pH-inducible, PhoPQ-dependent acid tolerance response protects Salmonella typhimurium against inorganic acid stress. J. Bacteriol. 180, 2409–2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benny E., Mesere K., Pavlin B. I., Yakam L., Ford R., Yoannes M., et al. (2014). A large outbreak of shigellosis commencing in an internally displaced population, Papua New Guinea, 2013. Western Pac. Surveill. Response J. 5, 18–21. 10.5365/wpsar.2014.5.2.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergen P. J., Bulman Z. P., Landersdorfer C. B., Smith N., Lenhard J. R., Bulitta J. B., et al. (2015). Optimizing polymyxin combinations against resistant gram-negative bacteria. Infect. Dis. Ther. 4, 391–415. 10.1007/s40121-015-0093-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini M. L., Fontaine A., Sansonetti P. J. (1990). The two-component regulatory system ompR-envZ controls the virulence of Shigella flexneri. J. Bacteriol. 172, 6274–6281. 10.1128/jb.172.11.6274-6281.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardini M. L., Mounier J., d'Hauteville H., Coquis-Rondon M., Sansonetti P. J. (1989). Identification of icsA, a plasmid locus of Shigella flexneri that governs bacterial intra- and intercellular spread through interaction with F-actin. Proc. Natl. Acad. Sci. U.S.A. 86, 3867–3871. 10.1073/pnas.86.10.3867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop R. E. (2005). The lipid A palmitoyltransferase PagP: molecular mechanisms and role in bacterial pathogenesis. Mol. Microbiol. 57, 900–912. 10.1111/j.1365-2958.2005.04711.x [DOI] [PubMed] [Google Scholar]
- Bozue J., Mou S., Moody K. L., Cote C. K., Trevino S., Fritz D., et al. (2011). The role of the phoPQ operon in the pathogenesis of the fully virulent CO92 strain of Yersinia pestis and the IP32953 strain of Yersinia pseudotuberculosis. Microb. Pathog. 50, 314–321. 10.1016/j.micpath.2011.02.005 [DOI] [PubMed] [Google Scholar]
- Brotcke Zumsteg A., Goosmann C., Brinkmann V., Morona R., Zychlinsky A. (2014). IcsA is a Shigella flexneri adhesin regulated by the type III secretion system and required for pathogenesis. Cell Host Microbe 15, 435–445. 10.1016/j.chom.2014.03.001 [DOI] [PubMed] [Google Scholar]
- Brown P., Dawson M. J. (2017). Development of new polymyxin derivatives for multi-drug resistant Gram-negative infections. J. Antibiot. 70, 386–394. 10.1038/ja.2016.146 [DOI] [PubMed] [Google Scholar]
- Cabeza M. L., Aguirre A., Soncini F. C., Vescovi E. G. (2007). Induction of RpoS degradation by the two-component system regulator RstA in Salmonella enterica. J. Bacteriol. 189, 7335–7342. 10.1128/JB.00801-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X., Zhang J., Chen M., Wu Y., Wang X., Chen J., et al. (2011). The effect of the potential PhoQ histidine kinase inhibitors on Shigella flexneri virulence. PLoS ONE 6:e23100. 10.1371/journal.pone.0023100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cano D. A., Martinez-Moya M., Pucciarelli M. G., Groisman E. A., Casadesus J., Garcia-Del Portillo F. (2001). Salmonella enterica serovar Typhimurium response involved in attenuation of pathogen intracellular proliferation. Infect. Immun. 69, 6463–6474. 10.1128/IAI.69.10.6463-6474.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang A. C., Cohen S. N. (1978). Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J. Bacteriol. 134, 1141–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang J., Chen J., Zhou D. (2005). Delineation and characterization of the actin nucleation and effector translocation activities of Salmonella SipC. Mol. Microbiol. 55, 1379–1389. 10.1111/j.1365-2958.2004.04480.x [DOI] [PubMed] [Google Scholar]
- Chen T., Leung R. K., Zhou Z., Liu R., Zhang X., Zhang L. (2014). Investigation of key interventions for shigellosis outbreak control in China. PLoS ONE 9:e95006. 10.1371/journal.pone.0095006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97, 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Detweiler C. S., Monack D. M., Brodsky I. E., Mathew H., Falkow S. (2003). virK, somA and rcsC are important for systemic Salmonella enterica serovar Typhimurium infection and cationic peptide resistance. Mol. Microbiol. 48, 385–400. 10.1046/j.1365-2958.2003.03455.x [DOI] [PubMed] [Google Scholar]
- D'Hauteville H., Khan S., Maskell D. J., Kussak A., Weintraub A., Mathison J., et al. (2002). Two msbB genes encoding maximal acylation of lipid A are required for invasive Shigella flexneri to mediate inflammatory rupture and destruction of the intestinal epithelium. J. Immunol. 168, 5240–5251. 10.4049/jimmunol.168.10.5240 [DOI] [PubMed] [Google Scholar]
- Flego D., Marits R., Eriksson A. R., Koiv V., Karlsson M. B., Heikinheimo R., et al. (2000). A two-component regulatory system, pehR-pehS, controls endopolygalacturonase production and virulence in the plant pathogen Erwinia carotovora subsp. carotovora. Mol. Plant Microbe Interact. 13, 447–455. 10.1094/MPMI.2000.13.4.447 [DOI] [PubMed] [Google Scholar]
- Gajiwala K. S., Burley S. K. (2000). HDEA, a periplasmic protein that supports acid resistance in pathogenic enteric bacteria. J. Mol. Biol. 295, 605–612. 10.1006/jmbi.1999.3347 [DOI] [PubMed] [Google Scholar]
- Gall A. R., Datsenko K. A., Figueroa-Bossi N., Bossi L., Masuda I., Hou Y. M., et al. (2016). Mg2+ regulates transcription of mgtA in Salmonella typhimurium via translation of proline codons during synthesis of the MgtL peptide. Proc. Natl. Acad. Sci. U.S.A. 113, 15096–15101. 10.1073/pnas.1612268113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Q., Ye Z., Wang X., Mu X., Gao S., Liu X. (2015). RstA is required for the virulence of an avian pathogenic Escherichia coli O2 strain E058. Infect. Genet. Evol. 29, 180–188. 10.1016/j.meegid.2014.11.022 [DOI] [PubMed] [Google Scholar]
- Garcia Vescovi E., Soncini F. C., Groisman E. A. (1996). Mg2+ as an extracellular signal: environmental regulation of Salmonella virulence. Cell 84, 165–174. 10.1016/S0092-8674(00)81003-X [DOI] [PubMed] [Google Scholar]
- Goldberg M. B., Theriot J. A. (1995). Shigella flexneri surface protein IcsA is sufficient to direct actin-based motility. Proc. Natl. Acad. Sci. U.S.A. 92, 6572–6576. 10.1073/pnas.92.14.6572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman S. R., Tu Y., Goldberg M. B. (2008). Differential regulation by magnesium of the two MsbB paralogs of Shigella flexneri. J. Bacteriol. 190, 3526–3537. 10.1128/JB.00151-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gooderham W. J., Gellatly S. L., Sanschagrin F., McPhee J. B., Bains M., Cosseau C., et al. (2009). The sensor kinase PhoQ mediates virulence in Pseudomonas aeruginosa. Microbiology 155(Pt 3), 699–711. 10.1099/mic.0.024554-0 [DOI] [PubMed] [Google Scholar]
- Gooderham W. J., Hancock R. E. (2009). Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 33, 279–294. 10.1111/j.1574-6976.2008.00135.x [DOI] [PubMed] [Google Scholar]
- Grabenstein J. P., Fukuto H. S., Palmer L. E., Bliska J. B. (2006). Characterization of phagosome trafficking and identification of PhoP-regulated genes important for survival of Yersinia pestis in macrophages. Infect. Immun. 74, 3727–3741. 10.1128/IAI.00255-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groisman E. A. (2001). The pleiotropic two-component regulatory system PhoP-PhoQ. J. Bacteriol. 183, 1835–1842. 10.1128/JB.183.6.1835-1842.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunn J. S., Miller S. I. (1996). PhoP-PhoQ activates transcription of pmrAB, encoding a two-component regulatory system involved in Salmonella typhimurium antimicrobial peptide resistance. J. Bacteriol. 178, 6857–6864. 10.1128/jb.178.23.6857-6864.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale T. L., Formal S. B. (1981). Protein synthesis in HeLa or Henle 407 cells infected with Shigella dysenteriae 1, Shigella flexneri 2a, or Salmonella typhimurium W118. Infect. Immun. 32, 137–144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon J., Kim H., Yun J., Ryu S., Groisman E. A., Shin D. (2008). RstA-promoted expression of the ferrous iron transporter FeoB under iron-replete conditions enhances Fur activity in Salmonella enterica. J. Bacteriol. 190, 7326–7334. 10.1128/JB.00903-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Q., Yuan Z., Xu J., Wang Y., Shen Y., Lu W., et al. (2002). Genome sequence of Shigella flexneri 2a: insights into pathogenicity through comparison with genomes of Escherichia coli K12 and O157. Nucleic Acids Res. 30, 4432–4441. 10.1093/nar/gkf566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson C. R., Newcombe J., Thorne S., Borde H. A., Eales-Reynolds L. J., Gorringe A. R., et al. (2001). Generation and characterization of a PhoP homologue mutant of Neisseria meningitidis. Mol. Microbiol. 39, 1345–1355. 10.1111/j.1365-2958.2001.02324.x [DOI] [PubMed] [Google Scholar]
- Kato A., Tanabe H., Utsumi R. (1999). Molecular characterization of the PhoP-PhoQ two-component system in Escherichia coli K-12: identification of extracellular Mg2+-responsive promoters. J. Bacteriol. 181, 5516–5520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lejona S., Aguirre A., Cabeza M. L., Vescovi E. G., Soncini F. C. (2003). Molecular characterization of the Mg2+-responsive PhoP-PhoQ regulon in Salmonella enterica. J. Bacteriol. 185, 6287–6294. 10.1128/JB.185.21.6287-6294.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leupold S., Busing P., Mas P. J., Hart D. J., Scrima A. (2017). Structural insights into the architecture of the Shigella flexneri virulence factor IcsA/VirG and motifs involved in polar distribution and secretion. J. Struct. Biol. 198, 19–27. 10.1016/j.jsb.2017.03.003 [DOI] [PubMed] [Google Scholar]
- Ma Z., Richard H., Tucker D. L., Conway T., Foster J. W. (2002). Collaborative regulation of Escherichia coli glutamate-dependent acid resistance by two AraC-like regulators, GadX and GadW (YhiW). J. Bacteriol. 184, 7001–7012. 10.1128/JB.184.24.7001-7012.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller S. I., Kukral A. M., Mekalanos J. J. (1989). A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. U.S.A. 86, 5054–5058. 10.1073/pnas.86.13.5054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minagawa S., Ogasawara H., Kato A., Yamamoto K., Eguchi Y., Oshima T., et al. (2003). Identification and molecular characterization of the Mg2+ stimulon of Escherichia coli. J. Bacteriol. 185, 3696–3702. 10.1128/JB.185.13.3696-3702.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss J. E., Fisher P. E., Vick B., Groisman E. A., Zychlinsky A. (2000). The regulatory protein PhoP controls susceptibility to the host inflammatory response in Shigella flexneri. Cell. Microbiol. 2, 443–452. 10.1046/j.1462-5822.2000.00065.x [DOI] [PubMed] [Google Scholar]
- Mounier J., Vasselon T., Hellio R., Lesourd M., Sansonetti P. J. (1992). Shigella flexneri enters human colonic Caco-2 epithelial cells through the basolateral pole. Infect. Immun. 60, 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakata N., Sasakawa C., Okada N., Tobe T., Fukuda I., Suzuki T., et al. (1992). Identification and characterization of virK, a virulence-associated large plasmid gene essential for intercellular spreading of Shigella flexneri. Mol. Microbiol. 6, 2387–2395. 10.1111/j.1365-2958.1992.tb01413.x [DOI] [PubMed] [Google Scholar]
- Ogawa M., Yoshimori T., Suzuki T., Sagara H., Mizushima N., Sasakawa C. (2005). Escape of intracellular Shigella from autophagy. Science 307, 727–731. 10.1126/science.1106036 [DOI] [PubMed] [Google Scholar]
- Oyston P. C., Dorrell N., Williams K., Li S. R., Green M., Titball R. W., et al. (2000). The response regulator PhoP is important for survival under conditions of macrophage-induced stress and virulence in Yersinia pestis. Infect. Immun. 68, 3419–3425. 10.1128/IAI.68.6.3419-3425.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez E., Samper S., Bordas Y., Guilhot C., Gicquel B., Martin C. (2001). An essential role for phoP in Mycobacterium tuberculosis virulence. Mol. Microbiol. 41, 179–187. 10.1046/j.1365-2958.2001.02500.x [DOI] [PubMed] [Google Scholar]
- Perez J. C., Shin D., Zwir I., Latifi T., Hadley T. J., Groisman E. A. (2009). Evolution of a bacterial regulon controlling virulence and Mg2+ homeostasis. PLoS Genet. 5:e1000428 10.1371/journal.pgen.1000428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilione M. R., Pishko E. J., Preston A., Maskell D. J., Harvill E. T. (2004). pagP is required for resistance to antibody-mediated complement lysis during Bordetella bronchiseptica respiratory infection. Infect. Immun. 72, 2837–2842. 10.1128/IAI.72.5.2837-2842.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plesa M., Hernalsteens J. P., Vandenbussche G., Ruysschaert J. M., Cornelis P. (2006). The SlyB outer membrane lipoprotein of Burkholderia multivorans contributes to membrane integrity. Res. Microbiol. 157, 582–592. 10.1016/j.resmic.2005.11.015 [DOI] [PubMed] [Google Scholar]
- Sereny B. (1957). Experimental keratoconjunctivitis shigella. Acta Microbiol. Acad. Sci. Hung. 4, 367–376. [PubMed] [Google Scholar]
- Shprung T., Peleg A., Rosenfeld Y., Trieu-Cuot P., Shai Y. (2012). Effect of PhoP-PhoQ activation by broad repertoire of antimicrobial peptides on bacterial resistance. J. Biol. Chem. 287, 4544–4551. 10.1074/jbc.M111.278523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh K., Senadheera D. B., Cvitkovitch D. G. (2014). An intimate link: two-component signal transduction systems and metal transport systems in bacteria. Future Microbiol. 9, 1283–1293. 10.2217/fmb.14.87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R. L., Kaczmarek M. T., Kucharski L. M., Maguire M. E. (1998). Magnesium transport in Salmonella typhimurium: regulation of mgtA and mgtCB during invasion of epithelial and macrophage cells. Microbiology 144(Pt 7), 1835–1843. 10.1099/00221287-144-7-1835 [DOI] [PubMed] [Google Scholar]
- Somerville J. E., Jr., Cassiano L., Darveau R. P. (1999). Escherichia coli msbB gene as a virulence factor and a therapeutic target. Infect. Immun. 67, 6583–6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teh M. Y., Morona R. (2013). Identification of Shigella flexneri IcsA residues affecting interaction with N-WASP, and evidence for IcsA-IcsA co-operative interaction. PLoS ONE 8:e55152. 10.1371/journal.pone.0055152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Cen X. F., Zhao G. P., Wang J. (2012). Characterization of a new GlnR binding box in the promoter of amtB in Streptomyces coelicolor inferred a PhoP/GlnR competitive binding mechanism for transcriptional regulation of amtB. J. Bacteriol. 194, 5237–5244. 10.1128/JB.00989-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe H. (1988). Genetics of virulence of Shigella species. Microbiol. Sci. 5, 307–310. [PubMed] [Google Scholar]
- Yao Z., Valvano M. A. (1994). Genetic analysis of the O-specific lipopolysaccharide biosynthesis region (rfb) of Escherichia coli K-12 W3110: identification of genes that confer group 6 specificity to Shigella flexneri serotypes Y and 4a. J. Bacteriol. 176, 4133–4143. 10.1128/jb.176.13.4133-4143.1994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C. L., Liu Q. Z., Wang J., Chu X., Shen L. M., Guo Y. Y. (2014). Epidemic and virulence characteristic of Shigella spp. with extended-spectrum cephalosporin resistance in Xiaoshan District, Hangzhou, China. BMC Infect. Dis. 14:260. 10.1186/1471-2334-14-260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zwir I., Shin D., Kato A., Nishino K., Latifi T., Solomon F., et al. (2005). Dissecting the PhoP regulatory network of Escherichia coli and Salmonella enterica. Proc. Natl. Acad. Sci. U.S.A. 102, 2862–2867. 10.1073/pnas.0408238102 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The phoPQ gene disruption strategy. H1 and H2 refer to the homology regions of phoPQ, P1 and P2 refer to the primers of kan resistance gene.
The invasion ability of ΔicsA, ΔyoaE, ΔyrbL, and ΔrstA to HeLa cells. The bacteria that grew to logarithmic phase were added into the cells for 30 min infection. Then gentamicin was added into the medium to kill extracellular bacteria. Colonies of lysates on LB plates were counted. The invasion rate refered to the number of intracellular bacteria divided by that of inoculated bacteria and multiplied by 10,000. Values are means ± standard deviations from 3 independent wells. **P < 0.01.
Primers used for qRT-PCR verification of differentially expressed genes in ΔphoPQ.
Homological analysis of Sf301 phoP/phoQ with other homologues.
Homological analysis of Sf301 PhoP/PhoQ with other homologues.
Primers used for construction of ΔicsA, ΔyoaE, ΔyrbL, ΔrstA and their complementation strain.
Differentially expressed genes of ΔphoPQ compared to Sf301 by microarray and qRT-PCR at early-stationary phase.
The transcriptional levels of phoPQ and its regulated genes in different concentrations of Mg2+ condition.
The transcriptional levels of phoPQ and its regulated genes in different pH condition.
The transcriptional levels of phoPQ and its regulated genes with or without polymyxin B condition.