Abstract
Cell death inducing DNA fragmentation factor-alpha-like A (Cidea) is a member of cell death-inducing DFF45-like effector (CIDE) protein. The initial function of CIDE is the promotion of cell death and DNA fragmentation in mammalian cells. Cidea was recently reported to play critical roles in the development of hepatic steatosis. The purpose of present study is to determine the effect of chronic alcohol intake on Cidea expression in the livers of mice with alcoholic fatty liver disease. Cidea expression was significantly increased in the liver of alcohol-induced fatty liver mice. While, knockdown of Cidea caused lipid droplets numbers reduction. Next, we detected the activity of ALDH2 reduction and the concentration of serum acetaldehyde accumulation in our alcohol-induced fatty liver mice. Cidea expression was elevated in AML12 cells exposed to 100uM acetaldehyde. Interestingly, Dual-luciferase reporter gene assay showed that 100 uM acetaldehyde led to the activation of Cidea reporter gene plasmid which containing SRE element. What’s more, the knockdown of SREBP1c suppressed acetaldehyde-induced Cidea expression. Overall, our findings suggest that Cidea is highly associated with alcoholic fatty liver disease and Cidea expression is specifically induced by acetaldehyde, and this up-regulation is most likely mediated by SREBP1c.
Introduction
Alcohol consumption is a major risk factor for many chronic disease, especially alcoholic liver disease (ALD)1. Alcoholic fatty liver disease is the initial stage of alcohol-induced liver disease (ALD), which is characterized by the excessive hepatic accumulation of triglycerides2. Alcoholic fatty liver is a worldwide health problem without effective therapeutic methods. The mechanisms by which alcohol leads to fatty liver appear to be complex, include the changes of the redox condition, transportation impairment of the synthesized lipid, inhibition of fatty acid oxidation, and the enhancement of the lipid genesis3. However, the currently underlying mechanisms which is on promotion of the alcoholic fatty liver is still not fully understand.
The cell death-inducing DNA fragmentation factor-alpha-like effector (CIDE) proteins contain three members (Cidea, Cideb, and Cidec) that are well known as apoptosis-inducing factors for mammalian cells4. However, abundant evidences indicate that CIDE play important roles in hepatic lipid metabolism5. Cidea is a lipid-coated protein involved in lipid droplet formation and storage that are expressed in an inducible manner6–9. In pathological conditions, Cidea is highly expressed in the liver of mice with hepatic steatosis fed a high fat diet (HFD)8–11, dystrophic mice with fatty livers12, obese mice or humans8,13 and mice with type 2 diabetes14. Hepatic overexpression of Cidea increases lipid accumulation and lipid droplets formation8,9. In contrast, Cidea−/− mice exhibit reduced hepatic lipid accumulation, and knockdown of Cidea in the livers of obese mice decreases hepatic triglyceride levels and lipid droplets formation8. Thus, Cidea plays critical roles in promoting hepatic lipid accumulation and in the development of hepatic steatosis. However, the molecular mechanism regulating Cidea expression in the development of alcoholic fatty liver disease remains unclear.
Acetaldehyde, as a key toxin involved in alcohol-induced liver injury, increases triglycerides accumulation in recombinant HepG2 cells15, enhances SREBP1c expression16,17 and may impair the ability of PPARα to promote hepatic fat accumulation18. Recent studies focus on the reduced oxidation of fatty acid and the enhancement of the do novo lipogenesis. There are two important nuclear transcriptions, peroxisome proliferator-activated receptor-α(PPARα)19 and sterol regulatory element-binding protein-1 (SREBP-1c)16, are proved to be involved in alcohol-induced fatty liver. That is to say, acetaldehyde may modulate hepatic lipid metabolism and homeostasis. However, the role of acetaldehyde to promote the development of alcoholic fatty liver is still unclear. Abundant evidences have shown that Cidea promoter regions contain sterol-regulatory elements (SRE)9,20, the expression of Cidea was induced in the presence of saturated fatty acids (FAs)8 or insulin20. Additionally, Cidea promoter regions contain peroxisome proliferator response elements (PPREs) that are activated by a PPAR agonist21. Cidea expression is also regulated by the PPARα transcriptional coactivator-1 alpha22. Thus, we formulate a hypothesis that acetaldehyde may promote the development of alcoholic fatty liver, and it is mediated by regulating Cidea expression.
Here, we demonstrated that Cidea expression is markedly increased in the livers of chronic alcohol-fed mice and is correlated with the development of alcoholic fatty liver disease. Furthermore, Cidea expression is specifically induced by acetaldehyde, and this up-regulation is likely mediated by SREBP1c in hepatocytes.
Results
Hepatic Cidea expression increases in alcohol-induced fatty liver in mice
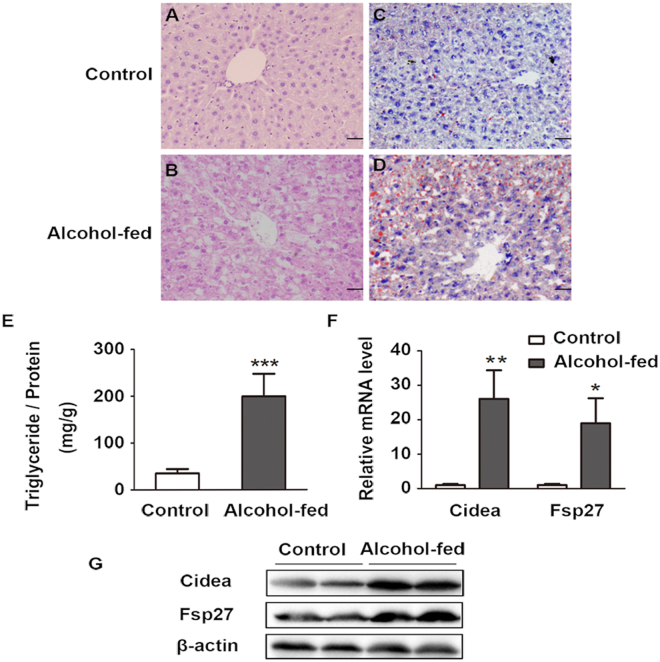
Recently, we have been established a new model mice of alcoholic fatty liver disease that represents a suitable model for studying the progression of AFLD23. As shown in Table 1, LDH, usually considered as a marker of common injury that is released during tissue damage, was significantly increased from 377.9 ± 106.62 U/L in control group to 580.9 ± 183.08 U/L in the alcohol-fed mice. ALT increased from 33 ± 6.57 U/L to 41.55 ± 9.7 U/L, and AST from 58.7 ± 11.78 U/L to 103.75 ± 61.86 U/L. Furthermore, chronic alcohol consumption increased the serum triglycerides from 0.6 ± 0.25 mg/dl in control group to 1.6 ± 0.7 mg/dl in alcohol-fed mice. HE staining and Oil Red O staining showed that hepatic lipid droplets increased in alcohol-fed mice (Fig. 1A–D). In addition, chronic alcohol consumption also significantly increased hepatic triglycerides levels (Fig. 1E). Interestingly, we found that Cidea mRNA and protein levels were markedly increased (Fig. 1F–G). Fsp27, another family member of CIDE, mRNA and protein levels were also enhanced in the alcohol-fed mice (Fig. 1F–G).
Table 1.
Serum biochemical indexes of 12-month-old mice were subjected to chronic alcohol feeding.
| Control group (n = 10) | Alcohol-fed (n = 12) | |
|---|---|---|
| LDH(U/L) | 377.9 ± 106.62 | 580.9 ± 183.08** |
| AST(U/L) | 58.7 ± 11.78 | 103.75 ± 61.86* |
| ALT(U/L) | 33 ± 6.57 | 41.55 ± 9.7* |
| ALB(g/L) | 31.71 ± 3.68 | 29.5 ± 5.76 |
| TP(g/L) | 51.21 ± 3.79 | 48.57 ± 5.46 |
| GLU(mmol/L) | 9.86 ± 1.97 | 7.09 ± 1.95** |
| TG(mmol/L) | 0.6 ± 0.25 | 1.6 ± 0.7*** |
| T-CHO(mmol/L) | 2.56 ± 0.66 | 2.97 ± 1.24 |
| HDL(mmol/L) | 2.43 ± 0.68 | 2.64 ± 1.19 |
| LDL(mmol/L) | 0.29 ± 0.15 | 0.43 ± 0.2 |
LDH, lactate dehydrogenase; AST, aspartate aminotransferase; ALT, alanineaminotransferase; ALB, albumin; TP, total protein; GLU, glucose; TG, triglycerides; T-CHO, total cholesterol; HDL-C, high-density lipoprotein cholesterol; LDL-C, low density lipoprotein cholesterol.
Means ± SD. Control group, n = 10; Alcohol-fed mice, n = 12 *p < 0.05; **p < 0.01; ***p < 0.001.
Figure 1.
Increased hepatic Cidea expression in alcohol-induced fatty liver in mice. (A,B) Sections of the liver from control group and alcohol-fed mice were subjected to H&E staining and analysis using OLYMPUS FSX100 with a 200× objective. (C,D) Oil red O-stained liver sections (X200). (E) Total Triglycerides level were detected from liver tissue. Means ± SD, n = 4–5 mice per group, ***p < 0.001 vs. control group. (F) Relative Cidea and Fsp27 mRNA levels in control group and alcohol-fed mice. Means ± SD, n = 4–5 mice per group, **p < 0.01, *p < 0.05 vs. control group. (G) Cidea and Fsp27 protein levels in the liver tissue, full-length blots are presented in Supplementary Figure S1.
Knockdown of Cidea reduces triglyceride accumulation in alcohol-exposed cells
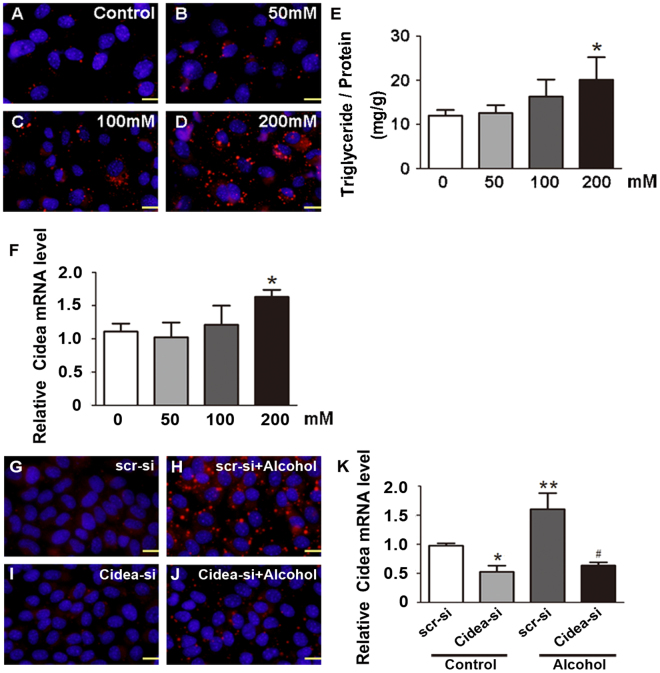
To evaluate the potential role of Cidea in development of alcoholic fatty liver disease, we treated AML12 cells with different alcohol concentrations. We found that alcohol increased the lipid droplets (LDs) numbers and sizes in the cells in a dose-dependent manner (Fig. 2A–D). Compared with control group, triglycerides (TG) (Fig. 2E) and Cidea expression (Fig. 2F) were increased by alcohol treatment. The LDs numbers and sizes markedly decreased in the Cidea-si + alcohol group compared with the alcohol group when Cidea was knocked down by transfecting the Cidea small interfering RNA (siRNA), at the same time, AML12 cells were treated with or without 200 mM alcohol (Fig. 2G–J). Cidea mRNA levels also decreased (Fig. 2K).
Figure 2.
Knockdown of Cidea reduces triglyceride accumulation in alcohol-exposed cells. AML12 cells were incubated with various concentrations of alcohol for 48 hours. (A–D) Evaluation of Nile red staining (X200). The triglyceride contents (E) and (F) the Cidea mRNA levels in AML12 cells. Means ± SD, n = 3, *p < 0.05 vs. control group. (G–J) Evaluation of Nile red staining (X200) and (K) the Cidea mRNA levels in AML12 cells with or without Cidea knockdown by small interfering RNA (siRNA) in the presence or absence of 200 mM alcohol. Means ± SD, n = 3, *p < 0.05; **p < 0.01 vs. scr-si group; #P < 0.05 vs. scr-si + Alcohol group. Scr-si, scrambled siRNA; Cidea-si, Cidea siRNA.
Cidea is directly induced by acetaldehyde not by alcohol
Next, we sought to understand the molecular mechanisms of Cidea expression in hepatocytes during alcohol intake. Previous reports found that chronic alcohol feeding can cause hepatic ALDH2 dysfunction and acetaldehyde accumulation in alcoholic hepatic steatosis mice and alcohol-treated hepatic cells24–26. Acetaldehyde modulates hepatic lipid metabolism and homeostasis27.
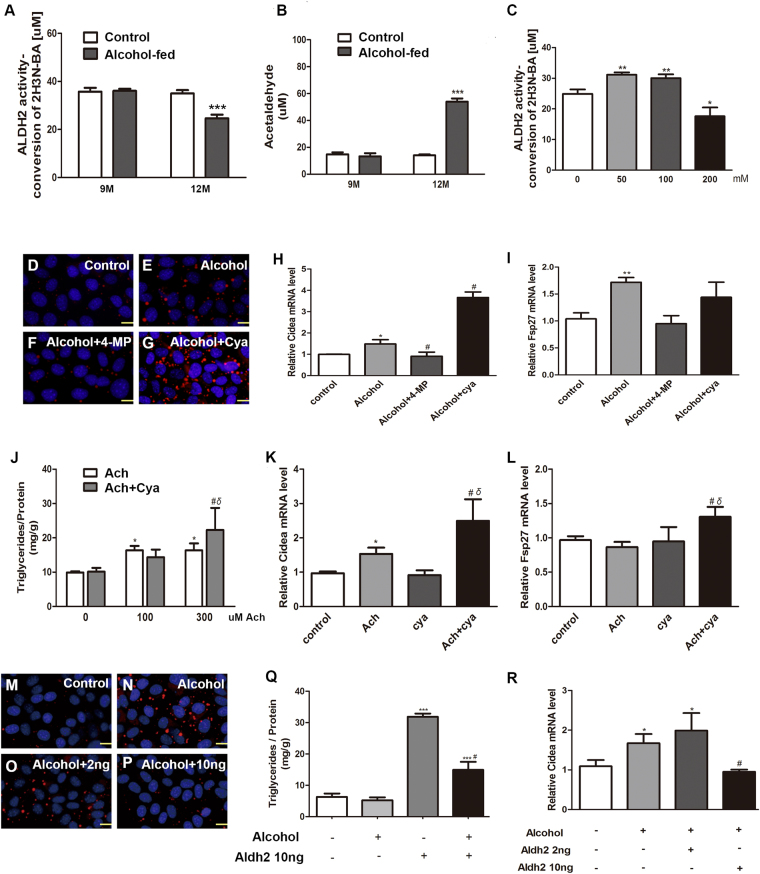
In our work, we assessed ALDH2 activity and acetaldehyde concentration in mice serum. Compared to control group, alcohol intake reduced ALDH2 activity in 12-month-old mice, while there was no change in 9-month-old mice (Fig. 3A). Acetaldehyde level significantly increased in alcohol-treated 12-month-old mice (Fig. 3B). Then we treated AML12 cells with different concentration of alcohol. ALDH2 activity increased in 50 mM and 100 mM alcohol, while 200 mM alcohol-exposed reduced ALDH2 activity compared with control group (Fig. 3C). Therefore, we hypothesized that the effect of alcohol on Cidea expression may directly due to its metabolic product acetaldehyde. As shown in Fig. 3D–G, 4-MP abolished the ability of alcohol to increase the LDs numbers and sizes, whereas cyanamide markedly augmented the alcohol effect. Similar to LDs numbers, Cidea mRNA level exhibited the same trend (Fig. 3H). Intriguingly, the Fsp27 mRNA level was not markedly induced by alcohol + cyanamide group compared with alcohol group (Fig. 3I). To further investigate whether acetaldehyde is directly a factor in the regulation of Cidea expression in alcoholic fatty liver disease, we treated the cells with 100 µM and 300 µM acetaldehyde with or without cyanamide. We found that triglyceride level is higher in the 300 µM acetaldehyde + cyanamide group than in 300 µM acetaldehyde group, and much higher than in control group (Fig. 3J). Next, we detected Cidea mRNA level, it increased in acetaldehyde-treated group, whereas cyanamide further enhanced the acetaldehyde effect (Fig. 3K). Interestingly, Fsp27 expression was not directly induced by acetaldehyde but Fsp27 increased in the acetaldehyde + cyanamide group (Fig. 3L). Therefore, we proposed a hypothesis that reducing acetaldehyde in alcohol metabolism could reduce Triglyceride level. We treated AML12 cells with alcohol and ALDH2 recombinant protein. Compared with the control group (Fig. 3M), the large LDs numbers significantly increased in the alcohol group (Fig. 3N); although 2 ng did not have an obvious effect (Fig. 3O), this effect was reversed by 10 ng of the ALDH2 recombinant protein (Fig. 3P), Triglycerides level increased, and the effect was reversed by 10 ng of the ALDH2 recombinant protein (Fig. 3Q). Cidea expression exhibited the same trend (Fig. 3R). These results suggest that acetaldehyde generated from alcohol metabolism is responsible for alcohol-induced Cidea expression.
Figure 3.
Cidea is directly induced by acetaldehyde but not by alcohol. (A) Activity of ALDH2 in liver tissue, ***p < 0.001 vs. control group. (B) Concentration of serum acetaldehyde, ***p < 0.001 vs. control group. (C) Activity of ALDH2 in AML12 cells were treated with different concentration alcohol, *p < 0.05; **p < 0.01. AML12 cells were treated with alcohol, 4-methylpyrazole (0.1 mM 4-MP), or cyanamide (0.1 mM Cya) for 48 h. (D–G) Nile red staining (X200). (H) Cidea mRNA level and (I) Fsp27 mRNA level were measured. Means ± SD, n = 3, *p < 0.05 vs. control group; **p < 0.01 vs. control group; #P < 0.05 vs. Alcohol group; NS, not significant. AML12 cells were treated with 100 uM and 300 uM of acetaldehyde or cyanamide (0.1 mM Cya). (J) Triglyceride contents were measured. Means ± SD, n = 3, *p < 0.05 vs. control group; #P < 0.05 vs. cya group; δ < 0.05 vs. 300 uM Ach group. AML12 cells were treated with 100 uM acetaldehyde or cyanamide (0.1 mM Cya) for 48 h. (K) Cidea mRNA level and (L) Fsp27 mRNA level were measured. Means ± SD, n = 3, *p < 0.05 vs. control group; #P < 0.05 vs. cya group; δ < 0.05 vs. Ach group. AML12 cells were treated with alcohol and 2 ng or 10 ng of the Aldh2 protein for 48 h. (M–P) Nile red staining (X200) and (Q) Triglyceride contents and (R) qPCR analysis of the Cidea mRNA level were measured. Means ± SD, n = 3, *P < 0.05 vs. control group; ***P < 0.001 vs. control group; #P < 0.05 vs. Alcohol group.
SREBP1c mediates acetaldehyde-induced Cidea expression
Acetaldehyde might be a significant factor that causes triglycerides accumulation and SREBP1c up-regulation in HepG2 cells15–17. In addition, Cidea promoter regions contain sterol-regulatory elements (SRE), and Cidea’s up-regulation is mediated by SREBP1c in the presence of saturated fatty acids (FAs) or insulin9,20.
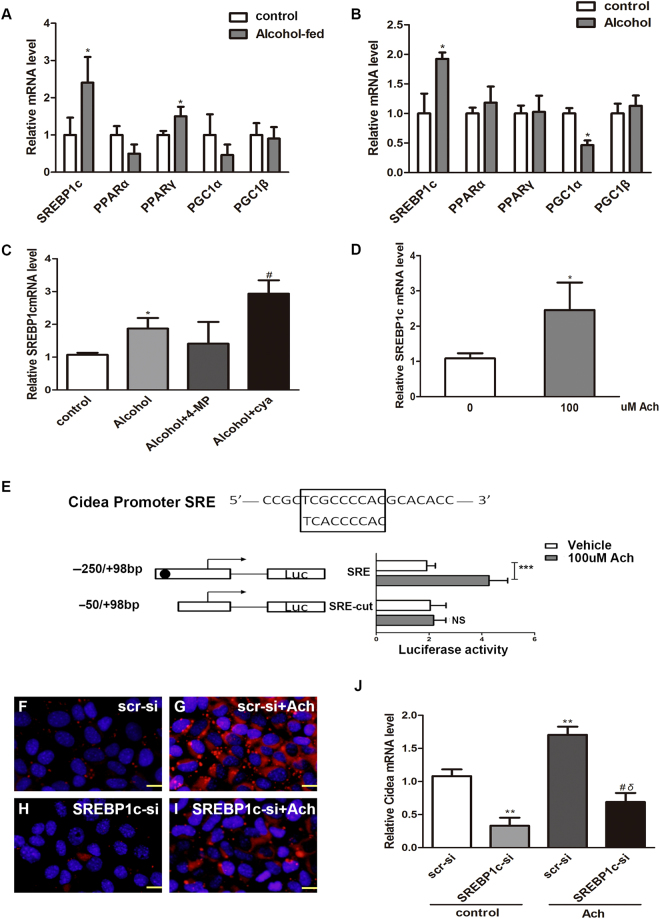
To define the specific transcription factor that mediates acetaldehyde-induced Cidea expression, we detected several key transcription factors that might regulate Cidea expression. Real-time PCR analysis of the gene levels showed a significant increase in SREBP1c and PPARα expression in the livers of AFLD mice (Fig. 4A); however, only SREBP1c expression was up-regulated in the AML12 cells (Fig. 4B). Then 4-MP abolished the ability of alcohol to increase SREBP1c mRNA level, whereas cyanamide markedly augmented the alcohol effect (Fig. 4C). Furthermore, SREBP1c expression increased in the acetaldehyde-treated group (Fig. 4D). To determine whether SREBP1c proteins were bound to the Cidea SRE element, we prepared the Luciferase reporter plasmid constructs of containing Cidea SRE element or completely deleting the SRE element of Cidea promoter and transfected into HepG2 cells. Dual-luciferase reporter assay results showed that 100 µM acetaldehyde led to the considerable activation of the Cidea reporter gene plasmid containing SRE element. However, when the reporter gene plasmid was further truncated completely deleting the SRE element, the activation was almost lost (Fig. 4E). To further verify the relationship between Cidea expression and transcription factor SREPB1c. AML12 cells were transfected with SREBP1c small interfering RNA (siRNA). The LDs numbers and sizes decreased in the SREBP1c-si + acetaldehyde group compared with the acetaldehyde group (Fig. 4F–I). Cidea expression, which was down-regulated by siRNA, was slightly elevated by acetaldehyde (Fig. 4J). Our subsequent series of results revealed that SREBP1c might serve as a transcription factor for acetaldehyde-induced Cidea expression.
Figure 4.
SREBP1cmediated acetaldehyde-induced Cidea expression. qPCR analysis of relative mRNA levels of key transcription factors levels in the livers of AF mice (A) and (B) Alcohol-treated AML12 cells were measured. Means ± SD, n = 4–5 mice per group, *p < 0.05; NS, not significant. (C) SREBP1c mRNA levels were measured in AML12 cells. Means ± SD, n = 3, *p < 0.05 vs. control group; #P < 0.05 vs. Alcohol group; NS, not significant. (D) AML12 cells were treated with 100 uM acetaldehyde for 48 h, SREBP1c mRNA levels were measured. Means ± SD, n = 3, *p < 0.05 vs. control group. (E) HepG2 cells were treated with 100uM acetaldehyde for 48 h. Then Luciferase reporter plasmid were transfected into HepG2 cells with Lipofectamine 2000. Means ± SD, n = 6, ***p < 0.001; NS, not significant. (F–I) Nile red staining (X200) and (J) Cidea mRNA and in AML12 cells with or without SREBP1c knockdown in the presence or absence of acetaldehyde. Means ± SD, n = 3, **p < 0.01 vs. scr-si group; #P < 0.05 vs. scr-si + Alcohol group; δ < 0.05 vs. SREBP1c-si group. scr-si, scrambled siRNA; SREBP1c-si, SREBP1c siRNA.
Discussion
Although Cidea mRNA is not detectable in the mouse liver21, Cidea has highly expressed in the liver under pathological conditions, such as HFD feeding, leptin deficiency or type 2 diabetes. High fat diet (HFD) feeding leads to hepatic steatosis as a result of higher plasma lipid levels from food intake. Unsaturated fatty acids (FAs) form the basis of the high fat diet, and Cidea expression is specifically induced by saturated FAs8. Interestingly, Cidea expression is an example of age-dependent augmented expression in HFD-fed mice11. Cidea is also reported to be markedly up-regulated in ob/ob mouse livers8,13, and its expression is induced by insulin in type 2 diabetic mouse livers14,20. Our findings showed that Cidea expression was highly increased in our alcohol-fed mice and alcohol-exposed AML12 cells. High Cidea expression resulted in significantly increased hepatic lipid accumulation and large LD numbers8,9. In contrast, Cidea−/− mice exhibited reduced hepatic lipid accumulation when fed the HFD, and knockdown of Cidea in the livers of ob/ob mice8 or in hepatocytes20 decreased hepatic triglyceride levels and LD formation. Moreover, Cidea expression was also reduced in several genetically modified animals that were resistant to hepatic steatosis28,29, and concomitant administration of eicosapentaenoic acid (EPA) with a high-fat diet ameliorated hepatic steatosis in mice and down-regulated Cidea expression9. These studies illustrate that Cidea may act as a liver marker for the appearance of hepatic steatosis. Our results suggest that Cidea knock-down leads to an obvious abrogation of alcohol-induced Cidea expression and large LD formation in AML12 cells.
Cidec (Fsp27 in mice) like Cidea, is also a lipid-coated protein involved in lipid droplet formation and storage that are expressed in an inducible manner6–9. Fsp27 expression markedly elevated in both HFD-fed and ob/ob mouse livers7,30. Furthermore, Fsp27 has been shown to be a regulator of PPARα-dependent hepatic steatosis7. Therefore, Fsp27 expression in the liver is correlated with the development of hepatic steatosis. Indeed, our findings showed that the Fsp27 mRNA was up-regulated in both the livers of alcohol-fed mice and alcohol-induced AML12 cells. Previous studies reported that Cidea expression was induced by saturated fatty acids (FAs), whereas Fsp27 expression was not affected by FAs but was significantly enhanced by a PPAR agonist8. Interestingly, our results showed that Fsp27 expression was not directly induced by acetaldehyde but the Fsp27 level was lightly increased in the acetaldehyde + cyanamide group. Acetaldehyde accumulation impaired the activity of PPARα resulted in severe abnormal lipid metabolism18. In addition, acetaldehyde sensitizes HepG2 cells to TNFa by impairing mGSH transport through an ER stress-mediated increase in cholesterol17. However, supporting the role of pharmacologic ER stress in up-regulating de novo lipogenesis, tunicamycin (TM) and thapsigargin (Tg) treatment which induce ER stress by pharmacologic reagents significantly increased accumulation of cytosolic lipid droplet formation and hepatic lipogenesis, among these lipogenesis gene including Fsp2731. Thus, the molecular mechanisms that regulate Cidea and Fsp27 expression in Alcoholic fatty liver disease may be different.
A recurring emergent theory in the alcohol field is that the reinforcing properties of alcohol are not produced by the alcohol molecule, but may depend upon the action of acetaldehyde within alcohol-associated disorders including alcoholic liver disease (ALD)32; alcoholic cardiomyopathy33; central nervous system (CNS)34; even carcinogenic35,36. As acetaldehyde is a toxic molecule which is a risk factor for pathogenesis of various alcohol-associated disorders including ALD. Accumulating evidence suggests that the development of liver fibrosis in alcoholics has been linked to the oxidation of alcohol to the highly reactive compound acetaldehyde37. Acetaldehyde is fibrogenic and induces expression of both COL1A1 and COL1A2 genes by a mechanism dependent on the generation of H2O238 and by a mechanism through Ca2+-independent PKC activation39. In fact, chronic Alcohol feeding can cause hepatic ALDH2 dysfunction and acetaldehyde accumulation in alcoholic fatty liver, that is the initial stage of alcohol-induced liver injury24. Interestingly, the concentration of serum acetaldehyde level in our model mice also increased. However, the effect of acetaldehyde in alcoholic fatty liver is not caused much attention by people. Previous studies suggested that Cidea promoter regions contain sterol-regulatory elements (SRE), and Cidea up-regulation is mediated by SREBP1c in the presence of saturated fatty acids (FAs) or insulin9,20. In addition, acetaldehyde enhanced SREBP-1c gene and protein expression, resulting in increasing lipogenic enzyme gene expression15,17. To further verify whether the effects of acetaldehyde on Cidea expression is directly due to SREBP1c mediated, we did Dual-luciferase reporter gene experiments, the results showed that 100 uM acetaldehyde led to the considerable activation of the Cidea reporter gene plasmid containing SRE element. Therefore, we found that SREBP1c directly mediates the effect of acetaldehyde on Cidea expression in hepatocytes.
ALDH2 is the second enzyme of the major oxidative pathway of alcohol metabolism, which is perhaps the most efficient mitochondrial ALDH isozyme24. However, chronic alcohol exposure caused oxidative stress may reduce the activity of ALDH224,25 and ALDH2 protein level27, this is leading to acetaldehyde accumulation significantly increase in the blood and liver24,26. Several researches have showed that ALDH2 overexpression transgenic mice40 or concomitant administration of Alda-124 with a chronic alcohol feeding can ameliorate alcoholic hepatic steatosis. Therefore, removal of excessive acetaldehyde loaded in the blood appears to be a new strategy for the treatment of alcoholic fatty liver disease. Recently, the ALDH2 recombinant protein was reported to play a protective role in the reduction of vascular contraction in AngII mice aorta41. Therefore, we treated AML12 cells with the ALDH2 recombinant protein and we found that Cidea expression and triglyceride contents decreased in the presence of alcohol. However, we didn’t verify the effect of ALDH2 recombinant protein on mice with alcoholic fatty liver disease. Further studies will be necessary to clarify whether the ALDH2 recombinant protein might ameliorate alcoholic fatty liver disease.
Materials and Methods
Animals
All animal experiments were approved by the Harbin Medical Univerisity’s Animal Care and Use Committee and conducted according to the National Institutes of Health guidelines. Male C57BL/6J mice at 8-weeks of age were housed in a room with a 12 h light/dark cycle and provided free access to food and water. The experimental group mice were fed 15% alcohol water (v/v) (alcohol-fed) and the wild-type (WT) mice were fed water for 10 months23.
Cell culture
Normal mouse hepatocyte cell line AML12(ATCC, CRL-2254) was cultured in DMEM/F12 medium (Gibco, Life Technologies) supplemented with 10% fetal bovine serum (FBS) (Gibco, Life Technologies), 100 ng/ml streptomycin, 63 ng/ml penicillin G, 0.1 uM dexamethasone, and insulin-transferrin-selenium (ITS; Gibco-BRL) in 37 °C humidified incubator containing 5% CO2.
Transfection of siRNA
Cells were transfected with the Cidea siRNA or SREBP1c siRNA using the X-treme GENE siRNA Transfection Reagent (Invitrogen)and treated with 200 mM alcohol or 100 uM acetaldehydes six hours after transfection.
Nile red staining
AML12 cells were stained with Nile Red solution (0.1 mg/ml) at 37 °C for 20 min, fixed with 4% formaldehyde for 10 min, and then counterstained with DAPI (1 ug/ml) for 10 min.
Measurement of triglycerides
The hepatic levels of triglycerides were detected using the Triglyceride Colorimetric Test kit (Ann Arbor, MI). The Cells were treated as described above.
Plasmid construction and transfection
Mouse Cidea cDNA was amplified from mouse liver tissue cDNA by polymerase chain reaction (PCR). Different deletion fragments were cloned by PCR using mouse liver cDNA as template and insert into the Nhel/Hindlll site of the pGL3basic vector. The Luciferase reporter plasmid were transfected into HepG2 cells which already have been induced by 100 uM acetaldehyde.
Determination of ALDH2 activity
The activity of ALDH2 is isolated from mitochondria of mouse liver tissue and AML12 cells. Conversion of 2-hydroxy-3-nitrobenzaldehyde to its benzoic acid product was followed by HPLC-based analysis by its absorbance at 340 nm as previously described29.
Measurement of serum acetaldehyde
Blood was drawn from the angular vein of the eyes and was subsequently incubated at room temperature for half an hour followed by centrifugation at 5000 rpm for 5 min. Samples were then analyzed by Colorimetric Aldehyde Assay Kit, Blue (Sigma-Aldrich).
Western blotting
Liver tissues and AML12 cell protein lysates were prepared using RIPA buffer containing complete EDTA-free protease inhibitor cocktail (Roche). A total of 30–50 ng of protein was subjected to 12% SDS-PAGE and transferred to a 0.45 um PVDF membrane (Millipore, Bedford, MA, USA). The membranes were probed with antibodies to Cidea (1:1000; ab8402, abcam) and Fsp27 (1:1000; ab77115, abcam) detected using chemiluminescence reagents.
Real-time PCR
Total RNA was prepared from liver tissues and AML12 cells using the TRIzol reagent according to the manufacturer’s protocol (Invitrogen). Total RNA was reverse transcribed into cDNA using the High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, CA, USA). Quantitative real-time PCR was performed using the SYBR Green kit with the Applied Biosystems’ 7500 Real-Time PCR System.
Data analysis
The results are from at least three independent experiments. Data are presented as the mean ± SD. The statistical analysis was calculated using Student’s t-test. Multiple comparisons were estimated by analysis of variance (ANOVA) followed by the Newman-Keuls test. Values of P < 0.05 were considered statistically significant.
Electronic supplementary material
Acknowledgements
This work was supported by the National Natural Science Foundation of China (81200406, 81570534), National Natural Science Foundation International (regional) cooperation and the exchange program of China (81611130072), the Scientific Research Foundation for the Returned Overseas Chinese Scholars Heilongjiang Province (C140202). Dr. Ying Liu was supported by the Scientific Research Foundation of Heilongjiang Province (ZD201419).
Author Contributions
Qi He, Yan Diao, and Tingting Zhao, wrote the main manuscript text. Baoyu Hou, Linel Darrel Ngokana, and Huan Liang, acquisition of data, analysis and interpretation of data. Junhui Nie, prepared Figure 1. Peizhu Tan, Hui Huang, prepared Figure 2. Yanze Li, Lin Qi, prepared Figure 3. Yuanyuan Zhao, Ying Liu, prepared Figure 4. Xu Gao, and Lingyun Zhou, study concept and design, obtained funding.
Competing Interests
The authors declare that they have no competing interests.
Footnotes
Qi He, Yan Diao and Tingting Zhao contributed equally to this work.
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-19466-z.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Xu Gao, Email: gaoxu_671227@163.com.
Lingyun Zhou, Email: zhoulingyun27@sina.com.
References
- 1.Purohit V, Gao B, Song BJ. Molecular Mechanisms of Alcoholic Fatty Liver. Alcohol Clin Exp Res. 2009;33(2):191–205. doi: 10.1111/j.1530-0277.2008.00827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu J. Ethanol and liver: recent insights into the mechanisms of ethanol-induced fatty liver. World J Gastroenterol. 2014;20(40):14672–85. doi: 10.3748/wjg.v20.i40.14672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zeng T, Xie KQ. Ethanol and liver: recent advances in the mechanisms of ethanol-induced hepatosteatosis. Arch Toxicol. 2009;83(12):1075–81. doi: 10.1007/s00204-009-0457-4. [DOI] [PubMed] [Google Scholar]
- 4.Inohara N, Koseki T, Chen S, Wu X. Núñez G. CIDE, a novel family of cell death activators with homology to the 45 kDa subunit of the DNA fragmentation factor. EMBO J. 1998;17(9):2526–33. doi: 10.1093/emboj/17.9.2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gong J, Sun Z, Li P. CIDE proteins and metabolic disorders. Curr Opin Lipidol. 2009;20(2):121–6. doi: 10.1097/MOL.0b013e328328d0bb. [DOI] [PubMed] [Google Scholar]
- 6.Puri V, et al. Fat-specific protein 27, a novel lipid droplet protein that enhances triglyceride storage. J Biol Chem. 2007;282(47):34213–8. doi: 10.1074/jbc.M707404200. [DOI] [PubMed] [Google Scholar]
- 7.Matsusue K, et al. Hepatic Steatosis in Leptin-Deficient Mice Is Promoted by the PPARγ Target Gene Fsp27. Cell Metab. 2008;7(4):302–11. doi: 10.1016/j.cmet.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Zhou L, et al. Cidea promotes hepatic steatosis by sensing dietary fatty acids. Hepatology. 2012;56(1):95–107. doi: 10.1002/hep.25611. [DOI] [PubMed] [Google Scholar]
- 9.Jinno Y, et al. Cide-a and Cide-c are induced in the progression of hepatic steatosis and inhibited by eicosapentaenoic acid. Prostaglandins Leukot Essent Fatty Acids. 2010;83(2):75–81. doi: 10.1016/j.plefa.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 10.Reynolds TH, et al. Effects of a High Fat Diet and Voluntary Wheel Running Exercise on Cidea and Cidec Expression in Liver and Adipose Tissue of Mice. PLoS One. 2015;10(7):e0130259. doi: 10.1371/journal.pone.0130259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Do GM, et al. Long term adaptation of global transcription and metabolism in the liver of high-fat diet-fed C57BL/6J mice. Mol Nutr Food Res. 2011;55(Suppl 2):S173–85. doi: 10.1002/mnfr.201100064. [DOI] [PubMed] [Google Scholar]
- 12.Hall AM, et al. Dynamic and differential regulation of proteins that coat lipid droplets in fatty liver dystrophic mice. J Lipid Res. 2010;51(3):554–63. doi: 10.1194/jlr.M000976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Montastier E, et al. Adipose Tissue CIDEA Is Associated, Independently of Weight Variation, to Change in Insulin Resistance during a Longitudinal Weight Control Dietary Program in Obese Individuals. PLoS One. 2014;9(7):e98707. doi: 10.1371/journal.pone.0098707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kelder, B. et al. CIDE-A is expressed in liver of old mice and in Type 2 diabetic mouse liver exhibiting steatosis. Comp Hepatol (2007). [DOI] [PMC free article] [PubMed]
- 15.Thomes PG, Osna NA, Davis JS, Donohue TM., Jr. Cellular steatosis in ethanol oxidizing-HepG2 cells is partially controlled by the transcription factor, early growth response-1. Int J Biochem Cell Biol. 2013;45(2):454–63. doi: 10.1016/j.biocel.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.You M, Fischer M, Deeg MA, Crabb DW. Ethanol induces fatty acid synthesis pathways by activation of sterol regulatory element-binding protein (SREBP) J Biol Chem. 2002;277:29342–29347. doi: 10.1074/jbc.M202411200. [DOI] [PubMed] [Google Scholar]
- 17.Lluis JM, Colell A, García-Ruiz C, Kaplowitz N, Fernández-Checa JC. Acetaldehyde impairs mitochondrial glutathione transport in HepG2 cells through endoplasmic reticulum stress. Gastroenterology. 2003;124(3):708–24. doi: 10.1053/gast.2003.50089. [DOI] [PubMed] [Google Scholar]
- 18.Galli A, et al. Activity of Peroxisome Proliferator-activated ReceptorαIs Inhibited by Ethanol Metabolism. J Biol Chem. 2001;276(1):68–75. doi: 10.1074/jbc.M008791200. [DOI] [PubMed] [Google Scholar]
- 19.Fischer M, You M, Matsumoto M, Crabb DW. Peroxisome Proliferator-activated Receptor (PPARalpha) Agonist Treatment Reverses PPARalpha Dysfunction and Abnormalities in Hepatic Lipid Metabolism in Ethanol-fed Mice. J Biol Chem. 2003;278(30):27997–8004. doi: 10.1074/jbc.M302140200. [DOI] [PubMed] [Google Scholar]
- 20.Wang R, et al. Sterol-regulatory-element-binding protein 1c mediates the effect of insulin on the expression of Cidea in mouse hepatocyte. Biochem J. 2010;430(2):245–54. doi: 10.1042/BJ20100701. [DOI] [PubMed] [Google Scholar]
- 21.Viswakarma N, et al. Transcriptional Regulation of Cidea, Mitochondrial Cell death-inducing DNA fragmentation factor alpha-like effector A, in mouse liver by peroxisome proliferator-activated receptor alpha and gamma. J Biol Chem. 2007;282(25):18613–24. doi: 10.1074/jbc.M701983200. [DOI] [PubMed] [Google Scholar]
- 22.Hallberg M, et al. A Functional Interaction between RIP140 and PGC-1 Regulates the expression of the lipid droplet protein CIDEA. Mol Cell Biol. 2008;28(22):6785–95. doi: 10.1128/MCB.00504-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tan P, et al. Establishment of an alcoholic fatty liver disease model in mice. Am J Drug Alcohol Abuse. 2017;43(1):61–68. doi: 10.1080/00952990.2016.1217539. [DOI] [PubMed] [Google Scholar]
- 24.Zhong W, et al. Pharmacological activation of aldehyde dehydrogenase 2 by Alda-1 reverses alcohol-induced hepatic steatosis and cell death in mice. J Hepatol.Jun. 2015;62(6):1375–81. doi: 10.1016/j.jhep.2014.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moon KH, et al. Inactivation of Oxidized and S-nitrosylated Mitochondrial Proteins in Alcoholic Fatty Liver of Rats. Hepatology. 2006;44(5):1218–30. doi: 10.1002/hep.21372. [DOI] [PubMed] [Google Scholar]
- 26.Kwon HJ, et al. Aldehyde dehydrogenase 2 deficiency ameliorates alcoholic fatty liver but worsens liver inflammation and fibrosis in mice. Hepatology. 2014;60(1):146–57. doi: 10.1002/hep.27036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Newton BW, Russell WK, Russell DH, Ramaiah SK, Jayaraman A. Liver Proteome Analysis in a Rodent Model of Alcoholic Steatosis. J Proteome Res. 2009;8(4):1663–71. doi: 10.1021/pr800905w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bai L, et al. Transcription Coactivator Mediator Subunit Med1 is Required for the Development of Fatty Liver in the Mouse. Hepatology. Apr. 2011;53(4):1164–74. doi: 10.1002/hep.24155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kang HS, et al. Nuclear Orphan Receptor TAK1_TR4-Deficient Mice Are Protected Against Obesity-Linked Inflammation, Hepatic Steatosis, and Insulin Resistance. Diabetes. 2011;60(1):177–88. doi: 10.2337/db10-0628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tanaka N, et al. Adipocyte-specific disruption of fat-specific protein 27 causes hepatosteatosis and insulin resistance in high-fat diet-fed mice. J Biol Chem. Jan 30. 2015;290(5):3092–105. doi: 10.1074/jbc.M114.605980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee JS, et al. Pharmacological ER stress promotes hepatic lipogenesis and lipid droplet formation. Am J Transl Res. 2012;4(1):102–13. [PMC free article] [PubMed] [Google Scholar]
- 32.Tilg H, Moschen AR, Kaneider NC. Pathways of liver injury in alcoholic liver disease. J Hepatol. 2011;55(5):1159–61. doi: 10.1016/j.jhep.2011.05.015. [DOI] [PubMed] [Google Scholar]
- 33.Cai L. Alcoholic cardiomyopathy: acetaldehyde, insulin in sensitization and ER stress. J Mol Cell Cardiol. 2008;44(6):979–82. doi: 10.1016/j.yjmcc.2008.03.018. [DOI] [PubMed] [Google Scholar]
- 34.Quertemont E, et al. Is ethanol a pro-drug Acetaldehyde contribution to brain ethanol effects. Alcohol Clin Exp Res. 2005;29(8):1514–21. doi: 10.1097/01.alc.0000175015.51329.45. [DOI] [PubMed] [Google Scholar]
- 35.Rusyn I, Bataller R. Reply to: The autophagic response to alcohol toxicity: the missing layer. J Hepatol. 2013;59(2):399–400. doi: 10.1016/j.jhep.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yukawa Y, et al. Impairment of aldehyde dehydrogenase 2 increases accumulation of acetaldehyde-derived DNA damage in the esophagus after ethanol ingestion. Am J Cancer Res. 2014;4(3):279–84. [PMC free article] [PubMed] [Google Scholar]
- 37.Reyes-Gordillo K, et al. Mechanisms of action of acetaldehyde in the up-regulation of the human α2(I) collagen gene in hepatic stellate cells: key roles of Ski, SMAD3, SMAD4, and SMAD7. Am J Pathol. 2014;184(5):1458–67. doi: 10.1016/j.ajpath.2014.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Svegliati-Baroni G, et al. Intracellular signaling pathways involved in acetaldehyde-induced collagen and fibronectin gene expression in human hepatic stellate cells. Hepatology. 2001;33(5):1130–40. doi: 10.1053/jhep.2001.23788. [DOI] [PubMed] [Google Scholar]
- 39.Greenwel, P. 1. et al. Hydrogen Peroxide: A Link Between Acetaldehyde-Eliciteda1(I) Collagen Gene Up-regulation and Oxidative Stress in Mouse Hepatic Stellate Cells. Hepatology. 31, 109–16 (2000). [DOI] [PubMed]
- 40.Rui G, et al. Aldehyde dedydrogenase-2 plays a beneficial role in ameliorating chronic alcohol-induced hepatic steatosis and inflammation through regulation of autophagy. J Hepatol. 2015;62(3):647–56. doi: 10.1016/j.jhep.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 41.Choi H, Tostes RC, Webb RC. Mitochondrial aldehyde dehydrogenase prevents ROS-induced vascular contraction in angiotensin-II hypertensive mice. J Am Soc Hypertens. 2011;5(3):154–60. doi: 10.1016/j.jash.2011.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.