Abstract
While several studies correlated increased expression of the histone code reader Spin1 with tumor formation or growth, little is known about physiological functions of the protein. We generated Spin1M5 mice with ablation of Spin1 in myoblast precursors using the Myf5-Cre deleter strain. Most Spin1M5 mice die shortly after birth displaying severe sarcomere disorganization and necrosis. Surviving Spin1M5 mice are growth-retarded and exhibit the most prominent defects in soleus, tibialis anterior, and diaphragm muscle. Transcriptome analyses of limb muscle at embryonic day (E) 15.5, E16.5, and at three weeks of age provided evidence for aberrant fetal myogenesis and identified deregulated skeletal muscle (SkM) functional networks. Determination of genome-wide chromatin occupancy in primary myoblast revealed direct Spin1 target genes and suggested that deregulated basic helix-loop-helix transcription factor networks account for developmental defects in Spin1M5 fetuses. Furthermore, correlating histological and transcriptome analyses, we show that aberrant expression of titin-associated proteins, abnormal glycogen metabolism, and neuromuscular junction defects contribute to SkM pathology in Spin1M5 mice. Together, we describe the first example of a histone code reader controlling SkM development in mice, which hints at Spin1 as a potential player in human SkM disease.
Spindlin1 (Spin1) is a histone code reader binding histone H3 trimethylated at lysine 4 (H3K4me3) with high affinity.1, 2, 3 H3K4me3 association is enhanced by the presence of asymmetrically dimethylated arginine 8 of histone H34. Spin1 is highly expressed in several types of tumors5, 6, 7 and affects cell cycle, chromatin segregation, apoptosis, and transformation of cell lines, as well as tumor formation in nude mice.6, 8, 9, 10, 11 While these studies suggest important roles in cancer, physiological functions of Spin1 have only been subject to initial investigation. Mouse oocytes deficient for maternal Spin1 undergo normal folliculogenesis, but fail to resume meiosis.12 Furthermore, mice with ubiquitous Spin1 ablation die shortly after birth.12 However, tissue-restricted defects accounting for postnatal death have not been reported.
Skeletal muscle (SkM) is the most abundant tissue in vertebrates mediating support and movement and contributing to overall metabolism. SkM development is orchestrated by key transcription factors including Pax3 and Pax7, which are also required for muscle stem cell specification,13, 14 and the myogenic regulatory factors (MRFs) Myf5, MyoD (Myod1), Mrf4 (Myf6), and myogenin (Myog).13, 14, 15 MRFs are tissue-specific basic helix-loop-helix (bHLH) transcription factors acting as homodimers or as heterodimers with other bHLH transcription factors such as the ubiquitously expressed E-proteins E12/E47 (Tcf3), E2-2/ITF2 (Tcf4), and HEB/HTF4 (Tcf12).14
SkM fiber formation in mice comprises three successive phases, an embryonic wave from around embryonic day (E) 10.5 to E12.5, a fetal wave from around E14.5 to E17.5, and a postnatal period during which adult fibers are established.14, 16, 17, 18 Adult myofibers exhibit distinct contractile properties (slow- or fast-twitch), patterns of innervation, and metabolic activities (oxidative or glycolytic), which correlate with the expression of specific myosin heavy chain (MHC) isoforms.19, 20 Limb muscle of adult mice is composed of type I (slow, oxidative), type IIa (fast, oxidative), type IIx (fast, glycolytic), and type IIb (fast, glycolytic) fibers.19, 20 SkM mass and functions become compromised in disease and numerous gene mutations causing myopathies or muscular dystrophies have been documented.21, 22, 23, 24, 25, 26, 27, 28 Interestingly, selected fiber or muscle types preferentially degenerate in certain disease states.29, 30
In this study, we crossed mice harboring conditional Spin1 alleles (Spin1p/p) with the Myf5-Cre deleter strain31 to ablate Spin1 in myogenic precursors. Most homozygous Spin1p/p Myf5-Cre (hereafter termed Spin1M5) mice die shortly after birth, while surviving mice display severe growth retardation. Histological, transcriptome, and cistrome analyses provide evidence for aberrant fetal myogenesis and deregulated basic bHLH transcription factor networks around the onset of SkM defects. Furthermore, our observations suggest that altered expression of titin-associated proteins, aberrant glycogen metabolism, and defective NMJs contribute to SkM pathology in Spin1M5 mice. In summary, our data reveal a severe developmental defect caused by ablation of a histone code reader in SkM and hint at Spin1 as a potential player in human SkM disease.
Results
Loss of Spin1 in SkM results in postnatal lethality
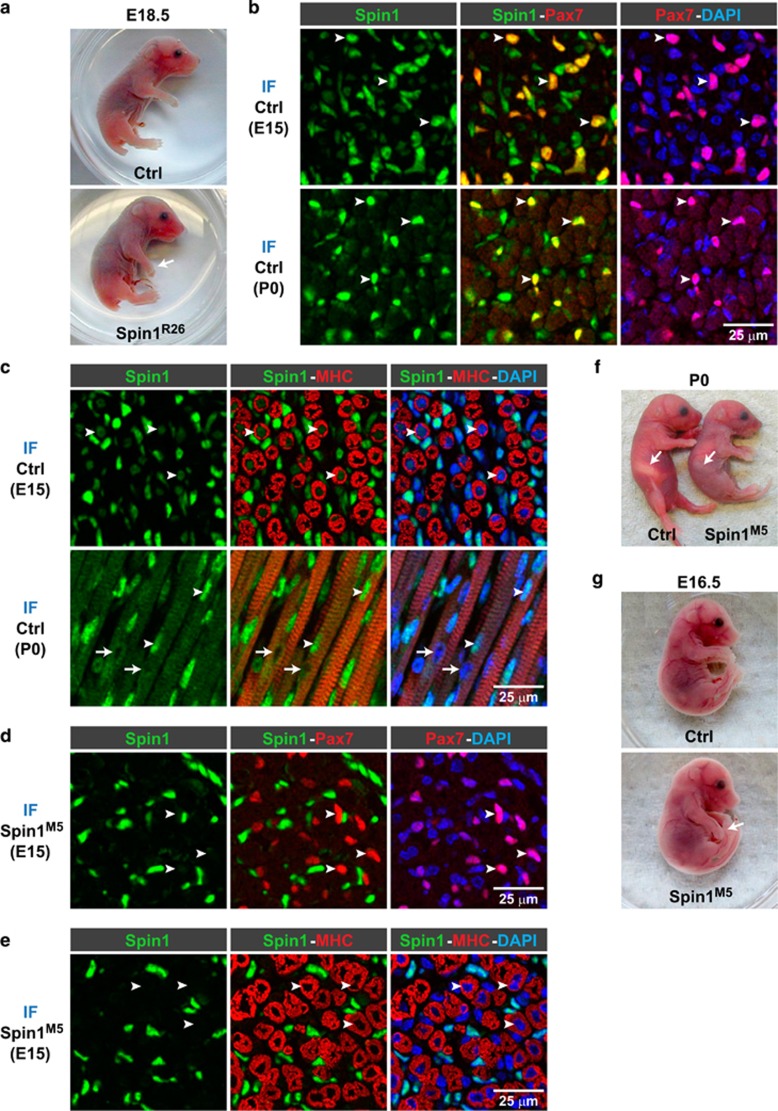
To investigate physiological functions of Spin1 in vivo, we generated ubiquitous knockout (Spin1R26) mice by crossing Spin1p/p mice with the Rosa26-Cre deleter strain32 (Supplementary Figure 1a). Spin1R26 mice were born, but died within one day after birth (Supplementary Figure 1b), which is in agreement with observations by others.12 Of note, at E18.5 Spin1R26 mice displayed dropping forelimbs (Figure 1a) indicating a neuromuscular defect.33
Figure 1.
Loss of Spin1 in SkM results in postnatal lethality. (a) Dropping forelimbs (arrow) indicating a neuromuscular defect observed in ubiquitous Spin1 knockout (Spin1R26) but not Spin1+/+ (Ctrl) fetuses. (b,c) Immunofluorescence (IF) staining of hind limb sections of control fetuses at E15 and of newborn mice (P0) with antibodies directed against Spin1 (green), Pax7 (red), or myosin heavy chain (MHC) (red). Arrowheads mark nuclei of myogenic precursors coexpressing Spin1 and Pax7 (b) or Spin1-positive nuclei in myofibers (c). Arrows depict nuclei devoid of Spin1 staining in myofibers at P0 (c). (d,e) IF staining of hind limb sections of Spin1M5 fetuses at E15 with the indicated antibodies. Loss of Spin1 expression in Pax7-positive nuclei of myogenic precursors (d) or myofibers (e) in Spin1M5 fetuses is highlighted with arrowheads. (b-e) Nuclei were visualized with DAPI (blue). (f) Absence of milk in the stomach (arrows) and an abnormal posture observed in newborn Spin1M5 mice but not control littermates. (g) Dropping forelimbs (arrow) observed in Spin1M5 but not control fetuses
To address potential functions of Spin1 in SkM, we analyzed Spin1 protein expression in hind limb sections of control mice at E15 and after birth (P0) by immunofluorescence. At both time points, we observed intense Spin1 staining in Pax7-positive myoblast precursors (Figure 1b (arrowheads)) and weaker staining in nuclei of myofibers (Figure 1c (arrowheads)). Of note, in newborn mice, Spin1 expression was undetectable in some nuclei of myofibers (Figure 1c, bottom row (arrows)).
Next, we deleted Spin1 in myoblast precursors by crossing Spin1p/p mice with the Myf5-Cre deleter strain31 resulting in Spin1M5 mice. Immunostaining confirmed the absence of Spin1 protein in nuclei of Pax7-positive myoblast precursors (Figure 1d (arrowheads); Supplementary Figure 1c) and myofibers of Spin1M5 fetuses (Figure 1e (arrowheads); Supplementary Figure 1d). Remaining Spin1 staining is due to expression in non-myogenic cells such as Tcf4-positive fibroblasts34 (Supplementary Figure 1e (arrowheads); see Materials and Methods and Supplementary Figure 1f, g for further characterization).
Homozygous Spin1M5 mice were obtained at the expected Mendelian ratio at birth (Supplementary Figure 1h). However, about 80% of Spin1M5 mice died within one day after birth. Newborn Spin1M5 mice could typically be distinguished from control littermates by an abnormal posture and the absence of milk in the stomach (Figure 1f). Moreover, at E16.5, we observed dropping forelimbs for Spin1M5 fetuses (Figure 1g). Together, our data show that ablation of Spin1 in SkM causes early postnatal death of the majority of mice.
SkM of Spin1M5 mice is characterized by necrosis and structural defects in non-necrotic fibers
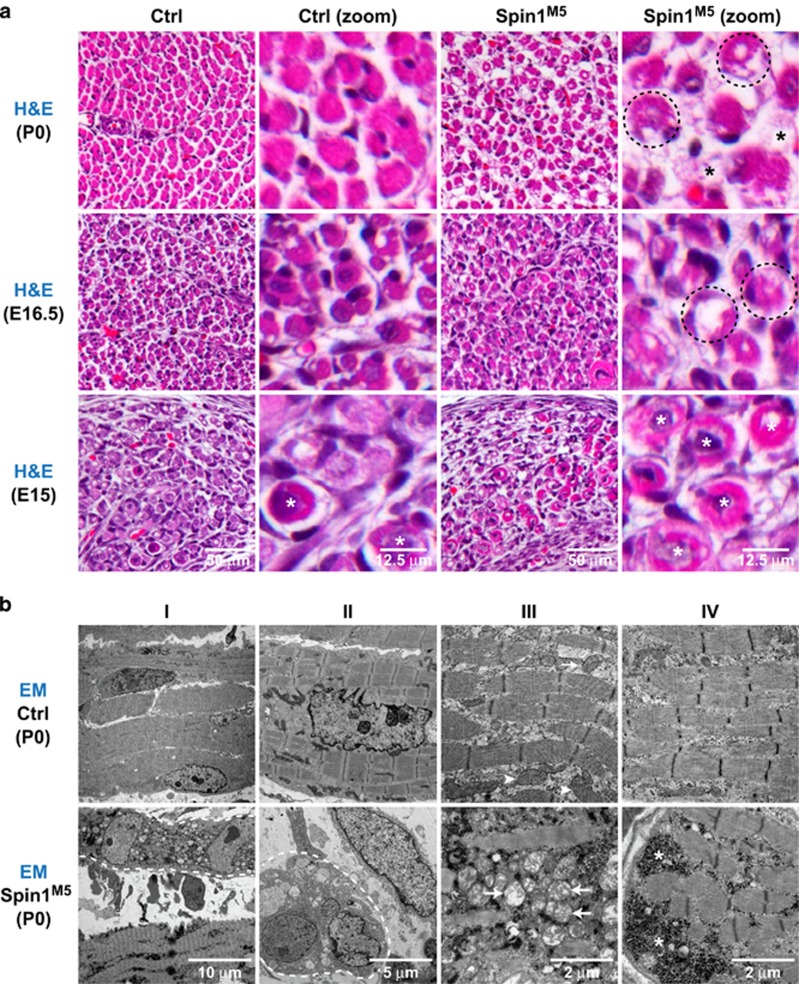
To characterize SkM defects in Spin1M5 mice, we inspected hematoxylin & eosin (H&E)-stained hind limb sections at different stages of development. Compared with control littermates, we observed in newborn Spin1M5 mice loss of fibers (Figure 2a, top row (black asterisks)) and numerous immature or degenerating fibers lacking contractile material (dashed circles). In Spin1M5 fetuses at E16.5, we also noted fibers with irregular H&E staining (Figure 2a, middle row (dashed circles)) and at E15 enlarged fibers, which were less abundant in control samples (Figure 2a, bottom row (white asterisks)).
Figure 2.
SkM of Spin1M5 mice is characterized by necrosis and structural defects in non-necrotic fibers. (a) Hematoxylin and eosin (H&E) staining of transversal tibialis anterior sections of Spin1M5 and control mice at P0, E16.5, and E15. Fiber loss in Spin1M5 mice is indicated by black asterisks (top row), fibers with irregular H&E staining are encircled, and unusually large fibers more frequently observed in Spin1M5 than in control fetuses at E15 are marked with white asterisks (bottom row). (b) Electron microscopy (EM) images of SkM samples of newborn (P0) Spin1M5 and control mice. Dashed lines demarcate degenerating, necrotic fibers (I–II, bottom), arrowheads mark normal mitochondria (III, top), arrows point at defective mitochondria (III, bottom), and asterisks indicate abnormal glycogen accumulation (IV, bottom)
Electron microscopy analyses of hind limb sections revealed for newborn Spin1M5 mice degenerating, necrotic fibers (Figure 2b, columns I–II (demarcated by dashed lines)), defective mitochondria (Figure 2b, column III (arrows)), and abnormal glycogen accumulation (Figure 2b, column IV (asterisks)). Similar defects were detected for Spin1M5 fetuses at E16.5 (Supplementary Figure 2). Furthermore, in non-necrotic fibers of Spin1M5 fetuses, we observed structural defects including a low density of contractile material and the lack of a clear M-line (Supplementary Figure 2, column III (triangles)). Together, our H&E and electron microscopy analyses uncovered necrotic and structurally defective fibers in Spin1M5 mice. In addition, the data suggested an onset of SkM defects before E16.5.
Transcriptome and histological analyses provide evidence for aberrant fetal myogenesis in Spin1M5 mice
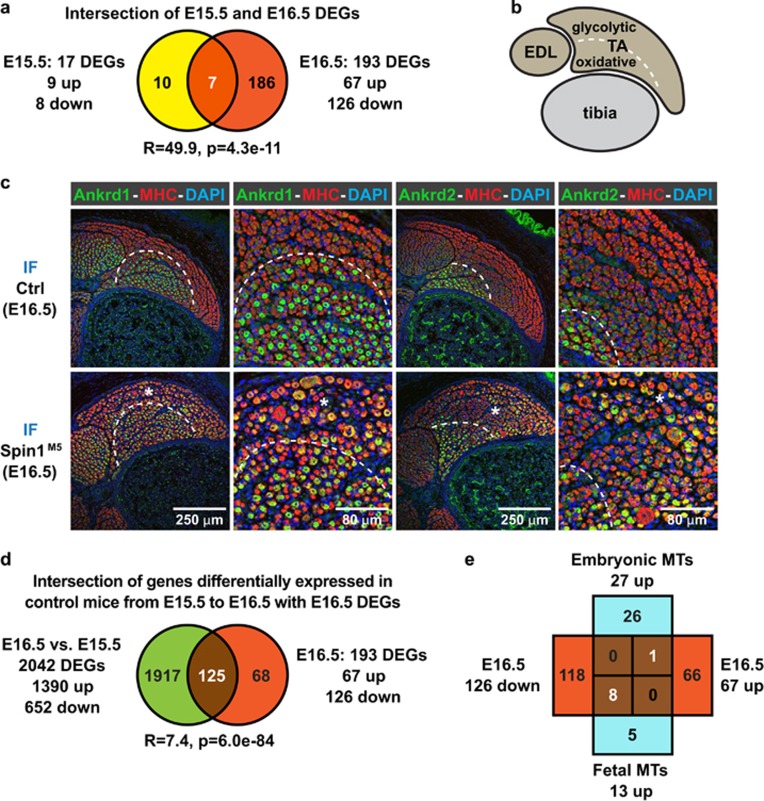
To investigate alterations of the transcriptome in SkM of Spin1M5 fetuses, we performed RNA sequencing (RNA-seq) analyses using RNA isolated from limb muscle. At E15.5, we observed only 17 differentially expressed genes (DEGs) (P≤1e−3; fold change ≥1.5) (Figure 3a; Supplementary Table 1a). In comparison, at E16.5 RNA-seq detected 193 DEGs, of which seven (Ankrd1, Ankrd2, Pmaip1, Scn4b (upregulated); Myf5, Msc, Nos1 (downregulated)) overlapped with the E15.5 DEGs (Figure 3a; Supplementary Table 1b). The strong increase in DEGs from E15.5 to E16.5 provided evidence for E15.5 as the approximate onset of SkM defects in Spin1M5 fetuses.
Figure 3.
Transcriptome and histological analyses provide evidence for aberrant fetal myogenesis in Spin1M5 mice. (a) Intersection of differentially expressed genes (DEGs) observed for SkM of Spin1M5 and control fetuses at E15.5 (yellow) and E16.5 (red). (R: enrichment factor; p: P-value calculated for the intersection.) (b) Schematic representation of the hind limb section depicting tibialis anterior (TA), extensor digitorum longus (EDL), and tibia analyzed in c. Prospective oxidative or glycolytic parts of the TA (demarcated by a dashed line) are located proximal or distal to the tibia, respectively. (c) Detection of Ankrd1 or Ankrd2 (green) by immunofluorescence (IF) staining in transversal hind limb sections of Spin1M5 and control fetuses at E16.5. Muscle fibers were visualized with myosin heavy chain (MHC) antibody (red), nuclei were stained with DAPI (blue). Dashed lines demarcate areas with or without Ankrd1 or Ankrd2 expression in control fetuses. Asterisks mark regions of the TA of Spin1M5 fetuses, in which Ankrd1 or Ankrd2 are aberrantly expressed. (d) Intersection of the E16.5 DEGs (red) with genes differentially expressed in control fetuses at E15.5 and E16.5 (green). (e) Comparison of the E16.5 DEGs (red) with transcripts reported to be more highly expressed in either embryonic or fetal myotubes (MTs, cyan)37
The common E15.5/E16.5 DEGs Ankrd1 (CARP) and Ankrd2 (ARPP) encode titin-associated regulators of sarcomere function, whose expression is often deregulated in SkM disease.35 We therefore investigated Ankrd1 and Ankrd2 protein levels in hind limb sections of E16.5 fetuses by immunofluorescence (Figures 3b and c). Most prominently, in control fetuses, expression of both proteins was restricted to the inner (prospective oxidative) part of the tibialis anterior (TA) neighboring the tibia (Figure 3c (demarcated by dashed lines)), whereas in Spin1M5 fetuses staining was also present in the outer (prospective glycolytic) part of the TA (Figure 3c (asterisks)). In comparison, at E15 Ankrd1 and Ankrd2 staining of hind limb muscle of Spin1M5 and control mice was similar (Supplementary Figures 3a and b). Thus, deregulation of Ankrd1 and Ankrd2 transcripts at E15.5 and E16.5 and aberrant protein expression at E16.5 are early markers of SkM defects in Spin1M5 fetuses.
To analyze which proportion of developmentally regulated genes is affected by the absence of Spin1, we compared the E16.5 DEGs with gene expression changes in control fetuses from E15.5 to E16.5. In control fetuses, we observed 2042 genes differentially expressed between E15.5 and E16.5 (Figure 3d; Supplementary Table 1c). Phenotype and pathway analyses for these 2042 DEGs using WebGestalt36 confirmed relevance of the identified genes for SkM development or function (Supplementary Figures 3c and d). Intersection with the 193 E16.5 DEGs revealed an overlap of 125 genes (Figure 3d). Therefore, the absence of Spin1 prevents adequate expression of a subset of genes regulated during the fetal phase of myogenesis in control mice.
To corroborate aberrant fetal myogenesis in Spin1M5 mice, we compared the E16.5 DEGs with transcripts previously observed to be differentially expressed in embryonic or fetal myotubes.37 Of the 27 transcripts more highly expressed in embryonic (compared with fetal) myotubes, only one (Tnnc1) overlapped with the E16.5 DEGs (Figure 3e). Importantly, eight out of thirteen transcripts more highly expressed in fetal myotubes (Myh8, Tnni2, Tnnt3, Atp2a1, Casq1, Pvalb, Ckm, and Eno3) were downregulated DEGs in Spin1M5 mice at E16.5 (Figure 3e). Together, our data provided evidence for an impaired progression of fetal myogenesis in Spin1M5 mice.
Identification of deregulated SkM functional networks in Spin1M5 fetuses
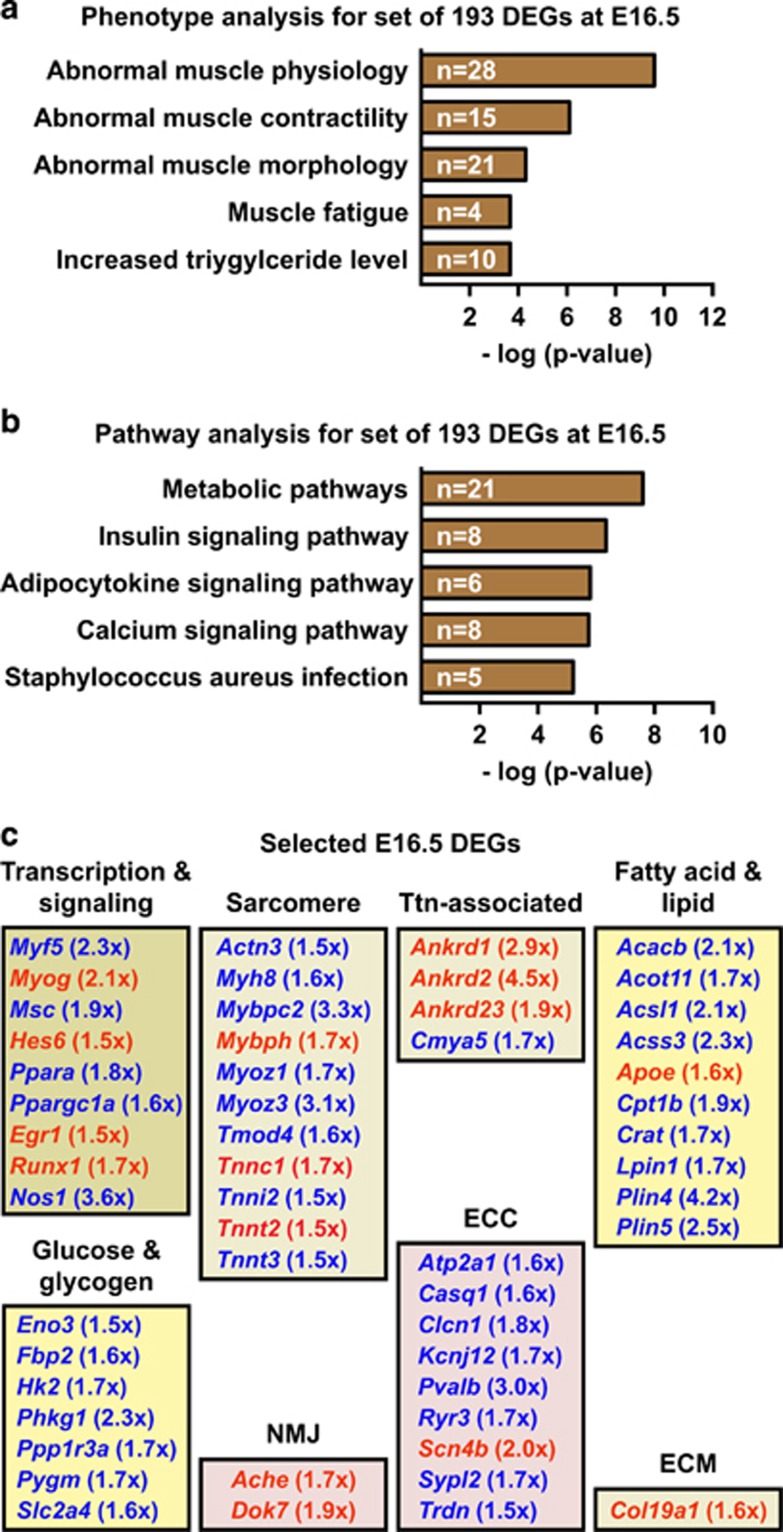
To identify genes accounting for SkM defects in Spin1M5 fetuses, we performed phenotype and pathway analyses for the E16.5 DEGs using WebGestalt36 (Figures 4a and b). These analyses revealed terms related to SkM function, metabolism, and pathology. We grouped the genes associated with the phenotype and pathway terms according to functional networks previously proposed in systems biology analyses for SkM38, 39 (Figure 4c). The deregulated SkM networks comprise genes encoding transcription factors and signaling molecules, proteins important for sarcomere function, titin (Ttn)-associated factors, metabolic enzymes, and proteins involved in the neuromuscular junction (NMJ) and excitation-contraction coupling (ECC) (Figure 4c). Deregulation of these functional networks likely causes SkM dysfunction in Spin1M5 fetuses.
Figure 4.
Identification of deregulated SkM functional networks in Spin1M5 fetuses. (a,b) Phenotype (a) and pathway (b) analysis for the set of 193 DEGs observed in SkM of Spin1M5 fetuses at E16.5. The number of genes in each category is indicated. For the phenotype analysis, the ‘top 5’ non-redundant terms are depicted. (c) Assignment of selected E16.5 DEGs identified by phenotype and pathway analyses to functional SkM networks. Upregulated DEGs are depicted in red, downregulated in blue color (fold change in brackets)
Deregulated basic helix-loop-helix transcription factor networks account for SkM defects in Spin1M5 fetuses
In addition to genes required for SkM structure or function, we found the myogenic bHLH transcription factor-encoding genes Myf5, Myog, Msc (musculin, MyoR), and Hes615, 40, 41, 42, 43 among the E16.5 DEGs (Figure 4c (network 'transcription and signaling')). Therefore, deregulated bHLH transcription factor-controlled gene expression may be an early event accounting for aberrant myogenesis in Spin1M5 fetuses. In support of this idea, Myf5 and Msc were also among the E15.5 DEGs (Supplementary Table 1a).
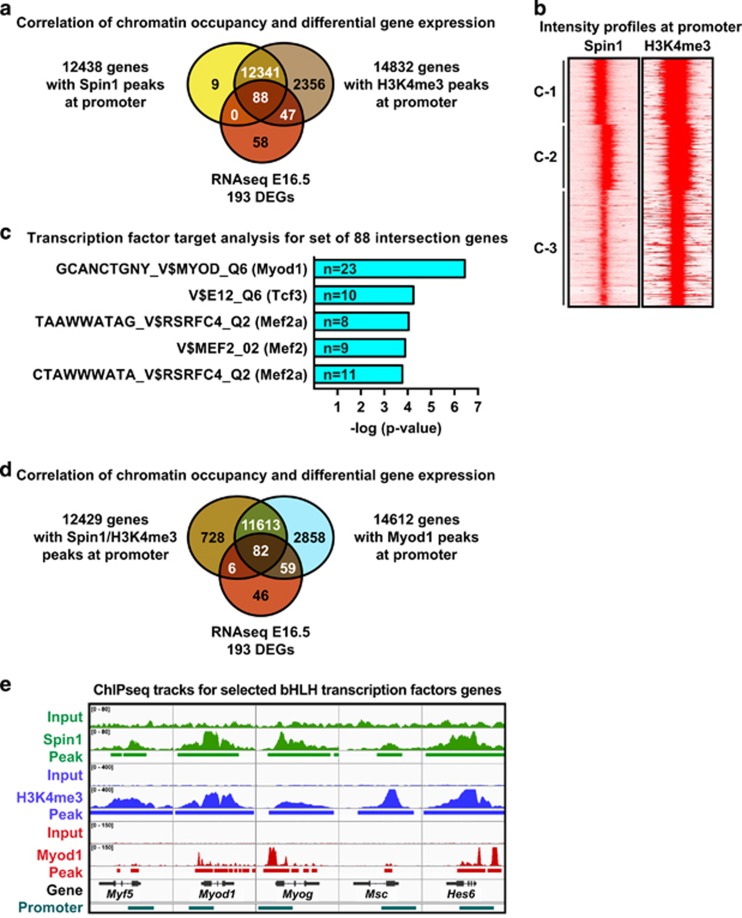
To correlate gene expression with Spin1 chromatin occupancy, we performed chromatin immunoprecipitation followed by massive parallel sequencing (ChIP-seq) using primary myoblasts. ChIP-seq with Spin1-specific antibody revealed 17106 peaks, of which the majority (61.2%) was located at the promoters (transcription start site (TSS)±1 kb) of 12438 genes (Figure 5a; Supplementary Figure 4a). Next, ChIP-seq analysis for H3K4me3 detected 37% of the peaks at the promoters of 14832 genes (Figure 5a; Supplementary Figure 4a). Further analyses confirmed the presence of H3K4me3 at almost all gene promoters occupied by Spin1 (Figure 5a) as well as overlapping intensity profiles (Figure 5b). Thus, in primary myoblasts Spin1 and H3K4me3 occupy a large portion of cellular gene promoters.
Figure 5.
Deregulated basic helix-loop-helix transcription factor networks account for SkM defects in Spin1M5 fetuses. (a) Intersection of genes with Spin1 peaks (yellow) or H3K4me3 peaks (brown) at the promoter (transcription start site (TSS)±1 kb) with the E16.5 DEGs (red). (b) Intensity profiles for Spin1 and H3K4me3 occupancy of 12 429 gene promoters (TSS±1 kb) (KMeans=3 clusters (C-1 to C-3)). (c) Transcription factor target analysis for 88 E16.5 DEGs with Spin1 and H3K4me3 peaks at the promoter observed by intersection of data sets shown in (a). (d) Intersection of genes with Spin1 and H3K4me3 peaks (brown) or Myod1 peaks (cyan) at the promoter (TSS±1 kb) with the E16.5 DEGs (red). (e) ChIP-seq tracks for selected genes encoding myogenic bHLH transcription factors. Spin1 (green) and H3K4me3 (blue) tracks were obtained by ChIP-seq analysis of primary myoblasts, Myod1 (red) tracks were obtained by analysis of previously deposited ChIP-seq data for C2C12 myoblasts (GEO data set GSE36024). Identified peaks and promoters (TSS±1 kb) are indicated by bars
Despite occupying thousands of gene promoters, Spin1 apparently only regulates a subset of target genes during fetal myogenesis. To further investigate Spin1-mediated gene regulation, we intersected the E16.5 DEGs with the cistrome observed in primary myoblasts. This analysis revealed the presence of Spin1 and H3K4me3 at the promoter of 88 E16.5 DEGs (Figure 5a). A transcription factor target analysis for these 88 genes using WebGestalt36 revealed a significant enrichment of potential Myod1 target genes followed by E12 (Tcf3) and Mef2 targets (Figure 5c).
To validate Myod1 promoter occupancy of Spin1 target genes, we analyzed a previously deposited Myod1 ChIP-seq data set for C2C12 myoblasts (GEO data set GSE36024), which uncovered Myod1 peaks at the promoters of 14612 genes (Figure 5d). Intersection with Spin1-occupied genes showed a large overlap of 11695 direct targets of Spin1 and Myod1 (Figure 5d). Comparison with the E16.5 DEGs identified 82 direct Spin1/Myod1 targets (Figure 5d). Inspection of ChIP-seq tracks confirmed the presence of Spin1, H3K4me3, and Myod1 at promoters of SkM functional, metabolic, and regulatory genes (Supplementary Figure 4b) as well as the promoters of Myf5, Myod1, Myog, Msc, and Hes6 (Figure 5e). Together, our data suggest that deregulated bHLH transcription factors affect Spin1/Myod1-dependent target gene transcription thereby accounting, at least in part, for SkM defects in Spin1M5 fetuses.
Surviving Spin1M5 mice exhibit major defects in soleus, tibialis anterior, and diaphragm
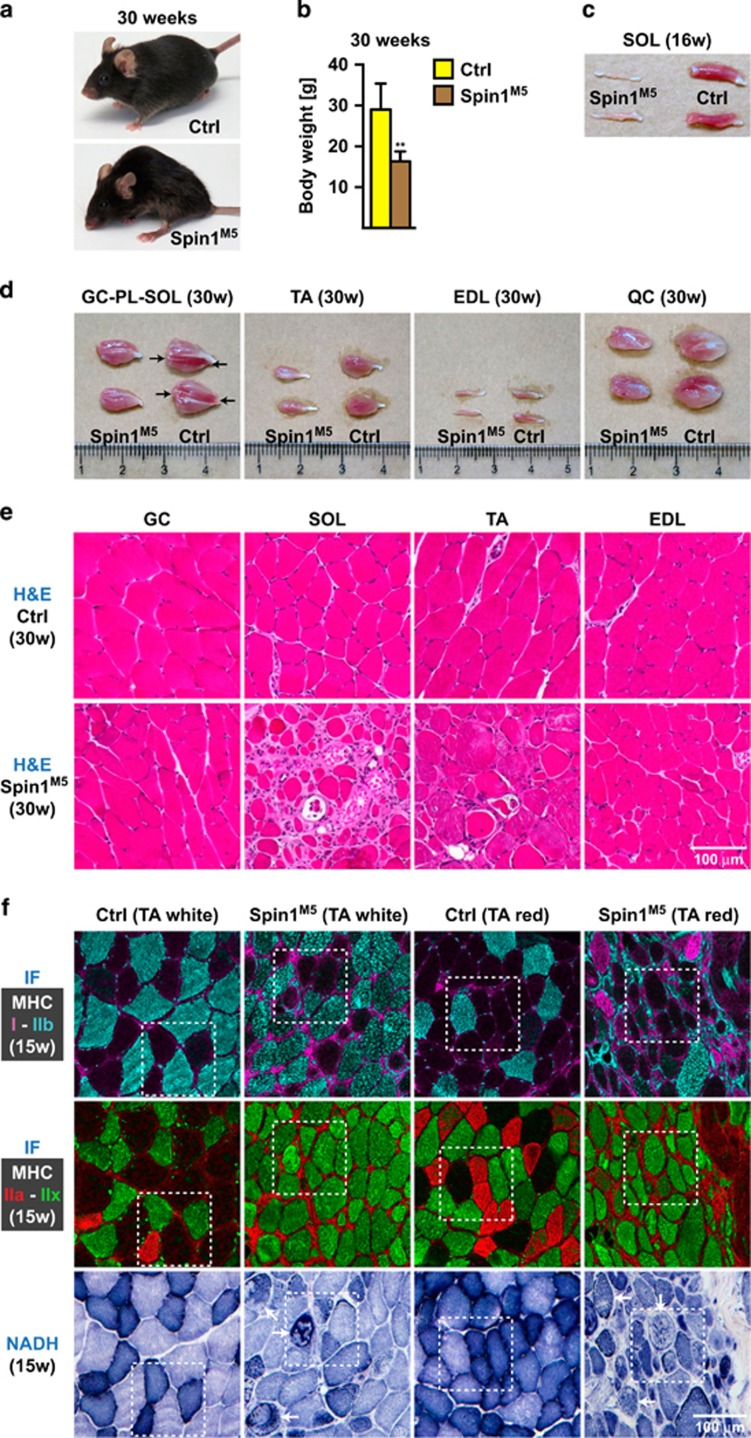
While the majority of Spin1M5 mice died shortly after birth, about 20% survived and reached adulthood. We next investigated the consequences of aberrant fetal myogenesis for SkM in surviving Spin1M5 mice between 15 and 30 weeks of age, the latter representing the oldest cohort obtained. Adult Spin1M5 mice exhibit a severe scoliosis, cachexia, and weigh only around 60% of control littermates (Figures 6a and b).
Figure 6.
Surviving Spin1M5 mice exhibit major defects in soleus, tibialis anterior, and diaphragm. (a,b) Appearance (a) and average body weight (b) of Spin1M5 and control mice (n=5 females in each category) at 30 weeks of age. Error bars represent +S.D., **P<0.01. (c) Degeneration of the soleus (SOL) muscle in adult Spin1M5 compared with control mice exemplified at 16 weeks of age. (d) Hind limb muscles (gastrocnemius (GC), plantaris (PL), soleus, tibialis anterior (TA), extensor digitorum longus (EDL), and quadriceps (QC)) of Spin1M5 and control mice at 30 weeks of age. Arrows point at the soleus embedded in gastrocnemius and plantaris, which is visible in control but degenerated in Spin1M5 mice. (e) Hematoxylin & eosin (H&E) staining of gastrocnemius, soleus, TA, and EDL muscle of Spin1M5 and control mice at 30 weeks of age. (f) Fiber types in glycolytic (white) or oxidative (red) parts of the TA of Spin1M5 and control mice at 15 weeks of age (top and middle rows) observed by immunofluorescence (IF) staining. Tissue sections were stained with selective antibody directed against MHC-I (purple), MHC-IIb (cyan), MHC-IIa (red), and MHC-IIx (green). For comparison, NADH staining was included (bottom row). Fibers with abnormal NADH staining are marked with arrows. Corresponding fibers in each column of images are squared
Inspection of hind limb muscles revealed as the most prominent feature an abnormally thin and pale soleus (SOL) muscle in Spin1M5 mice (Figure 6c). Other hind limb muscles (gastrocnemius (GC), plantaris (PL), TA, extensor digitorum longus (EDL), and quadriceps (QC)) also displayed a reduced mass relative to controls (Figure 6d; Supplementary Figure 5a). The muscle mass reduction was more pronounced (about 60%) for TA than for gastrocnemius-plantaris, EDL, or quadriceps (about 30%) (Supplementary Figure 5a). Counting showed that the number of TA fibers was reduced by about 50%, but did not uncover a significant difference for the EDL of Spin1M5 compared with control mice (Supplementary Figure 5b).
H&E staining confirmed degeneration of the soleus as evidenced by rounded fibers, enormous differences in fiber diameters, and the presence of inflammatory cells (Figure 6e). Major defects were also observed in the TA, while gastrocnemius and EDL appeared largely normal (Figure 6e). Also, the diaphragm (DP) of Spin1M5 mice displayed severe muscle fiber degeneration (Supplementary Figure 5c). Gomori staining detected abnormal collagen deposition indicating fibrosis mainly in the soleus and TA muscle of Spin1M5 mice (Supplementary Figure 5d). Together, our observations revealed severe defects in soleus, TA, and diaphragm.
To investigate whether degenerative changes affected specific fiber types, we analyzed SkM sections by immunofluorescence. The TA of control mice exhibited typical fiber type compositions, that is, mainly type IIb fibers in white (primarily glycolytic) TA and type IIa and IIx fibers in red (primarily oxidative) TA (Figure 6f, top and middle rows). NADH staining correlated with the glycolytic or oxidative properties of the fiber types (Figure 6f, bottom row). In comparison, in the TA of some Spin1M5 mice, fiber types could not be clearly distinguished since most fibers expressed varying levels of MHC-IIx, in part together with MHC-IIb (Figure 6f). We rarely observed type IIa fibers in the TA of these animals (Figure 6f). However, in the TA of other Spin1M5 mice type IIa fibers were present (Supplementary Figure 5e, right column). Furthermore, all expected fiber types were observed in the degenerating soleus and the neighboring plantaris of Spin1M5 mice (Supplementary Figure 5e, left and middle columns), although defective fibers could be identified by irregular fiber size, shape, or NADH staining (Figure 6f; Supplementary Figure 5e, bottom row (arrows)). Overall, these observations argue against selective fiber type degeneration in Spin1M5 mice, but rather suggest that certain muscles including soleus, TA, and diaphragm degenerate.
Identification of deregulated SkM functional networks in surviving Spin1M5 mice
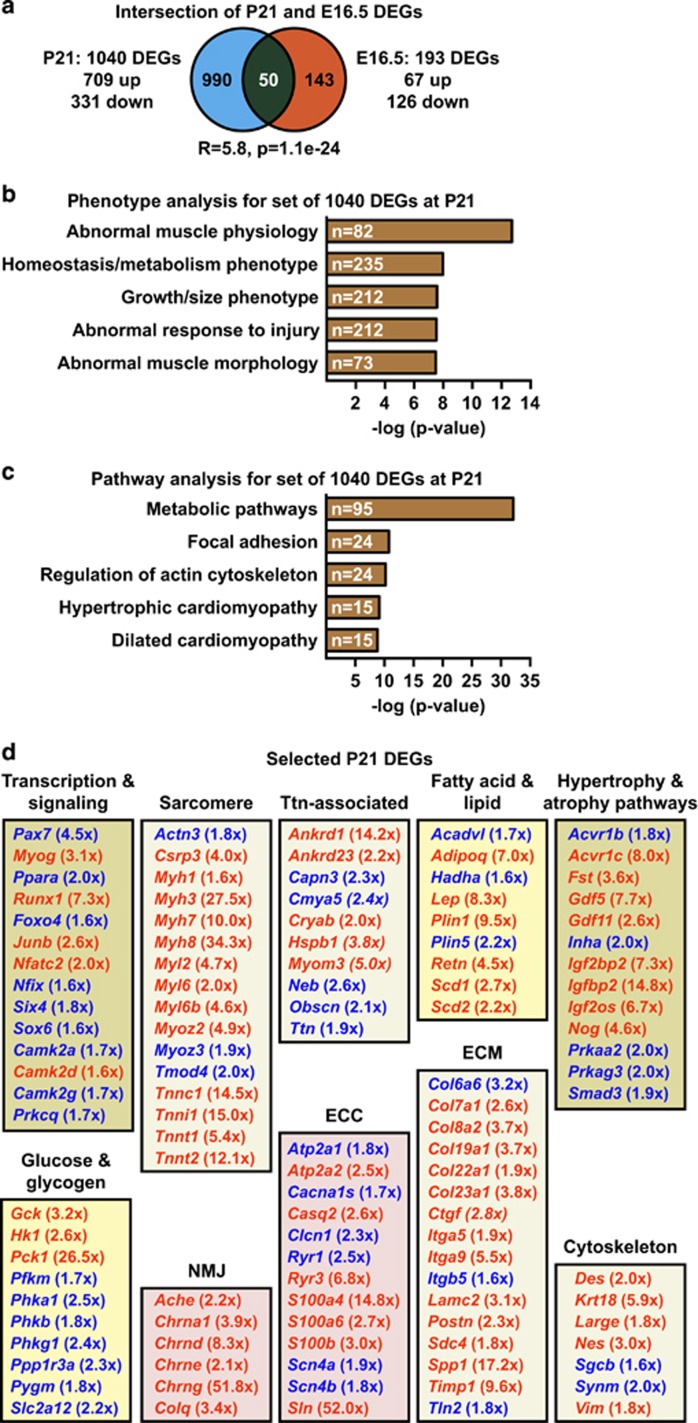
We next analyzed the transcriptomes of TA of Spin1M5 and control mice at three weeks of age (P21), which revealed 1040 DEGs (P≤1e−5; fold change ≥1.5; Figure 7a; Supplementary Table 1d). Intersection with the E16.5 DEGs detected a highly significant overlap of 50 genes (Figure 7a) suggesting that defects in surviving Spin1M5 mice are related to differential gene expression at early stages of disease.
Figure 7.
Identification of deregulated SkM functional networks in surviving Spin1M5 mice. (a) Intersection of DEGs observed for TA of Spin1M5 mice at P21 (blue) and SkM at E16.5 (red). (R: enrichment factor; p: P-value calculated for the intersection.) (b,c) Phenotype (b) and pathway (c) analysis for the set of 1040 DEGs observed in the TA of Spin1M5 mice at P21. For the phenotype analysis, the ‘top 5’ non-redundant terms are depicted. (d) Assignment of selected P21 DEGs identified by phenotype and pathway analyses to functional SkM networks. Upregulated DEGs are depicted in red, downregulated in blue color (fold change in brackets)
Phenotype and pathway analyses for the P21 DEGs uncovered terms related to muscle function and disease, as well as metabolism (Figures 7b and c). In addition, we noted terms such as ‘focal adhesion’ and ‘regulation of actin cytoskeleton’, which were not identified at E16.5 (Figures 4a and b). Grouping of the P21 DEGs according to systems biology classifications38, 39 revealed the same deregulated SkM networks (Figure 7d) as for the E16.5 DEGs (Figure 4c). At P21, however, the networks contained more DEGs than at E16.5, and we identified additional networks ('hypertrophy & atrophy pathways' and 'cytoskeleton'). Of note, the strong increase in deregulated genes including collagen isoforms in the network 'extracellular matrix (ECM)' (Figure 7d) correlates with fibrosis detected in the TA of adult Spin1M5 mice (Supplementary Figure 5d). Together, this analysis provided a comprehensive disease signature for the TA of surviving Spin1M5 mice.
Differential gene expression in surviving Spin1M5 mice correlates with SkM disease patterns
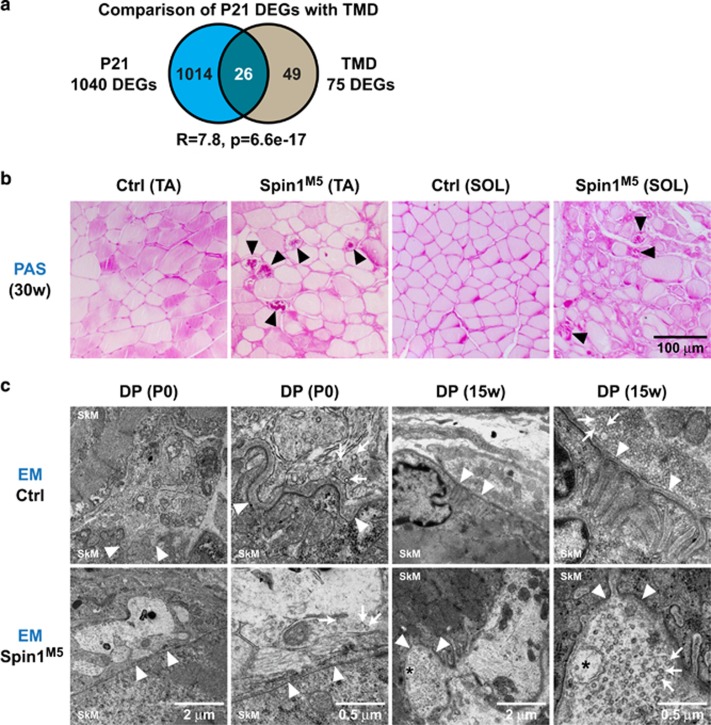
In the final set of experiments, we aimed to correlate differential gene expression in surviving Spin1M5 mice with SkM disease patterns. Since several P21 DEGs encode titin-associated proteins21, 22 (Figure 7d, network 'Ttn-associated'), we hypothesized similarity with diseases caused by Ttn mutations such as tibial muscular dystrophy (TMD).44, 45 Muscular dystrophy with myositis (mdm) mice (serving as a model of TMD) express a titin mutant lacking the N2A region, which binds Ankrd1, Ankrd2, and Ankrd23. Accordingly, defective Ankrd1 signaling has been implicated in TMD in mdm mice.46 Intersection of the P21 DEGs with 75 DEGs previously reported in mdm mice46 revealed a significant overlap of 26 genes (Figure 8a). Given that Ankrd1 and Ankrd2 are already deregulated in Spin1M5 fetuses at E16.5 (Figures 3c and 4c), aberrant expression of titin-associated proteins may account for SkM defects in Spin1M5 mice.
Figure 8.
Differential gene expression in surviving Spin1M5 mice correlates with SkM disease patterns. (a) Intersection of DEGs observed for the TA of Spin1M5 mice at P21 (blue) and SkM of mdm mice46 (gray) representing a model for tibial muscular dystrophy (TMD) (R: enrichment factor; p: P-value calculated for the intersection). (b) Periodic acid–Schiff (PAS) staining for glycogen accumulation in TA and soleus muscle (SOL) of Spin1M5 and control mice at 30 weeks of age. Fibers showing abnormal glycogen accumulation are marked with black triangles. (c) Electron microscopy (EM) images of neuromuscular junctions in the diaphragm (DP) of newborn and 15-week-old Spin1M5 and control mice. Membranes at the synaptic cleft are marked with white triangles, synaptic vesicles with arrows, and vacuoles observed at neuromuscular junctions of Spin1M5 mice with asterisks
Next, we observed downregulation of genes involved in glycogen metabolism including Pygm, Pfkm, Phka1, Phkg1, Prkaa2, and Prkag3 (Figure 7d, networks 'glucose & glycogen' metabolism and 'hypertrophy & atrophy pathways'). Mutations of these genes are known to occur in glycogen storage diseases (glycogenoses).28 Periodic acid–Schiff (PAS) staining revealed abnormal glycogen deposits in TA and soleus fibers of Spin1M5 mice (Figure 8b, black triangles). Although these deposits are limited to individual fibers, which differs from typical glycogenoses, our observation suggests that defective glycogen metabolism contributes to SkM disease in adult Spin1M5 mice.
Finally, we noted strong deregulation of acetylcholine receptor subunits (Chrna1, Chrnd, Chrne, Chrng) and several genes involved in excitation-contraction coupling (Figure 7d, networks 'NMJ' and 'ECC') hinting at defective neuromuscular junctions and/or excitation-contraction coupling. Therefore, we analyzed neuromuscular junctions in the diaphragm of newborn and adult Spin1M5 mice by electron microscopy (Figure 8c). At both time points, the analysis revealed defects of the synaptic membrane (white triangles) and an abnormal appearance as well as a reduced number of synaptic vesicles (arrows) in Spin1M5 mice (Figure 8c). Furthermore, we observed the presence of vacuoles at nerve terminals of adult Spin1M5 mice (Figure 8c, right columns (asterisks)). These data provide evidence for neuromuscular junction damage and abnormal excitation-contraction coupling in Spin1M5 mice.
Discussion
In this study, we ablated the H3K4me3 reader Spin1 in myoblast precursors resulting in abnormal fetal myogenesis, early postnatal death, or severe SkM defects in few surviving Spin1M5 mice. Our analyses suggest that numerous E16.5 DEGs are direct target genes of Spin1 and Myod1. Furthermore, aberrant expression of Myf5, Myog, Msc, and Hes6 in Spin1M5 fetuses may indirectly affect Myod1-dependent gene regulation. Msc, for example, can repress transcription and antagonize the action of Myod1 in undifferentiated myoblasts by heterodimerization with E-proteins.40 Hes6 was reported to inhibit expression of Msc thereby enhancing myoblast differentiation.43 Therefore, deregulated bHLH transcription factor networks appear to determine, at least in part, aberrant gene expression in Spin1M5 fetuses.
The preferential degeneration of soleus, TA, and diaphragm in adult Spin1M5 mice raised the question whether this is caused by fiber type- or muscle type-selective mechanisms. Currently, we favor the latter hypothesis. While degeneration of type I and type II fibers might account for soleus defects, it would not convincingly explain the severe TA damage since this muscle contains only about 1 and 10% of these fibers types, respectively.47 Furthermore, at E15.5 or E16.5, we did not identify DEGs involved in the regulation of fiber type identity or plasticity (e.g., Six1, Six4, Eya1, Sox6).30
One possible determinant of muscle type-selective degeneration in Spin1M5 mice could be the deregulation of titin-associated proteins such as Ankrd1, which has been linked with TMD in mdm mice.46 However, despite some similarity with TMD, the SkM phenotype of Spin1M5 mice is apparently more complex. We exemplarily correlated the P21 DEGs with abnormal glycogen accumulation in individual TA and soleus fibers as well as neuromuscular junction defects in Spin1M5 mice. Due to the high number of DEGs observed in the TA of Spin1M5 mice at P21, these examples only provide an initial characterization of deregulated SkM networks.
Compared with transcription factors, signaling molecules, SkM structural proteins, or metabolic enzymes, the roles of epigenetic regulators in SkM physiology and disease have only more recently become a focus of research. However, while epigenetic writers, erasers, and non-coding RNAs have received considerable attention,48, 49 little is known about potential functions of histone code readers in SkM. Few readers (Brd4, Ing2, Sfmbt1, Dpf3) have been implicated in myogenesis using the C2C12 cell culture model50, 51, 52, 53 or in zebrafish.54 However, in these cases either knockout mouse models have not been reported, or in mice no apparent SkM phenotype was observed.55, 56, 57 Thus, to the best of our knowledge, Spin1 is the first histone code reader, for which ablation in SkM has fatal consequences in mice. Together, our histological and transcriptome analyses provide insight into severe SkM defects in Spin1M5 fetuses and surviving adult mice, hinting at Spin1 as a potential player in human SkM disease.
Materials and methods
Mouse studies
All mice were housed in the pathogen-free barrier facility of the University Medical Center Freiburg in accordance with institutional guidelines and approved by the regional board. Mice were maintained under temperature- and humidity-controlled conditions with a 12-h light/dark cycle, free access to water, and a standard rodent chow (3807, Provimi Kliba). Animals were killed by cervical dislocation and tissues immediately processed for further analyses.
Generation and validation of Spin1R26 and Spin1M5 mice
The targeting strategy for generation of a conditional Spin1 allele is outlined in Supplementary Figure 1a. Details are available upon request. The targeting construct was electroporated into C57BL/6 N Tac embryonic stem (ES) cells (Taconic), and neomycin-and puromycin-resistant clones were expanded. Selected ES cells were injected into blastocysts of C57BL/6 N mice. Resulting mice were bred to Rosa26-Flp mice to remove NeoR and PuroR selection markers. Offspring harbored a conditional Spin1 allele, in which exon 4 of Spin1 was flanked by loxP sites. C57/Bl6N mice homozygous for the conditional allele (Spin1p/p) were crossed with the Rosa26-Cre deleter strain32 to generate ubiquitous knockout (Spin1R26) mice or with the Myf5-Cre deleter strain31 to selectively ablate Spin1 in myoblast and brown adipocyte precursors (Spin1M5 mice). Mice were maintained on the C57/Bl6N background. In phenotypic analyses of Spin1M5 mice, Spin1+/p or Spin1p/p littermates served as controls. Mice were genotyped by PCR amplification using GoTaq G2 Flexi DNA polymerase (Promega) of genomic DNA extracted from tail biopsies with the NucleoSpin 96 tissue kit (Macherey Nagel). The following genotyping primers were used: forward 5-ATAGGCTCTCTGGCATGGTG-3 / reverse 5-ACAGCGTGACACATCAAAGC-3 detecting the wild-type (177 bp) and the conditional (337 bp) allele and forward 5′-GGAGGAAGACACCTAATAGCACC-3′ /reverse 5′-AAGGCAAAACGGAGACAGC-3′ detecting the deleted allele (395 bp).
Efficient Spin1 depletion in myogenic cells and remaining Spin1 expression in non-myogenic cells of Spin1M5 mice was validated as follows. Spin1M5 and control mice were crossed with mice harboring a green fluorescent protein/nuclear lacZ (GNZ) reporter.58 In mice harboring the reporter, GNZ expression is Cre dependent. GNZ-positive myogenic cells in Spin1M5 mice did not express Spin1, whereas Spin1-positive cells were GNZ-negative and therefore non-myogenic (Supplementary Figure 1f). Part of these Spin1-expressing, non-myogenic cells were Tcf4-positive fibroblasts34 (Supplementary Figure 1e). Reduction of Spin1 mRNA in hind limb muscle of newborn Spin1M5 mice was confirmed by quantitative RT-PCR (Supplementary Figure 1g).
Quantitative RT-PCR and RNA sequencing
SkM tissue was homogenized in TRIzol reagent (Life Technologies, Darmstadt, Germany) using a Minilys personal homogenizer (Bertin, Montigny, France) and 0.5 or 2.0 ml CK14 lysing kits (Precellys, Montigny, France). RNA was isolated using a standard phenol/chloroform extraction protocol. cDNA was prepared by reverse transcription of total RNA using SuperScript II (Life Technologies) and oligo(dT) primer according to the supplier’s protocol. Quantitative RT-PCR was performed with a Lightcycler 480 II (Roche, Mannheim, Germany) using Absolute SYBR green ROX Mix (Thermo Scientific, Scherte, Germany) and the following primers: Spin1 (exon 4, forward) 5′-CAGGTGCCTGTGAATCCTTC-3′, Spin1 (exon 5, reverse) 5′-ACATGTGCTCCACTGCTTTG-3′, Tbp (forward) 5′-CCCCTTGTACCCTTCACCAAT-3′, Tbp (reverse) GAAGCTGCGGTACAATTCCAG-3′, Hprt (forward) 5′-GTTAAGCAGTACAGCCCCAAA-3′, Hprt (reverse) 5′-AGGGCATATCCAACAACAAACTT-3′, Polr2a (forward) 5′-CACCCCAGCTTCTCCCAAAT-3′, and Polr2a (reverse) 5′-AGTATGTCGGGGAGGTTGGA-3′. Data were analyzed using the 2(-Delta Delta C(T)) method.59
RNA-seq analyses for E15.5 and E16.5 were performed with SkM dissected from front and hind limb of three Spin1M5 and four control fetuses. For RNA-seq analysis of mice at three weeks of age (P21), TA isolated from four Spin1M5 and four control mice was used. RNA was isolated as described above, except that minced tissue in TRIzol was further homogenized with QIA shredder spin columns (Qiagen, Hilden, Germany). RNA quality was determined using the RNA 6000 Nano Kit (Agilent, Waldbronn, Germany) on an Agilent 2100 Bioanalyzer. RNA with a RIN above 8.0 was sequenced at the DKFZ core facility (Heidelberg, Germany) or the Deep Sequencing Unit (Max-Planck-Institute of Immunology and Epigenetics, Freiburg) using the standard Illumina protocol. Paired-end reads were mapped to Ensemble annotation NCBIm38/mm10 with TopHat260 using default parameters. The aligned reads were counted with HOMER software61 and DEGs calculated with edgeR.62 Overrepresentation analyses for the identified DEGs (P≤1e−3 (E15.5 and E16.5), P≤1e−5 (P21); fold change ≥1.5; ≥50 reads in all Spin1M5 or all control samples] were performed using WebGestalt36 with 'genome' as reference set.
Of note, none of the RNA-seq analyses identified Spin1 as differentially expressed gene. Examination of the normalized reads at exons 1 to 6 of the Spin1 gene explained this observation (Supplementary Table 1e). Reads were significantly decreased at exon 4 (which is excised in a Myf5-Cre-dependent manner in myogenic cells), but not at other exons in Spin1M5 relative to control mice. Thus, in myogenic cells of Spin1M5 mice a truncated transcript (lacking exon 4) is expressed, which results in translation of a truncated Spin1 peptide (containing 56 N-terminal amino acids lacking any known functional domain) due to the presence of a premature STOP codon in exon 5. Normal Spin1 transcript (containing exon 4) present in SkM of Spin1M5 mice is most likely produced by non-myogenic cells (Supplementary Figures 1e and f). Since mapped reads at all exons contribute to the overall count, differential expression of Spin1 is not detected in RNA-seq analyses.
Antibodies
The generation and validation of anti-SPIN1 antibodies 5865 and 5867 was described previously.6 Antibodies SPIN1(5867) and H3K4me3 (Diagenode, Oxford, UK, C15410003) were used for ChIP. For immunofluorescence staining the following primary antibodies were used: SPIN1(5865) 1 μg/ml; Pax7 (PAX7, DSHB, batch 7/2/15) 2 μg/ml; Tcf4 (6H5-3, Millipore, Darmstadt, Germany, 05-511, batch 2155406) 10 μg/ml; Ankrd1 (Proteintech Group, Manchester, UK, 11427-1-AP, batch 1951) 1:100; Ankrd2 (Proteintech Group, 11821-1-AP, batch 7649) 1:100; dystrophin (Abcam, Cambridge, UK, ab15277, batch GR226781-6) 1:500; MHC-I (NOQ7.5.4D, Sigma, Munich, Germany, M8421, batch 035M4792V) 1:2000; MHC-IIa (SC-71, DSHB, batch 4/7/16) 1:10; MHC-IIx (6H1, DSHB, batch 3/3/16) 1:6; MHC-IIb (BF-F3, DSHB, batch 5/12/16) 1:20; MCH, skeletal, fast (MY-32, Sigma, M4276, batch 083M4790V) 1:1000; GNZ (anti-GFP, Abcam, ab13970, batch GR236651) 1:1000; normal rabbit IgG (Santa Cruz, Heidelberg, Germany, sc-2027). The following secondary antibodies were used: Alexa Fluor 488 (goat anti-mouse IgM, Molecular Probes, Karlsruhe, Germany, A21042, batch 1387726) 1:600; Alexa Fluor 488 (goat anti-mouse IgG, Molecular Probes, A11029, batch 1306597) 1:600; Alexa Fluor 546 (goat anti-mouse IgG, Molecular Probes, 11030, batch 517979) 1:600; Alexa Fluor 594 (goat anti-chicken IgG, Molecular Probes, A11042, batch 762712) 1:600; donkey anti-rabbit IgG-HRP (Santa Cruz, sc-2313) 1:500.
Immunofluorescence staining
SkM tissues were either fixed in 4% paraformaldehyde/PBS or flash-frozen in 2-methylbutane (Sigma). Paraffin sections (5 μm) were deparaffinized, heated in antigen retrieval solution (20 mM Tris (pH9.0)) for 20 min in a pressure cooker, and blocked for 1 h at room temperature in 3% skim milk powder/PBS containing 0.1% Tween (PBST). Cryosections (10 μm) were blocked with 5% fetal calf serum/PBST. Sections were incubated overnight with primary antibody at 4 °C, washed with PBST, incubated with secondary antibody for 1 h at room temperature, and then washed with PBST. For Spin1 staining, signal amplification using the TSA fluorescein system (Perkin Elmer, NEL701A001KT) was applied according to the manufacturer’s instructions. Briefly, tissue sections were blocked in 0.5% TNB blocking buffer, labeled with primary and secondary antibody, and then incubated with TSA reagent for 15 minutes at room temperature followed by two washing steps with PBST. Finally, nuclei were stained with DAPI (1 μg/ml) followed by two washing steps with PBST, and sections were mounted using Fluoromount-G (SouthernBiotech). Images were recorded with a confocal microscope (Leica TCS SP2 AOBS). Counting was done with ImageJ63 followed by visual validation of the results. Pax7-positive nuclei or nuclei of myofibers (expressing Spin1 or being Spin1-negative) were quantified using confocal images with dimensions of 187.5 μm × 187.5 μm or 375 μm × 375 μm, respectively.
Isolation of primary myoblasts and ChIP sequencing
Primary myoblasts were isolated from 10- day-old C57BL/6 N mice using an established preplating protocol.64 Briefly, limb muscles were collected from front and hind leg, minced, digested for 1 h in 0.2% collagenase type I (Sigma, C0130), and filtered through 100 μm cell strainers (Falcon, 352360). Cells were collected by centrifugation, resuspended in DMEM medium (Gibco, Schwerte, Germany, 11995-065) supplemented with 10% FCS, 10% horse serum, and 1.25% chicken embryo extract (Seralab, CE-650-J), and cultivated for 1 h on 6 cm tissue culture dishes to allow for attachment of fibroblasts. Supernatants were transferred to 6 cm dishes, again incubated for one hour, and then transferred to 10 cm tissue culture dishes coated with collagen (Gibco, A10483-01). Myoblasts were cultivated for 3 to 4 days until a confluency of about 70% was reached. Cells were fixed with 1% formaldehyde for 5 minutes, quenched in glycine (1.25 M), washed with PBS buffer, collected, and snap-frozen in liquid nitrogen.
Chromatin was prepared using the NEXSON procedure.65 Briefly, nuclei were extracted by sonication with a Covaris E220 sonicator (75 W peak power, 2% duty factor, 200 cycles/burst, 60 s). Nuclei were pelleted, resuspended in 1 ml of shearing buffer, and sonicated for 12 min (140 W peak power, 5% duty factor, 200 cycles/burst). Chromatin was diluted 1:2 in buffer H (Diagenode auto histone ChIP-seq kit (C01010022)) before ChIP. For ChIP, one tenth of the chromatin was incubated with 1 μg of H3K4me3 antibody (Diagenode, C15410003). The remaining chromatin (from ~1.2 million cells) was incubated with 5 μg of SPIN1(5867) antibody. ChIP was performed using the automated platform SX-8G IP-Star (Diagenode) and the program 'ChIP indirect method'. Chromatin was incubated with antibody for 10 h followed by 3 h of incubation with protein A-conjugated beads. 1% of the original chromatin was used as input. After elution from beads, ChIP and input samples were reverse crosslinked, and DNA purified with MinElute columns (Qiagen, 28004). Libraries were prepared using the NEBNext Ultra DNA library preparation kit (NEB, E7370S) according to the manufacturer’s instruction and without size selection. Adapter-ligated fragments were amplified with 12 PCR cycles and sequenced on the Illumina HiSeq 2500 platform by the Deep Sequencing Unit of the Max-Planck-Institute of Immunology and Epigenetics (Freiburg).
Paired-end reads were mapped to the mouse reference genome (mm10) using bowtie 266 with default parameters. Data were further analyzed using the peak finding algorithm of MACS 1.4.267 using input as control. All peaks with a false discovery rate >1% were excluded from further analyses. The uniquely mapped reads were used to generate genome-wide intensity profiles, which were visualized using the integrative genomics viewer (IGV).68 Peaks were annotated and overlaps between different peak files were calculated with HOMER.61 The genomic features (promoter, exon, intron, 5′ or 3′ untranslated region, and intergenic region) were defined and calculated using Refseq and HOMER. Myod1 chromatin association in C2C12 myoblasts was analyzed using a previously deposited GEO data set (GSE36024). Venn diagrams were generated with the help of Venny.69 Intensity profiles for Spin1 and H3K4me3 gene promoter occupancy were analyzed with SeqMINER.70
GEO data availability
The RNA and DNA sequencing data discussed in this publication have been deposited in NCBI's Gene Expression Omnibus71 and are accessible through GEO Series accession number GSE92539.
Electron microscopy
Muscle samples were fixed by immersion in 2.5% glutaraldehyde and 2.5% paraformaldehyde in cacodylate buffer (0.1 M, pH 7.4) and then washed in cacodylate buffer for further 30 min. The samples were post-fixed in 1% osmium tetroxide in 0.1 M cacodylate buffer for 1 h at 4 °C, and dehydrated in an ascending ethanol gradient (50, 70, 90, and 100%) and propylene oxide for 30 min each. Samples were embedded in Epon 812 substitute (Sigma-Aldrich). Semi-thin sections cut at 2 μm and ultra-thin sections cut at 70 nm were contrasted with uranyl acetate and lead citrate and examined at 70 kV with a Morgagni 268D electron microscope. Images were digitally captured by Mega View III camera (Soft Imaging System).
Hematoxylin & eosin staining
Deparaffinized and rehydrated or flash-frozen tissue sections (5 or 10 μm, respectively) were stained according to a standard protocol with hematoxylin (Gill No. 3, Sigma, GHS332) and eosin Y solution (Sigma, HT110332) and mounted using Roti-Histokitt (Roth).
Gomori trichrome staining
Deparaffinized and rehydrated tissue sections (5 μm) were stained using Bouin’s solution (Sigma, HT10132), hematoxylin-Weigert’s iron kit (Dianova, HWI-2), and trichrome stain (blue) solution (Dianova, TGB500) according to the supplier's instructions and mounted using Roti-Histokitt (Roth).
NADH staining
Cryosections (10 μm) were incubated with staining solution (0.2 M Tris (pH 7.4), 1.5 mM NADH (Roche, 10128015001), 1.5 mM nitroblue tetrazolium (Sigma, N-6876)), dehydrated in an ascending ethanol gradient, incubated twice with xylene, and mounted using Roti-Histokitt (Roth).
Periodic acid–Schiff staining
Deparaffinized and rehydrated tissue sections (5 μm) were treated with 0.5% periodic acid solution (Sigma, 3951), stained with Schiff’s reagent (Sigma, 3952016), dehydrated, and mounted using Roti-Histokitt (Roth).
Statistics
Transcriptome and cistrome data were analyzed as described above. Statistical significance of gene set intersections was evaluated by a hypergeometric test using the program 'R' (http://www.R-project.org) [phyper (N12-1, N1, N-N1, N2, lower.tail=FALSE) with N1 (genes in set 1), N2 (genes in set 2), N12 (genes in intersection), and N (genome size)]. The enrichment factor (R) was calculated according to R=(N × N12)/(N1 × N2). Other data are presented as the mean value or percentage change +S.D. Comparisons between two data sets were made using the two-tailed Student's t-test for parametric data and the Wilcoxon signed-rank test for nonparametric data. A P-value of less than 0.05 was considered statistically significant. Statistical significance is indicated as follows: *P<0.05, **P<0.01, ***P<0.001.
Acknowledgments
This work was supported by grants from the European Research Council (ERC AdGrant 322844 to R.S.) and Deutsche Forschungsgemeinschaft (SFB 992, 850, and 746 to R.S.). L.A. and T.M. received funding from the German Epigenome Programme DEEP (BMBF 01KU1216G). The authors thank Ines Steinhauser, Jean-Luc Weickert, and Josiane Hergueux for providing excellent technical assistance and Jin Rong Ow for critical reading of the manuscript and helpful discussions. The authors are grateful to Ulrike Bönisch and members of the Deep Sequencing Unit at the Max-Planck-Institute of Immunology and Epigenetics (Freiburg). The authors are indebted to Hans-Henning Arnold for providing Myf5-Cre mice.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by M Agostini
The authors declare no conflict of interest.
Supplementary Material
References
- Bartke T, Vermeulen M, Xhemalce B, Robson SC, Mann M, Kouzarides T. Nucleosome-interacting proteins regulated by DNA and histone methylation. Cell 2010; 143: 470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W, Chen Z, Mao Z, Zhang H, Ding X, Chen S et al. Nucleolar protein Spindlin1 recognizes H3K4 methylation and stimulates the expression of rRNA genes. EMBO Rep 2011; 12: 1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N, Wang W, Wang Y, Wang M, Zhao Q, Rao Z et al. Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1. Proc Natl Acad Sci USA 2012; 109: 17954–17959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su X, Zhu G, Ding X, Lee SY, Dou Y, Zhu B et al. Molecular basis underlying histone H3 lysine-arginine methylation pattern readout by Spin/Ssty repeats of Spindlin1. Genes Dev 2014; 28: 622–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue W, Sun LY, Li CH, Zhang LX, Pei XT. Screening and identification of ovarian carcinomas related genes. Ai Zheng 2004; 23: 141–145. [PubMed] [Google Scholar]
- Franz H, Greschik H, Willmann D, Ozretic L, Jilg CA, Wardelmann E et al. The histone code reader SPIN1 controls RET signaling in liposarcoma. Oncotarget 2015; 6: 4773–4789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang YW, Xing AY, Xiang S, Shi DB, Liu L et al. Suppression of SPIN1-mediated PI3K-Akt pathway by miR-489 increases chemosensitivity in breast cancer. J Pathol 2016; 239: 459–472. [DOI] [PubMed] [Google Scholar]
- Gao Y, Yue W, Zhang P, Li L, Xie X, Yuan H et al. Spindlin1, a novel nuclear protein with a role in the transformation of NIH3T3 cells. Biochem Biophys Res Commun 2005; 335: 343–350. [DOI] [PubMed] [Google Scholar]
- Wang JX, Zeng Q, Chen L, Du JC, Yan XL, Yuan HF et al. SPINDLIN1 promotes cancer cell proliferation through activation of WNT/TCF-4 signaling. Mol Cancer Res 2012; 10: 326–335. [DOI] [PubMed] [Google Scholar]
- Zhang P, Cong B, Yuan H, Chen L, Lv Y, Bai C et al. Overexpression of spindlin1 induces metaphase arrest and chromosomal instability. J Cell Physiol 2008; 217: 400–408. [DOI] [PubMed] [Google Scholar]
- Yuan H, Zhang P, Qin L, Chen L, Shi S, Lu Y et al. Overexpression of SPINDLIN1 induces cellular senescence, multinucleation and apoptosis. Gene 2008; 410: 67–74. [DOI] [PubMed] [Google Scholar]
- Chew TG, Peaston A, Lim AK, Lorthongpanich C, Knowles BB, Solter D. A tudor domain protein SPINDLIN1 interacts with the mRNA-binding protein SERBP1 and is involved in mouse oocyte meiotic resumption. PLoS One 2013; 8: e69764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckingham M, Relaix F. PAX3 and PAX7 as upstream regulators of myogenesis. Semin Cell Dev Biol 2015; 44: 115–125. [DOI] [PubMed] [Google Scholar]
- Comai G, Tajbakhsh S. Molecular and cellular regulation of skeletal myogenesis. Curr Top Dev Biol 2014; 110: 1–73. [DOI] [PubMed] [Google Scholar]
- Almada AE, Wagers AJ. Molecular circuitry of stem cell fate in skeletal muscle regeneration, ageing and disease. Nat Rev Mol Cell Biol 2016; 17: 267–279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi G, Messina G. Comparative myogenesis in teleosts and mammals. Cell Mol Life Sci 2014; 71: 3081–3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White RB, Bierinx AS, Gnocchi VF, Zammit PS. Dynamics of muscle fibre growth during postnatal mouse development. BMC Dev Biol 2010; 10: 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun T, Gautel M. Transcriptional mechanisms regulating skeletal muscle differentiation, growth and homeostasis. Nat Rev Mol Cell Biol 2011; 12: 349–361. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Reggiani C. Fiber types in mammalian skeletal muscles. Physiol Rev 2011; 91: 1447–1531. [DOI] [PubMed] [Google Scholar]
- Greising SM, Gransee HM, Mantilla CB, Sieck GC. Systems biology of skeletal muscle: fiber type as an organizing principle. Wiley Interdiscip Rev Syst Biol Med 2012; 4: 457–473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauveau C, Rowell J, Ferreiro A. A rising titan: TTN review and mutation update. Hum Mutat 2014; 35: 1046–1059. [DOI] [PubMed] [Google Scholar]
- Savarese M, Sarparanta J, Vihola A, Udd B, Hackman P. Increasing role of titin mutations in neuromuscular disorders. J Neuromuscul Dis 2016; 3: 293–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravenscroft G, Laing NG, Bonnemann CG. Pathophysiological concepts in the congenital myopathies: blurring the boundaries, sharpening the focus. Brain 2015; 138: 246–268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jungbluth H, Ochala J, Treves S, Gautel M. Current and future therapeutic approaches to the congenital myopathies. Semin Cell Dev Biol 2017; 64: 191–200. [DOI] [PubMed] [Google Scholar]
- Mercuri E, Muntoni F. Muscular dystrophies. Lancet 2013; 381: 845–860. [DOI] [PubMed] [Google Scholar]
- Dowling JJ, Lawlor MW, Dirksen RT. Triadopathies: an emerging class of skeletal muscle diseases. Neurotherapeutics 2014; 11: 773–785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suetterlin K, Mannikko R, Hanna MG. Muscle channelopathies: recent advances in genetics, pathophysiology and therapy. Curr Opin Neurol 2014; 27: 583–590. [DOI] [PubMed] [Google Scholar]
- Godfrey R, Quinlivan R. Skeletal muscle disorders of glycogenolysis and glycolysis. Nat Rev Neurol 2016; 12: 393–402. [DOI] [PubMed] [Google Scholar]
- Ciciliot S, Rossi AC, Dyar KA, Blaauw B, Schiaffino S. Muscle type and fiber type specificity in muscle wasting. Int J Biochem Cell Biol 2013; 45: 2191–2199. [DOI] [PubMed] [Google Scholar]
- Talbot J, Maves L. Skeletal muscle fiber type: using insights from muscle developmental biology to dissect targets for susceptibility and resistance to muscle disease. Wiley Interdiscip Rev Dev Biol 2016; 5: 518–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tallquist MD, Weismann KE, Hellstrom M, Soriano P. Early myotome specification regulates PDGFA expression and axial skeleton development. Development 2000; 127: 5059–5070. [DOI] [PubMed] [Google Scholar]
- Schwenk F, Baron U, Rajewsky K. A cre-transgenic mouse strain for the ubiquitous deletion of loxP-flanked gene segments including deletion in germ cells. Nucleic Acids Res 1995; 23: 5080–5081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turgeon B, Meloche S. Interpreting neonatal lethal phenotypes in mouse mutants: insights into gene function and human diseases. Physiol Rev 2009; 89: 1–26. [DOI] [PubMed] [Google Scholar]
- Mathew SJ, Hansen JM, Merrell AJ, Murphy MM, Lawson JA, Hutcheson DA et al. Connective tissue fibroblasts and Tcf4 regulate myogenesis. Development 2011; 138: 371–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojic S, Radojkovic D, Faulkner G. Muscle ankyrin repeat proteins: their role in striated muscle function in health and disease. Crit Rev Clin Lab Sci 2011; 48: 269–294. [DOI] [PubMed] [Google Scholar]
- Wang J, Duncan D, Shi Z, Zhang B. WEB-based GEne SeT AnaLysis Toolkit (WebGestalt): update 2013. Nucleic Acids Res 2013; 41: W77–W83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biressi S, Tagliafico E, Lamorte G, Monteverde S, Tenedini E, Roncaglia E et al. Intrinsic phenotypic diversity of embryonic and fetal myoblasts is revealed by genome-wide gene expression analysis on purified cells. Dev Biol 2007; 304: 633–651. [DOI] [PubMed] [Google Scholar]
- Wang Y, Winters J, Subramaniam S. Functional classification of skeletal muscle networks. I. Normal physiology. J Appl Physiol 2012; 113: 1884–1901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith LR, Meyer G, Lieber RL. Systems analysis of biological networks in skeletal muscle function. Wiley Interdiscip Rev Syst Biol Med 2013; 5: 55–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu J, Webb R, Richardson JA, Olson EN. MyoR: a muscle-restricted basic helix-loop-helix transcription factor that antagonizes the actions of MyoD. Proc Natl Acad Sci USA 1999; 96: 552–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu JR, Bassel-Duby R, Hawkins A, Chang P, Valdez R, Wu H et al. Control of facial muscle development by MyoR and capsulin. Science 2002; 298: 2378–2381. [DOI] [PubMed] [Google Scholar]
- Cossins J, Vernon AE, Zhang Y, Philpott A, Jones PH. Hes6 regulates myogenic differentiation. Development 2002; 129: 2195–2207. [DOI] [PubMed] [Google Scholar]
- Gao X, Chandra T, Gratton MO, Quelo I, Prud'homme J, Stifani S et al. HES6 acts as a transcriptional repressor in myoblasts and can induce the myogenic differentiation program. J Cell Biol 2001; 154: 1161–1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvey SM, Rajan C, Lerner AP, Frankel WN, Cox GA. The muscular dystrophy with myositis (mdm) mouse mutation disrupts a skeletal muscle-specific domain of titin. Genomics 2002; 79: 146–149. [DOI] [PubMed] [Google Scholar]
- Hackman P, Vihola A, Haravuori H, Marchand S, Sarparanta J, De Seze J et al. Tibial muscular dystrophy is a titinopathy caused by mutations in TTN, the gene encoding the giant skeletal-muscle protein titin. Am J Hum Genet 2002; 71: 492–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witt CC, Ono Y, Puschmann E, McNabb M, Wu Y, Gotthardt M et al. Induction and myofibrillar targeting of CARP, and suppression of the Nkx2.5 pathway in the MDM mouse with impaired titin-based signaling. J Mol Biol 2004; 336: 145–154. [DOI] [PubMed] [Google Scholar]
- Bloemberg D, Quadrilatero J. Rapid determination of myosin heavy chain expression in rat, mouse, and human skeletal muscle using multicolor immunofluorescence analysis. PLoS One 2012; 7: e35273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sincennes MC, Brun CE, Rudnicki MA. Concise review: epigenetic regulation of myogenesis in health and disease. Stem Cells Transl Med 2016; 5: 282–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moresi V, Marroncelli N, Coletti D, Adamo S. Regulation of skeletal muscle development and homeostasis by gene imprinting, histone acetylation and microRNA. Biochim Biophys Acta 2015; 1849: 309–316. [DOI] [PubMed] [Google Scholar]
- Lin S, Shen H, Li JL, Tang S, Gu Y, Chen Z et al. Proteomic and functional analyses reveal the role of chromatin reader SFMBT1 in regulating epigenetic silencing and the myogenic gene program. J Biol Chem 2013; 288: 6238–6247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eapen SA, Netherton SJ, Sarker KP, Deng L, Chan A, Riabowol K et al. Identification of a novel function for the chromatin remodeling protein ING2 in muscle differentiation. PLoS One 2012; 7: e40684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proserpio V, Fittipaldi R, Ryall JG, Sartorelli V, Caretti G. The methyltransferase SMYD3 mediates the recruitment of transcriptional cofactors at the myostatin and c-Met genes and regulates skeletal muscle atrophy. Genes Dev 2013; 27: 1299–1312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du C, Jin YQ, Qi JJ, Ji ZX, Li SY, An GS et al. Effects of myogenin on expression of late muscle genes through MyoD-dependent chromatin remodeling ability of myogenin. Mol Cells 2012; 34: 133–142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange M, Kaynak B, Forster UB, Tonjes M, Fischer JJ, Grimm C et al. Regulation of muscle development by DPF3, a novel histone acetylation and methylation reader of the BAF chromatin remodeling complex. Genes Dev 2008; 22: 2370–2384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mertsalov IB, Ninkina NN, Wanless JS, Buchman VL, Korochkin LI, Kulikova DA. Generation of mutant mice with targeted disruption of two members of the d4 gene family: neuro-d4 and cer-d4. Dokl Biochem Biophys 2008; 419: 65–68. [DOI] [PubMed] [Google Scholar]
- Saito M, Kumamoto K, Robles AI, Horikawa I, Furusato B, Okamura S et al. Targeted disruption of Ing2 results in defective spermatogenesis and development of soft-tissue sarcomas. PLoS One 2010; 5: e15541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kichina JV, Zeremski M, Aris L, Gurova KV, Walker E, Franks R et al. Targeted disruption of the mouse ing1 locus results in reduced body size, hypersensitivity to radiation and elevated incidence of lymphomas. Oncogene 2006; 25: 857–866. [DOI] [PubMed] [Google Scholar]
- Stoller JZ, Degenhardt KR, Huang L, Zhou DD, Lu MM, Epstein JA. Cre reporter mouse expressing a nuclear localized fusion of GFP and beta-galactosidase reveals new derivatives of Pax3-expressing precursors. Genesis 2008; 46: 200–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 2013; 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 2010; 38: 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010; 26: 139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider CA, Rasband WS, Eliceiri KW NIH Image to ImageJ: 25 years of image analysis Nat Methods 2012; 9: 671–675. [DOI] [PMC free article] [PubMed]
- Rando TA, Blau HM. Primary mouse myoblast purification, characterization, and transplantation for cell-mediated gene therapy. J Cell Biol 1994; 125: 1275–1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrigoni L, Richter AS, Betancourt E, Bruder K, Diehl S, Manke T et al. Standardizing chromatin research: a simple and universal method for ChIP-seq. Nucleic Acids Res 2016; 44: e67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 2009; 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol 2008; 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G et al. Integrative genomics viewer. Nat Biotechnol 2011; 29: 24–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliveros JC Venny. An interactive tool for comparing lists with Venn's diagrams. Available at http://bioinfogp.cnb.csic.es/tools/venny/index.html2007-2015.
- Ye T, Krebs AR, Choukrallah MA, Keime C, Plewniak F, Davidson I et al. seqMINER: an integrated ChIP-seq data interpretation platform. Nucleic Acids Res 2011; 39: e35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002; 30: 207–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.