Summary
The RNA polymerase II-associated protein 1 (RPAP1) is conserved across metazoa and required for stem cell differentiation in plants; however, very little is known about its mechanism of action or its role in mammalian cells. Here, we report that RPAP1 is essential for the expression of cell identity genes and for cell viability. Depletion of RPAP1 triggers cell de-differentiation, facilitates reprogramming toward pluripotency, and impairs differentiation. Mechanistically, we show that RPAP1 is essential for the interaction between RNA polymerase II (RNA Pol II) and Mediator, as well as for the recruitment of important regulators, such as the Mediator-specific RNA Pol II factor Gdown1 and the C-terminal domain (CTD) phosphatase RPAP2. In agreement, depletion of RPAP1 diminishes the loading of total and Ser5-phosphorylated RNA Pol II on many genes, with super-enhancer-driven genes among the most significantly downregulated. We conclude that Mediator/RPAP1/RNA Pol II is an ancient module, conserved from plants to mammals, critical for establishing and maintaining cell identity.
Keywords: transcription, RNA polymerase II, Mediator, cell identity, differentiation, interactome, enhancer
Graphical Abstract
Highlights
-
•
RPAP1 is an RNA Pol II regulator, conserved from plants to mammals
-
•
RPAP1 depletion erases cell identity gene expression, triggering de-differentiation
-
•
Mechanistically, RPAP1 is critical for the Mediator-RNA Pol II interaction
-
•
RPAP1 preferentially contributes to enhancer-driven gene transcription
Lynch et al. report a regulator of RNA Pol II called RPAP1, displaying functional conservation from plants to mammals. RPAP1 is required to establish and maintain cell identity. Mechanistically, RPAP1 is critical for the Mediator-RNA Pol II interaction, thereby preserving normal transcription at enhancer-driven genes.
Introduction
Coordinated regulation of RNA polymerase II (RNA Pol II) transcription is central to cell identity transitions and reflects a common developmental principle across the plant-animal divide (Gaillochet and Lohmann, 2015, Levine, 2011, Meyerowitz, 2002). High-throughput studies have recently revealed a set of conserved Pol-II-associated proteins (RPAP1, 2, 3, and 4) sharing multiple interactions among themselves (Jeronimo et al., 2004, Jeronimo et al., 2007). RPAP2 is an atypical phosphatase that targets Ser5P on the RNA Pol II C-terminal domain (CTD) (Egloff et al., 2012a, Mosley et al., 2009), and RPAP2, RPAP3, and RPAP4 all have essential roles as nuclear transport chaperones for the RNA Pol II complex (Boulon et al., 2010, Forget et al., 2010, Forget et al., 2013). In contrast, the function of RPAP1 remains uncharacterized in mammals.
RPAP1 is a large (153-kDa) multidomain protein with a high degree of conservation across species (Jeronimo et al., 2004, Jeronimo et al., 2007, Sanmartín et al., 2011). Studies in plants, yeasts, and mammals indicate that RPAP1 interacts with the RPB3 (official name POLR2C) and RPB11 (POLR2J) subunits of the RNA Pol II complex (Giaever et al., 2002, Ito et al., 2001, Jeronimo et al., 2004, Jeronimo et al., 2007, Sanmartín et al., 2011). Importantly, the heterodimer RPB3/RPB11 provides a critical interface of RNA Pol II with the Mediator complex (Allen and Taatjes, 2015, Davis et al., 2002). Indeed, a high-throughput screen in yeast indicated that depletion of RPAP1 results in dramatic gene expression changes that were similar to depletion of the RNA Pol II subunit RPB11, although these changes were not characterized further (Jeronimo et al., 2004, Jeronimo et al., 2007).
The multiprotein Mediator complex associates with transcriptional enhancers through protein-protein interactions, being critical for enhancer-promoter looping (Allen and Taatjes, 2015, Jeronimo and Robert, 2017). The largest accumulations of Mediator are in super-enhancers, and super-enhancer target genes are typically the most important for defining cell identity and the most heavily dependent on Mediator to drive their transcription by RNA Pol II (Allen and Taatjes, 2015, Hnisz et al., 2013, Kagey et al., 2010, Whyte et al., 2013).
RPAP1 was recently identified in plants as a critical factor for differentiation by promoting developmental gene expression (Muñoz et al., 2017, Sanmartín et al., 2011). Specifically, in Arabidopsis, RPAP1 was necessary and rate limiting to initiate stem cell differentiation (Sanmartín et al., 2011, Sanmartín et al., 2012). Based on this, we hypothesized that mammalian RPAP1 may also coordinate gene expression and cell identity at a global level. Here, we characterize the mammalian homolog of RPAP1 to investigate putative roles in mammalian transcription and differentiation and reveal a mechanism involving direct RNA Pol II regulation through interaction with Mediator.
Results
Mammalian RPAP1 Expression
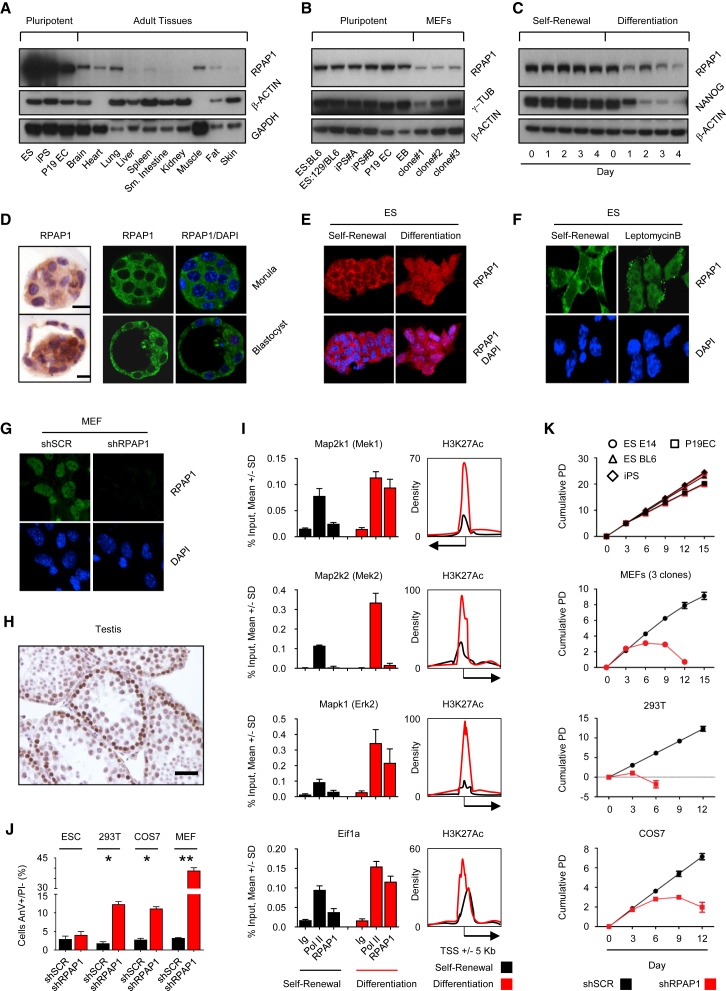
The plant homolog of RPAP1 is highly expressed in stem cells compared to differentiated cells (Sanmartín et al., 2011). Based on this, we began by examining RPAP1 expression in pluripotent and differentiated mouse cells. Compared to adult tissues or mouse embryonic fibroblasts (MEFs), RPAP1 protein levels were high in embryonic stem cells (ESCs), induced pluripotent stem cells (iPSCs), embryo carcinoma (P19EC) cells, and embryoid bodies (EBs) (Figures 1A and 1B). Moreover, RPAP1 expression levels decreased during in vitro differentiation of ESCs by leukemia inhibitory factor (LIF) removal and retinoic acid addition (Figures 1C and S1A). In the case of plants, RPAP1 in stem cells is cytoplasmic and only enters into the nucleus upon differentiation, suggesting that RPAP1 functions as a differentiation switch (Sanmartín et al., 2011). Interestingly, we observed a similar behavior in mouse cells. In particular, RPAP1 was mostly cytoplasmic in the morula and blastocyst (Figure 1D), as well as in ESCs undergoing self-renewal (Figures 1E and 1F). However, RPAP1 became partly nuclear upon ESC differentiation (Figure 1E) and completely nuclear in differentiated cells and tissues (Figures 1G, 1H, and S1B). Indeed, around gene promoters that become activated soon after launching differentiation, we could detect enrichment of RPAP1 coincident with an increase in H3K27Ac (Figure 1I). Moreover, treatment of ESCs with the nuclear export inhibitor leptomycin B produced rapid nuclear accumulation of RPAP1 (Figure 1F), which, similar to plants, is consistent with active nuclear export of RPAP1 during stem cell self-renewal. Therefore, mammalian RPAP1 shares similar expression and subcellular localization dynamics as observed in plants during the switch between self-renewal and differentiation.
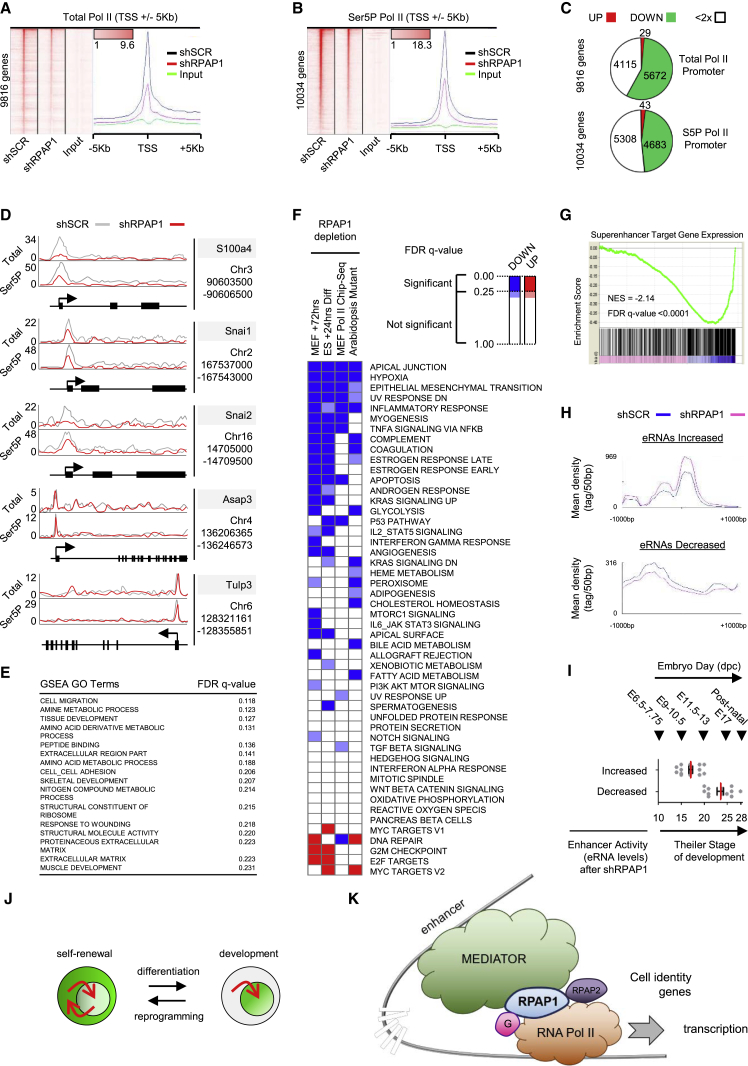
Figure 1.
RPAP1 Expression, Localization, and Requirement for Cell Viability
(A and B) Western blot of RPAP1 expression in a range of pluripotent cell types versus adult tissues (A) or MEFs (B).
(C) Western blot of RPAP1 expression and the ESC marker NANOG during a time course of ESC differentiation by LIF removal and retinoic acid addition.
(D) Immunohistochemical and immunofluorescence staining for RPAP1 in mouse E3.0 morula (upper panel) or E4.0 blastocyst (lower panel). The scale bars represent 20 μm.
(E–G) Immunofluorescence for RPAP1.
(E) ESCs undergoing self-renewal versus 24 hr differentiation by LIF removal.
(F) ESCs exposed to leptomycin B for 3 hr.
(G) MEFs at day 3 after lentiviral transduction with non-targeting control (shSCR) or with RPAP1 targeting (shRPAP1) shRNAs.
(H) Immunohistochemical staining for RPAP1 in mouse adult testis. The scale bar represents 30 μm.
(I) Left panels show ChIP-qPCR for RNA Pol II or RPAP1 enrichment at promoters of the indicated genes. Analysis was performed on ESCs maintained in self-renewal conditions or after 24 hr of differentiation by LIF withdrawal and addition of retinoic acid (diff). See resources table in Supplemental Experimental Procedures for ChIP-qPCR primers. Panels on the right show the enhancer histone mark H3K27Ac enrichment surrounding each promoter (Creyghton et al., 2010), where the enhancer is present in ESCs undergoing self-renewal and becomes activated after differentiation. Mean ± SD; n = 3 technical replicates.
(J) Quantification of apoptosis by annexin V/propidium iodide co-staining and fluorescence-activated cell sorting (FACS) of the indicated cell lines at day 6 after lentiviral transduction with non-targeting control (shSCR) or with RPAP1 targeting (shRPAP1) shRNAs. Mean ± SEM; n = 3 replicates; ∗p < 0.05; ∗∗p < 0.01.
(K) Proliferation curves (shown by cumulative population doubling) treated by control (shSCR) or lentiviral shRNA against RPAP1 in the indicated cell lines. Mean ± SD; n = 3 biological replicates.
See also Figure S1.
RPAP1 Is Essential for Cell Viability
To assess the relevance of RPAP1 in cells, we first identified short hairpin RNAs (shRNAs) that efficiently downregulated RPAP1 both in mouse and human cells (Figures 1G and S1C; see also below Figures 4 and S4). RPAP1 knockdown in non-pluripotent cells, such as human 293T, monkey COS7, various human cancer cell lines, murine MEFs, and immortalized primary hepatocytes, severely attenuated proliferation, induced senescence, and triggered apoptosis, typically with a delay of 2–6 days (Figures 1J, 1K, and S1D–S1G). These observations were recapitulated using a total of three different shRNAs against murine Rpap1 mRNA (Figure S1F). In contrast to the above cell types, knockdown of RPAP1 expression had no effect on ESC viability during self-renewal (Figures 1J, 1K, S1C, S1D, and S1H). Considering the high levels of RPAP1 in ESCs, we wondered if shRNA-mediated depletion was not sufficient to reveal an essential role of RPAP1 on ESC viability. Indeed, we were unable to obtain viable ESC clones with complete Rpap1 elimination using CRISPR technology. It is important to note that we successfully targeted the mouse and human RPAP1-encoding gene using multiple independent CRISPR delivery systems (transient, constitutive, or inducible), guide RNAs, and several wild-type mouse ESC lines or a haploid human cancer cell line (HAP1). In particular, we obtained many ESC clones where RPAP1 suffered small deletions but never a complete loss. Also, when using an ESC line with a LacZ reporter knocked in within intron 8 of the Rpap1 gene, we were able to efficiently eliminate LacZ expression using guide RNAs against the first 7 exons of Rpap1; however, we never obtained clones with elimination of the remaining wild-type Rpap1 allele (Figures S1I–S1L; see Experimental Procedures). Taken together, the data suggest that RPAP1 performs an essential function in all the cell types tested, including ESCs.
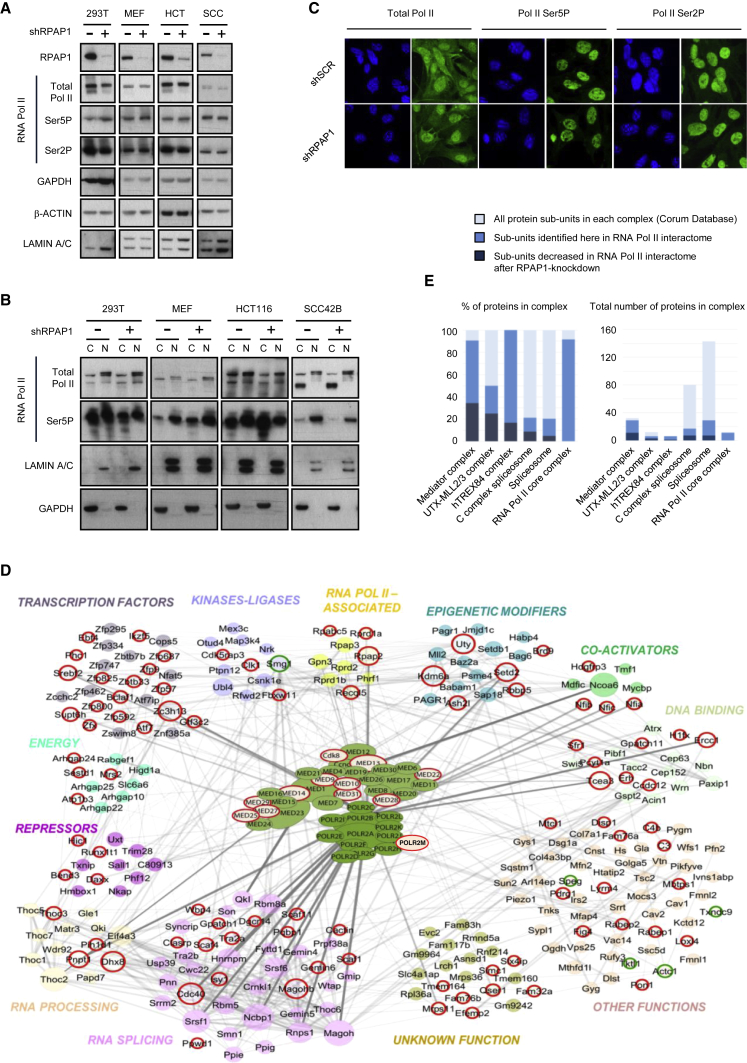
Figure 4.
RPAP1 Regulates the RNA Pol II Interactome
(A) Western blots of RNA Pol II total (RPB1), Ser5P, or Ser2P expression in whole-cell lysates from a range of cell lines at day 3 after RPAP1 depletion. GAPDH, β-ACTIN, and LAMIN A/C were used as internal controls.
(B) Western blots of RNA Pol II total (RPB1), Ser5P, or Ser2P expression in nuclear/cytoplasmic fractions from a range of cell lines at day 3 after RPAP1 depletion. GAPDH and LAMIN A/C were used as indicators of fraction separation. C, cytoplasmic fraction; N, nuclear fraction.
(C) Immunofluorescence of RNA Pol II total (RPB1), Ser5P, or Ser2P in MEFs at day 3 after lentiviral transduction with non-targeting control (shSCR) or with RPAP1 targeting (shRPAP1) shRNAs. Nuclei were stained with DAPI.
(D) Schematic of the 294 specific interactors of POLR2A/RPB1 detected in primary MEFs in this study by RNA Pol II immunoprecipitation and mass spectrometry analysis (see also Table S4). Interactors were displayed as a network using Cytoscape and grouped manually by their known physical interactions and general primary function, wherein the thickness and intensity of the connecting edges indicate the strength of their known interactions in the STRING database. Following RPAP1 depletion, the RNA Pol II interactors reduced (circled in red) and RNA Pol II interactors gained (circled in green) are indicated. The Mediator complex is depicted centrally and in full color based on the data in Figure 4E, below.
(E) RNA Pol II interactors that were decreased following RPAP1 depletion were assigned to all 3,000 known protein complexes in the Corum database (see Supplemental Experimental Procedures). On left, complexes are ranked according to the highest percentage of proteins whose interactions were decreased upon RPAP1 depletion from the cells. On right, the total number of subunits per complex is indicated, together with the number of subunits detected in this study and the number of subunits decreased following RPAP1 depletion.
RPAP1 Depletion Impairs ESC Differentiation
Whereas strong shRNA depletion of RPAP1 did not affect pluripotent cells under self-renewal conditions (see above), we wondered if we could affect the activation of differentiation programs. For this, we first assessed differentiation by LIF removal for 24 or 72 hr (Savatier et al., 1996). We observed that RPAP1-depleted ESCs presented a delayed differentiation based on the expression of pluripotency markers and morphological changes, followed by an increase in apoptosis (Figures S2A and S2B). Differentiation of ESCs to EBs by hanging-drop culture constitutes a longer term and more complex in vitro differentiation assay. RPAP1 depletion in ESCs followed by EB differentiation resulted in severely reduced efficiency of cardiac center development (formation of beating cell clusters) in EBs (Figure 2A). In agreement, analyses of RNA expression also revealed a delay in the loss of pluripotency markers and delayed induction of cardiac muscle differentiation markers associated with RPAP1-depleted EBs (Figures 2B and S2C), suggesting that a decrease in RPAP1 expression is incompatible with development. Impaired cardiac center formation by RPAP1-depleted ESCs may reflect their reduced capacity to differentiate and/or the accumulation of dying or dysfunctional cells. Consistent with a developmental defect, Rpap1(+/−) ESCs displayed weak contribution to chimeric offspring (10 from 254 micro-injected embryos; Figure S2D). Furthermore, when chimeric mice were crossed to look for germline transmission, we did not obtain mice that were Rpap1(+/−) or Rpap1(−/−) (0 out of 156 pups born; Figure S2D).
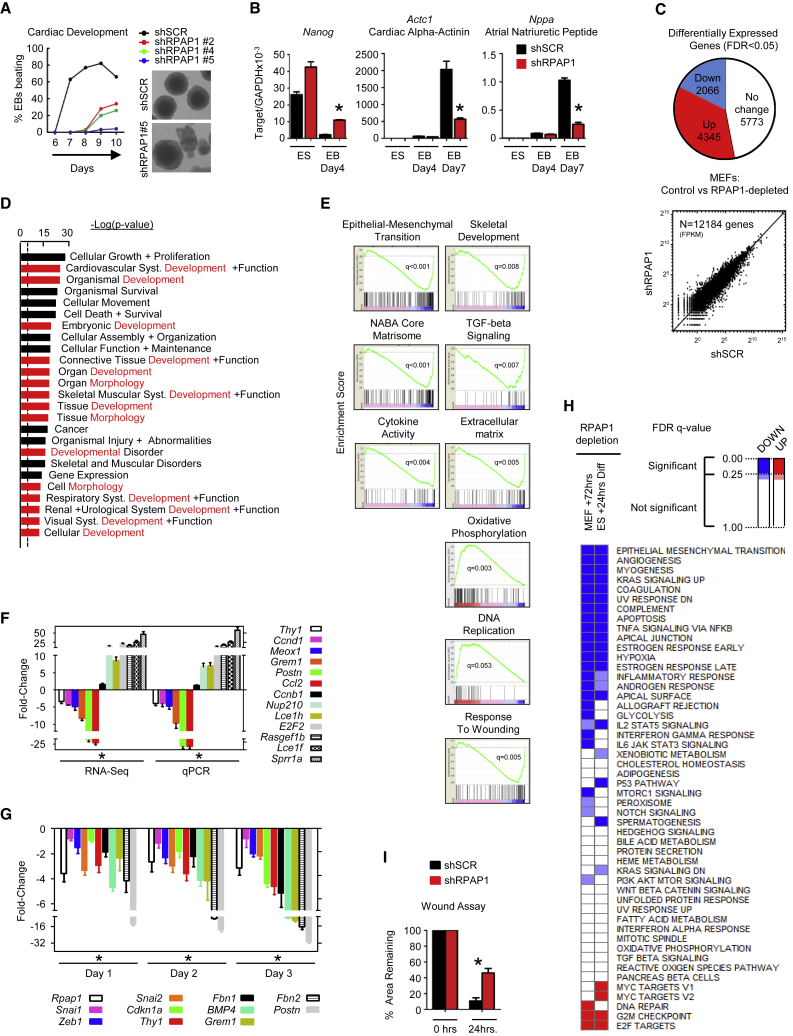
Figure 2.
RPAP1 Is Required for the Establishment and Maintenance of Cell Identity
(A) Effect of RPAP1 depletion on embryoid body (EB) cardiac center development. EBs were scored daily by microscopy for the appearance of clusters of actively beating cells indicative of cardiac muscle development. The graph shows the kinetics over several days. Representative pictures of EBs are shown.
(B) qPCR analyses of pluripotency or cardiac development markers at the indicated time points from the EB differentiation assay in (A). Mean ± SD; n = 3 replicates; ∗p < 0.05.
(C) Overview of RNA-seq transcriptome analyses summarizing differential gene expression (FDR q < 0.05) in MEFs at day 3 after RPAP1 depletion. (Upper panel) Proportional representation pie chart of significantly differentially expressed genes is shown. (Lower panel) Dot plot of FPKM values for all genes shows that many genes of high and low expression level remain unchanged.
(D) Ingenuity pathway analysis showing the top 25 most significantly enriched GO terms among those genes that were significantly downregulated at day 3 after RPAP1 depletion in MEFs (FDR q < 0.01). Terms highlighted in red contain “development” or “morphogenesis.” Dotted line indicates the basal threshold of significance.
(E) Examples of the most significantly up- or downregulated gene sets identified by GSEA analysis in RNA-seq data at day 3 after RPAP1 depletion in MEFs (FDR q < 0.01). See also Figures S3J and S3K and Table S2.
(F) qPCR validation of RNA-seq data. Mesenchymal, fibroblast, and epithelial marker mRNA expression levels were assessed by RNA-seq (left) or qPCR (right) at day 3 after RPAP1 knockdown in MEFs. Data indicate fold change relative to control shSCR. Mean ± SD; n = 3 independent MEF lines; ∗p < 0.05.
(G) qPCR measurement of mesenchymal and fibroblast marker mRNA levels during days 1–3 lentiviral transduction of MEFs with non-targeting control (shSCR) or with RPAP1 targeting (shRPAP1) shRNAs. Data indicate fold change relative to control. Mean ± SD; n = 3 independent MEF lines; ∗p < 0.05. See also Figure S2L.
(H) Heatmap summarizing the most significantly up- or downregulated hallmark gene sets identified by GSEA analysis among all gene expression at day 3 after RPAP1 depletion in MEFs (FDR q < 0.01; left column; see Tables S2 and S3) or in ESCs 24 hr after triggering differentiation (FDR q < 0.05; right column; see Tables S1 and S2). Hallmark gene sets with FDR q < 0.25 are significant. Also highlighted in the heatmap are borderline gene sets (where FDR q = 0.35–0.25).
(I) Wound assay scratch test recovery. Graph shows the percent damaged area remaining at +24 hr. Mean ± SD; n = 3 independent MEF lines with 12 replicates each; ∗p < 0.05.
To characterize the influence of RPAP1 on early events during the pluripotency-to-differentiation transition, we performed RNA sequencing (RNA-seq) analyses in ESCs, both control and RPAP1 depleted, after 24 hr of differentiation (LIF removal). Of 12,827 transcripts detected, 899 (7.1%) were significantly differentially expressed in RPAP1-depleted cells (Figure S2E; Table S1). Global investigation via gene set enrichment analysis (GSEA) and supervised network analyses indicated that, following RPAP1 depletion, differentiating ESCs maintained proliferation pathways (Myc and E2F-regulated gene sets were significantly higher) and had an attenuated induction of mesenchymal identity (epithelial-mesenchymal transition [EMT]-related gene sets were lower) compared to the controls, including key mesenchymal genes, such as Ctgf, Mest, and Col4a2 (Figures S2F–S2I; Tables S1 and S2). This is consistent with the delayed loss of pluripotency markers and morphological changes observed upon differentiation of RPAP1-depleted ESCs (Figures 2A, 2B, and S2C). Thus, RPAP1 depletion delayed ESC differentiation, suggesting that high levels of RPAP1 endow ESCs with the ability to rapidly differentiate, whereas reduced levels of RPAP1 dramatically slow differentiation.
RPAP1 Depletion Induces Loss of Differentiated Cell Identity
Because RPAP1 depletion impaired ESC differentiation, we investigated the role of RPAP1 in differentiated cells. Following RPAP1 depletion in MEFs, cells proliferated and still appeared morphologically normal during days 1–3, prior to the defects that subsequently emerged at days 4–6 (see above Figures 1I, 1J, and S1D). Thus, RNA-seq was performed at day 3 in control or RPAP1-depleted MEFs to assess the transcriptome while avoiding death-related secondary effects. Nevertheless, transcriptomic alterations were dramatic, with >52% of the 12,249 genes detected displaying significantly altered expression (false discovery rate [FDR] q < 0.05; Figure 2C; Table S3). Using multiple approaches to assess gene expression, including GSEA, gene ontology, and supervised network analysis, we observed that RPAP1 triggered a rapid and pronounced loss of multiple developmental processes and robust erasure of fibroblast identity within 3 days (Figures 2F–2H, S2J, and S2K; Tables S2 and S3), a sequence that was initiated after 24 hr, as confirmed by qRT-PCR for multiple mesenchymal/fibroblastic identity markers (Figures 2G and S2L). Notably, there was a remarkable parallel between the gene sets that were downregulated in MEFs by RPAP1 loss and the gene sets that failed to be upregulated in differentiating RPAP1-depleted ESCs (Figure 2H). Moreover, there was significant overlap in the mRNAs differentially expressed due to RPAP1 depletion in the two contexts (hypergeometric overlap p < 10−6). Lastly, a defining feature of mesenchymal cell identity is a high capacity for cell migration. Consistent with the above gene expression profile, RPAP1 depletion followed by a wound healing scratch assay revealed an attenuation of MEF migration capacity (Figure 2I). In summary, after RPAP1 depletion, MEFs display rapid de-differentiation via loss of mesenchymal-fibroblastic identity.
RPAP1 Depletion Favors Reprogramming
Because RPAP1 is important for maintaining the mesenchymal cell identity of MEFs, we hypothesized that RPAP1 depletion may recapitulate an early stage of reprogramming to iPSC. Previous investigators have found that, during reprogramming, there is an initial de-differentiation wave followed by a transient intermediate state, which is resolved by a second wave of transcriptional changes, leading to pluripotency (Polo et al., 2012). Interestingly, the gene expression profile induced by RPAP1 depletion was significantly similar to the intermediate state of reprogramming (Figures 3A, S3A, and S3B). This was supported by validation with markers of the intermediate state (Polo et al., 2012), including downregulation of Meox1 and Meox2 and upregulation of Nup210 (Figure 3B). This suggested that RPAP1 knockdown phenocopies the de-differentiation and loss of mesenchymal identity observed in the first wave of transcriptional changes during iPSC reprogramming. Consistent with this, prior knockdown of RPAP1 for 2 days in MEFs led to significantly enhanced iPSC reprogramming with the four Yamanaka factors (Oct4, Sox2, Klf4, and Myc, abbreviated as OSKM; Figures 3C and 3D). Importantly, therefore, the lethality of RPAP1 depletion in MEFs was rescued by reprogramming to pluripotent iPSCs, suggesting that RPAP1-depleted MEFs at days 2 or 3 may represent de-differentiated cells without a defined identity, which subsequently collapse unless rescued by reprogramming into pluripotency.
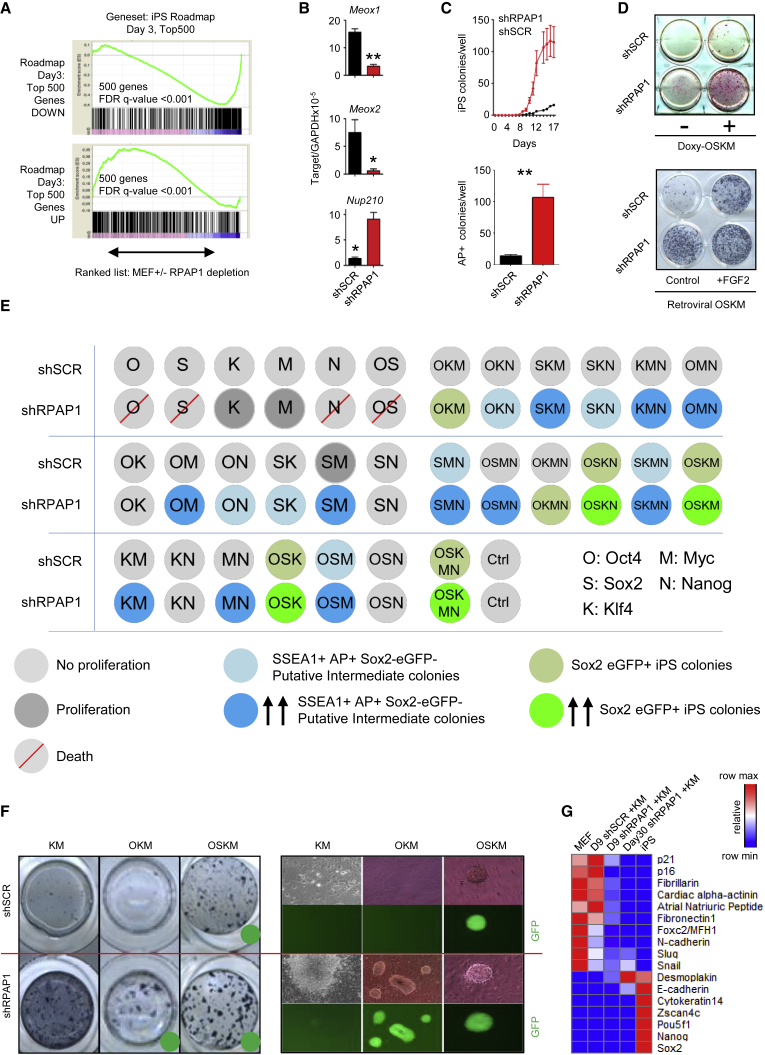
Figure 3.
Rpap1 Knockdown Favors De-differentiation and Reprogramming
(A) Comparison of gene expression at day 3 after RPAP1 depletion in MEFs versus a published iPSC roadmap gene expression profile (Polo et al., 2012). Panels show GSEA comparison of the published top 500 genes up- or downregulated at day 3 of the iPSC roadmap versus a ranked list of the gene expression profile in the current study at day 3 after RPAP1 depletion in MEFs (x axis). See Supplemental Experimental Procedures for assessment of the iPSC roadmap data from parental MEFs verses Thy1-negative cells at day 3 of iPSC reprogramming. FDR q < 0.25 is significant. See also Figures S3A and S3B.
(B) qPCR measurement of selected genes at day 3 after RPAP1 depletion in MEFs. Downregulation of Meox1 and Meox2 and upregulation of Nup210 were reported to correlate with cell gene expression during the intermediate stages of iPSC reprogramming (Hansson et al., 2012, Polo et al., 2012). Mean ± SD; n = 3 independent MEF lines; ∗p < 0.05; ∗∗p < 0.01.
(C and D) MEF to iPSC reprogramming after RPAP1 depletion. Expression of the OSKM reprogramming factors was initiated at day 2 after lentiviral transduction of MEFs with non-targeting control (shSCR) or with RPAP1 targeting (shRPAP1) shRNAs. In (C), top panel, kinetics of iPSC colony appearance during doxycycline-induced reprogramming of i4F MEFs that express the four Yamanaka factors is shown (see Experimental Procedures). A profile representative of three independent i4F MEF lines is shown (mean ± SD; 3 technical replicates). In (C), bottom panel, quantification of iPSC colony yield at day 14 of doxycycline-induced 4F reprogramming is shown. Mean ± SD; n = 3 independent MEF lines; ∗∗p < 0.01. In (D), examples of alkaline phosphatase staining to indicate iPSC colonies formed at day 12 of i4F-MEF doxycycline-induced-OSKM iPSC reprogramming (top panel) or retroviral delivery of the OSKM factors (bottom panel) are shown. FGF2 was added to stimulate reprogramming efficiency.
(E) Summary of outcomes from 32 combinations of OSKMN Yamanaka transcription factors. Sox2-eGFP MEFs at day 2 after control or RPAP1 depletion received the indicated factors by retroviral delivery, followed by culture in standard iPSC reprogramming media. Progress of iPSC reprogramming was assessed by cell proliferation rate, morphology changes, colony formation, staining for alkaline phosphatase, SSEA1 expression, and Sox2-eGFP levels. Sox2-eGFP-positive cells forming typical iPSC colonies were scored as successfully reprogrammed iPSCs. Rapidly proliferating cells that initiated colony formation and that were positive for alkaline phosphatase and SSEA1 but negative for Sox2-eGFP were scored as putative intermediate stages of reprogramming.
(F) Examples of alkaline phosphatase staining to indicate formation rates of iPSC colonies and putative intermediate cell types at day 14 of MEF reprogramming with the indicated combinations of Yamanaka factors ± RPAP1 depletion. Green dot indicates those combinations that produced Sox2-eGFP-positive full reprogrammed iPSC colonies.
(G) qPCR measurement of mesenchymal, epithelial, and pluripotency marker mRNA expression levels. Data were converted to heatmap format to highlight the intermediate nature of marker expression displayed by the cells that were generated by shRPAP1+Klf4/Myc.
See also Figure S3.
To explore the minimal complement of the Yamanaka factors sufficient to rescue lethality of RPAP1 depletion and/or confer pluripotency, we tested all possible combinations of Oct4, Sox2, Klf4, Myc, and Nanog (OSKMN) (32 combinations; Figure 3E), in combination with a panel of media supplements reported to enhance reprogramming (15 media cocktails; Figure S3C). Four interesting features emerged: (1) RPAP1 knockdown plus several of the transcription factor combinations, including Klf4 or Myc, were sufficient to rescue cell survival; in particular, shRPAP1 with Klf4/Myc together converted the majority of MEFs to putative intermediates of reprogramming, that is, rapidly proliferating colony-forming cells that were also positive for markers of the early stages of the reprogramming process, including alkaline phosphatase and SSEA1 cell surface expression, but were Sox2-eGFP negative (Figures 3E–3G and S3D); (2) RPAP1 depletion increased the efficiency of all successful reprogramming combinations (Figure 3E); (3) RPAP1 depletion can replace Sox2 in combination with OKM or OKMN (Figures 3E–3G); and (4) pharmacological inhibition of transforming growth factor β (TGFβ) signaling, which is known to replace Sox2 (Li et al., 2010), cooperated with RPAP1 depletion in the OKM or OKMN reprogramming (Figure S3E). Taken together, phenotypic and expression data suggest that RPAP1 depletion induces a de-differentiated state that can be stabilized by Klf4/Myc and can be converted into full pluripotency if Oct4 is included.
RPAP1 Regulates the RNA Pol II Interactome
To understand the mechanism by which RPAP1 is required for somatic cell proliferation, we first wondered whether RPAP1 could affect the stability and localization of RNA Pol II. The RNA Pol II complex is formed by 12 subunits (RPB1–12), where RPB1 (official name POL2R2A) is the largest and catalytic subunit, and 4 additional associated proteins (RPAP1–4; Wild and Cramer, 2012). The full complex is assembled in the cytoplasm, and remarkably, individual depletion of the subunits RPB2–12 or RPAP2–4 prevents nuclear import of the catalytic subunit RPB1 (Boulon et al., 2010, Forget et al., 2010, Forget et al., 2013, Wild and Cramer, 2012), whereas the effect of RPAP1 depletion has not been reported. Therefore, we assessed the effect of RPAP1 knockdown on RNA Pol II expression and localization in five different cell lines. RPAP1 depletion did not affect RNA Pol II expression levels (using as surrogate the catalytic subunit RPB1), or the phosphorylation levels on serine 5 (Ser5P) or serine 2 (Ser2P) of RPB1 (Figures 4A, S4A, and S4B). In contrast to all the other subunits of the RNA Pol II complex, depletion of RPAP1 did not affect RNA Pol II (RPB1) nuclear localization (Figures 4B and S4C). This was confirmed by immunofluorescence (Figures 4C and S4D). These observations rule out RNA Pol II destabilization and/or mislocalization as an explanation for the essential role of RPAP1 in the survival of differentiated cells.
We further investigated the mechanism by which RPAP1 might regulate RNA Pol II. Because RPAP1 is a large protein directly associated with RNA Pol II (see Introduction), we compared the RNA Pol II protein interactome of control versus Rpap1 knockdown MEFs. Immunoprecipitation of the largest and core RNA Pol II subunit (RPB1), followed by mass spectrometry revealed 294 specific interactor proteins (Figure 4D; see Experimental Procedures), with a clear enrichment for transcription-related factors, including, for example, all 12 subunits of the RNA Pol II complex and almost all (28 out of 30) of Mediator subunits, illustrating the depth and specificity of this interactome analysis (Table S4). Importantly, Rpap1 knockdown did not affect the integrity of the RNA Pol II complex itself but it resulted in a significant reduction of 104 RNA Pol II interactors (red circles in Figure 4D; see also Table S4), whereas 5 new interactors were found (green circles). Among the RNA Pol II interactors significantly affected by RPAP1 depletion, the Mediator complex was ranked the highest in terms of proportion of affected subunits (Figure 4E; Table S4), suggesting an important alteration in the functions controlled by this complex. Furthermore, we observed that depletion of RPAP1 led to the loss of Gdown1 (official name POLR2M) from RNA Pol II complexes. Gdown1 is a recently discovered protein that tightly binds approximately half of RNA Pol II in cells, forming the so-called RNA Pol II(G) complex (Hu et al., 2006, Jishage et al., 2012). Importantly, RNA Pol II(G) complexes are known to contain RPAP1 (Jishage et al., 2012). Finally, it is relevant to note that Gdown1 is recruited by Mediator and associates with RNA Pol II on Mediator-regulated target genes (Cheng et al., 2012, Hu et al., 2006, Jishage et al., 2012, Li and Price, 2012). Altogether, we conclude that RPAP1 is a critical ingredient for Mediator-competent RNA Pol II.
RPAP1 Is Required for Transcription of Identity and Developmental Genes
Because Mediator has a critical role recruiting RNA Pol II to genes controlling cell identity and development (Allen and Taatjes, 2015, D’Alessio et al., 2009, Hnisz et al., 2013, Whyte et al., 2013), we next investigated the global effect of RPAP1 depletion on RNA Pol II binding to chromatin. For this, we performed chromatin immunoprecipitation sequencing (ChIP-seq) for total RNA Pol II and for Ser5P RNA Pol II, the latter reflecting active RNA Pol II (Egloff et al., 2012b, Hsin and Manley, 2012). Knockdown of RPAP1 in MEFs reduced the abundance of both total and Ser5P RNA Pol II at about 50% of detected genes, whereas very few genes (<0.5%) displayed an increase (Figures 5A–5D; Table S5). Interestingly, GSEA and leading edge analyses revealed that mesenchymal regulators and related developmental processes were among the gene sets (GSEA) and genes (leading edge) with the most significant loss of RNA Pol II (Figures 5E, 5F, and S5A; Table S2).
Figure 5.
RPAP1 Is Required for RNA Pol II Transcription in MEFs, Particularly on Developmental and Mesenchymal Genes
(A and B) ChIP-seq enrichment data after lentiviral transduction of MEFs with non-targeting control (shSCR) or with RPAP1 targeting (shRPAP1) shRNAs. Data are plotted as heatmaps of RNA Pol II total (A) or RNA Pol II Ser5P (B) occupancy around the transcriptional start site (TSS) region ± 5 Kb. Rows are sorted by decreasing RNA Pol II occupancy at the promoter (−100 to +300 bp) in the shSCR control. Color-scaled intensities are in units of reads per million mapped reads (rpm) (see Supplemental Experimental Procedures—ChIP-seq analysis).
(C) Proportional representation of ChIP data, classifying genes according to the changes in abundance of RNA Pol II total (upper panel) or Ser5P (lower panel) at the promoter (−100 to +300 bp) following RPAP1 depletion in MEFs for 3 days.
(D) Schematics of RNA Pol II total and Ser5P abundance on selected genes, showing examples of RNA Pol II depletion (S100a4, Snai1, and Snai2) or minimal effects (Asap3 and Tulp3).
(E) Table summarizing the most significantly up- or downregulated GO term gene sets identified by GSEA among the genes with >2×-fold decrease in RNA Pol II following RPAP1 depletion in MEFs (see also Tables S2 and S5). Gene sets with FDR q value < 0.25 were considered significant.
(F) Summary heatmap displaying the overlay of significant GSEA hallmark gene sets across 4 experiments (columns 1 and 2: see also Figure 2H). Column 1: GSEA on the ranked list of differential mRNA expression in MEFs at day 3 ± RPAP1 depletion is shown. Column 2: GSEA on the ranked list of differential mRNA expression in ESCs at +24 hr after inducing differentiation ± RPAP1 depletion is shown. Column 3: GSEA on the ranked list of differential RNA Pol II abundance at all promoters in MEFs at day 3 ± RPAP1 depletion is shown. Column 4: GSEA on the ranked list of differential gene expression in Arabidopsis thaliana plant tissues ± RPAP1 mutation is shown (Sanmartín et al., 2011), following conversion to the nearest mouse homolog based on protein sequence conservation (see Supplemental Experimental Procedures, conversion of plant to mouse homologs, and Table S2).
(G) GSEA to assess mRNA expression levels of MEF super-enhancer target genes (n = 661; defined by GREAT analysis as described; see Supplemental Experimental Procedures) within the transcriptome of primary MEFs at day 3 after Rpap1 knockdown. Compare with housekeeper gene expression in Figure S5G.
(H) Plots show the average eRNA levels within two groups of MEF super-enhancers regions: those which were increased (n = 63 enhancers; top panel) or decreased (n = 64 enhancers; bottom panel) in MEFs at day 3 after RPAP1 knockdown.
(I) GREAT analysis was used to identify target genes for each of the two super-enhancer groups identified in (H) (see Supplemental Experimental Procedures). These two sets of target genes (with increased eRNAs or with decreased eRNAs, respectively) were compared with gene groups associated to particular developmental Theiler stages (these gene groups are detailed in Figures S5H and S5I). The Theiler stage of those gene groups with high similarity to our target genes of interest is represented (gray dots) together with the average (red) and SEM (black).
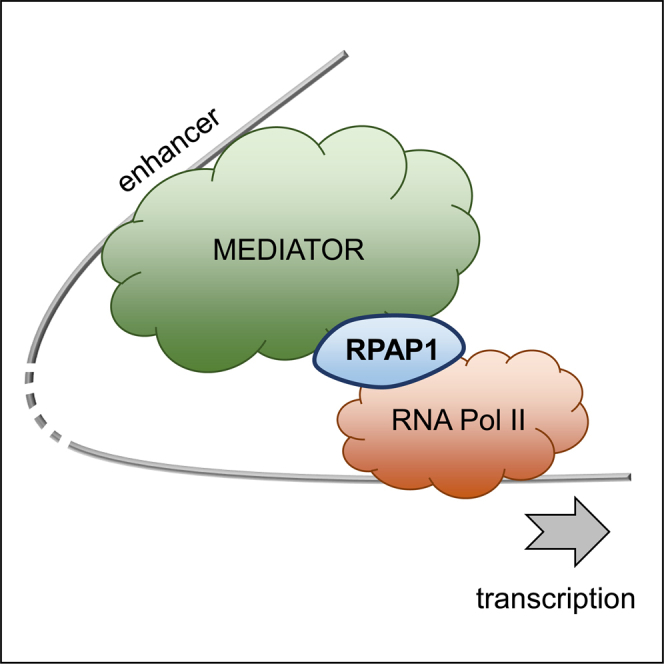
(J and K) Model for RPAP1 function in the mechanism for triggering development.
(J) In self-renewing ESCs, RPAP1 (in green) is abundantly expressed and predominantly cytoplasmic. Upon differentiation, nuclear accumulation of RPAP1 permits increased transcriptional regulation and activation of developmental programs. Depletion of RPAP1 in differentiated cells results in loss of cell identity and enhanced susceptibility for reprogramming toward pluripotency.
(K) Taken together, our data suggest a model where RPAP1 exists in a complex with RNA Pol II and plays an essential role in the Mediator-RNA Pol II regulatory axis. Thus, loss of RPAP1 triggers a decrease in the association between Mediator and RNA Pol II (including the key regulators Gdown1 [G] and the Ser5P phosphatase RPAP2), preferentially affecting the ability of enhancers to activate Mediator target genes, which are known to include key markers and regulators of cell identity. In somatic cells, such as MEFs, this leads to de-differentiation, as expression of fibroblastic, mesenchymal, and developmental markers is erased.
RNA Pol II regulation at individual genes is often more complex than overall abundance, particularly in relation to two critical steps, namely RNA Pol II loading at promoters and transitioning into productive elongation (Chen et al., 2015, Liu et al., 2015, Rahl et al., 2010). Hence, we compared RNA Pol II abundance at promoter versus gene body by calculating the promoter-to-body ratio (also referred to as the “pausing index”) as described (Chen et al., 2015, Rahl et al., 2010; Figure S5B). Overall, in those genes with reliable RNA Pol II signal, we observed that 84% of the genes in MEFs had a promoter/body ratio >2.0 (Figures S5C and S5D; Table S5), which is similar to published data in mouse ESCs (91%; Rahl et al., 2010) or human cancer cells (90%; Chen et al., 2015). Following RPAP1 depletion, the promoter/body ratio was altered in many genes; in some cases, it was increased and in others it was decreased (Figure S5E). Interestingly, whereas no significant gene sets were enriched among those genes with decreased promoter/body ratios, gene sets corresponding to regulators of cell identity and development were significantly present among the genes with increased ratios (Figure S5E; Table S5).
To investigate whether RPAP1 depletion altered RNA Pol II activity and abundance through altered Ser5P levels, we calculated the Ser5P/total RNA Pol II ratio (also known as “Ser5P density”) for all genes at the promoters and gene bodies. We detected widespread changes in Ser5P density (Figure S5F), a phenomenon that has been observed before when RNA Pol II elongation is blocked (Allepuz-Fuster et al., 2014). Notably, these changes were more pronounced at promoter regions than at gene bodies (Figure S5F). Moreover, GSEA analyses revealed that, upon RPAP1 depletion, the only significantly enriched gene sets were associated with increased Ser5P density at promoters, and these included gene sets and gene ontology (GO) terms, such as TNFα signaling via NFκB, cell migration, locomotory behavior, and genes in which RNA levels are also downregulated (Tables S2 and S5), such as Snai2, Tgfb1i1, Tgfb3, Tgfbrap, Lox, Loxl1, Tlr2, Tlr3, Vegfa, Myo6, Smad6, Ccl7, S100a4 (fibroblast-specific protein1), and S100a6 (Figure S5F; Table S5). Taken together, this suggests that RPAP1 depletion affects RNA Pol II transcription, including the levels of Ser5P, and this preferentially perturbs the expression of cell identity and developmental regulators.
In summary, upon Rpap1 knockdown in MEFs, the genes and gene sets linked to the regulation of fibroblastic/mesenchymal identity or closely related developmental processes were the most significantly enriched in four key categories: (1) genes with the most significantly downregulated mRNA expression; (2) genes with the greatest overall depletion of RNA Pol II; (3) genes with selective RNA Pol II depletion from their gene body; and (4) genes with the most enhanced Ser5P density at their promoters.
Conservation of RPAP1 Function from Plants to Mammals
Previously, it was shown that mutations of the RPAP1 homolog in plants inhibited cell differentiation, and microarray analyses showed a specific defect on developmental gene expression (Sanmartín et al., 2011). In order to directly compare the mouse and plant functional overlap, we converted the published plant differential gene expression data to the nearest mammalian protein homolog where possible (see Experimental Procedures and Table S2). Interestingly, conversion of the plant expression data to mouse homologs also revealed significant downregulation of developmental processes (Figure 5F; Table S2). This suggests that loss of RPAP1 function in mice and plants downregulates similar developmental processes, including lineage specifiers and regulators of cell identity, such as hypoxia, cell polarity, extra-cellular matrix, and chemokine signaling.
RPAP1 Preferentially Regulates Mediator-Driven Gene Expression
Mediator physically links enhancers with target genes and then recruits RNA Pol II for their transcriptional activation (Allen and Taatjes, 2015). This process is especially critical to maintain transcription of genes regulated by super-enhancers, which typically encode key markers and regulators of cell identity (Allen and Taatjes, 2015, Hnisz et al., 2013, Whyte et al., 2013). Given our observations above that RPAP1 depletion triggered both a decrease in RNA Pol II interaction with the Mediator complex and selective loss of cell identity gene expression, we next assessed the transcription of super-enhancer-driven genes. We found that following RPAP1 depletion in MEFs, the mRNA levels of genes proximal to super-enhancers were significantly decreased (Figure 5G), whereas highly expressed housekeeper genes were not affected (Figure S5G). Expression levels of enhancer RNAs (eRNAs) are proportional to their enhancer activity (Andersson et al., 2014, Li et al., 2016). In our RNA-seq, we detected eRNA expression in ∼20% of super-enhancers, and we divided those enhancers into two groups, those with increased or decreased eRNA levels (Figure 5H). Interestingly, after RPAP1 depletion, enhancers with decreased eRNA levels (decreased activity) had target genes associated with Theiler stages 20–25 (embryonic day 11.5 [E11.5]–17), whereas enhancers with increased eRNA levels (increased activity) had target genes associated with Theiler stages 14–20 (E8–13; Figures 5I and S5H). Because MEFs arise from E13.5 embryos, the data suggest that enhancers of this embryo stage are decreased in activity, whereas enhancers of earlier embryo stages are activated. This is consistent with the de-differentiation effects that we observed above in MEFs after RPAP1 depletion. Taken together, this suggests that RPAP1 depletion affects RNA Pol II transcription by disruption of the Mediator/RNA Pol II interaction, and this preferentially reduces the expression of super-enhancer-driven cell identity and developmental regulators (Figures 5J and 5K).
Discussion
We have characterized the function of mammalian RPAP1 and observed prominent parallels with its plant homolog in terms of subcellular localization, developmental expression patterns, regulation of RNA polymerase II transcription, and a requirement to establish and maintain differentiated cell identity. Based on this, we propose that this is an ancient mechanism to trigger the transition from pluripotency to differentiation.
RPAP1 Nucleo-cytoplasmic Shuttling
We found that RPAP1 protein is very abundant and largely cytoplasmic in pluripotent cells, which is consistent with the apparent lack of effect of RPAP1 depletion on self-renewing pluripotent cells. However, we cannot exclude the possibility that Rpap1 knockdown has some impact on self-renewal pathways, and we note that we were unable to isolate Rpap1 knockout (KO) ESCs, suggesting that ESCs require a small fraction of RPAP1 either for an essential function or to maintain fast proliferation under self-renewal conditions. Interestingly, we observed rapid nuclear accumulation of RPAP1 by blocking nuclear export, implying a continuous cycle of RPAP1 in/out of pluripotent cell nuclei. In contrast, the onset of differentiation coincided with RPAP1 nuclear accumulation, observed both in vitro and in vivo, and recruitment to promoters together with RNA Pol II. In fact, this developmental switch in nucleo-cytoplasmic shuttling is similar to the behavior of the RPAP1 plant homolog (Muñoz et al., 2017, Sanmartín et al., 2011, Sanmartín et al., 2012). This is also consistent with the existence of multiple conserved nuclear localization signals (NLS)/nuclear export signal (NES) sequences on RPAP1 and an armadillo superfamily repeat region (ARM), a motif associated with nucleo-cytoplasmic shuttling, that is highly conserved in RPAP1 homologs of Saccharomyces, Drosophila, and mammals (Jeronimo et al., 2004). Together, this suggests a conserved model for RPAP1 in the mechanism for triggering development (Figure 5J).
RPAP1 Is Required to Establish and Maintain Cell Identity
During development, new cell identity can arise through a series of reversible epithelial-to-mesenchymal transitions (EMTs) (Thiery et al., 2009). RPAP1 expression was required during ESC differentiation, including toward cardiac muscle development, a path containing several EMT transitions (Thiery et al., 2009). Consistent with this, we failed to obtain homozygous Rpap1-null mice. Moreover, RPAP1 depletion resulted in a striking loss of the mesenchymal identity of MEFs and subsequent cell death. Similarly, all tested cell lines (a total of 8) died several days after RPAP1 depletion or attempted CRISPR knockout. Taken together, these data suggest a role for RPAP1 in establishment and maintenance of cell identity and this explains why its complete elimination is incompatible with cell viability.
RPAP1 Depletion Permits De-differentiation and Reprogramming
RPAP1 depletion in MEFs induced loss of the mesenchymal/fibroblastic identity. Strikingly, however, such de-differentiation complemented the early stages of reprogramming to pluripotent iPSCs, and thus, RPAP1 depletion enhanced the efficiency of recapturing pluripotency. Therefore, reprogramming with OSKM rescued the lethality of RPAP1 depletion, a phenomenon we found could be attributed to the overexpression of Klf4 plus Myc in particular. We hypothesize that Klf4/Myc dual overexpression may revert or compensate the lethal effects of RPAP1 depletion because MYC amplifies active RNA Pol II transcription (Lin et al., 2012, van Riggelen et al., 2010), whereas the ectodermal lineage specifier KLF4 may help to specify a new epithelial identity. In this way, RPAP1 depletion plus Klf4/Myc overexpression may stabilize a highly proliferative reprogramming intermediate.
RPAP1 Acts at the Interface between RNA Pol II and Mediator
RPAP1 is a large (153-kDa) multidomain protein that has been reported to bind a number of interesting RNA Pol II regulators, most notably the RPB3/11 heterodimer, and this is well substantiated in plants, yeasts, and mammals (Giaever et al., 2002, Hazbun et al., 2003, Ito et al., 2001, Jeronimo et al., 2004, Jeronimo et al., 2007, Sanmartín et al., 2011). Indeed, loss of RPAP1 in yeast produces global changes in gene expression that resemble those produced by loss of RPB11 (Jeronimo et al., 2004). The RPB3/RPB11 heterodimer provides the interface between RNA Pol II and the Mediator complex (Davis et al., 2002). Importantly, Mediator plays a critical role in establishing cell identity (Allen and Taatjes, 2015, Hnisz et al., 2013, Jeronimo and Robert, 2017, Whyte et al., 2013), and RPB3 is reported to specify muscle identity (Corbi et al., 2002). Here, we detected a major disruption of the RNA Pol II interactome following RPAP1 depletion, and most notably, out of 3,000 known protein complexes in the Corum database, the complex most heavily affected was the Mediator complex. Therefore, our current findings suggest a model whereby RPAP1 operates at the interface between RNA Pol II and Mediator to direct the transcription of cell identity genes.
RPAP1 Is Required for RNA Pol II Transcription at Cell Identity Genes
Consistent with the pivotal role of RPAP1 in the Mediator/RNA Pol II axis, we observed widespread transcriptional changes in RPAP1-depleted MEFs, with significantly altered gene expression in 52% of all detectable mRNAs and decreased RNA Pol II loading in 50%–60% of all genes. However, we also observed that about 40% of genes displayed minimal changes in RNA Pol II abundance (Figure 5C), and many highly expressed mRNAs remained unaffected (Figures 2C and S5G), arguing against a non-specific defect in RNA Pol II transcription. Furthermore, upon RPAP1 knockdown in MEFs, genes regulating developmental processes and fibroblastic/mesenchymal identity were the most significantly affected according to four criteria: (1) downregulated mRNA expression; (2) greatest overall depletion of RNA Pol II; (3) increased Ser5P RNA Pol II density at promoters; and (4) depletion of RNA Pol II within gene bodies relative to promoters. These features are consistent with RPAP1 deletion affecting RNA Pol II loading on promoters and promoter escape into gene bodies. Remarkably, these aspects mirror Mediator's best known functions (Jeronimo and Robert, 2017).
Our proteomic data provide mechanistic explanations for the relative increase in Ser5P RNA Pol II at promoters and for the relative reduction of RNA Pol II from gene bodies. In particular, RPAP1 has conserved interactions with the Ser5P phosphatase RPAP2 in plants and mammals (Egloff et al., 2012a, Jeronimo et al., 2007, Mosley et al., 2009, Muñoz et al., 2017). We observed that RPAP2 phosphatase was depleted from the Pol II interactome upon knockdown of RPAP1, and this may explain the relative accumulation of Ser5P RNA Pol II at promoters. Meanwhile, Gdown1 (official name POLR2M) is a recently discovered protein, often referred to as “the 13th subunit,” that tightly binds approximately half of the RNA Pol II complexes in cells, forming RNA Pol II(G) (Hu et al., 2006, Jishage et al., 2012). Specifically, Gdown1 is recruited by Mediator and associates with RNA Pol II on Mediator-regulated target genes (Cheng et al., 2012, Hu et al., 2006, Jishage et al., 2012, Li and Price, 2012). It has been reported that RNA Pol II(G) contains RPAP1 (Jishage et al., 2012), and here, we show that depletion of RPAP1 leads to the loss of Gdown1 from RNA Pol II complexes. Therefore, RPAP1 acts as a critical ingredient for Mediator-competent RNA Pol II.
Mediator is most abundant in super-enhancers, and super-enhancer target genes are typically the most important for defining cell identity and the most heavily dependent on Mediator to drive their transcription by RNA Pol II (Allen and Taatjes, 2015, Hnisz et al., 2013, Jeronimo and Robert, 2017, Whyte et al., 2013). In agreement, the gene expression of super-enhancer target genes was preferentially decreased following RPAP1 depletion in MEFs, and this pattern of gene expression correlates closely with the first 3 days of iPSC reprogramming, constituting a de-differentiation effect. Consistent with a de-differentiation effect, we observed that the activity of enhancers, measured by their eRNA levels, shifted from the developmental stage of MEFs toward an earlier developmental stage. This is consistent with recent evidence that, during cell identity transitions, coordinated changes in enhancer activity lead the re-organization of transcriptional networks (Arner et al., 2015, Factor et al., 2014). Taken together, the data point toward a primary role for RPAP1 in maintaining the expression of identity regulators through the Mediator/RNA Pol II axis.
Conclusions
Collectively, our data point toward a developmental requirement for mammalian RPAP1, both in establishing and maintaining cell identity, through direct regulation of RNA polymerase II transcription. Mechanistically, we present evidence suggesting a unified model whereby RPAP1 operates by coordinating the communication between Mediator and RNA Pol II, particularly on super-enhancer-driven genes.
Experimental Procedures
Further details and an outline of resources used in this work can be found in Supplemental Experimental Procedures.
Animal Experimentation
Experiments with mice at the CNIO, Madrid, were performed according to protocols approved by the CNIO-ISCIII Ethics Committee for Research and Animal Welfare (CEIyBA).
Cell Culture, RPAP1 Knockdown, and CRISPR-Cas9 Knockout
Primary MEFs (wild-type; passage 2) were obtained at E13.5 from pure inbred C57BL6 background mice. Immortalized primary mouse hepatocytes HEP cells have been previously described (Lopez-Guadamillas et al., 2016). Mouse P19EC cells, monkey COS7 cells, and the human cell lines 293T, HCT116, SCC42B, and H226 were from ATCC. All the above-mentioned cells were maintained in DMEM medium with 10% fetal bovine serum (FBS) (Gibco) with antibiotics (penicillin/streptomycin 100 U/mL). For ESC culture and iPSC reprogramming conditions, see the Supplemental Experimental Procedures for full details. For shRNA knockdown or overexpression methods with retroviral and lentiviral vectors and for a detailed description of the CRISPR-Cas9 strategies used here, see the Supplemental Experimental Procedures for full details.
RNA Pol II Protein Interactome and Protein Expression Analysis
RNA Pol II immunoprecipitation, interactome analysis, and liquid chromatography (LC)/LC mass spectrometry was performed on day +2 after lentiviral shRNA knockdown of RPAP1 in primary MEFs. See the Supplemental Experimental Procedures for full details. For analysis of protein expression by western blot, immunofluorescence, cytometry, and histopathology, see the Supplemental Experimental Procedures for full details.
RNA Isolation and Gene Expression Analyses by qRT-PCR or RNA-Seq
Total RNA was extracted from cells on column by RNeasy kit with DNA digestion following provider’s recommendations (QIAGEN no. 74104) and retro-transcribed into cDNA following manufacturer's protocol with Superscript Reverse Transcriptase (Life Technologies). Real-time qPCR was performed using Syber Green Power PCR Master Mix (Applied Biosystems) in an ABI PRISM 7700 thermocycler (Applied Biosystems). Primers are listed in the resource table in Supplemental Experimental Procedures. For RNA-seq transcriptomic analyses, see the Supplemental Experimental Procedures for full details.
Chromatin Immunoprecipitation: ChIP-qPCR and ChIP-Seq
ChIP-qPCR was performed with antibodies for total RNA Pol II and RPAP1 and with primers listed in the resource tables in Supplemental Experimental Procedures. ChIP-seq for RNA Pol II was performed as described (Rahl et al., 2010). See the Supplemental Experimental Procedures for full details of ChIP-seq methods, analyses, and definition of promoter and enhancer regions
Quantification and Statistical Analysis
Unless otherwise specified, quantitative data are presented as mean ± SD and significance was assessed by the two-tailed Student’s t test; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.
Acknowledgments
We are grateful to Elisa Varela for assistance with morula and blastocyst fixation. Work in the laboratory of M.S. is funded by the CNIO and the IRB and by grants from the Spanish Ministry of Economy co-funded by the European Regional Development Fund (ERDF) (SAF2013-48256-R), the European Research Council (ERC-2014-AdG/669622), the Regional Government of Madrid co-funded by the European Social Fund (ReCaRe project), the European Union (RISK-IR project), the Botin Foundation and Banco Santander (Santander Universities Global Division), the Ramon Areces Foundation, and the AXA Foundation. S.R. was funded by a contract from the Ramon y Cajal Program(RYC-2011-09242) and by the Spanish Ministry of Economy co-funded by the ERDF (SAF2013-49147-P and SAF2016-80874-P).
Author Contributions
C.J.L. performed most of the experiments, contributed to experimental design and data analysis, and co-wrote the manuscript; R.B., I.C., S.N.-P., S.R., and N.I. contributed to experimental work; C.J.L., A.M.-V., O.G.-C., G.G.-L., and E.A.-L. contributed to bioinformatic analyses; V.E.A. and A.d.S. performed supervised network analyses; S.O. provided reagants, contributed to experimental design, and supervised embryo work; and E.R., O.F.-C., and J.M. provided reagents, discussion, and revisions. M.S. designed and supervised the study, secured funding, analyzed the data, and co-wrote the manuscript. All authors discussed the results and commented on the manuscript.
Declaration of Interests
The authors declare no competing interests.
Published: January 9, 2018
Footnotes
Supplemental Information includes Supplemental Experimental Procedures, five figures, and five tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2017.12.062.
Data and Software Availability
The accession number for the three datasets (two RNA-seq and one ChIP-seq experiments) reported in this paper is GEO: GSE78795. The accession number for the mass spectrometry proteomics data reported in this paper is ProteomeXchange: PXD007114.
Supplemental Information
Sheets 1-3: List of all normalized mRNA expression data generated by RNA-seq in mouse ES cells after lentiviral transduction with non-targeting control (shSCR) or with RPAP1 targeting (shRPAP1) shRNAs, followed by 24 hr of differentiation by LIF-removal. Genes are ranked from most upregulated to most downregulated by significance after correction for multiple testing (FDR q-value). Significant genes are highlighted in yellow (FDR < 0.05). Sheets 4-5: mRNA transcriptome ranked by Log2 fold-change, for use in GSEA analysis. Sheets 6-10: Supervised Network analysis of the GO terms significantly up- or downregulated (Z-score > 4) among the significantly differentially expressed genes (FDR q < 0.01) in mouse ES cells after lentiviral shSCR control or shRPAP1, followed by 24 hr of differentiation by LIF-removal. GO Terms are ranked by significance after correction for multiple testing (z-value). FDR z-value < 0.05 indicates significance. See also Supplemental Experimental Procedures, section on Network Analysis.
Sheets 1-3: GSEA results. Sheets 4-7: Processing of Arabidopsis RPAP1-mutation data (Sanmartín et al., 2011) to the nearest mouse homolog for use as a ranked list in GSEA analysis (see also: Supplemental Experimental Procedures). In sheets 1-3, the GSEA results include the Normalized Enrichment score (NES), the p value, and the FDR (False Discovery Rate) q-value for each geneset. Genesets are ranked by significance after correction for multiple hypothesis testing (FDR q-value). FDR q-value < 0.25 indicates significance. Sheet 1: Hallmark genesets. Sheet 2: C5-GO term genesets. Sheet 3: C2-Curated genesets (here only genesets which are significant are shown; FDR q-value < 0.25). Sheet 4: original Arabidopsis data for mRNA expression profile of RPAP1-mutant plant tissues versus wild type plant tissues (31,200 mRNAs). Data are ranked by log2 fold change. Sheet 5: Conversion of differential plant mRNA expression to nearest protein homolog in mouse (21,080 homologous mouse genes; see: Supplemental Experimental Procedures). Data are ranked by log2 fold change. Sheet 6: Ranked list of 21,080 homologs used for GSEA analyses. Sheet 7: Two lists of Arabidopsis genes which were filtered out during the conversion process (see: Experimental Procedures). “Not Found”: 250 genes in the Arabidopsis dataset that did not map (neither in TAIR 10 database nor in EnsEMBL). “Not aligned”: 1933 Arabidopsis genes without homology/orthology calculation. A description is listed, when available, to see their function. The majority of these are transposons.
Sheets 1-4: List of all normalized mRNA expression data generated by RNA-seq in MEF cells at day 3 after lentiviral transduction with non-targeting control (shSCR) or with RPAP1 targeting (shRPAP1) shRNAs. Genes are ranked from most upregulated to most downregulated by significance after correction for multiple testing (FDR q-value). Significant genes are highlighted in yellow (FDR < 0.05). Sheet 5: mRNA transcriptome ranked by Log2 fold-change, for use in GSEA analysis. Sheets 6-10: Supervised Network analysis of the GO terms significantly up- or downregulated (Z-score > 4) among the significantly differentially expressed genes (FDR q < 0.01) at day 3 after RPAP1 depletion in MEFs. GO Terms are ranked by significance after correction for multiple testing (z-value). FDR z-value < 0.05 indicates significance. See also Supplemental Experimental Procedures, section on Network Analysis.
Sheet 1: Summary of RNA Pol II interactome in MEFs at day 2 in control (shSCR) or after RPAP1 depletion (shRPAP1). Sheet 2: Magnified image of Figure 4D, showing the RNA Pol II interactome identified in this study. Schematic of the 294 specific interactors of RPB1 (official name POLR2A) detected in primary MEFs in this study by RNA Pol II immunoprecipitation and mass spectrometry analysis. Interactors were displayed as a network using Cytoscape, and grouped manually by their known physical interactions and general primary function, wherein the thickness and intensity of the connecting edges indicates the strength of their known interactions in the STRING database. Following RPAP1-depletion, the RNA Pol II-interactors reduced (circled in red) and RNA Pol II-interactors gained (circled in green) are indicated. The Mediator complex is depicted centrally and in full color based on the data in Figure 4E. Sheet 3: Summary of CORUM database analysis. RNA Pol II interactors which were decreased following RPAP1 depletion were assigned to all 3,000 known protein complexes in the Corum database (see Supplemental Experimental Procedures). In Column B, known complexes are ranked according to the highest number of proteins whose interaction were decreased upon RPAP1-depletion from the cells, listed in Column F (see: Figure 4E). Sheet 4: Data calculation for the CORUM plots of the data in Figure 4E.
Sheets 1-8: Summary tables of RNA Pol II quantification, calculations, and GSEA analyses on ChIP-seq data in this study, related to Figures 5 and S5. See also Supplemental Experimental Procedures in section describing the ChIP-seq data analyses.
References
- Allen B.L., Taatjes D.J. The Mediator complex: a central integrator of transcription. Nat. Rev. Mol. Cell Biol. 2015;16:155–166. doi: 10.1038/nrm3951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allepuz-Fuster P., Martínez-Fernández V., Garrido-Godino A.I., Alonso-Aguado S., Hanes S.D., Navarro F., Calvo O. Rpb4/7 facilitates RNA polymerase II CTD dephosphorylation. Nucleic Acids Res. 2014;42:13674–13688. doi: 10.1093/nar/gku1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson R., Gebhard C., Miguel-Escalada I., Hoof I., Bornholdt J., Boyd M., Chen Y., Zhao X., Schmidl C., Suzuki T. An atlas of active enhancers across human cell types and tissues. Nature. 2014;507:455–461. doi: 10.1038/nature12787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arner E., Daub C.O., Vitting-Seerup K., Andersson R., Lilje B., Drabløs F., Lennartsson A., Rönnerblad M., Hrydziuszko O., Vitezic M., FANTOM Consortium Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science. 2015;347:1010–1014. doi: 10.1126/science.1259418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S., Pradet-Balade B., Verheggen C., Molle D., Boireau S., Georgieva M., Azzag K., Robert M.C., Ahmad Y., Neel H. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell. 2010;39:912–924. doi: 10.1016/j.molcel.2010.08.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F.X., Woodfin A.R., Gardini A., Rickels R.A., Marshall S.A., Smith E.R., Shiekhattar R., Shilatifard A. PAF1, a molecular regulator of promoter-proximal pausing by RNA Polymerase II. Cell. 2015;162:1003–1015. doi: 10.1016/j.cell.2015.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng B., Li T., Rahl P.B., Adamson T.E., Loudas N.B., Guo J., Varzavand K., Cooper J.J., Hu X., Gnatt A. Functional association of Gdown1 with RNA polymerase II poised on human genes. Mol. Cell. 2012;45:38–50. doi: 10.1016/j.molcel.2011.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbi N., Di Padova M., De Angelis R., Bruno T., Libri V., Iezzi S., Floridi A., Fanciulli M., Passananti C. The alpha-like RNA polymerase II core subunit 3 (RPB3) is involved in tissue-specific transcription and muscle differentiation via interaction with the myogenic factor myogenin. FASEB J. 2002;16:1639–1641. doi: 10.1096/fj.02-0123fje. [DOI] [PubMed] [Google Scholar]
- Creyghton M.P., Cheng A.W., Welstead G.G., Kooistra T., Carey B.W., Steine E.J., Hanna J., Lodato M.A., Frampton G.M., Sharp P.A. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA. 2010;107:21931–21936. doi: 10.1073/pnas.1016071107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Alessio J.A., Wright K.J., Tjian R. Shifting players and paradigms in cell-specific transcription. Mol. Cell. 2009;36:924–931. doi: 10.1016/j.molcel.2009.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis J.A., Takagi Y., Kornberg R.D., Asturias F.A. Structure of the yeast RNA polymerase II holoenzyme: Mediator conformation and polymerase interaction. Mol. Cell. 2002;10:409–415. doi: 10.1016/s1097-2765(02)00598-1. [DOI] [PubMed] [Google Scholar]
- Egloff S., Zaborowska J., Laitem C., Kiss T., Murphy S. Ser7 phosphorylation of the CTD recruits the RPAP2 Ser5 phosphatase to snRNA genes. Mol. Cell. 2012;45:111–122. doi: 10.1016/j.molcel.2011.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egloff S., Dienstbier M., Murphy S. Updating the RNA polymerase CTD code: adding gene-specific layers. Trends Genet. 2012;28:333–341. doi: 10.1016/j.tig.2012.03.007. [DOI] [PubMed] [Google Scholar]
- Factor D.C., Corradin O., Zentner G.E., Saiakhova A., Song L., Chenoweth J.G., McKay R.D., Crawford G.E., Scacheri P.C., Tesar P.J. Epigenomic comparison reveals activation of “seed” enhancers during transition from naive to primed pluripotency. Cell Stem Cell. 2014;14:854–863. doi: 10.1016/j.stem.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget D., Lacombe A.A., Cloutier P., Al-Khoury R., Bouchard A., Lavallée-Adam M., Faubert D., Jeronimo C., Blanchette M., Coulombe B. The protein interaction network of the human transcription machinery reveals a role for the conserved GTPase RPAP4/GPN1 and microtubule assembly in nuclear import and biogenesis of RNA polymerase II. Mol. Cell. Proteomics. 2010;9:2827–2839. doi: 10.1074/mcp.M110.003616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forget D., Lacombe A.A., Cloutier P., Lavallée-Adam M., Blanchette M., Coulombe B. Nuclear import of RNA polymerase II is coupled with nucleocytoplasmic shuttling of the RNA polymerase II-associated protein 2. Nucleic Acids Res. 2013;41:6881–6891. doi: 10.1093/nar/gkt455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaillochet C., Lohmann J.U. The never-ending story: from pluripotency to plant developmental plasticity. Development. 2015;142:2237–2249. doi: 10.1242/dev.117614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giaever G., Chu A.M., Ni L., Connelly C., Riles L., Véronneau S., Dow S., Lucau-Danila A., Anderson K., André B. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- Hansson J., Rafiee M.R., Reiland S., Polo J.M., Gehring J., Okawa S., Huber W., Hochedlinger K., Krijgsveld J. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2012;2:1579–1592. doi: 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazbun T.R., Malmström L., Anderson S., Graczyk B.J., Fox B., Riffle M., Sundin B.A., Aranda J.D., McDonald W.H., Chiu C.-H. Assigning function to yeast proteins by integration of technologies. Mol. Cell. 2003;12:1353–1365. doi: 10.1016/s1097-2765(03)00476-3. [DOI] [PubMed] [Google Scholar]
- Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A., Hoke H.A., Young R.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsin J.P., Manley J.L. The RNA polymerase II CTD coordinates transcription and RNA processing. Genes Dev. 2012;26:2119–2137. doi: 10.1101/gad.200303.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu X., Malik S., Negroiu C.C., Hubbard K., Velalar C.N., Hampton B., Grosu D., Catalano J., Roeder R.G., Gnatt A. A Mediator-responsive form of metazoan RNA polymerase II. Proc. Natl. Acad. Sci. USA. 2006;103:9506–9511. doi: 10.1073/pnas.0603702103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T., Chiba T., Ozawa R., Yoshida M., Hattori M., Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc. Natl. Acad. Sci. USA. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C., Robert F. The Mediator complex: at the nexus of RNA Polymerase II transcription. Trends Cell Biol. 2017;27:765–783. doi: 10.1016/j.tcb.2017.07.001. [DOI] [PubMed] [Google Scholar]
- Jeronimo C., Langelier M.-F., Zeghouf M., Cojocaru M., Bergeron D., Baali D., Forget D., Mnaimneh S., Davierwala A.P., Pootoolal J. RPAP1, a novel human RNA polymerase II-associated protein affinity purified with recombinant wild-type and mutated polymerase subunits. Mol. Cell. Biol. 2004;24:7043–7058. doi: 10.1128/MCB.24.16.7043-7058.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeronimo C., Forget D., Bouchard A., Li Q., Chua G., Poitras C., Thérien C., Bergeron D., Bourassa S., Greenblatt J. Systematic analysis of the protein interaction network for the human transcription machinery reveals the identity of the 7SK capping enzyme. Mol. Cell. 2007;27:262–274. doi: 10.1016/j.molcel.2007.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M., Malik S., Wagner U., Uberheide B., Ishihama Y., Hu X., Chait B.T., Gnatt A., Ren B., Roeder R.G. Transcriptional regulation by Pol II(G) involving mediator and competitive interactions of Gdown1 and TFIIF with Pol II. Mol. Cell. 2012;45:51–63. doi: 10.1016/j.molcel.2011.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey M.H., Newman J.J., Bilodeau S., Zhan Y., Orlando D.A., van Berkum N.L., Ebmeier C.C., Goossens J., Rahl P.B., Levine S.S. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine M. Paused RNA polymerase II as a developmental checkpoint. Cell. 2011;145:502–511. doi: 10.1016/j.cell.2011.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T., Price D. Gdown1: making a link between mediator and RNA polymerase II elongation control. Transcription. 2012;3:177–180. doi: 10.4161/trns.20600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63. doi: 10.1016/j.stem.2010.04.014. [DOI] [PubMed] [Google Scholar]
- Li W., Notani D., Rosenfeld M.G. Enhancers as non-coding RNA transcription units: recent insights and future perspectives. Nat. Rev. Genet. 2016;17:207–223. doi: 10.1038/nrg.2016.4. [DOI] [PubMed] [Google Scholar]
- Lin C.Y., Lovén J., Rahl P.B., Paranal R.M., Burge C.B., Bradner J.E., Lee T.I., Young R.A. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151:56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Kraus W.L., Bai X. Ready, pause, go: regulation of RNA polymerase II pausing and release by cellular signaling pathways. Trends Biochem. Sci. 2015;40:516–525. doi: 10.1016/j.tibs.2015.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Guadamillas E., Fernandez-Marcos P.J., Pantoja C., Muñoz-Martin M., Martínez D., Gómez-López G., Campos-Olivas R., Valverde A.M., Serrano M. p21Cip1 plays a critical role in the physiological adaptation to fasting through activation of PPARα. Sci. Rep. 2016;6:34542. doi: 10.1038/srep34542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerowitz E.M. Plants compared to animals: the broadest comparative study of development. Science. 2002;295:1482–1485. doi: 10.1126/science.1066609. [DOI] [PubMed] [Google Scholar]
- Mosley A.L., Pattenden S.G., Carey M., Venkatesh S., Gilmore J.M., Florens L., Workman J.L., Washburn M.P. Rtr1 is a CTD phosphatase that regulates RNA polymerase II during the transition from serine 5 to serine 2 phosphorylation. Mol. Cell. 2009;34:168–178. doi: 10.1016/j.molcel.2009.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muñoz A., Mangano S., González-García M.P., Contreras R., Sauer M., De Rybel B., Weijers D., Sánchez-Serrano J.J., Sanmartín M., Rojo E. RIMA-dependent nuclear accumulation of IYO triggers auxin-irreversible cell differentiation in Arabidopsis. Plant Cell. 2017;29:575–588. doi: 10.1105/tpc.16.00791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polo J.M., Anderssen E., Walsh R.M., Schwarz B.A., Nefzger C.M., Lim S.M., Borkent M., Apostolou E., Alaei S., Cloutier J. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–1632. doi: 10.1016/j.cell.2012.11.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahl P.B., Lin C.Y., Seila A.C., Flynn R.A., McCuine S., Burge C.B., Sharp P.A., Young R.A. c-Myc regulates transcriptional pause release. Cell. 2010;141:432–445. doi: 10.1016/j.cell.2010.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanmartín M., Sauer M., Muñoz A., Zouhar J., Ordóñez A., van de Ven W.T.G., Caro E., de la Paz Sánchez M., Raikhel N.V., Gutiérrez C. A molecular switch for initiating cell differentiation in Arabidopsis. Curr. Biol. 2011;21:999–1008. doi: 10.1016/j.cub.2011.04.041. [DOI] [PubMed] [Google Scholar]
- Sanmartín M., Sauer M., Muñoz A., Rojo E. MINIYO and transcriptional obligation: lifting the roadblock to differentiation. Transcription. 2012;3:25–28. doi: 10.4161/trns.3.1.19303. [DOI] [PubMed] [Google Scholar]
- Savatier P., Lapillonne H., van Grunsven L.A., Rudkin B.B., Samarut J. Withdrawal of differentiation inhibitory activity/leukemia inhibitory factor up-regulates D-type cyclins and cyclin-dependent kinase inhibitors in mouse embryonic stem cells. Oncogene. 1996;12:309–322. [PubMed] [Google Scholar]
- Thiery J.P., Acloque H., Huang R.Y.J., Nieto M.A. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- van Riggelen J., Yetil A., Felsher D.W. MYC as a regulator of ribosome biogenesis and protein synthesis. Nat. Rev. Cancer. 2010;10:301–309. doi: 10.1038/nrc2819. [DOI] [PubMed] [Google Scholar]
- Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H., Rahl P.B., Lee T.I., Young R.A. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild T., Cramer P. Biogenesis of multisubunit RNA polymerases. Trends Biochem. Sci. 2012;37:99–105. doi: 10.1016/j.tibs.2011.12.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Sheets 1-3: List of all normalized mRNA expression data generated by RNA-seq in mouse ES cells after lentiviral transduction with non-targeting control (shSCR) or with RPAP1 targeting (shRPAP1) shRNAs, followed by 24 hr of differentiation by LIF-removal. Genes are ranked from most upregulated to most downregulated by significance after correction for multiple testing (FDR q-value). Significant genes are highlighted in yellow (FDR < 0.05). Sheets 4-5: mRNA transcriptome ranked by Log2 fold-change, for use in GSEA analysis. Sheets 6-10: Supervised Network analysis of the GO terms significantly up- or downregulated (Z-score > 4) among the significantly differentially expressed genes (FDR q < 0.01) in mouse ES cells after lentiviral shSCR control or shRPAP1, followed by 24 hr of differentiation by LIF-removal. GO Terms are ranked by significance after correction for multiple testing (z-value). FDR z-value < 0.05 indicates significance. See also Supplemental Experimental Procedures, section on Network Analysis.
Sheets 1-3: GSEA results. Sheets 4-7: Processing of Arabidopsis RPAP1-mutation data (Sanmartín et al., 2011) to the nearest mouse homolog for use as a ranked list in GSEA analysis (see also: Supplemental Experimental Procedures). In sheets 1-3, the GSEA results include the Normalized Enrichment score (NES), the p value, and the FDR (False Discovery Rate) q-value for each geneset. Genesets are ranked by significance after correction for multiple hypothesis testing (FDR q-value). FDR q-value < 0.25 indicates significance. Sheet 1: Hallmark genesets. Sheet 2: C5-GO term genesets. Sheet 3: C2-Curated genesets (here only genesets which are significant are shown; FDR q-value < 0.25). Sheet 4: original Arabidopsis data for mRNA expression profile of RPAP1-mutant plant tissues versus wild type plant tissues (31,200 mRNAs). Data are ranked by log2 fold change. Sheet 5: Conversion of differential plant mRNA expression to nearest protein homolog in mouse (21,080 homologous mouse genes; see: Supplemental Experimental Procedures). Data are ranked by log2 fold change. Sheet 6: Ranked list of 21,080 homologs used for GSEA analyses. Sheet 7: Two lists of Arabidopsis genes which were filtered out during the conversion process (see: Experimental Procedures). “Not Found”: 250 genes in the Arabidopsis dataset that did not map (neither in TAIR 10 database nor in EnsEMBL). “Not aligned”: 1933 Arabidopsis genes without homology/orthology calculation. A description is listed, when available, to see their function. The majority of these are transposons.
Sheets 1-4: List of all normalized mRNA expression data generated by RNA-seq in MEF cells at day 3 after lentiviral transduction with non-targeting control (shSCR) or with RPAP1 targeting (shRPAP1) shRNAs. Genes are ranked from most upregulated to most downregulated by significance after correction for multiple testing (FDR q-value). Significant genes are highlighted in yellow (FDR < 0.05). Sheet 5: mRNA transcriptome ranked by Log2 fold-change, for use in GSEA analysis. Sheets 6-10: Supervised Network analysis of the GO terms significantly up- or downregulated (Z-score > 4) among the significantly differentially expressed genes (FDR q < 0.01) at day 3 after RPAP1 depletion in MEFs. GO Terms are ranked by significance after correction for multiple testing (z-value). FDR z-value < 0.05 indicates significance. See also Supplemental Experimental Procedures, section on Network Analysis.
Sheet 1: Summary of RNA Pol II interactome in MEFs at day 2 in control (shSCR) or after RPAP1 depletion (shRPAP1). Sheet 2: Magnified image of Figure 4D, showing the RNA Pol II interactome identified in this study. Schematic of the 294 specific interactors of RPB1 (official name POLR2A) detected in primary MEFs in this study by RNA Pol II immunoprecipitation and mass spectrometry analysis. Interactors were displayed as a network using Cytoscape, and grouped manually by their known physical interactions and general primary function, wherein the thickness and intensity of the connecting edges indicates the strength of their known interactions in the STRING database. Following RPAP1-depletion, the RNA Pol II-interactors reduced (circled in red) and RNA Pol II-interactors gained (circled in green) are indicated. The Mediator complex is depicted centrally and in full color based on the data in Figure 4E. Sheet 3: Summary of CORUM database analysis. RNA Pol II interactors which were decreased following RPAP1 depletion were assigned to all 3,000 known protein complexes in the Corum database (see Supplemental Experimental Procedures). In Column B, known complexes are ranked according to the highest number of proteins whose interaction were decreased upon RPAP1-depletion from the cells, listed in Column F (see: Figure 4E). Sheet 4: Data calculation for the CORUM plots of the data in Figure 4E.
Sheets 1-8: Summary tables of RNA Pol II quantification, calculations, and GSEA analyses on ChIP-seq data in this study, related to Figures 5 and S5. See also Supplemental Experimental Procedures in section describing the ChIP-seq data analyses.