Abstract
We have previously described a novel taxon of the genus Ehrlichia (type strain WisconsinT), closely related to Ehrlichia muris, that causes human ehrlichiosis among patients with exposures to ticks in the upper midwestern USA. DNA from this bacterium was also detected in Ixodes scapularis and Peromyscus leucopus collected in Minnesota and Wisconsin. To determine the relationship between the E. muris-like agent (EMLA) and other species of the genus Ehrlichia phenotypic, genotypic and epidemiologic comparisons were undertaken, including sequence analysis of eight gene loci (3906 nucleotides) for 39 EMLA DNA samples and the type strain of E. muris AS145T. Three loci were also sequenced from DNA of nine strains of E. muris from mouse spleens from Japan. All sequences from E. muris were distinct from homologous EMLA sequences, but differences between them were less than those observed among other species of the genus Ehrlichia. Phenotypic comparison of EMLA and E. muris revealed similar culture and electron microscopic characteristics, but important differences were noted in their geographic distribution, ecological associations and behavior in mouse models of infection. Based on these comparisons, we propose that type strain WisconsinT represents a novel subspecies, Ehrlichia murissubsp. eauclairensis,subsp. nov. This strain is available through the Centers for Disease Control and Prevention Rickettsial Isolate Reference Collection (CRIRC EMU002T) and through the Collection de Souches de l’Unité des Rickettsies (CSURP2883 T). The subspecies Ehrlichia murissubsp. muris subsp. nov. is automatically created and the type strain AS145T is also available through the same collections (CRIRC EMU001T, CSUR E2T). Included is an emended description of E. muris.
Keywords: tick-borne, vector-borne, Rickettsiales, Anaplasmataceae
Abbreviations
CDC, Centers for Disease Control and Prevention; EMLA, Ehrlichia muris-like agent; IFA, immunofluorescence assay; EM, electron microscopy.
The genus Ehrlichia includes multiple species of Gram-negative, non-motile, coccoid to ellipsoidal, obligate intracellular tick-borne bacteria that reside within cytoplasmic vacuoles of haematopoietic or endothelial cells in mammals [1]. Ehrlichiae are not cultivable in cell-free media but most can be isolated in one or more haematopoietic, endothelial or tick-derived cell lines [1]. There are currently five species with validly published names in this genus including the type species Ehrlichia canis [2, 3], Ehrlichia chaffeensis [4], Ehrlichia ewingii (Anderson et al., 1992), Ehrlichia muris [5] and Ehrlichia ruminantium, [1, 6, 7].
We have reported previously the detection of a novel taxon of the genus Ehrlichia, type strain WisconsinT, in whole blood specimens from four febrile patients with histories of tick exposures in the upper midwestern United States using culture, serology and PCR [8]. DNA from this bacterium was detected in Ixodes scapularis (black-legged ticks) [8–10] and blood of Peromyscus leucopus collected in Minnesota and Wisconsin [11].
WisconsinT was originally described as representing a novel species of the genus Ehrlichia based on analyses of the partial groEL and 16S rRNA gene sequences [8]. These genes exhibited approximately 98 % sequence identity to homologous regions of the genome of Ehrlichia muris [8] thereby leading to the commonly applied moniker ‘E. muris-like agent (EMLA)’ that has been used to describe this bacterium since its initial characterization. While this member of the genus Ehrlichia has only been detected in the upper midwestern USA, E. muris is considered to be an Old World pathogen, found in Eastern Europe and Japan [12–17]. Recently, genotypic, phenotypic and epidemiological comparisons of EMLA WisconsinT, E. muris and other species of the genus Ehrlichia prompted us to re-examine our prior conclusion that WisconsinT represented a novel species of the genus Ehrlichia. This communication describes the results of this comparison and proposes that WisconsinT represents a novel subspecies, E. muris subsp. eauclairensis subsp. nov., and that the organism represented by the type strain AS145T be renamed E. muris subsp. muris, subsp. nov.
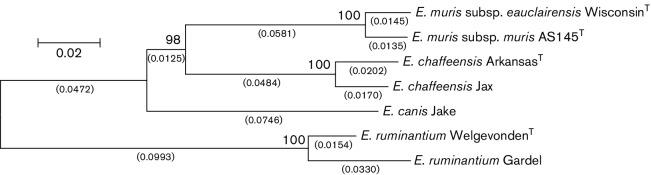
To better understand the degree of genetic variability found between and within species of the genus Ehrlichia, we analyzed sequences of eight gene loci (gltA, groEL, nadA, dsb, fbpA, p13, p28-14, p28-19) comprising 3906 nucleotides from EMLA-positive DNA samples, including WisconsinT, E. muris AS145T (ATCC, VR-1411) and other species of the genus Ehrlichia (Table 1 and S1, available in the online Supplementary Material), [9, 18–21]. EMLA-positive DNA extracts were obtained from 21 humans, 17 I. scapularis and 1 P. leucopus. Three loci (16S, groEL and p28-14; 1559 nt) were also sequenced for nine E. muris-positive DNA extracts obtained from mouse spleens at Osaka Prefecture University, Japan and compared with the E. muris type strain, AS145T. PCR products were sequenced in both directions, and sequencing reads were assembled using Sequencher 5.1 (Gene Codes). Sequences were aligned using mega 5.1 (www.megasoftware.net/) and percentage identity was determined using blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi). An estimation of the evolutionary history of E. muris subsp. eauclairensis compared with other established ehrlichial species was inferred using mega 5.1 (www.megasoftware.net/) by trimming the PCR primer sequences and concatenating the sequences for gltA, groEL, nadA, dsb, fbpA, p28-14 and p28-19. Indels were removed using the simple indel coding method [22]. EMLA WisconsinT was found to be closely related to E. muris AS145T (Fig. 1), with a shorter branch length than found between existing ehrlichial species. Diversity within EMLA samples was seen for only one locus (p28-14), in which one EMLA-positive sample (UW-M7) was indistinguishable from E. muris (Genbank accession DQ335244). Similarly, of the three E. muris loci, no diversity was found among the nine samples tested and all were identical to E. muris AS145T. However, all E. muris sequences were distinct from the homologous sequences of EMLA. The level of genetic similarity between EMLA and E. muris AS145T at these eight loci was compared with those observed among four distinct species of the genus Ehrlichia at the same loci (Table 1). From these data, we determined that the genetic differences between EMLA and strains of E. muris at these loci were less than those observed among multiple recognized species of the genus Ehrlichia.
Table 1. Comparison of strain WisconsinT to strains of other species of the genus Ehrlichia.
Amplicon lengths are shown in the online Supplementary Material. The following GenBank files were used in this analysis (E. chaffeensis Arkansas genome NC_007799, E. canis Jake genome NC_007354, E. ewingii gltA DQ365879, groEL AF195273, dsb KM458249, p28-14 EF116932 and p28-19 EF116932 and E. ruminantium WelgevondenT genome NC_005295.2).
| Percentage identity of strain WisconsinT to other species of the genus Ehrlichia | ||||||||
|---|---|---|---|---|---|---|---|---|
| gltA | groEL | nadA | dsb | fbpA | p13 | p28-14 | p28-19 | |
| E. muris AS145T | 98 | 98 | 98 | 96 | 96 | 90 | 95 | 95 |
| E. chaffeensis ArkansasT | 88 | 94 | 89 | 87 | 91 | * | 83 | 80 |
| E. canis Jake | 87 | 93 | 89 | 82 | 88 | * | 82 | 77 |
| E. ewingii | 83 | 91 | * | 78 | * | * | 76 | 81 |
| E. ruminantium WelgevondenT | 79 | 88 | 84 | 79 | 82 | * | 78 | 77 |
*Indicates genetic data unavailable in GenBank.
Fig. 1.
The evolutionary history was inferred using the neighbor-joining method [34]. The optimal tree with the sum of branch lengths=0.45356965 is shown. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown above the branches [35]. The tree is drawn to scale with branch lengths, shown under the branches (in parentheses), in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Maximum Composite Likelihood method [36] and are in the units of the number of base substitutions per site. The analysis involved seven nucleotide sequences and all positions with less than 95 % site coverage were eliminated. There were a total of 3781 positions in the final dataset. Evolutionary analyses were conducted in mega5 [37]. The following GenBank files were used in this analysis (E. chaffeensis ArkansasT genome NC_007799, E. chaffeensis Jax genome NZ_CP007475.1, E. canis Jake genome NC_007354, E. ruminantium WelgevondenT genome NC_005295.2, and E. ruminantium Gardel genome NC_006831.1).
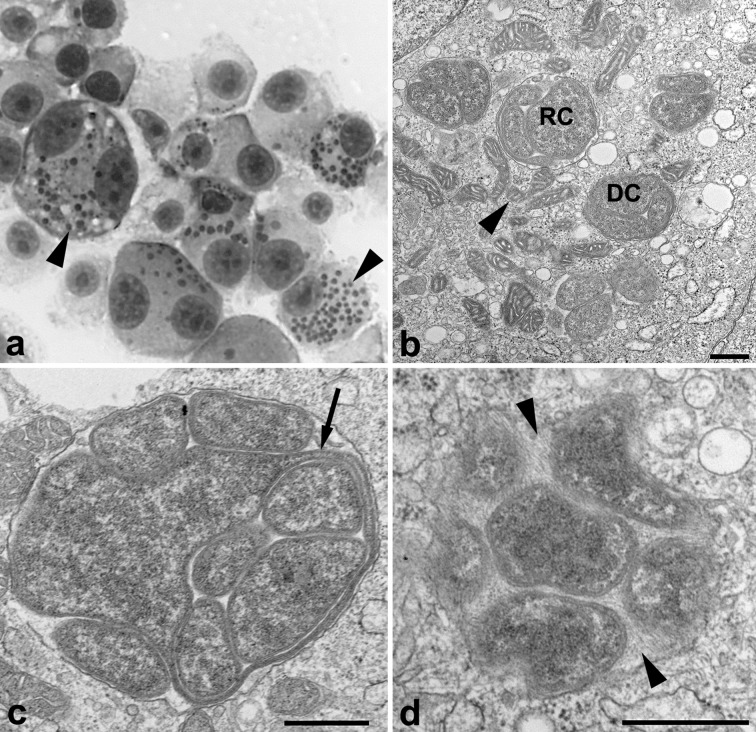
A phenotypic comparison of EMLA WisconsinT and E. muris AS145T was also performed using cell culture, electron microscopy (EM) and animal inoculation. EMLA WisconsinT and E. muris AS145T [5] were propagated in DH82 canine macrophage-like cells (American Type Culture Collection number CRL-10389) at 37 °C in a 5 % CO2 atmosphere. The cells were fed with minimal essential medium (MEM) (Gibco) supplemented with 0.1 mM MEM non-essential amino acids (Gibco), 10 mM HEPES buffer (Gibco), 2 mM l-glutamine (Gibco), 10 mM sodium pyruvate (Gibco) and 10 % heat-inactivated fetal bovine serum (Atlanta Biologicals). When grown in DH82 canine macrophage cells and stained with the Jorvet Dip Quick stain (Jorgensen Laboratories), EMLA appears as small clusters of bacteria, known as morulae, within vacuoles in the host cell cytoplasm (Fig. 2a), and no growth differences were observed between EMLA WisconsinT and E. muris AS145T.
Fig. 2.
EMLA in cultured cells. (a) Dip Quick stain of EMLA infecting DH82 canine macrophage cells (×100) showing bacterial morulae (arrow heads). (b–d) Electron microscopic analysis of EMLA infecting DH82 cells, scale bars, 500 nm. (b) EMLA exhibits two cell types, dense-cored cells (DC) and reticulate cells (RC). Host cell mitochondria (arrowhead) are also found in close association with the morulae. (c) Long projection of RC ehrlichial cell membrane (arrow) that completely surrounds other RC. (d) A fibrillary matrix (arrow heads) of varying densities is often observed in EMLA morulae.
To fix cells for transmission EM analysis, an infected DH82 monolayer was washed in 0.1 M phosphate buffer, pH 7.3 and fixed in buffered 2.5 % glutaraldehyde for 5 min at 4 °C. Cells were detached from the monolayer by using a cell scraper, and then centrifuged at 2500 r.p.m. for 5 min at 4 °C and allowed to fix for 10 min. The glutaraldehyde was removed and fresh phosphate buffer was layered onto the pellet, which was stored at 4 °C. The cells were post-fixed in 1 % buffered osmium tetroxide, stained in 4 % uranyl acetate, dehydrated through a graded series of alcohols and acetone and embedded in a mixture of Epon-substitute and Araldite [23]. Thin sections were stained with 4 % uranyl acetate and Reynold’s lead citrate. As described for other species of the genus Ehrlichia, intracellular ELMA bacteria exist as two forms, reticulate cells (RC) and dense-cored cells (DC), [24] (Fig. 2b). Both cell types appear slightly oblong, with dense-cored cells averaging 700×403 nm while the reticulate cells are a little larger, with an average cell size of 964×458 nm. The morulae averaged 1.42×1.27 µm and usually consisted of a homogeneous population of either cell type. Morulae contained a fibrillary matrix of varying density (Fig. 2d) and host cell mitochondria were often found in proximity to morulae (Fig. 2b). Reticulate cells occasionally demonstrated long projections of the bacterial cell membrane that invested other ehrlichial cells in morulae (Fig. 2c). Similar findings were observed for E. muris AS145T (data not shown) and have been previously described when AS145T was grown in other cell lines [24].
Despite similarities in culture and EM characteristics, EMLA WisconsinT and E. muris AS145T demonstrated pathogenic differences in mouse models of infection. E. muris AS145T causes a sub-lethal infection in C57BL/6 mice [25–27]. EMLA WisconsinT can lead to either a lethal or persistent infection in the same mouse strain, depending on the route of infection [28, 29]. Additionally, EMLA WisconsinT that was transmitted by ticks has also been shown to cause mouse mortality [29].
Finally, ecological analysis of EMLA WisconsinT and E. muris AS145T reveals differences between the geographic distributions and host associations of these organisms. E. muris has been found in Japan [17], Russia [14], Slovakia [12] and Korea [16], while EMLA strains, including WisconsinT, have thus far been found only in Minnesota and Wisconsin in the Western Hemisphere [8–10, 30]. Ixodes persulcatus and Haemaphysalis flava ticks serve as vectors of E. muris [12, 31], whereas EMLA WisconsinT is transmitted by I. scapularis [28, 29, 32]. It is also apparent that EMLA causes human disease, while it is not known whether E. muris AS145T is a human pathogen, despite serological evidence of human exposure [31].
We conclude that EMLA WisconsinT represents a novel subspecies of E. muris and propose naming it E. muris subspecies eauclairensis based on the geographic origin of the original isolate.
Description of Ehrlichia muris subsp. muris subsp. nov.
Ehrlichia muris subsp. muris (mu′ris. L. gen. n. muris, of a mouse; the subspecies was first isolated from a mouse).
Ehrlichia muris subsp. muris [5] has been found in Ixodes persulcatus and Haemaphysalis flava hard-bodied ticks, wild mice and sika deer (Cervus nippon yesoensis) in regions of Eastern Europe and Japan [13, 15, 20, 25, 31, 33]. There is also serological evidence of human, boar, dog, deer, bear and monkey infections in Japan [31], although it is difficult to determine if these antibodies result from E. muris or other ehrlichial agents described in that region. The natural history of E. muris subsp. muris is incompletely characterized but probably involves small rodent hosts; wild caught specimens of infected Eothenomys kageus [26], Apodemus flavicollis [12], and Apodemus speciosus, and Apodemus argenteus [31] have been identified. In laboratory settings E. muris subsp. muris causes sub-lethal infections in BALB/c, DBA/2, C57BL/6, C3H, ICR, CBA and ddY [17] AKR [25], and BALB/c mice [26]. Protection against infection in mice appears to be mediated through a combination of CD4 and CD8 T lymphocytes, antibodies, tumor necrosis factor and interferon gamma, with lethal infection observed in CD4 and CD8 lymphocyte-depleted mice [25]. Infection is associated with a short-lived clinical illness and persists for the life of the mouse [25, 26]. The target cell(s) in naturally infected vertebrate hosts is unknown; however, ehrlichiae can be found in mononuclear cells of various organs and tissues and occasional hepatocytes in AKR and C57BL/6 mice experimentally infected with this organism [25].
The type strain, AS145T, is available through the Centers for Disease Control and Prevention Rickettsial Isolate Reference Collection (CRIRC EMU001T) and through the Collection de Souches de l′Unité des Rickettsies (CSUR E2 T).
Description of Ehrlichia muris subsp. eauclairensis subsp. nov.
Ehrlichia muris subsp. eauclairensis (eau.clair.en′sis. N.L. fem. adj. eauclairensis, from Eau Claire; the type strain was isolated from a patient from Eau Claire, Wisconsin, in 2009).
To date, all infected tick and vertebrate hosts have originated from Minnesota and Wisconsin. Human infection with E. muris subsp. eauclairensis causes an illness characterized by fever, headache, myalgias, lymphopenia and thrombocytopenia [8, 30]. Ehrlichia muris subsp. eauclairensis is serologically cross-reactive with E. chaffeensis as determined by IFA [8]. The target cell(s) in naturally infected vertebrate hosts is unknown; however, ehrlichiae can be found in mononuclear and endothelial cells of various organs and tissues in mice experimentally infected with this organism [28] E. muris subsp. eauclairensis is passaged transstadially in and transmitted by I. scapularis ticks [28, 29, 32] and the bacterium has been detected in or isolated from nymphal and adult stages [8–10, 32]. In the tick, E. muris subsp. eauclairensis infects multiple cell types, particularly epithelial cells of the salivary glands, tracheae and male accessory glands, as well as neuronal cells of the synganglion [32]. The natural history of E. muris subsp. eauclairensis is incompletely characterized but probably involves small rodent hosts; wild-caught specimens of infected Peromyscus leucopus have been identified [11]. In the laboratory setting E. muris subsp. eauclairensis has been shown to infect C57BL/6 mice, and is capable of causing a lethal infection in a dose-dependent manner. Although bacteremia only occurs for a short time, multiple organs are infected, including the lungs, liver and spleen [28, 29]. The bacteremia is sufficient for ehrlichial transmission to I. scapularis ticks through feeding in both C57BL/6 mice [28, 29] and Syrian hamsters (Mesocricetus auratus) [32] and tick transmission may lead to mouse mortality [29]. A draft genome assembly for strain EmCRT is available (NCBI NZ_LANU01000000) consisting of 1.15 mb with 29.8 % GC.
The type strain, WisconsinT, is available through the Centers for Disease Control and Prevention Rickettsial Isolate Reference Collection (CRIRC EMU002T) and through the Collection de Souches de l’Unité des Rickettsies (CSURP2883T).
Funding information
This work received no specific grant from any funding agency.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Ethical statement
Patient follow-up and DNA sequencing of clinical specimens was approved by the Mayo Clinic institutional review board. All animal experiments were performed in accordance with Nagoya Environmental Health Institute Institutional Animal Care and Use Committee guidelines and approved.
Supplementary Data
References
- 1.Dumler JS, Barbet AF, Bekker CP, Dasch GA, Palmer GH, et al. Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and 'HGE agent' as subjective synonyms of Ehrlichia phagocytophila. Int J Syst Evol Microbiol. 2001;51:2145–2165. doi: 10.1099/00207713-51-6-2145. [DOI] [PubMed] [Google Scholar]
- 2.Donatien A, Lestoquard F. Existence en Algerie d'une Rickettsia du chien. Bull Soc Pathol Exot. 1935;28:418–419. [Google Scholar]
- 3.Moshkovski SD. Cytotropic inducers of infection and the classification of the Rickettsiae with Chlamydozoa. Advances in Modern Biology. 1945;19:1–44. [Google Scholar]
- 4.Anderson BE, Greene CE, Jones DC, Dawson JE. Ehrlichia ewingii sp. nov., the etiologic agent of canine granulocytic ehrlichiosis. Int J Syst Bacteriol. 1992;42:299–302. doi: 10.1099/00207713-42-2-299. [DOI] [PubMed] [Google Scholar]
- 5.Wen B, Rikihisa Y, Mott J, Fuerst PA, Kawahara M, et al. Ehrlichia muris sp. nov., identified on the basis of 16S rRNA base sequences and serological, morphological, and biological characteristics. Int J Syst Bacteriol. 1995;45:250–254. doi: 10.1099/00207713-45-2-250. [DOI] [PubMed] [Google Scholar]
- 6.Cowdry EV. Studies on the etiology of heartwater 1. Observation of a rickettsia, Rickettsia ruminantium (n. sp.), in the tissues of infected animals. J Exp Med. 1925;42:231–252. doi: 10.1084/jem.42.2.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cowdry EV. Studies on the etiology of heartwater 2. Rickettsia ruminantium (n.sp.) in the tissues of ticks transmitting the disease. J Exp Med. 1925;42:253–274. doi: 10.1084/jem.42.2.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pritt BS, Sloan LM, Johnson DK, Munderloh UG, Paskewitz SM, et al. Emergence of a new pathogenic Ehrlichia species, Wisconsin and Minnesota, 2009. N Engl J Med. 2011;365:422–429. doi: 10.1056/NEJMoa1010493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Telford III SR, Goethert HK, Cunningham JA. Prevalence of Ehrlichia muris in Wisconsin deer ticks collected during the mid 1990s. Open Microbiol J. 2011;5:18–20. doi: 10.2174/1874285801105010018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stromdahl E, Hamer S, Jenkins S, Sloan L, Williamson P, et al. Comparison of phenology and pathogen prevalence, including infection with the Ehrlichia muris-like (EML) agent, of Ixodes scapularis removed from soldiers in the midwestern and the northeastern United States over a 15 year period (1997–2012) Parasit Vectors. 2014;7:553. doi: 10.1186/s13071-014-0553-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Castillo CG, Eremeeva ME, Paskewitz SM, Sloan LM, Lee X, et al. Detection of human pathogenic Ehrlichia muris-like agent in Peromyscus leucopus. Ticks Tick Borne Dis. 2015;6:155–157. doi: 10.1016/j.ttbdis.2014.11.006. [DOI] [PubMed] [Google Scholar]
- 12.Smetanová K, Boldis V, Kocianová E, Spitalská E. Detection of Ehrlichia muris in a yellow-necked mouse (Apodemus flavicollis) in Central Slovakia. Acta Virol. 2007;51:69–71. [PubMed] [Google Scholar]
- 13.Spitalská E, Boldis V, Kostanová Z, Kocianová E, Stefanidesová K. Incidence of various tick-borne microorganisms in rodents and ticks of central Slovakia. Acta Virol. 2008;52:175–179. [PubMed] [Google Scholar]
- 14.Alekseev AN, Dubinina HV, van de Pol I, Schouls LM. Identification of Ehrlichia spp. and Borrelia burgdorferi in Ixodes ticks in the Baltic regions of Russia. J Clin Microbiol. 2001;39:2237–2242. doi: 10.1128/JCM.39.6.2237-2242.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eremeeva ME, Oliveira A, Moriarity J, Robinson JB, Tokarevich NK, et al. Detection and identification of bacterial agents in Ixodes persulcatus Schulze ticks from the north western region of Russia. Vector Borne Zoonotic Dis. 2007;7:426–436. doi: 10.1089/vbz.2007.0112. [DOI] [PubMed] [Google Scholar]
- 16.Kang SW, Doan HT, Choe SE, Noh JH, Yoo MS, et al. Molecular investigation of tick-borne pathogens in ticks from grazing cattle in Korea. Parasitol Int. 2013;62:276–282. doi: 10.1016/j.parint.2013.02.002. [DOI] [PubMed] [Google Scholar]
- 17.Kawahara M, Suto C, Rikihisa Y, Yamamoto S, Tsuboi Y. Characterization of ehrlichial organisms isolated from a wild mouse. J Clin Microbiol. 1993;31:89–96. doi: 10.1128/jcm.31.1.89-96.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Doyle CK, Labruna MB, Breitschwerdt EB, Tang YW, Corstvet RE, et al. Detection of medically important Ehrlichia by quantitative multicolor TaqMan real-time polymerase chain reaction of the dsb gene. J Mol Diagn. 2005;7:504–510. doi: 10.1016/S1525-1578(10)60581-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Doyle CK, Zhang X, Popov VL, McBride JW. An immunoreactive 38-kilodalton protein of Ehrlichia canis shares structural homology and iron-binding capacity with the ferric ion-binding protein family. Infect Immun. 2005;73:62–69. doi: 10.1128/IAI.73.1.62-69.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tamamoto C, Seino N, Suzuki M, Kaji K, Takahashi H, et al. Detection of Ehrlichia muris DNA from sika deer (Cervus nippon yesoensis) in Hokkaido, Japan. Vet Parasitol. 2007;150:370–373. doi: 10.1016/j.vetpar.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 21.Yu XJ, Walker DH. Sequence and characterization of an Ehrlichia chaffeensis gene encoding 314 amino acids highly homologous to the NAD A enzyme. FEMS Microbiol Lett. 1997;154:53–58. doi: 10.1111/j.1574-6968.1997.tb12623.x. [DOI] [PubMed] [Google Scholar]
- 22.Ogden TH, Rosenberg MS. How should gaps be treated in parsimony? A comparison of approaches using simulation. Mol Phylogenet Evol. 2007;42:817–826. doi: 10.1016/j.ympev.2006.07.021. [DOI] [PubMed] [Google Scholar]
- 23.Mollenhauer HH. Plastic embedding mixtures for use in electron microscopy. Stain Technol. 1964;39:111–114. [PubMed] [Google Scholar]
- 24.Popov VL, Han VC, Chen SM, Dumler JS, Feng HM, et al. Ultrastructural differentiation of the genogroups in the genus Ehrlichia. J Med Microbiol. 1998;47:235–251. doi: 10.1099/00222615-47-3-235. [DOI] [PubMed] [Google Scholar]
- 25.Olano JP, Wen G, Feng HM, McBride JW, Walker DH. Histologic, serologic, and molecular analysis of persistent ehrlichiosis in a murine model. Am J Pathol. 2004;165:997–1006. doi: 10.1016/S0002-9440(10)63361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kawahara M, Suto C, Shibata S, Futohashi M, Rikihisa Y. Impaired antigen specific responses and enhanced polyclonal stimulation in mice infected with Ehrlichia muris. Microbiol Immunol. 1996;40:575–581. doi: 10.1111/j.1348-0421.1996.tb01111.x. [DOI] [PubMed] [Google Scholar]
- 27.Borjesson D, Macnamara K, Johns J, Winslow G. Anaplasma phagocytophilum and Ehrlichia muris induce cytopenias and global defects in hematopoiesis. Clin Microbiol Infect. 2009;15:66–67. doi: 10.1111/j.1469-0691.2008.02182.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Saito TB, Thirumalapura NR, Shelite TR, Rockx-Brouwer D, Popov VL, et al. An animal model of a newly emerging human ehrlichiosis. J Infect Dis. 2015;211:452–461. doi: 10.1093/infdis/jiu372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karpathy SE, Allerdice ME, Sheth M, Dasch GA, Levin ML. Co-feeding transmission of the Ehrlichia muris-like agent to mice (Mus musculus) Vector Borne Zoonotic Dis. 2016;16:145–150. doi: 10.1089/vbz.2015.1878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson DK, Schiffman EK, Davis JP, Neitzel DF, Sloan LM, et al. Human infection with Ehrlichia muris-like pathogen, United States, 2007-2013(1) Emerg Infect Dis. 2015;21:1794–1799. doi: 10.3201/eid2110.150143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kawahara M, Ito T, Suto C, Shibata S, Rikihisa Y, et al. Comparison of Ehrlichia muris strains isolated from wild mice and ticks and serologic survey of humans and animals with E. muris as antigen. J Clin Microbiol. 1999;37:1123–1129. doi: 10.1128/jcm.37.4.1123-1129.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lynn GE, Oliver JD, Nelson CM, Felsheim RF, Kurtti TJ, et al. Tissue distribution of the Ehrlichia muris-like agent in a tick vector. PLoS One. 2015;10:e0122007. doi: 10.1371/journal.pone.0122007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rar VA, Fomenko NV, Dobrotvorsky AK, Livanova NN, Rudakova SA, et al. Tickborne pathogen detection, Western Siberia, Russia. Emerg Infect Dis. 2005;11:1708–1715. doi: 10.3201/eid1111.041195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 35.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.2307/2408678. [DOI] [PubMed] [Google Scholar]
- 36.Tamura K, Nei M, Kumar S. Prospects for inferring very large phylogenies by using the neighbor-joining method. Proc Natl Acad Sci USA. 2004;101:11030–11035. doi: 10.1073/pnas.0404206101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.