Abstract
The EepR transcription factor positively regulates secondary metabolites and tissue-damaging metalloproteases. To gain insight into mechanisms by which EepR regulates pigment and co-regulated factors, genetic suppressor analysis was performed. Suppressor mutations that restored pigment to the non-pigmented ∆eepR mutant mapped to the hexS ORF. Mutation of hexS also restored haemolysis, swarming motility and protease production to the eepR mutant. HexS is a known direct and negative regulator of secondary metabolites in Serratia marcescens and is a LysR family regulator and an orthologue of LrhA. Here, we demonstrate that HexS directly controls eepR and the serralysin gene prtS. EepR was shown to directly regulate eepR expression but indirectly regulate hexS expression. Together, these data indicate that EepR and HexS oppose each other in controlling stationary phase-associated molecules and enzymes.
Keywords: biosurfactant, prodigiosin, motility, secondary metabolism, transcriptional regulation
Introduction
In stationary phase, the Gram-negative bacterium and opportunistic pathogen Serratia marcescens synthesizes a number of secondary metabolites and secreted enzymes. Generation of these factors is highly regulated by a number of transcription factors including negative regulators CopA [1], CRP [2], HexS [3, 4], RssAB [5] and SpnR [6] and positive regulators EepR [7, 8], PigP [3] and SmaI [9].
The EepR putative response regulator is a direct positive regulator of several compounds including the biologically active pigment prodigiosin, the antibiotic biosurfactant serratamolide and the cytotoxic metalloprotease serralysin (PrtS) [7, 8]. The eepR gene is also important in positive regulation of chitinases and chitin binding protein Cbp21, as well as other proteins such as the SlpB protease and S-layer protein SlaA [7]. EepR-like regulators have been found in other medically relevant organisms including Burkholderia species [10]. The coordinated interplay between EepR and other transcriptional regulators that govern secondary metabolites and virulence factors has not been determined.
In this study, suppressor analysis was used to gain insight into the regulatory network of the EepR transcription factor. Transposon mutations that restored pigmentation to a ∆eepR mutant mapped to the hexS transcription factor and upstream of the eepR ORF. Subsequent analysis supports that HexS directly binds to and inhibits eepR expression and that EepR inhibits hexS expression. Together, the data presented here suggest that EepR and HexS are key regulators that oppose one another in control of secondary metabolites and the cytotoxic metalloprotease serralysin.
Methods
Microbiological growth conditions and media
Escherichia coli and S. marcescens strains are listed in Table 1 and were grown in lysogeny broth (LB) [11, 12] at 30 °C. Growth in liquid medium was performed with aeration using a tissue culture roller (TC-7). Swarming motility plates were composed of LB with 0.6 % agar, and swimming motility plates were LB with 0.3 % agar. Haemolysis detection plates consisted of tryptic soy agar with 5 % sheep erythrocytes. Antibiotics used were gentamicin at 10 µg ml−1, kanamycin at 50–100 µg ml−1 and tetracycline at 10 µg ml−1.
Table 1. Strains and plasmids used in this study.
| Strain or plasmid | Description | Reference or source* |
|---|---|---|
| Saccharomyces cerevisiae | ||
| InvSc1 | Uracil auxotroph for in vivo cloning | Invitrogen |
| E. coli | ||
| SM10 λpir | thi thr leu tonA lacY Sup.E recA :: RP4-2Tc ::Mu pir | [19] |
| S17-1 λpir | thi pro hsdR hsdM+ ∆recA RP4-2 ::TcMu-Km ::Tn7 pir | [19] |
| EC100D pir-116 | Cloning strain | Epicentre |
| S. marcescens | ||
| PIC3611 | Wild-type ‘parental strain’ | Presque Isle Cultures |
| CMS2922 | PIC3611 ∆hexS | [17] |
| CMS2097 | PIC3611 ∆eepR | [8] |
| CMS2204 | PIC3611 ∆eepR ∆hexS | This study |
| Nima | Wild-type ‘parental strain’ | Pryce Haddix |
| CMS2089 | Nima ∆eepR | [8] |
| CMS2320 | Nima ∆eepR ∆hexS | This study |
| CMS3125 | CMS2922 with pMQ294 integrated at hexS | [17] |
| Plasmids | ||
| pMQ131 | oripBBR1 aphA-3 oriT URA3 CEN6/ARSH4 | [18] |
| pMQ236 | oriR6K nptII rpsL oriT URA3 CEN6/ARSH4 I-SceI site | [18] |
| pMQ240 | oripSC101tsaacC1 oriT Plac-I-SceI URA3 CEN6/ARSH4 | [18] |
| pMQ248 | pMQ131 with PflhDC-lacZ | [3] |
| pMQ292 | pMQ131 with hexS | [17] |
| pMQ294 | pMQ236 with hexS wild-type allele | [17] |
| pMQ296 | pMQ236 with hexS-∆1 mutant allele | [17] |
| pMQ361 | pMQ131 with PnptII-tdtomato | [21] |
| pMQ407 | pMQ131 with lrhA from E. coli | This study |
| pMQ412 | pMQ361 with PeepR-tdtomato | [8] |
*Invitrogen, Waltham, MA, USA; Epicentre Biotechnologies, Madison, WI, USA; Presque Isle Cutlures, Erie, PA, USA; Pryce Haddix, Auburn University at Montgomery, Montgomery, AL, USA.
Mutagenesis and genetic manipulations
Transposon mutagenesis was performed as previously described [13] using Himar1 delivery plasmids pBT20 [14] and pSC189 [15]. Transposons were mapped by arbitrary PCR [16] or marker rescue [15]. After eight mutations were mapped to the hexS gene, primers that amplify the hexS ORF were used to screen transposon mutants with desired phenotypes. The primer sequences were GTTATTCTTCTTCGTCCACCAGGCTGG and ATGACAACTGCAAATCGTCCGATACTTAATCTCG (all primer sequences are shown 5′ to 3′).
The hexS gene was mutated by allelic replacement as previously described using plasmid pMQ296 [17]. The pMQ296 plasmid was introduced into strains CMS2089 and CMS2097 by conjugation and was resolved using pMQ240, an I-SceI delivery plasmid [18]. The hexS mutation was screened for by hyper-pigment phenotype, followed by PCR amplification and sequencing of the hexS gene to verify the hexS-∆1 mutation. This mutation deletes one base pair of the hexS ORF causing a frameshift mutation and a null allele [17].
The lrhA gene was amplified from E. coli strain S17-1λpir [19] using Phusion high-fidelity polymerase (New England Biolabs) and primers cgacggccagtgccaagcttgcatgcctgcaggtcgacT-TACTCGATATCCCTTTCAATC and gtggaattgtgagcggataacaatttcacacggaaacagATGATAAGTGCAAATCGTCC. The lower-case nucleotides target recombination and the upper-case letters direct amplification of the lrhA ORF, which was placed under control of the E. coli lac promoter on pMQ131 using yeast recombineering techniques [18, 20]. The resulting plasmid pMQ407 was introduced into S. marcescens by conjugation.
Mass spectrometry
Serratamolide analysis was performed as described previously [8, 21]. Bacteria were grown in LB medium for 20 h in 10×5 ml cultures per genotype and pooled. Cultures were centrifuged for 10 min at 10 000 g and 50 ml of the supernatant was extracted three times with an equal volume of ethyl acetate. The extract was dried over sodium sulphate and evaporated in vacuo and the residue was dissolved in methanol and analysed by HPLC-MS (Shimadzu LCMS-2020) equipped with a DIONEX Acclaim 120C18 column (3 µm particle size, 120 Å pore size, 2.1×150 mm dimensions). A previously described [8], mobile-phase gradient was used along with a column flow rate of 0.2 ml min−1 at 40 °C. Serratamolide was monitored at m/z=515 with an ES-MS detector at positive mode, and purified serratamolide [21] was used as a positive control. The experiment was performed three times using independent bacterial cultures.
Gene expression analysis and electrophoretic mobility shift assays
β-Galactosidase assay: Bacteria with a plasmid-borne flhDC-lacZ transcriptional reporter, pMQ248, were grown in LB with kanamycin (100 µg ml−1) overnight and then subcultured 1 : 100 into the same medium. After 20 h, samples were taken and the OD600 reading was determined with a spectrophotometer (Spectronic 200, Thermo Scientific). β-Galactosidase activity was determined as described by Griffith and Wolf [22].
Tdtomato assay: Bacteria with a plasmid-based eepR promoter fusion to tdtomato, pMQ412, were grown under the same experimental conditions described for the β-galactosidase assays noted above, and Tdtomato fluorescence was read as previously described [21] with a plate reader (Biotek, Synergy 2).
RNA preparation and quantitative reverse transcriptase PCR (qRT-PCR) were performed as previously described [8]. Primers for eepR (GGATTGGAAAACGTCAGCAT and CACGAAAAAGATGGCATCAC) and hexS (CGTTAAAGCGCAGGATCTTC and AAGAACCTTTGTTGCGGTTG) were designed to amplify DNA from the deletion alleles (all primers are listed as 5′ to 3′). Primer sequences for 16S and prtS analysis were noted in Brothers et al. [7]. Electrophoretic mobility shift assay (EMSA) reactions were performed with a commercial EMSA kit (Lightshift Chemiluminescent EMSA kit, Pierce) using previously described reagents (purified protein and promoter regions) and conditions [3, 8, 23]. The hexS promoter region was amplified using primers CCCGCGTTCTATAAGCACC and GCTCTAATCGCTGCATTTGTTG. The amplicon is 345 bp in length and includes 194 bp upstream of the hexS ORF that contains a predicted promoter determined using Softberry BPROM promoter prediction software. The eepR, flhDC and prtS promoter regions were as described previously [3, 8, 23]. Each EMSA experiment was performed three to six times with consistent results.
Statistical analysis
GraphPad Prism software was used for statistical analysis with significance set to P<0.05. Mann–Whitney U-tests were used for gene expression comparison and ANOVA with Tukey’s post-test was used for other experiments as noted.
Results
Suppressor analysis of the eepR mutant pigment defect
To gain insight into mechanisms by which EepR regulates pigment and co-regulated factors, suppressor analysis was performed. Random transposon mutations were introduced into a previously described ∆eepR mutant strain [8] that is pigmentless. Eighteen pigmented colonies were isolated from among 32 independent mutagenesis pools representing approximately 50 400 mutant colonies. A sample mutant screen plate is shown in Fig. 1(a). Pigmented strains were noted as red eepR suppressors (reep). A maximum of one pigmented colony was taken from each mutagenesis pool to eliminate sibling mutant colonies.
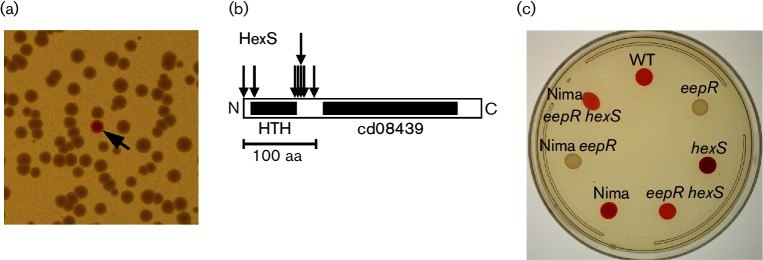
Fig. 1.
Genetic screen for eepR suppressor mutations. Transposon mutations were introduced into the CMS2097 strain (∆eepR) to find pigmented suppressor mutants. (a) A portion of one plate is shown with one red suppressor (reep) mutant (black arrow) visible among the pigmentless ∆eepR colonies. (b) Location of ∆eepR suppressor mutations (vertical arrows) in the hexS gene (horizontal bar). Of the 12 insertions, 8 are shown, the other 4 are in the hexS ORF but not mapped. (c) Prodigiosin pigmentation of strains grown on LB agar for 20 h at 30 °C. WT refers to parental strain PIC3611; eepR, to CMS2097; hexS, to CMS2922; eepR hexS, to CMS2204; Nima, to parental strain CMS1787; Nima eepR, to CMS2089; and Nima eepR hexS, to CMS2320.
Transposon insertion sites were mapped in the majority of the reep strains. The mutations mapped to one of two locations: in the hexS ORF and upstream of the eepR ORF. This manuscript will describe the genetic interactions and transcriptional regulation of eepR and hexS. The mutations upstream of the eepR ORF will be described in a separate study.
Eight mutants had transposon insertions in the hexS gene at base pairs 1, 45, 210, 213, 214, 214, 265 and 292. Four more had mutations in the hexS ORF whose specific insertion sites were not mapped, as noted in Methods. The specifically mapped mutations clustered near the N-terminus of the HexS protein proximal to a helix–turn–helix domain, whereas none was isolated in a predicted cd08439 (substrate-binding domain) in the C-terminus. HexS is a LysR family transcription factor that directly and negatively regulates prodigiosin and serratamolide production by S. marcescens [3, 4, 17]. HexS is closely related to the LrhA protein of E. coli [24]. LrhA homologues, found in a variety of micro-organisms including E. coli (LrhA), Erwinia species (HexA and PecT), Serratia ATCC 39006 (PigU) and Yersinia pseudotuberculosis (RovM), are involved in regulation of secreted enzymes, motility and virulence [25–29].
The robust pigment phenotypes observed in this screen suggest a regulatory relationship between EepR and HexS in the coordination of secondary metabolite biosynthesis. We investigated whether this relationship went beyond prodigiosin, as previous studies demonstrate that EepR positively and HexS negatively regulates biosynthesis of the secondary metabolite serratamolide and proteases [4, 8].
Opposing control of serratamolide and serralysin biosynthesis by EepR and HexS
An eepR hexS double mutant strain was built incorporating the previously described hexS-∆1 null mutation [17] by allelic exchange into the ∆eepR strain background. The double mutant strain was used for epistasis analysis to explore the relationship between EepR and HexS in coordinated regulation of secondary metabolites and secreted enzymes.
Introduction of the hexS mutation into the ∆eepR strain suppressed the pigment-defective phenotype of the eepR mutant (Fig. 1c). There were no obvious phenotypic differences observed between the reep mutants or between the reep mutants and the directed eepR hexS double mutant (CMS2204). Serratamolide is required for haemolysis and swarming phenotypes of many strains of S. marcescens, and PIC3611 harbouring the ∆eepR mutation (strain CMS2097) is unable to accomplish either phenotype due to a severe deficiency in serratamolide biosynthesis [8]. Both swarming ability and haemolysis were restored in the eepR hexS double mutant indicating that the double mutant synthesizes serratamolide (Fig. 2a, b). Swarming zone radii measured at 24 h for strains in the PIC3611 strain background were observed to be 3.0±1.7 mm for wild-type, 19.5±2.3 mm for ∆hexS, 0±0 for ∆eepR and 4.8±1.9 mm for ∆eepR ∆hexS. Importantly, these strains grew at similar rates indicating that the difference in motility is not due to altered growth by the mutant strains (Fig. 3a).
Fig. 2.
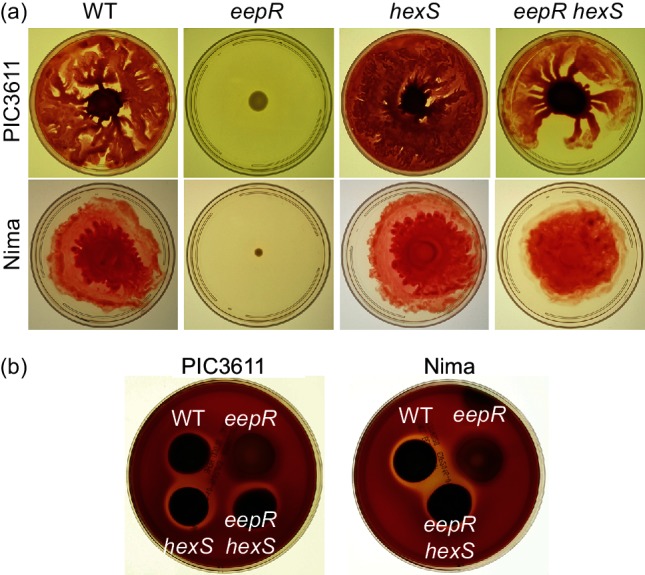
Genetic evidence suggests coordinated swarming and haemolysis regulation by EepR and HexS. (a) Swarming motility after incubation for 20–48 h on LB medium with 0.6 % agar. (b) Haemolysis phenotype after 4 days of growth on tryptic soy agar +5 % sheep erythrocytes.
Fig. 3.
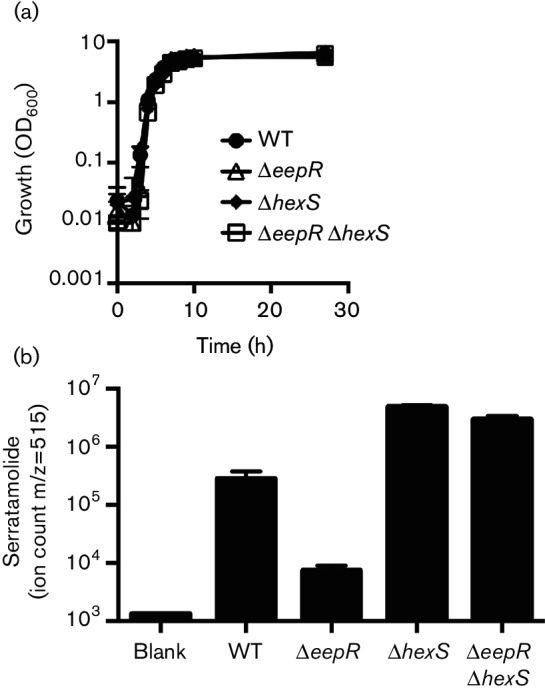
Growth and MS analysis of serratamolide production by the eepR hexS double mutant. (a) Growth curve analysis of the wild-type strain PIC3611, ∆eepR, ∆hexS and ∆eepR ∆hexS in LB medium. (b) MS analysis of serratamolide biosynthesis of cultures grown for 24 h and normalized to OD600 2.0. Means and standard deviations are shown, n=3 independent samples. PIC3611-derived strains were used.
MS analysis was used to measure serratamolide production in the wild-type (PIC3611) and derived strains. Compared to the wild-type, increased levels of serratamolide were measured in the hexS and double mutant, and reduced serratamolide in the eepR mutant (Fig. 3b). The eepR hexS double mutant produced serratamolide similarly to the hexS mutant, both significantly more than the ∆eepR mutant (P>0.001, ANOVA with Tukey’s post-test). These data indicate epistasis of the hexS mutant phenotype over the eepR mutant phenotype for serratamolide biosynthesis.
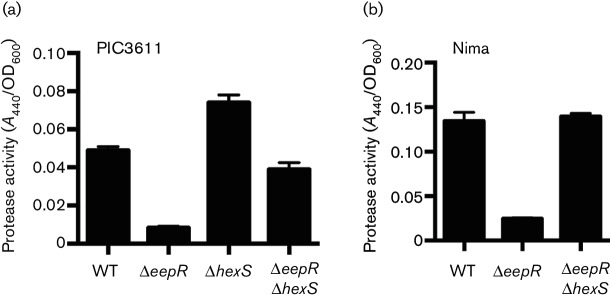
Since EepR positively regulates secreted enzymes such as the cytotoxic metalloprotease serralysin (PrtS) [7] and HexS has been reported to regulate undetermined secreted protease(s) [4], we tested whether the overlapping regulons of HexS and EepR extend to secreted protease activity. Azocasein was used as a quantitative substrate to detect proteases in normalized stationary-phase culture filtrates. Similar to a study by Tanikawa et al. [4] who used a different strain background, the hexS mutant version of strain PIC3611 exhibited elevated protease activity. Unlike the eepR mutant, the eepR hexS double mutant produced protease activity similar to wild-type (Fig. 4a).
Fig. 4.
Genetic evidence suggests coordinated protease regulation by EepR and HexS. Protease activity in supernatants from stationary-phase bacterial cultures normalized to OD600 2.0. Azocasein was used as a colorimetric protease substrate. Means and standard deviations are shown, n=3 independent experiments, each with three biological replicates. (a) Protease activity from the PIC3611 background. (b) Protease activity from the Nima strain background.
To ensure that this genetic interaction was not specific to strain PIC3611, we generated an eepR hexS double mutant variant of strain Nima [30]. Nima and PIC3611 are of different biotypes [31]. Pigment, serratamolide and protease activity were restored in a manner similar to strain PIC3611 and indicate that the relationship between HexS and EepR is not strain specific (Figs 1c, 2 and 4b).
Analysis of gene regulation by EepR and HexS
The suppression of several eepR mutant phenotypes by mutation of hexS led us to test whether HexS regulates eepR gene expression. The expression of eepR was increased 2.4-fold in the ∆hexS mutant compared to the wild-type (P<0.001, Fig. 5a). This trend of increased eepR expression was reversed when hexS was added to the chromosome in cis using pMQ294 as previously described [17] (P<0.01, ANOVA with Tukey’s post-test; Fig. 5a). As a second way to measure the impact of hexS mutation on eepR expression, a PeepR–tdtomato fusion was employed. Significant 4.8-fold and 3.1-fold increases in eepR expression were measured in the ∆hexS mutant relative to wild-type when measured at OD600 1.2 and 3, respectively (P<0.05), data not shown. These results suggest that HexS inhibits eepR gene expression.
Fig. 5.
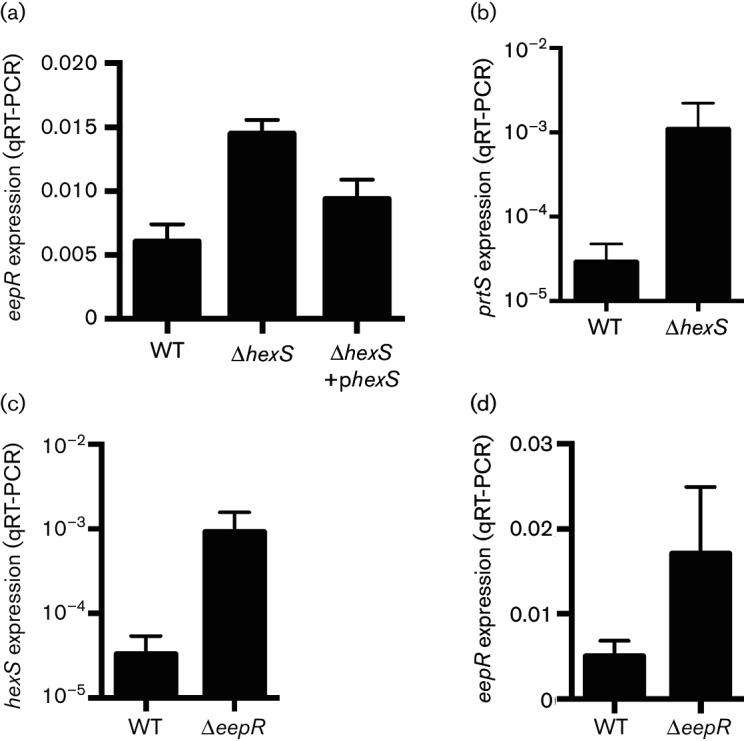
Genes regulated by HexS and EepR. (a–d) qRT-PCR analysis of gene expression using RNA from strains grown to OD600 3. (a, b) Analysis of eepR and prtS expression in WT compared to the ∆hexS mutant; (c, d) analysis of hexS and eepR expression in WT compared to the ∆eepR mutant. Gene expression was determined using PIC3611-derived strains. The phexS plasmid refers to pMQ294. Means and standard deviations are shown. At least three independent replicates were used for each experiment.
Experiments were carried out using qRT-PCR to test whether HexS mediates hexS expression. There was a non-significant twofold decrease in expression of the hexS gene in the hexS mutant strain compared to the wild-type strain when measured at OD600 3 (0.029±0.012 for the wild-type and 0.014±0.002 for the hexS mutant, P=0.10).
A hexS mutant is known to produce more extracellular protease [4, 8], but the specific protease was not determined. EepR is known to regulate the serralysin protease, coded for by the prtS gene [7]. Given the potential overlap of the EepR and HexS regulons, we tested whether mutation of hexS changes prtS expression. A 37-fold increase in prtS expression (P=0.009) was measured by qRT-PCR from the ∆hexS mutant compared to the wild-type at OD600 3 (Fig. 5b).
The role of EepR in transcriptional regulation of hexS and eepR was also tested. The hexS gene was elevated in expression 28-fold in the eepR mutant compared to the wild-type at OD600 3 (Fig. 5c, P=0.0006). The eepR gene was also elevated in the eepR mutant, but only by 3.4-fold (P=0.016, Fig. 5d).
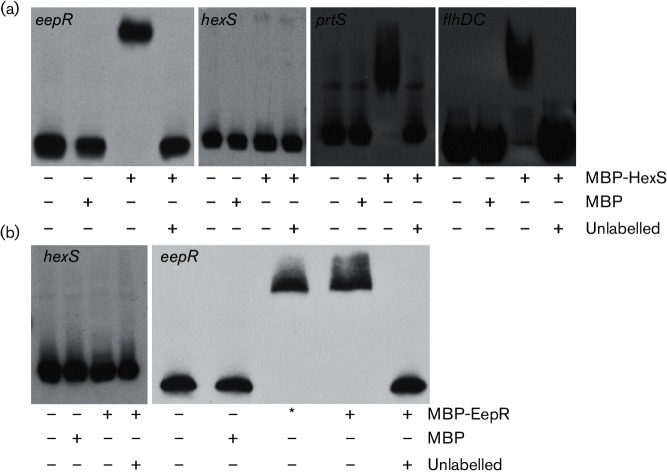
EMSA analysis was used to examine whether HexS and EepR directly regulated the genes tested for expression above. Formerly described maltose binding protein (MBP) protein fusions to EepR and HexS were used at previously optimized concentrations in promoter binding experiments; the MBP domain was used to affinity purify the fusion proteins and complementation analysis indicated that the fusion proteins retained functionality [3, 7, 8, 17]. Whereas MBP itself did not bind to the eepR promoter, the MBP–HexS fusion caused a gel shift of the biotinylated eepR promoter that could be inhibited by an excess of unlabelled eepR promoter DNA (Fig. 6a). Recombinant HexS did not bind to the hexS promoter region, suggesting that HexS does not directly regulate expression of the hexS gene (Fig. 6a). The absence of MBP–HexS binding to the hexS promoter also serves as a control for HexS binding specificity. Since HexS regulates secreted protease activity, we tested whether HexS bound to the prtS promoter, and we found evidence indicating that HexS could bind to the prtS promoter in vitro (Fig. 6a). Recombinant EepR bound to the eepR promoter, but not the hexS promoter region (Fig. 6b). The binding of EepR to eepR appears to be specific as the binding could be outcompeted with unlabelled eepR promoter sequence and recombinant EepR did not bind to the hexS promoter DNA. These data suggest that HexS directly regulates eepR and prtS, but not hexS expression, and that EepR directly regulates eepR expression but indirectly regulates hexS transcription.
Fig. 6.
EepR and HexS promoter binding analyses. EMSA analysis with biotinylated promoter DNA. Labelled promoter DNA was used in each reaction at 2 ng per reaction. Specific promoters are noted in the upper left-hand corner of each panel. MBP and MBP–HexS were used at 38 µM and 28 µM, respectively. MBP–EepR was used at 17.5 µM (*) or 35 µM (+); 500 ng of poly-dIdC was added per reaction to prevent non-specific protein–DNA interactions, and unlabelled promoters were used at 500 ng. –, indicates no addition of a particular reagent. (a) Representative EMSA using recombinant MBP or MBP–HexS. (b) Representative EMSA using recombinant MBP or MBP–EepR. Each EMSA experiment had a consistent result in at least three independent experiments.
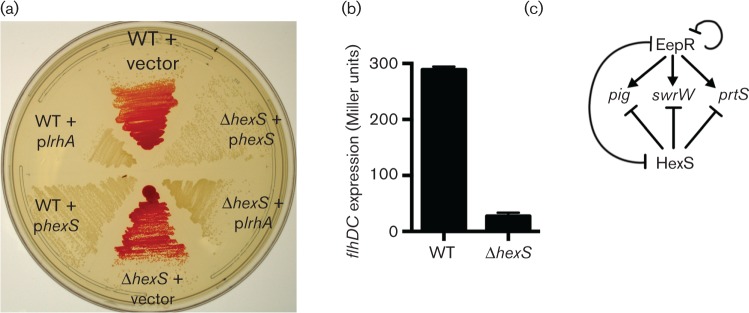
Mulitcopy expressions of hexS and lrhA inhibit flagellum and prodigiosin biosynthesis and reveal a functional conservation
As noted above, HexS is similar to LrhA from E. coli. blastp analysis [32] indicates a 69 % amino acid sequence identity between HexS and LrhA. To test whether the HexS protein and LrhA are functionally related, we cloned the lrhA gene under control of the E. coli Plac promoter and moved it into S. marcescens. Wild-type S. marcescens bearing hexS and lrhA on a medium-copy plasmid were both able to completely inhibit prodigiosin pigment production, whereas the vector control did not (Fig. 7a). This suggested remarkably conserved function as E. coli does not have the prodigiosin biosynthetic operon and yet multicopy expression of lrhA could impair pigment production similar to multicopy expression of hexS. Thus, former studies performed with LrhA in E. coli may give insight into other roles of HexS in S. marcescens. For example, LrhA regulates flhDC expression in E. coli [33–35]. FlhD and FlhC are the master regulators of flagellum biosynthesis and control biosynthesis of phospholipase and other metabolites in Serratia species [23, 36, 37]. Therefore, we tested whether HexS also regulates flhDC. A plasmid-borne flhDC promoter–lacZ fusion transcriptional reporter construct was introduced into the hexS mutant and isogenic wild-type strain. β-Galactosidase activity was >10-fold higher at OD600 3 in the wild-type compared to the hexS mutant suggesting positive regulation of flhDC by HexS (Fig. 7b). EMSA analysis supports direct regulation of the flhDC promoter by HexS (Fig. 6a). However, this reduction in flhDC expression did not result in a corresponding loss in swimming motility under the tested conditions: the wild-type had a 46±7 mm swim diameter and the hexS mutant had a 44±7 mm swim zone in 24 h (P=0.58, Student’s t-test).
Fig. 7.
Multicopy expressions of lhrA and hexS inhibit prodigiosin biosynthesis and flhDC expression in the hexS mutant. (a) Prodigiosin pigmentation of strains grown on LB agar with kanamycin for 20 h at 30 °C. Plasmids with the hexS or lhrA genes under control of the Plac promoter inhibit pigmentation in both the wild-type and the ∆hexS mutant. Experiments were performed with the PIC3611 strain background and vector indicates pMQ131. (b) β-Galactosidase activity produced by WT and ∆hexS strains bearing a plasmid-borne flhDC-lacZ transcriptional reporter after growth in LB medium to OD600 3. A representative experiment with three independent biological replicates is shown. Mean and standard deviation is shown. (c) Model, described in Discussion, for coordinated regulation of secondary metabolite biosynthetic genes (pigA-N and swrW) and the prtS protease gene by EepR and HexS. The line between EepR and HexS indicates that each inhibits transcription of the other.
Discussion
The EepR regulator of S. marcescens is a global positive regulator of secreted enzymes and secondary metabolites and is necessary for wild-type levels of virulence in a rabbit keratitis model [7, 8]. The goal of this study was to use genetic suppressor analysis to find other regulatory factors that coordinate with EepR in control of the EepR transcriptional regulon. Suppressor mutations of the ∆eepR mutant pigment phenotype mapped to the hexS ORF.
The results presented here indicate that the hexS mutations not only suppressed the pigment defect of the eepR mutation but also were able to reverse other eepR mutant defects including loss of protease production, serratamolide biosynthesis and associated phenotypes, haemolysis and swarming motility. Notably, this study demonstrates that the elevated protease activity due to undetermined protease(s) generated by the hexS mutants is due, at least in part, to elevated production of PrtS. Importantly, suppression of the eepR mutant defects by mutation of hexS was consistent in two different strain backgrounds suggesting that EepR and HexS have a conserved relationship in control of the tested phenotypes.
Transcriptional and EMSA analyses suggest that EepR directly and negatively regulates expression of the eepR gene but indirectly regulates hexS expression in a strong negative manner. Evidence presented here supports the model that HexS negatively regulates expression of the eepR and prtS, but HexS did not bind to the hexS promoter and mutation of hexS did not cause a significant change in hexS transcript. The observation that, in the hexS mutant, eepR expression is elevated suggests that the derepression of the eepR promoter by hexS mutation is dominant compared to the negative regulation imparted by increased EepR. Together, these data suggest a model, Fig. 7(c), in which EepR and HexS oppose one another in transcriptional control of secondary metabolites prodigiosin and serratamolide and of the cytotoxic protease serralysin (PrtS). In the absence of HexS, EepR is predicted to be made at higher levels, leading to increased pigmentation, serratamolide production and protease production. The opposite is also true where, in the absence of EepR, HexS is expected to be made at higher levels leading to a lack of secondary metabolite and protease production, as is seen in the eepR mutant. At this point, the signals stimulating EepR and HexS are unknown. EepR also appears to weakly inhibit the expression of the eepR gene, perhaps as a way to prevent the overproduction of energetically costly secondary metabolites, as overexpression of eepR has been shown to stimulate prodigiosin production [7, 8].
Lastly, the similarity between HexS and LrhA led to the surprising result that multicopy expression of lrhA was able to inhibit pigmentation in a similar manner to multicopy expression of hexS. This suggests that the two proteins have a highly conserved binding site and that genes controlled by LhrA in E. coli are likely to be controlled by HexS in S. marcescens. Unfortunately, the predicted LrhA binding site (AT-N9-AT) [34] is common in the S. marcescens genome. Nevertheless, as an example of how we can take advantage of this similarity, we observed that the flhDC operon is regulated by HexS in S. marescens and a similar manner by LhrA in E. coli. However, the flhDC expression deficit in the hexS mutant did not result in a reduction in swimming motility through semisolid agar or in a reduction in swarming motility; this may be due to differences in liquid versus solid medium conditions. It is known that S. marcescens without flagella can swarm under certain conditions [38]. Importantly, these results indicate that the EepR-HexS regulon extends to FlhDC-regulated genes including flagella and phospholipase A [36], all of which may contribute to a bacterium’s success in interspecies competition and pathogenesis.
Funding information
This work was funded by National Institutes of Health grants AI085570 (R. M. Q. S.) and EY024785 (K. M. B.), P30 grant EY08098, the Eye and Ear Foundation of Pittsburgh and unrestricted funds from Research to Prevent Blindness.
Acknowledgements
The authors thank James Fender and Denise Polaski at the University of Pittsburgh for technical assistance.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: EMSA, electrophoretic mobility shift assay; LB, lysogeny broth; MBP; maltose binding protein; qRT-PCR,quantitative reverse transcriptase PCR
Edited by: H. Gramajo and S. V. Gordon
References
- 1.Williamson NR, Simonsen HT, Harris AK, Leeper FJ, Salmond GP. Disruption of the copper efflux pump (CopA) of Serratia marcescens ATCC 274 pleiotropically affects copper sensitivity and production of the tripyrrole secondary metabolite, prodigiosin. J Ind Microbiol Biotechnol. 2006;33:151–158. doi: 10.1007/s10295-005-0040-9. [DOI] [PubMed] [Google Scholar]
- 2.Kalivoda EJ, Stella NA, Aston MA, Fender JE, Thompson PP, et al. Cyclic AMP negatively regulates prodigiosin production by Serratia marcescens. Res Microbiol. 2010;161:158–167. doi: 10.1016/j.resmic.2009.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shanks RM, Lahr RM, Stella NA, Arena KE, Brothers KM, et al. A Serratia marcescens PigP homolog controls prodigiosin biosynthesis, swarming motility and hemolysis and is regulated by cAMP-CRP and HexS. PLoS One. 2013;8:e57634. doi: 10.1371/journal.pone.0057634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tanikawa T, Nakagawa Y, Matsuyama T. Transcriptional downregulator hexS controlling prodigiosin and serrawettin W1 biosynthesis in Serratia marcescens. Microbiol Immunol. 2006;50:587–596. doi: 10.1111/j.1348-0421.2006.tb03833.x. [DOI] [PubMed] [Google Scholar]
- 5.Horng YT, Chang KC, Liu YN, Lai HC, Soo PC. The RssB/RssA two-component system regulates biosynthesis of the tripyrrole antibiotic, prodigiosin, in Serratia marcescens. Int J Med Microbiol. 2010;300:304–312. doi: 10.1016/j.ijmm.2010.01.003. [DOI] [PubMed] [Google Scholar]
- 6.Horng YT, Deng SC, Daykin M, Soo PC, Wei JR, et al. The LuxR family protein SpnR functions as a negative regulator of N-acylhomoserine lactone-dependent quorum sensing in Serratia marcescens. Mol Microbiol. 2002;45:1655–1671. doi: 10.1046/j.1365-2958.2002.03117.x. [DOI] [PubMed] [Google Scholar]
- 7.Brothers KM, Stella NA, Romanowski EG, Kowalski RP, Shanks RM. EepR mediates secreted-protein production, desiccation survival, and proliferation in a corneal infection model. Infect Immun. 2015;83:4373–4382. doi: 10.1128/IAI.00466-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stella NA, Lahr RM, Brothers KM, Kalivoda EJ, Hunt KM, et al. Serratia marcescens cyclic AMP receptor protein controls transcription of EepR, a novel regulator of antimicrobial secondary metabolites. J Bacteriol. 2015;197:2468–2478. doi: 10.1128/JB.00136-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Coulthurst SJ, Williamson NR, Harris AK, Spring DR, Salmond GP. Metabolic and regulatory engineering of Serratia marcescens: mimicking phage-mediated horizontal acquisition of antibiotic biosynthesis and quorum-sensing capacities. Microbiology. 2006;152:1899–1911. doi: 10.1099/mic.0.28803-0. [DOI] [PubMed] [Google Scholar]
- 10.Aubert DF, O'Grady EP, Hamad MA, Sokol PA, Valvano MA. The Burkholderia cenocepacia sensor kinase hybrid AtsR is a global regulator modulating quorum-sensing signalling. Environ Microbiol. 2013;15:372–385. doi: 10.1111/j.1462-2920.2012.02828.x. [DOI] [PubMed] [Google Scholar]
- 11.Bertani G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951;62:293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bertani G. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J Bacteriol. 2004;186:595–600. doi: 10.1128/JB.186.3.595-600.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shanks RM, Stella NA, Kalivoda EJ, Doe MR, O'Dee DM, et al. A Serratia marcescens OxyR homolog mediates surface attachment and biofilm formation. J Bacteriol. 2007;189:7262–7272. doi: 10.1128/JB.00859-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kulasekara HD, Ventre I, Kulasekara BR, Lazdunski A, Filloux A, et al. A novel two-component system controls the expression of Pseudomonas aeruginosa fimbrial cup genes. Mol Microbiol. 2005;55:368–380. doi: 10.1111/j.1365-2958.2004.04402.x. [DOI] [PubMed] [Google Scholar]
- 15.Chiang SL, Rubin EJ. Construction of a mariner-based transposon for epitope-tagging and genomic targeting. Gene. 2002;296:179–185. doi: 10.1016/s0378-1119(02)00856-9. [DOI] [PubMed] [Google Scholar]
- 16.O'Toole GA, Pratt LA, Watnick PI, Newman DK, Weaver VB, et al. Genetic approaches to study of biofilms. Methods Enzymol. 1999;310:91–109. doi: 10.1016/s0076-6879(99)10008-9. [DOI] [PubMed] [Google Scholar]
- 17.Stella NA, Fender JE, Lahr RM, Kalivoda EJ, Shanks RM. The LysR transcription factor, HexS, is required for glucose inhibition of prodigiosin production by Serratia marcescens. Adv Microbiol. 2012;2:511–517. doi: 10.4236/aim.2012.24065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shanks RM, Kadouri DE, Maceachran DP, O'Toole GA. New yeast recombineering tools for bacteria. Plasmid. 2009;62:88–97. doi: 10.1016/j.plasmid.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller VL, Mekalanos JJ. A novel suicide vector and its use in construction of insertion mutations: osmoregulation of outer membrane proteins and virulence determinants in Vibrio cholerae requires toxR. J Bacteriol. 1988;170:2575. doi: 10.1128/jb.170.6.2575-2583.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shanks RM, Caiazza NC, Hinsa SM, Toutain CM, O'Toole GA. Saccharomyces cerevisiae-based molecular tool kit for manipulation of genes from Gram-negative bacteria. Appl Environ Microbiol. 2006;72:5027–5036. doi: 10.1128/AEM.00682-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shanks RM, Stella NA, Lahr RM, Wang S, Veverka TI, et al. Serratamolide is a hemolytic factor produced by Serratia marcescens. PLoS One. 2012;7:e36398. doi: 10.1371/journal.pone.0036398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Griffith KL, Wolf RE. Measuring beta-galactosidase activity in bacteria: cell growth, permeabilization, and enzyme assays in 96-well arrays. Biochem Biophys Res Commun. 2002;290:397–402. doi: 10.1006/bbrc.2001.6152. [DOI] [PubMed] [Google Scholar]
- 23.Stella NA, Kalivoda EJ, O'Dee DM, Nau GJ, Shanks RM. Catabolite repression control of flagellum production by Serratia marcescens. Res Microbiol. 2008;159:562–568. doi: 10.1016/j.resmic.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gibson KE, Silhavy TJ. The LysR homolog LrhA promotes RpoS degradation by modulating activity of the response regulator sprE. J Bacteriol. 1999;181:563–571. doi: 10.1128/jb.181.2.563-571.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Castillo A, Reverchon S. Characterization of the pecT control region from Erwinia chrysanthemi 3937. J Bacteriol. 1997;179:4909–4918. doi: 10.1128/jb.179.15.4909-4918.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fineran PC, Slater H, Everson L, Hughes K, Salmond GP. Biosynthesis of tripyrrole and beta-lactam secondary metabolites in Serratia: integration of quorum sensing with multiple new regulatory components in the control of prodigiosin and carbapenem antibiotic production. Mol Microbiol. 2005;56:1495–1517. doi: 10.1111/j.1365-2958.2005.04660.x. [DOI] [PubMed] [Google Scholar]
- 27.Harris SJ, Shih YL, Bentley SD, Salmond GP. The hexA gene of Erwinia carotovora encodes a LysR homologue and regulates motility and the expression of multiple virulence determinants. Mol Microbiol. 1998;28:705–717. doi: 10.1046/j.1365-2958.1998.00825.x. [DOI] [PubMed] [Google Scholar]
- 28.Heroven AK, Dersch P. RovM, a novel LysR-type regulator of the virulence activator gene rovA, controls cell invasion, virulence and motility of Yersinia pseudotuberculosis. Mol Microbiol. 2006;62:1469–1483. doi: 10.1111/j.1365-2958.2006.05458.x. [DOI] [PubMed] [Google Scholar]
- 29.Surgey N, Robert-Baudouy J, Condemine G. The Erwinia chrysanthemi pecT gene regulates pectinase gene expression. J Bacteriol. 1996;178:1593–1599. doi: 10.1128/jb.178.6.1593-1599.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Green JA, Rappoport DA, Williams RP. Studies on pigmentation of Serratia marcescens. II. Characterization of the blue and the combined red pigments of prodigiosin. J Bacteriol. 1956;72:483–487. doi: 10.1128/jb.72.4.483-487.1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fender JE, Bender CM, Stella NA, Lahr RM, Kalivoda EJ, et al. Serratia marcescens quinoprotein glucose dehydrogenase activity mediates medium acidification and inhibition of prodigiosin production by glucose. Appl Environ Microbiol. 2012;78:6225–6235. doi: 10.1128/AEM.01778-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 33.Lee C, Park C. Mutations upregulating the flhDC operon of Escherichia coli K-12. J Microbiol. 2013;51:140–144. doi: 10.1007/s12275-013-2212-z. [DOI] [PubMed] [Google Scholar]
- 34.Lehnen D, Blumer C, Polen T, Wackwitz B, Wendisch VF, et al. LrhA as a new transcriptional key regulator of flagella, motility and chemotaxis genes in Escherichia coli. Mol Microbiol. 2002;45:521–532. doi: 10.1046/j.1365-2958.2002.03032.x. [DOI] [PubMed] [Google Scholar]
- 35.Mouslim C, Hughes KT. The effect of cell growth phase on the regulatory cross-talk between flagellar and Spi1 virulence gene expression. PLoS Pathog. 2014;10:e1003987. doi: 10.1371/journal.ppat.1003987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Givskov M, Eberl L, Christiansen G, Benedik MJ, Molin S. Induction of phospholipase- and flagellar synthesis in Serratia liquefaciens is controlled by expression of the flagellar master operon flhD. Mol Microbiol. 1995;15:445–454. doi: 10.1111/j.1365-2958.1995.tb02258.x. [DOI] [PubMed] [Google Scholar]
- 37.Hampton HG, Mcneil MB, Paterson TJ, Ney B, Williamson NR, et al. CRISPR-Cas gene-editing reveals RsmA and RsmC act through FlhDC to repress the SdhE flavinylation factor and control motility and prodigiosin production in Serratia. Microbiology. 2016;162:1047–1058. doi: 10.1099/mic.0.000283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Matsuyama T, Bhasin A, Harshey RM. Mutational analysis of flagellum-independent surface spreading of Serratia marcescens 274 on a low-agar medium. J Bacteriol. 1995;177:987–991. doi: 10.1128/jb.177.4.987-991.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]