Abstract
The flagellar system in Escherichia coli K12 is expressed under the control of the flhDC-encoded master regulator FlhDC. Transposition of insertion sequence (IS) elements to the upstream flhDC promoter region up-regulates transcription of this operon, resulting in a more rapid motility. Wang and Wood (ISME J 2011;5:1517–1525) provided evidence that insertion of IS5 into upstream activating sites occurs at higher rates in semi-solid agar media in which swarming behaviour is allowed as compared with liquid or solid media where swarming cannot occur. We confirm this conclusion and show that three IS elements, IS1, IS3 and IS5, transpose to multiple upstream sites within a 370 bp region of the flhDC operon control region. Hot spots for IS insertion correlate with positions of stress-induced DNA duplex destabilization (SIDD). We show that IS insertion occurs at maximal rates in 0.24 % agar, with rates decreasing dramatically with increasing or decreasing agar concentrations. In mixed cultures, we show that these mutations preferentially arise from the wild-type parent at frequencies of up to 3×10−3 cell−1 day−1 when the inoculated parental and co-existing IS-activated mutant cells are entering the stationary growth phase. We rigorously show that the apparent increased mutation frequencies cannot be accounted for by increased swimming or by increased growth under the selective conditions used. Thus, our data are consistent with the possibility that appropriate environmental conditions, namely those that permit but hinder flagellar rotation, result in the activation of a mutational pathway that involves IS element insertion upstream of the flhDC operon.
Keywords: IS-mediated directed mutation, flagellar master regulator, FlhDC, transposon, gene activation, SIDD
Introduction
A transposon, a jumping gene, can hop from one location to another in the genome of an organism [1]. Transposons thereby induce mutations, and the consequences can be beneficial or detrimental to the host cell [2–4]. Transposition occurs by a conservative or replicative mechanism and can be regulated by host DNA-binding proteins [5–8]. Reversible transposon insertion/deletion can result in phase variation of phenotypic properties such as metabolism, virulence and biofilm formation [9–11].
The smallest bacterial transposons, about 1 kb long, are the so-called insertion sequences or ‘IS elements’. Multiple IS elements found in Escherichia coli and other prokaryotes often play important roles in molecular and organismal evolution [4, 12]. They sometimes modify the expression of global regulatory networks and even change mutation rates [13]. Earlier studies had shown that IS insertions can activate genes adjacent to and usually downstream of the insertion sites by a number of distinct mechanisms [14–17]. For example, IS5 has been observed to be inserted in response to nutritional stress upstream of the E. coli fuc [18], bgl [19], flhDC [20] and glpFK operons [21–25].
Flagellar motility allows bacteria to reach more nutritive environments and escape from toxic or otherwise deleterious environments [26]. Flagellar synthesis and function are energetically expensive; thus, the flagellar system, encoded by more than 50 genes, is highly regulated. Flagellar operons and their promoters in E. coli are categorized into three classes, Class I, Class II and Class III, according to their expression timing [27]. In several bacteria including E. coli, the heterohexameric transcription factor, FlhD4C2 (hereafter referred to as FlhDC), is the only Class I protein and is the master regulator of the entire flagellar cascade [28].
Transcription of flhDC is regulated by many transcription factors; in fact, no other bacterial operon has been shown to be subject to as complex a control network. Protein transcription factors reported to control flhDC expression include RssAB, H-NS, AtoSC, CsgD, GlgS, MatA, YdiV, FmrA, YiiQ, LrhA, HdfR, OmpR, PapX, RcsB, CysB, PhoB, NtrC and the cAMP-Crp complex [16, 17, 27–37]. Small RNAs, ArcZ, OmrA, OmrB and OxyS, negatively regulate and McaS positively regulates flhDC expression and cell motility in Hfq-dependent processes by binding to the 5′ untranslated region of the flhDC mRNA [38, 39]. flhDC operon expression is also autoregulated by FlhDC [40].
Fahrner and Berg [28] isolated several mutations that enhanced motility and flhDC expression, showing that both IS and kanamycin resistance gene insertions into the upstream region can cause increased motility. None of these mutations eliminated the cAMP-Crp requirement, although several did eliminate the need for H-NS. These investigators concluded that the upstream region into which IS elements insert must represent a negative control region for flhDC expression.
Motility and flagellar synthesis are differentially regulated in a complex way during biofilm development [41]. FlhDC influences biofilm formation and virulence [42], although the FlhDC regulon is surprisingly restricted with respect to the types of genes included [27]. Thus, flagellar biosynthesis and function are sensitive to many external and internal conditions, serving several related functions.
As shown in Fig. S1 (available in the online Supplementary Material), the chromosome of E. coli K12 strain BW25113 has ten identical copies of IS5 (1195 bp long), six copies of IS1 (768 bp) and five copies of IS3 (1258 bp) [9]. When any one of these three IS elements transposes into the upstream region of flhDC, transcription of the operon is up-regulated, resulting in an increased transcription of Class II and Class III genes and a consequent increased motility (see Refs [17, 20, 26] and the present report). Usually, IS elements closest to the target site on the chromosome are involved in the transposition event.
On solid agar (1.5 %) plates, increased flagellar gene expression due to IS insertion upstream of the flhDC promoter does not allow E. coli to swarm, while in agitated liquid media, cells do not need flagella to move. In soft agar (<0.4 %) plates, however, wild-type cells migrate slowly through the gel [43], and these benefit from higher flagellar expression because the cells with more flagella have greater access to nutrients and can escape toxic end products of metabolism [17, 28, 44]. In this article, we shall refer to the rate of migration from the point of inoculation outward as the ‘colony expansion rate’ or ‘swarming rate’.
In their 2011 paper, Wang and Wood demonstrated the effects of different environmental conditions on the occurrence of IS5 hopping into sites upstream of the flhDC operon. First, they measured flhDC-activating IS5 hopping frequencies in soft agar plates (0.3 %). The E. coli K12 BW25113 strain, weakly motile without an IS element upstream of the flhDC promoter, was inoculated into soft agar plates, and the appearance of swarming zones was recorded. The insertion frequencies on solid agar plates (1.5 % agar) as well as in LB and minimal nutrient liquid media remained low compared to the frequencies observed in soft agar. Second, they tested the conditions essential for IS5 hopping. A flhD-deleted strain as well as motA and flgK mutants showed no swarming halo on soft agar, and no IS5 insertions in the flhD upstream region were identified. Third, biofilm formation and growth rates for the strains with or without IS5 upstream of flhD were measured. The IS5-activating insertion mutants gained a greater capacity for biofilm formation than the wild-type strain. In fact, IS5 hopped into the upstream region of flhD during biofilm formation, resulting in a subpopulation of more motile cells. Finally, evidence was presented suggesting that IS5 insertions upstream of flhD probably were not accompanied by a general increase in IS hopping into random insertion sites.
The observations reported by Wang and Wood [44] are consistent with the possibility that environmental conditions influence the rates of beneficial IS insertion mutations, and we sought to confirm and extend these results. We show here that, in soft agar plates, new insertions of IS elements preferentially arise as growth of the parental cells slows down and ceases. We further show that the IS insertional mutation frequencies are greatly enhanced in soft agar. The optimal agar concentration for promotion of IS insertional activation of flhDC proved to be 0.24 % with decreasing mutational frequencies at higher and lower concentrations. In a carefully controlled study, we found that new IS5 insertional mutants arise as a wild-type culture simultaneously inoculated with an IS5 insertional mutant approached stationary phase. Moreover, using an assay that does not depend on motility, we showed that mutations that eliminate flagellar structure or motor function prevent IS insertion upstream of the flhDC operon, in agreement with the results of Wang and Wood [44]. These findings argue against the possibility that the increased frequency of IS-activated colony expansion mutations is an artefact due to an increase in growth and/or motility.
Methods
Construction of bacterial strains
Strains and DNA oligonucleotides used in this study are described in Tables S1 and S2, respectively. All the strains were derived from E. coli K12 strain BW25113 [45]. To make the flhDC reporter strain, we first fused a promoter-less cat (encoding chloramphenicol acetyltransferase) structural gene (plus its own ribosomal binding site) to the 3′ end of the flhC gene. The resultant operon, flhDC : cat, is under the control of the flhDC promoter. To do this, the region containing the cat gene and the downstream FRT-flanking kanamycin resistance gene (kmr) in pKD-cat [23, 24] were amplified using oligos, FlhC.cat-P1 and FlhC.cat-P2 (Table S2). FlhC.cat-P1 is composed of a 20 bp region at the 3′ end that is complementary to the beginning of cat and a 50 bp region at the 5′ end that is homologous to the 3′ end of flhC. FlhC.cat-P2 is composed of a 20 bp region at the 3′ end that is complementary to the FRT-flanking kmr sequence and a 50 bp region at the 5′ end that is homologous to the flhC/motA intergenic region. The PCR products (i.e. ‘cat:kmr’) were gel purified and electroporated into BW25113 cells expressing the lambda Red proteins encoded by plasmid pKD46 [45]. The cells were applied onto LB+Km plates, and Kmr colonies were confirmed for the replacement of the flhC/motA intergenic region (−326 to −1 relative to the start codon of motA) by the ‘cat:kmr’ fragment by colony PCR, followed by DNA sequencing. The FRT-flanking kmr gene was removed by first transforming the temperature-sensitive pCP20 plasmid and then growing a transformant at 40 °C overnight. The resultant strain was named CAT_△AB (swarming negative).
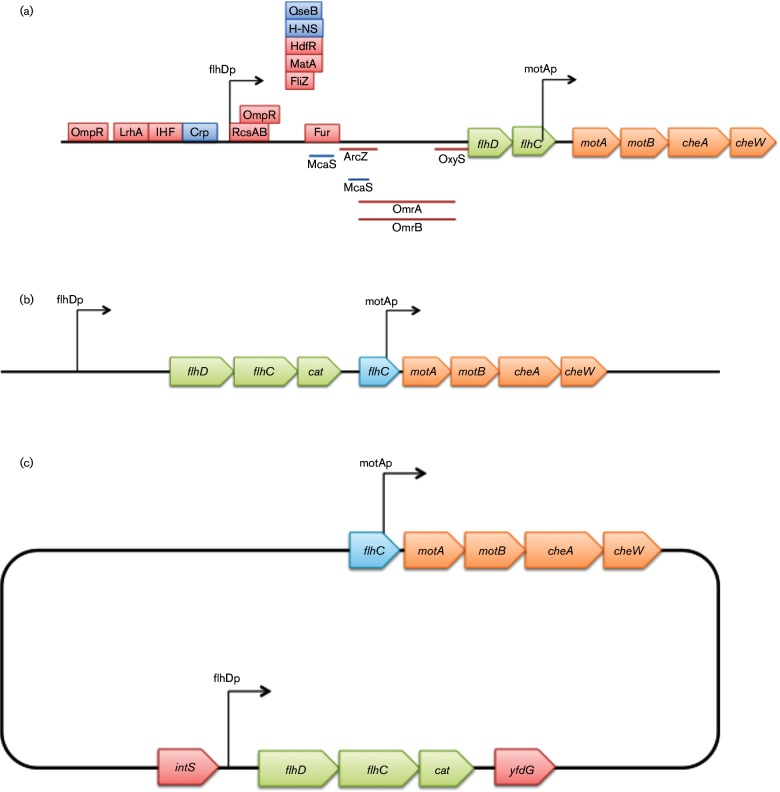
As shown in Fig. 1(a), the promoter region of the motAB operon is located within the 3′ end of flhC. The presence of cat in the flhC/motA intergenic region appears to abolish expression of the downstream motAB operon, since its promoter is destroyed. This strain, CAT_△AB, would be expected to be non-motile. To restore motility, the motAB promoter region was added back at its proper location in front of motA. To do this, a DNA fragment (PmotAB), containing the 3′ end of flhC and the flhC/motA intergenic region, was cloned into the XhoI/BamHI sites of pKDT, yielding pKDT-PmotAB. Present in this recombinant plasmid, the region (kmr : rrnBT : PmotAB) carrying kmr, the rrnB terminator (rrnBT) and PmotAB, was amplified using the primers PmotAB-P1 and PmotAB-P2 (Table S2). The amplified fragment, kmr : rrnBT : PmotAB, was electroporated into CAT_△AB cells to replace the region between the 3′ end of cat and the 5′ end of motA. The transformants were selected for Km resistance. The Kmr colonies were confirmed for such replacement by colony PCR and subsequently by DNA sequencing. The kmr gene was removed as above. This yielded the reporter strain, CAT, in which flhD, flhC and cat form a single operon that is under the control of the flhDC promoter, with motAB expression under the control of its own promoter (Fig. 1b). To create a second non-motile strain, the fliC mutation in strain JW1908 (The Coli Genetic Stock Center #9586) was transferred into CAT by P1 transduction, yielding strain CAT_ΔfliC.
Fig. 1.
Schematic diagram of the flhDC and motAB/cheAW operons and constructs used in the present study. (a) Schematic diagram of the wild-type flhDC and motAB/cheAW operons. The flhD and flhC genes are transcribed from one promoter while the motA, motB, cheA and cheW genes are transcribed from a second promoter located in the 3′ region of flhC. Red rectangles indicate binding sites for some negative regulators, and blue rectangles indicate the same for certain positive regulatory proteins. The lines below the gene depictions denote binding sites for small RNAs. The two arrows indicate the promoters. (b) Schematic diagram of the constructs used to understand the regulated expression of the flhDC operon in the CAT strain. The cat gene is fused immediately after the flhC gene, yielding the flhDC : cat operon that is under the control of the native flhDC promoter. The 3′ region of the flhC gene bearing the motA promoter was fused to the motA gene. This promoter is therefore between the cat and motA genes so that the downstream genes are expressed under the control of the native promoter. (c) Schematic diagram of the CAT2 strain. The region containing the flhDC promoter and the flhDC:cat operon were moved into the intS/yfdG intergenic region (bottom). The original flhDC promoter, flhD and the 5′ part of flhC were deleted (top).
In some cases, we needed to distinguish IS insertional mutants that arose after plating from the insertional mutants initially added. For this purpose, to facilitate isolation of IS insertional mutants, we inserted a kmr marker into the ycaC/ycaD intergenic region. The kmr gene was amplified from pKD4 using primers ycaD-P1 and ycaD-P2 (Table S2). The PCR products were purified and inserted into the ycaC/ycaD intergenic region of CAT. The resultant strain, which is kanamycin resistant, was named CAT_KmR.
As an alternative, we made a second flhDC reporter strain in which the flhD/flhC/cat operon, under the control of the native flhDC promoter (PflhDC-flhDC :cat), was moved to another chromosomal location. To do this, the region containing the native flhDC promoter, flhD and the 5′ end of flhC (but not the 3′ end of flhC containing the motAB promoter region), was deleted. flhDC:cat was amplified from the genomic DNA of the first reporter strain CAT using primers PflhDC-Xho-L and Cat-Sal-R (Table S2), digested with XhoI and SalI, then ligated into the same sites of pKDT, yielding pKDT-PflhDC-flhDC:cat. Using this plasmid as template, the region carrying kmr:rrnBT:PflhDC-flhDC:cat was amplified using primers IntC-P1 and IntC-P2. The PCR products were purified and then electroporated into BW25113 cells. The integration of ‘kmr:rrnBT:PflhDC-flhDC:cat’ into the intS/yfdG intergenic region was confirmed by colony PCR and DNA sequencing. The resultant strain was named CAT2, in which the native flhDC promoter, flhD and the 5′ part of flhC were deleted, and a new copy of the same promoter driving the flhDC:cat operon was located in the intS/yfdG intergenic region (Fig. 1c).
Swarming (colony expansion) rate measurements
A total of 2 wild-type strains and 13 IS insertional mutant strains were each grown in 3 ml of liquid medium (0.5 % tryptone and 0.5 % NaCl) at 30 °C overnight and diluted with an M9 salts solution to an optical density of 1.0. Then 1.5 µl of each culture was used to inoculate the centre of a soft agar plate (0.5 % tryptone, 0.5 % NaCl and 0.3 % agar). The plates were incubated at room temperature, and the diameters of the swarming zones were measured from 8 to 17 h after inoculation. Graphs of the diameters were plotted as a function of time, and the colony expansion (swarming) rates were calculated from the slopes.
The experiment was repeated with glucose (0.5 %) present in the medium. Before inoculating the soft agar plates, the strains were grown in liquid media (0.5 % tryptone, 0.5 % NaCl and 0.5 % glucose). The soft agar plates used were made with 0.5 % tryptone, 0.5 % NaCl, 0.5 % glucose and 0.3 % agar. These plates were treated in the same way as for the experiments without glucose. The slopes were used to determine the rates of colony expansion for each strain, with and without glucose.
Growth rates
Growth rates of the E. coli BW25113 flhDC:cat:‘flhC:motAB (CAT) strain as well as CAT_IS1 and CAT_IS5 were measured in liquid media (0.5 % tryptone, 0.5 % NaCl and 0 % agar), on solid agar plates (0.5 % tryptone, 0.5 % NaCl and 1.5 % agar) and on soft agar plates (0.5 % tryptone, 0.5 % NaCl and 0.3 % agar). Each strain was grown in 3 ml of liquid medium (0.5 % tryptone and 0.5 % NaCl) at 30 °C overnight, and the cultures were used for growth rate assays.
For liquid media, 5–10 µl of the overnight culture was added to 5 ml of medium. The tubes were incubated in a 30 °C shaking water bath for up to 24 h. Every 3 h, 100 µl of the cultures was removed, and the OD600 was measured to plot the growth curves.
For solid agar plate assays, 4 µl of the overnight culture (optical density of 1.0) was inoculated in a single streak across the plate and incubated in a 30 °C incubator. At every sample point, the solid agar plates were washed with 3 ml of 1× M9 buffer and the cell slurry was collected in a microcentrifuge tube. Then the cell culture was diluted with the same buffer to spread on LB plates. These plates were incubated at 37 °C before the colonies were counted.
The soft agar plates were inoculated in the same way as the solid agar plates. Every 3 h, the gel was scraped off into a large test tube, each plate was rinsed with 3 ml of M9 buffer, and the wash buffer was added to the same tube. The gel was vortexed for 2 min to mix the preparation, and the resultant suspension was centrifuged at 800 r.p.m. for 30 s to separate the gel from the buffer. Such a centrifugation procedure was shown to remove only a small fraction (<10 %) of the bacterial cells. The cells were collected in a microcentrifuge tube and diluted to plate onto LB plates. The plates were incubated at 37 °C to measure the cell populations.
Using soft agar plates, the growth rates of CAT_KmR (wild-type) and CAT_ IS5 were measured when these two types of cells were mixed before inoculation. The overnight cultures were diluted with M9 buffer, and CAT_KmR (2×109 c.f.u. ml−1) and CAT_ IS5 (2×106 c.f.u. ml−1) were mixed in equal volumes. Then, the mixture (4 µl) was inoculated in a single streak across the centre of the soft agar plates and incubated at 30 °C. Every 3 h, the cell samples were prepared as described above for the single-strain soft agar assay. They were appropriately diluted before being applied to LB plates, LB+Cm plates (LB with 6 µg ml−1 chloramphenicol) and LB+Cm+Km plates (LB with 6 µg ml−1 chloramphenicol and 25 µg ml−1 kanamycin). These plates were incubated in a 37 °C incubator before counting colonies. The numbers of colonies on LB plates gave the total population, that on LB+Cm plates gave the total population of the inoculated insertion mutant and that on LB+Cm+Km gave the population of the newly emerging mutants. The growth rate of the IS5 insertional mutant that was added at the beginning was calculated by subtracting the newly emerging population from the total insertion mutant population.
Mutant ratios
Mutant ratios were measured for three media, the same as for the growth rate experiments. The CAT strain was grown in 3 ml of liquid medium in a 30 °C water shaker overnight and diluted with M9 buffer to an optical density of 1.0.
For liquid media, 5–10 µl of the overnight culture was inoculated into 5 ml of medium and incubated in a 30 °C shaker. At four different time points, the culture was plated onto LB plates (diluted with M9 buffer) and LB+Cm plates (undiluted). The mutant ratio was calculated by dividing the mutant population by the wild-type population.
For solid agar plates, 4 µl of an overnight wild-type culture (optical density of 1.0) was plated in a single streak across the plate and incubated in a 30 °C incubator. At four different time points, each plate was washed with 3 ml of M9 buffer and plated onto LB plates (diluted with 1× M9 buffer) and LB+Cm plates and was incubated in a 37 °C incubator. The colonies were counted, and the ratios of the total population of wild-type to mutant cells were determined.
Soft agar plates were inoculated and incubated in the same way as solid agar plates. Every 3 h, the agar was collected in a test tube and the plates were washed with 3 ml of M9 buffer. The collected agar was mixed by vortexing and centrifuged as described above, and the cultures were diluted to plate onto LB and LB+Cm plates.
Mutant ratios were determined for another wild-type strain, CAT2, using these three media at two temperatures, 30 °C and 37 °C. The strain was grown overnight in 3 ml of liquid medium, diluted with M9 buffer to an optical density of 1 and used to inoculate fresh media and incubate as described above. For this strain, a higher concentration of chloramphenicol (LB with 14 µg ml−1 chloramphenicol) was used for mutant isolation.
Mutant ratios were determined for another two non-motile strains, CAT_ΔAB and CAT_ΔfliC, using soft agar plates [0.4 % nutrient broth (NB)+0.3 % agar]. The strains were cultured in 0.4 % NB overnight and diluted to OD600 of 1.0, and 1.4 µl of the diluted cells was streaked onto agar plates. The plates were incubated at 30 °C. After 18 h, the total and mutant populations were determined as described above.
To determine whether agar concentrations affected the appearance of swarming mutants, the cells of strain CAT were inoculated onto NB agar plates that contained 0.4 % NB and 0–1.2 % agar. After the plates were incubated at 30 °C for 18 h, the total and mutant populations were determined. For the plates with 0–0.4 % agar, the populations were determined as described for soft agar plate assays as above. For the plates with 0.6 % agar and above, the cells were washed off with 3 ml of M9 buffer without breaking the agar. The total populations and mutant populations were determined using LB and LB+Cm plates as above. Three plates were used for each concentration of agar.
Original locations of IS elements in the chromosome
NCBI blast was used to identify the locations and directions of original IS elements on the chromosome of E. coli K12 strain BW25113. Then, the relative locations were plotted on the circular diagram of the chromosome. In Fig. S1, the blue arrows indicate the IS1s; purple, the IS3s; and green, the IS5s. The arrowheads indicate the direction of transcription of the transposase gene.
Determining DNA destabilization energy plots for the flhDC upstream region
Destabilization energy G(x) is the incremental energy (kcal mol−1) needed to guarantee separation of base pair x under defined superhelical stress. Highly destabilized DNA regions (referred to as SIDD for superhelicity-induced DNA duplex destabilization) have lower values of G(x), and more stable regions have higher G(x) values [46]. Destabilization energy calculations were made by using the SIST programmes provided by Professor C. J. Benham [47] using default settings to be described elsewhere (M. Z. Humayun, Z. Zhang and M. H. Saier, unpublished) for a 4.4 kb DNA segment containing flhD flanked by 2000 bp upstream of the start codon for flhD and 2000 bp downstream of the stop codon for the flhD gene (base pairs 1 970 104 to 1 974 454 from the E. coli BW25513 genomic sequence, NCBI Reference NZ_CP009273.1). For clarity, Fig. 5 shows the plot for a 2.4 kb portion of the sequence.
Fig. 5.
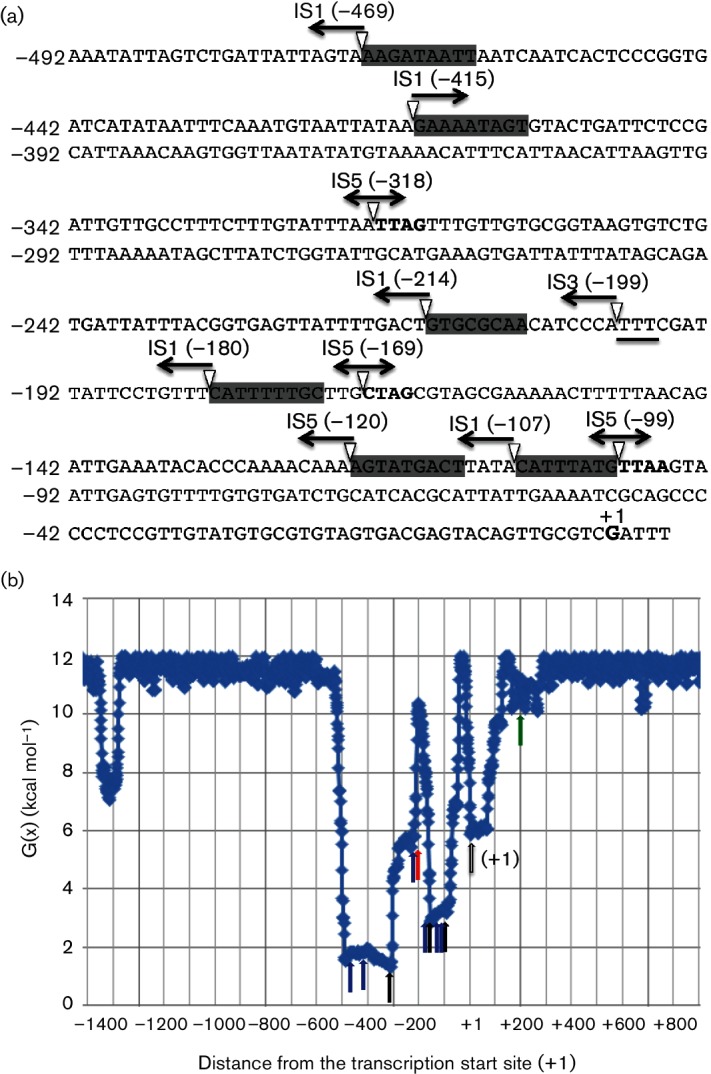
The upstream flhDC promoter region showing IS insertion sites and the orientations of the elements characterized in this study (a), as well as depiction of the SIDDs that overlap the 370 bp region into which IS elements insert in the flhDC upstream region (b). (a) The positions of the insertion sites and their orientations for the newly inserted IS1, IS3 and IS5 elements in the upstream region of the flhD promoter reported in this study are presented. The transcriptional start site is indicated as +1, and other locations, in parentheses, were assigned relative to this start site. The directions of the arrows correspond to the orientations of the insertions. IS1 has six different locations; IS3, one location; and IS5, three different locations. IS1, IS3 and IS5 insertion sites are grey highlighted, boldfaced and underlined, respectively. Single-headed arrows indicate the direction of transcription of the transposase gene. Double-headed arrows for IS5 indicate that this IS element was inserted in both orientations at each of its three sites. (b). Destabilization energy plot for a DNA segment that includes the flhD/uspC intergenic region (see Methods). Destabilization energy G(x) is the incremental energy (kcal mol−1) needed to guarantee separation of base pair x under defined superhelical stress. Highly destabilized (SIDD) DNA regions have lower values of G(x), and stable regions have higher G(x) values [46]. The green arrow shows the start codon of the flhD gene, the open arrow shows the transcriptional start site (+1) of the flhDC operon, the black arrows correspond to the IS5 insertion sites, blue arrows show the IS1 insertion sites and the red arrow reveals the IS3 insertion site.
Results
Original locations of IS elements on the chromosome
IS elements are present in most E. coli K12 chromosomes, but the numbers of copies and their locations differ from strain to strain. In this study, E. coli K12 BW25113 was used as the parental wild-type and the numbers and locations of three IS elements, IS1, IS3 and IS5, were determined using NCBI blast. There were six copies of IS1 in the chromosome, five copies of IS3 and ten copies of IS5 (Fig. S1). Some of these were in the direct orientation while others were in the reverse orientation as shown. One IS1 and three IS5s were located near the target flhDC promoter, but which IS elements serve as the sources for insertion upstream of the flhDC promoter are unknown.
Key strains used in this study
BW25113 is the parental strain used to make other strains. This strain has no IS element present in the flhDC regulatory region and is therefore a poor swarmer. Strain CAT carries a promotor-less cat gene that is fused immediately downstream of flhC so that flhD, flhC and cat form a single operon under the control of the flhDC promoter. Strain CAT2 is the same strain as CAT, except that the flhDC regulatory region and its downstream flhD : flhC : cat operon were moved to the intS/yfdG intergenic region while the original promoter, flhD and the first half of flhC were deleted. As alternative parental strains, CAT and CAT2 are sensitive to Cm at 3 µg ml−1 while all IS mutants are resistant to Cm at 6.5 µg ml−1.
Strain CAT_Kmr is the same as CAT except that it carries a Km resistance gene in the chromosome. This strain is resistant to Km at 25 µg ml−1. All IS insertional mutants derived from this strain are resistant to both Cm and Km. Strains CAT_IS1 and CAT_IS5 are two insertional mutants that contain IS1 (at −107) and IS5 (at −99), respectively, in the flhDC regulatory region. Both strains are resistant to Cm at 6.5 µg ml−1. Strain CAT_△AB is the same as CAT, except that the motAB operon is not expressed due to the lack of its promoter. CAT_△fliC is the same as CAT, except that the fliC gene is deleted. Both CAT_△AB and CAT_△fliC are non-motile and sensitive to Cm at 3 µg ml−1.
Swarm colony expansion
When an IS element was inserted upstream of the flhDC promoter, operon expression was increased, causing the cells to display increased motility [44]. Including two parental ‘wild-type’ E. coli K12 strains (BW25113 and CAT), 15 strains were tested for colony expansion (swarming) rates when glucose was absent from the media (Fig. 2a). All the IS mutants migrated at higher rates than the wild-type cells, while both types of wild-type colony migrated at similar rates (1–1.14 mm h−1). Thus, the slowest mutant showed a 1.9-fold increase while the fastest mutant showed a 2.7-fold increase over wild-type cells.
Fig. 2.
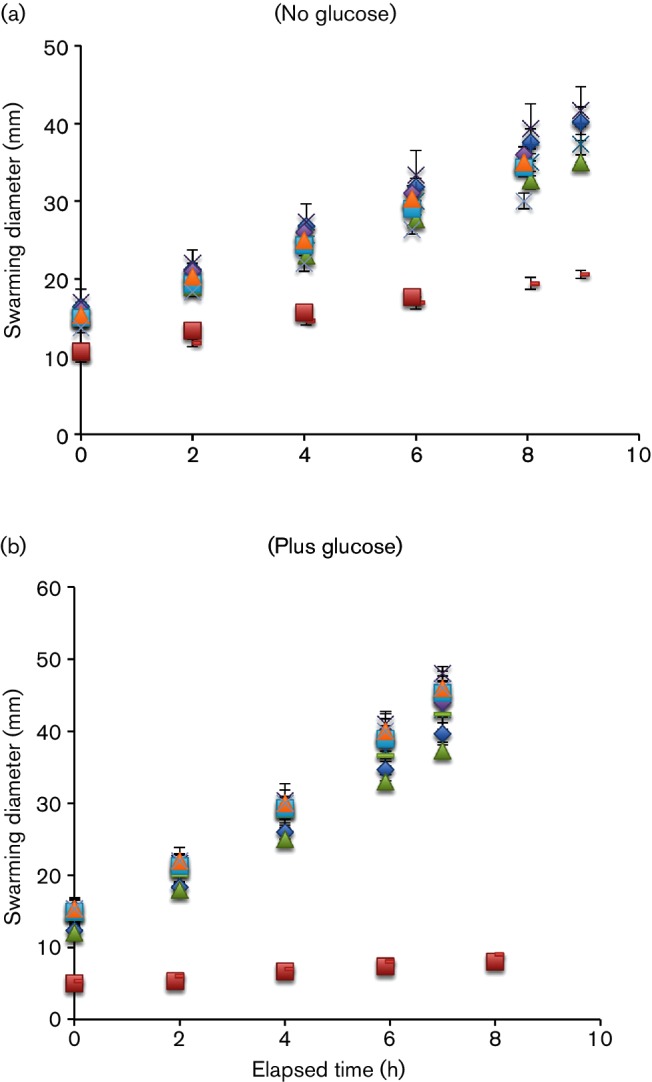
Swarming rates of wild-type (CAT) and IS mutants without (a) or with (b) glucose. Diameters of swarming zones of IS mutants and the wild-type strain when plated on soft agar without (a) or with (b) glucose (0.5 %) in the medium as a function of time. All plates were incubated at room temperature. The experiments were conducted in triplicate, and the error bars (not always visible) represent the sd of the three measurements. The error was always less than 10 % of the measured values. The strains examined are those listed in Table 1, and the raw numbers plotted here are those presented in that table.
The colony expansion rates for these strains were measured again with glucose (0.5 %) present in the media. In the presence of this nutrient, all of the mutant strains migrated at 40–60 % faster rates relative to the rates in the absence of glucose, whereas the wild-type strain virtually lost its swarming ability in the presence of glucose (Fig. 2b, Table 1).
Table 1. Colony expansion swarming rates of wild-type and IS mutants with and without glucose*.
| Swarming rate (mm h−1) | |||
|---|---|---|---|
| Strains | Without glucose | With glucose | Ratio |
| IS1(−469 rev) | 2.20 | 3.60 | 1.6 |
| IS1(−415 dir) | 2.50 | 3.90 | 1.6 |
| IS1(−214 rev) | 2.75 | 3.83 | 1.4 |
| IS1(−180 rev) | 2.80 | 3.92 | 1.4 |
| IS1(−120 rev) | 2.90 | 4.35 | 1.5 |
| IS1(−107 rev) | 3.05 | 4.30 | 1.4 |
| IS3(−199 rev) | 2.70 | 4.20 | 1.5 |
| IS5(−318) | 2.40 | 3.80 | 1.6 |
| IS5(−318 rev) | 2.30 | 3.90 | 1.7 |
| IS5(−169) | 2.35 | 3.80 | 1.7 |
| IS5(−169 rev) | 2.50 | 3.90 | 1.6 |
| IS5(−99) | 2.70 | 4.20 | 1.5 |
| IS5(−99 rev) | 2.60 | 4.10 | 1.5 |
| BW25113 (wild-type)† | 1.14 | 0.50 | 0.4 |
| CAT (wild-type)† | 1.00 | 0.40 | 0.4 |
*The diameters of the swarming zones were measured in soft agar plates (0.5 % tryptone, 0.5 % NaCl and 0.3 % agar) with or without 0.5 % glucose. Each strain was grown in a liquid medium for 10–12 h before inoculation, and 1.5 µl of diluted culture was inserted into the soft agar in the middle of the plates. The plates were incubated at room temperature. The diameters of the swarms were measured every 2 h to calculate the swarming rates (mm h−1), n=3. The three determinations were averaged. The error, expressed in sd of these determinations, was <0.4 mm h−1.
†The sizes of the colonies were measured for wild-type and CAT (wild-type) instead of the swarming zone because of their minimal motility. Thus, the ratio±glucose represents a maximal value.
Growth rates for wild-type and mutants when separately inoculated
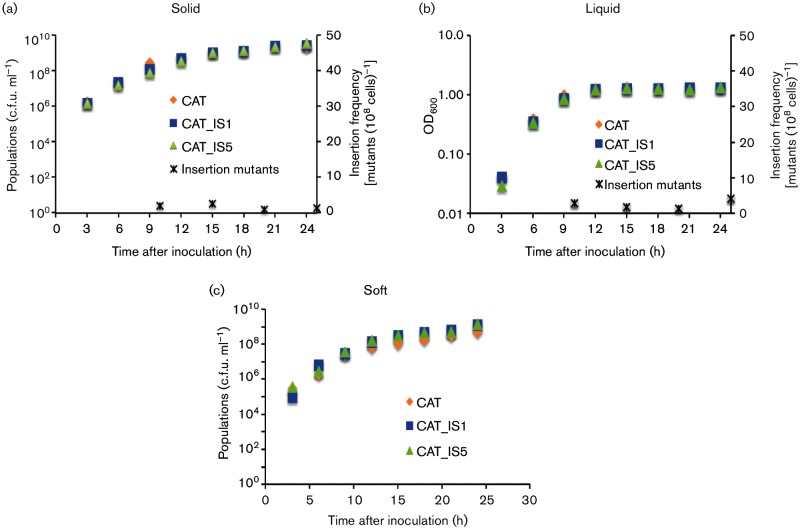
Three strains, CAT (wild-type, in which flhDC and cat form a single operon under the control of the flhDC promoter), CAT_IS1 (an IS1 insertion mutant) and CAT_IS5 (an IS5 insertion mutant) (Table S1), were inoculated into three different types of media. The media differed only in the concentration of agar, which was 0, 0.3 and 1.5 % for liquid media, soft agar plates and solid agar plates, respectively. For liquid media, growth rates were determined by measuring the optical densities (OD600) and plotting OD600 values in a semi-log format. For 0.3 and 1.5 % agar media, growth rates were determined by measuring absolute viable cell numbers and plotting the data in linear formats. The growth curves for these three strains overlapped in all three media up to 24 h, and no significant difference was identified (Fig 3a–c). It should be noted, however, that these experiments would not allow detection of small growth rate differences (see below). For example, if the growth data shown in Fig 3(c) are plotted in a semi-log format, CAT_IS1 and CAT_IS5 had slightly greater populations than wild-type cells after 15 h of growth.
Fig. 3.
Growth and mutant appearance frequencies on solid agar plates (a), in liquid medium (b) and in soft (0.3 %) agar (c). (a) CAT (orange rhombuses), CAT_IS1 (blue squares),and CAT_IS5 (green triangles) were inoculated separately on solid agar plates (0.5 % tryptone, 0.5 % NaCl and 1.5 % agar) and incubated at 30 °C for 25 h. Cells were washed off the plates, diluted, plated and grown for colony counting. The growth curves were plotted for each strain as a function of time. These three curves use the left y-axis. Insertion mutant populations were determined by plating onto LB+Cm plates. The insertion mutant ratios were determined at four time points throughout the 25 h period. The ratio is recorded on the right y-axis. Error, expressed in sd values, was less than 5 % of the measured values in (a), (b) and (c). (b) CAT, CAT_IS1 and CAT_IS5 strains were inoculated separately into liquid media (0.5 % tryptone, 0.5 % NaCl and 0 % agar) at 30 °C for 24 h, and cultures were rotated at 250 r.p.m. The growth curves were plotted for each strain. Insertion frequencies in liquid media from the CAT strain were measured, and the insertion mutant ratios were measured at four time points through the 25 h period. Format of presentation is as for (a). (c) CAT, CAT_IS1 and CAT_IS5 strains were inoculated into soft agar plates (0.5 % tryptone, 0.5 % NaCl and 0.3 % agar) separately and incubated at 30 °C for 24 h. The populations were measured every 3 h. Format of presentation is as for (a). Insertion frequencies are presented in Fig. 4.
Growth rates for wild-type and mutant strains when inoculated together
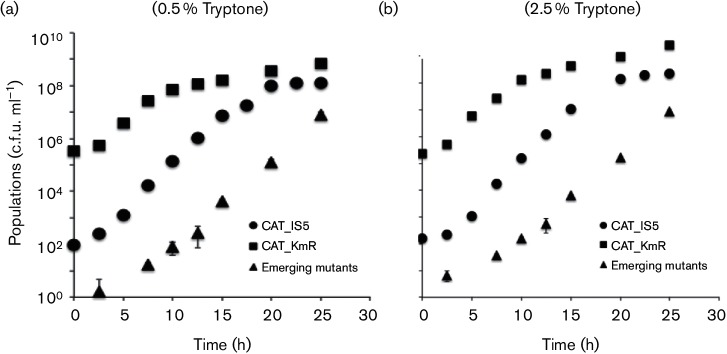
When the mixture of CAT_KmR (wild-type) and CAT_IS5 cells grew together in a soft agar medium (0.5 % tryptone, 0.5 % NaCl and 0.3 % agar), the IS5 strain showed a slight increase in growth rate where as the doubling time of wild-type was 62 min and that of the IS5 mutant was ~49 min (Table 2). This difference was nearly the same when the tryptone concentration in the medium was increased fivefold. The wild-type had a doubling time of 57 min, and the mutant doubled in 47 min (Table 2). The ‘wild-type’ strain (CAT_KmR) is resistant only to Km. Cells only become resistant to Cm when the flhDC operon was activated by IS insertion. This allowed us to distinguish between the originally inoculated IS5 mutant, CAT_IS5, resistant only to chloramphenicol (CmR KmS), and the newly derived mutants from CAT_KmR, which were resistant to both chloramphenicol and kanamycin (CmR KmR). This gave us more accurate growth curves for the wild-type and IS5 strains. In both 0.5 % tryptone and 2.5 % tryptone soft agar plates, all strains reached stationary phase around 15 h after inoculation (Fig. 4a, b). For these experiments, growth rates were determined by measuring absolute viable cell numbers over time and plotting the data in linear format.
Table 2. Doubling time comparisons in 0.5 % (top) and 2.5 % (bottom) tryptone soft agar plates.
| Apparent growth rate* | CAT_KmR | CAT_IS5 | Emerging mutants |
|---|---|---|---|
| Maximal (0.5 % tryptone; 1–15 h) | 62 | 49 | 65 |
| Average (0.5 % tryptone; 15–25 h) | 288 | 866 | 60 |
| Maximal (2.5 % tryptone; 1–15 h) | 57 | 47 | 60 |
| Average (2.5 % tryptone; 15–25 h) | 238 | 492 | 56 |
*Doubling times for CAT_KmR (wild-type), CAT_IS5 and emerging insertion mutants in 0.5 % tryptone soft agar plates (top) and 2.5 % tryptone soft agar plates (bottom). The maximal growth rate for the 1–15 h interval was the steepest slope before the 15 h time point, and the apparent average growth rate for the subsequent time period was the slope between 15 and 25 h. Values are expressed in minutes (min). Note that the values for the emerging mutants represent a sum of their appearance rates and their growth rates, with the former presumably predominating.
Fig. 4.
Growth curves and mutant appearance frequencies in soft agar plates containing 0.5 % tryptone (a) or 2.5 % tryptone (b). For (a), overnight cultures of CAT_KmR (wild-type, KmR, 2×109 c.f.u. ml−1; squares) and a CAT_ IS5 mutant (CmR, 2×106 c.f.u. ml−1; circles) were mixed and inoculated into soft agar plates (0.5 % tryptone, 0.5 % NaCl and 0.3 % agar) at 30 °C. For (b), the same strains were inoculated into soft agar with 2.5 % tryptone plus NaCl and 0.3 % agar. All soft agar plates were incubated at 30 °C. Every 3 h, the cells were collected and plated on LB and LB+Cm plates to measure the two inoculated populations. The emerging insertion mutant populations (CmR KmR, triangles) were determined on LB+Cm+Km plates. All measurements were conducted in triplicate, and the error, expressed in sd, was less than 10 % of the measured values. Error bars are often obscured by the symbols representing the averages of the three determinations.
To determine whether the newly derived mutants were IS insertion mutants, 30 CmRKmR mutants isolated from 0.5 % tryptone soft agar plates (incubated for 15 h) were purified, and their flhDC regulatory regions were amplified by PCR. All mutants were found to carry an IS element. DNA sequencing showed that these mutants remained as the IS5, IS1 or IS3 insertional mutant as present originally (data not shown).
Insertion sites
IS insertion sites were identified by PCR amplification of the intergenic region between flhD and the upstream gene, yecG (uspC), and subsequent DNA sequencing of the upstream region of the flhDC promoter. IS1 was found at six different locations (five in the reverse orientation between nucleotides −469 and −470, –214 and −215, –180 and −181, –120 and −121 and −107 and −108 and one in the direct orientation between −415 and −416) (Fig. 5a). A single IS3 insertion site was identified between −199 and −200 in the reverse orientation. IS5 was found to insert at three sites, between −318 and −319, –169 and −170 and −99 and −100. At all three of these insertion sites, IS5 was inserted in both orientations. Thus, while IS1 was never observed in both orientations at a particular site, IS5 was found in both orientations at all three sites. All insertion sites were upstream of the promoter, with none found downstream of the transcriptional start site of the flhDC promoter. Further analyses showed that all identified IS insertion sites are located between, not within, known operator sites in the flhDC upstream control region.
We observed that all insertions were in a 370 bp region (−100 to −470) that constitutes a strong SIDD region (Fig. 5b). SIDD sequences are susceptible to strand separation under negative superhelical stress [48–50]. As considered in greater detail elsewhere, IS elements preferentially insert at SIDD sequences (M. Z. Humayun, Z. Zhang and M. H. Saier, unpublished results).
Mutant ratios obtained in liquid media and on solid agar plates
CAT was used as the wild-type strain in this experiment. A chloramphenicol resistance gene was fused at the end of the flhC gene so that it was under the control of the flhDC promoter, and was up-regulated whenever the flhDC promoter was activated by an IS insertion upstream of flhDC. This allowed us to measure the populations of wild-type and IS mutant strains on LB plates and LB+Cm plates, respectively.
On solid agar plates (0.5 % tryptone, 0.5 % NaCl and 1.5 % agar), the mutant ratio was low (<5 mutants per 108 cells, 25 h after plating) (Fig. 3a). In liquid media (0.5 % tryptone, 0.5 % NaCl and 0 % agar), the insertion frequencies also remained low (<5 mutants per 108 cells) (Fig. 5b). CAT, CAT_IS1 and CAT_IS5 had the same growth rates in liquid media within experimental error when these strains were cultured separately (Fig. 3b).
Both in liquid media and on solid agar plates, up-regulating flhDC expression is not expected to benefit cells. On the contrary, the biosynthesis and function of the flagellar system would be expected to be detrimental because of the high energy consumption resulting from flagellar biosynthesis and function. Wang and Wood [44], in fact, reported slower growth rates for an IS5 insertional mutant compared to its wild-type strain when cells were grown in liquid LB medium, presumably reflecting the increased energetic burden in mutant cells. However, under these experimental conditions, there was little difference in growth rates of wild-type cells and insertion mutants in liquid media and on solid agar (however, see the next section).
IS insertion frequencies obtained in soft agar
Soft agar plates were tested with two different concentrations of tryptone in order to evaluate the effect of nutrient deficiency on mutation frequency. While wild-type (CmS KmR) and mutant (CmR KmS) cells entered the stationary growth phase about 15 h after inoculation, the emerging insertion mutants (CmR KmR) began to appear after about 5 h and continued to increase exponentially when both inoculated cell types (wild-type and IS5 insertional mutant cell) were barely growing (Fig. 6a, b).
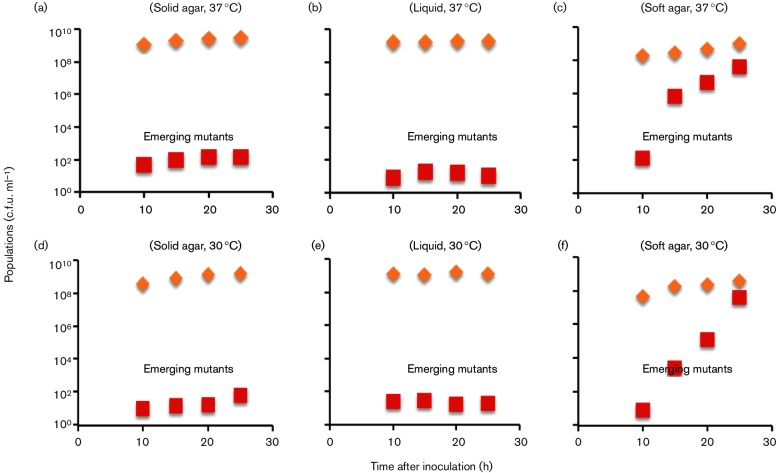
Fig. 6.
Mutant appearances from CAT2 [(a, b and c, 37 °C) and (d, e and f, 30 °C)]. An overnight culture of CAT2 (1×109 c.f.u. ml−1) was inoculated onto solid agar plates (0.5 % tryptone, 0.5 % NaCl and 1.5 % agar) (a and d), into liquid medium (0 % agar) (b and e) and into soft agar plates (0.3 % agar) (c and f). The solid agar and soft agar plates were incubated at 37 °C (a and c, repectively) or 30 °C (d and f, respectively) as specified, while the liquid media were incubated with rotation (250 r.p.m.) at 37 °C (b) or 30 °C (e). The total populations (diamonds) and the emerging insertion mutant populations (squares) were determined in LB and LB+Cm media, respectively. Error for triplicate determinations was within 10 % of the measured values.
Table 2 compares the doubling times for the CAT_KmR (wild-type), CAT_IS5 and emerging insertion mutants in 0.5 and 2.5 % tryptone soft agar plates. For 0.5 % tryptone soft agar plates, between 1 and 15 h, these three populations showed apparent doubling times of 62, 49 and 65 min, respectively. Thus, the IS mutants had a faster doubling time than the wild-type. Between 15 and 25 h, growth of both the initially inoculated wild-type and IS mutant cells approached the stationary phase when the doubling times were roughly 288 and 866 min, respectively (Fig. 4a, Table 2). The new mutant population, however, kept increasing as it maintained its apparent doubling time of 60 min, presumably reflecting the appearance of new insertional mutants rather than the rapid growth of this population. The average rate of appearance of these mutants, calculated from the slope of the curve, was about 3×10−3 cell−1 day−1.
This trend was the same in 2.5 % tryptone soft agar media (Fig. 4b, Table 2). For the first 15 h after inoculation, the doubling times for CAT_KmR, CAT_ IS5 and the emerging insertion mutants were 57, 47 and 60 min, respectively. When both the wild-type and IS5 strains initially plated approached the stationary phase, the emerging mutant population kept increasing with an apparent doubling time of 56 min. It should be noted that the apparent doubling times for the newly appearing mutants represent a sum of the insertion rate and the doubling rate for these cells alone. Since both the wild-type and initially inoculated IS5 mutant were barely growing, we assume that the increased numbers of newly emerging cells are primarily due to the appearance of new mutants, not to growth. This experiment, with a five fold increase in all nutrients, suggests that nutrient limitation was not the cause of growth cessation observed for the parent and co-inoculated IS5 insertion mutant.
The frequencies of mutant appearance were also determined with another wild-type strain, CAT2, in which the flhDC : cat operon plus its upstream regulatory region was moved to another (ectopic) chromosomal location. The experiments were conducted at both 30 and 37 °C, and the trends were the same for these two conditions (Fig. 6a, b). The mutant population remained low for solid agar and liquid media during the entire 25 h period. By contrast, emerging mutant cells kept increasing exponentially throughout this time period in soft agar, even after the parental wild-type cells had reached the stationary phase.
IS insertion frequencies of non-motile cells in soft agar
To determine whether IS insertional mutations occur in non-motile cells, we measured the IS insertion frequencies using strains CAT_△AB and CAT_△fliC that lack motility. As seen in Fig. 7, almost no IS mutants arose in these non-motile strains. As expected, insertional mutations arose at a high frequency from wild-type cells under the same soft agar conditions. These observations are consistent with the results of Wang and Wood [44], who deleted the flhD, motA or flgK gene and did not observe the appearance of IS5 insertional mutations.
Fig. 7.
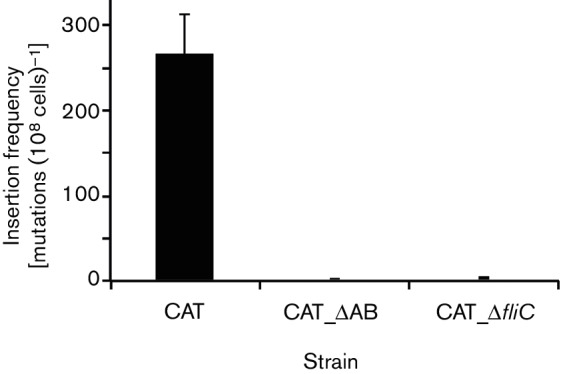
Frequencies of appearance of insertion mutants from wild-type and non-motile cells in soft agar Overnight LB cultures of strains CAT, CAT_ΔAB and CAT_ΔfliC were washed once using 1x M9 salts and then diluted to an OD600 of 1. Four microlitres was streaked onto soft NB agar plates (0.4 % NB+0.3 % agar), and the plates were incubated at 30 °C. After 18 h, the total populations and IS insertional mutant populations were determined as described in the legend to Fig. 4. Three soft agar plates were used for each strain. CAT_ΔAB and CAT_ΔfliC are non-motile.
Effects of agar concentration on the appearance of IS insertional mutants
The pore size of agar is inversely proportional to its concentration. To determine how the agar concentration affects the rates of IS insertion, we measured the frequencies of IS insertional mutations by incubating strain CAT in media containing various concentrations of agar, ranging from 0 to 1.2 %. As shown in Fig. 8, 0.2 % agar gave rise to IS insertion mutants with the highest frequency. When the agar concentration was 0.1 %, the mutation frequency was far less than when it was at 0.2 %. When agar was absent, almost no mutants arose. Similarly, the mutation frequencies decreased greatly as agar concentrations increased from 0.3 to 0.5 %. When the agar concentration was 0.6 % or above, the mutational frequencies were negligible. The pore diameters of agar are roughly 1.6, 0.7 and 0.2 µm in 0.25 % agar, 0.5 % agar and 1.0 % agar, respectively (51; Z. Zhang and M. H. Saier, unpublished data). When the pore diameters are smaller than 1 µm, cells can hardly migrate inside the agar. Our data with different concentrations of agar provide the first quantitative evidence showing the effect of agar concentration (and thus, pore size) on mutation rate. It should be noted, however, that increased agar concentration also results in an increase of viscosity that could play a role in determining the rate of mutation. We propose that inhibition of flagellar rotation is the signal promoting IS insertion.
Fig. 8.
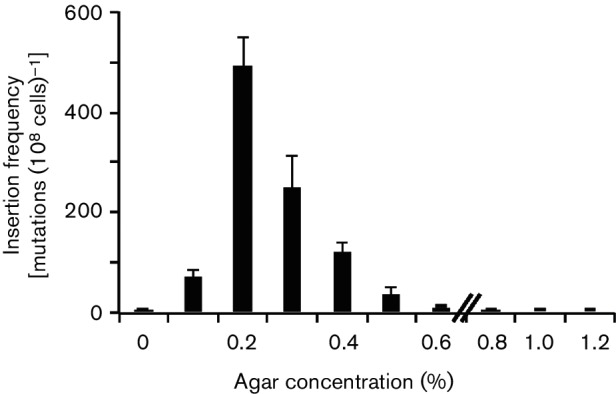
Effects of agar concentration on the appearance of insertional mutants. An overnight LB culture of CAT was washed once and diluted to OD600 of 1. Four microlitres of diluted culture was streaked onto NB agar plates (0.4 % NB+0–1.2 % agar), and the plates were incubated at 30 °C. After 18 h, the total populations and mutant populations in the plates with 0–0.4 % agar were determined as described in the legend to Fig. 4. For the plates with 0.6 % agar or above, the cells were washed off the plates without breaking the agar, and the total populations and mutant populations were determined as described in the legend to Fig. 4. These experiments were conducted in triplicate, and error bars represent sd.
Discussion
Activation of the flhDC operon by insertional mutations
Barker et al. [20] were the first to report a connection between IS element insertion upstream of the flhDC promoter and increased motility of E.coli K12, an observation that has been confirmed and extended by several investigators [17, 27, 28]. Many other types of mutations can also activate flhDC expression, as expected, since expression is regulated by over two dozen transcription factors (see Introduction). Though the mechanism(s) of activation of the operon downstream of the insertion sites remain(s) unknown, transposon insertion increased motility. Three IS elements (IS1, IS3 and IS5) hop into the regulatory region of the operon, increasing expression of the downstream genes [20, 27, 52]. Increased flagellar gene expression following IS insertion allows E. coli K12 to migrate more rapidly through soft agar (0.2–0.3 %), but not through solid agar (1.5 %), and flagella are not useful in agitated liquid media where the medium is of a uniform consistency. Wang and Wood [44] reported that IS5 insertional mutations upstream of the flhDC operon arose at high rates only in soft agar, precisely the condition where the consequences of the mutations were beneficial.
In this study, we confirm and extend the provocative results of Wang and Wood [44], showing that the frequency of IS element insertional events in the flhDC promoter increases during exposure to soft agar, but not to solid agar or liquid medium. We showed that the concentration that optimally promotes IS insertion is about 0.24 %, a concentration correlating with a pore size in the agar equivalent to that just barely allowing entry of the cells into the agar channels, presumably inhibiting motility by reducing the flagellar rotation rate. Further, we showed that this increase cannot simply be an artefact of a post-insertional growth advantage. We constructed two reporter systems, based on transcriptional fusion of a cat gene downstream of the flhC gene, such that IS activation of the flhDC operon leads to chloramphenicol resistance. This system provided a way to track both the emergence of novel mutants and expansion of the newly emerging mutant population in the presence of the inoculated wild-type and an inoculated IS-flhDC-activated mutant. While the assay system does not allow for discrimination between mutation and selection, the collection of experiments performed does yield compelling evidence that the growth and motility advantages are not sufficient to account for the accumulation of the newly arising more motile/chloramphenicol-resistant mutants.
The role of environment on insertional activation of the flhDC operon
The finding that IS insertions only occurred after growth in soft agar raises the possibility that an environmental trigger activated (or facilitated) insertion into flhDC-activating sites. Presumably, cells in the wild with more flagella swarm faster through soft agar and are capable of reaching nutrients ahead of cells with fewer flagella. Both Wang and Wood [44] and we noted that the increase in the insertion mutant population could not be accounted for simply by faster growth. Wang and Wood [44] reported slower growth rates for an insertion mutant when the cells were grown in liquid LB medium, but they did not measure these rates in soft agar. The results from our mixing experiment provide the most conclusive demonstration that growth rates cannot explain the apparent rapid increase in the emerging insertion mutant population on soft agar. Our results showed that the growth rate in soft agar differed by ~20 % when cells were in the log phase of growth, whereas the newly emerging mutant population increased dramatically only after most cells (both wild-type and the co-inoculated IS5 insertional mutant tested) had reached stationary phase. Between 15 and 25 h, the ratio of the wild-type and the originally inoculated IS insertion strains did not differ appreciably, but the numbers of emerging IS mutants increased dramatically when the cells were approaching the stationary growth phase (Fig. 4a, b). This result suggests that the rates of insertional mutant appearance are up-regulated when the inoculated cells are entering the stationary phase in soft agar. This trend was the same for the other parental wild-type strain examined (CAT2, data not shown).
Our observations, together with those of Wang and Wood [44], indicate that higher expression of the flagellar genes may confer an advantage to cells in soft agar plates, but not in liquid media. The advantage is presumed to be the ability to reach a new nutrient-rich niche or the ability to escape growth-inhibitory metabolic waste products, both of which are only feasible in soft agar. This appears to be an example of a mechanism whereby gene expression regulation provides adaptive evolutionary benefit in overcoming stress [53].
Effect of environment on insertional mutagenesis in other experimental systems
At present, molecular mechanisms by which the environment is sensed to enhance insertion frequency upstream of the flhDC operon are unknown. However, this behaviour is reminiscent of, and could be mechanistically related to, the swarming behaviour of Vibrio parahaemolyticus which induces a second, lateral, swarming flagellar system in response to rotational inhibition of the first polar one [54–56]. In the case of E. coli IS-mediated flhDC activation studied here, weak interference with flagellar rotation could provide the signal for IS insertion. One possibility is that flagellar rotation senses the viscosity of the medium.
In a different respect, this work is also reminiscent of the work of Vandecraen et al. [57] who reported that the presence of toxic levels of zinc in the growth medium induced transposition and insertion of several IS elements in the bacterium Cupriavidus metallidurans, increasing its adaptation to growth in the presence of zinc. This situation resembles the situation described here in that multiple IS elements insert at multiple sites to activate gene expression.
In its responsiveness to environmental conditions, the flhDC insertional mutagenesis system is also reminiscent of our previous extensive work on IS5 insertional activation of the glycerol utilization glpFK operon [21–25]. The glpFK system, which is mechanistically much better described than any other insertional mutagenesis system, is based on the inability of Crp− or CyaA− cells to utilize glycerol as a carbon source (i.e. Crp− and CyaA− cells have a Glp− phenotype). Prolonged incubation of these cells on minimal agar plates with glycerol as the sole carbon source, however, leads to an accumulation of promoter-activating IS5 insertional mutations upstream of the glpFK promoter that render the cells Glp+. The insertional mutagenesis appears to be specific to the glpFK region under these conditions, as no other examined sites showed elevated insertional mutagenesis [24]. We have recently proposed a plausible mechanism that connects glycerol starvation stress to localized mutagenesis of the glpFK promoter region, mediated by two DNA-binding proteins [25].
Effect of IS insertion events on flhDC expression
IS insertion into the flhDC operon upstream region of E. coli always leads to faster migration rates relative to parental cells (Fig. 2a, b). The mechanisms by which transposition increased expression of the downstream flhDC operon in E. coli is however, not yet clear (but see Refs [17, 23, 28, 44]). Activation may be accomplished by one of the following four mechanisms: (1) the IS element could directly activate an existing chromosomal promoter; (2) it could use a promoter already on the transposon to activate expression; (3) it could create a new hybrid promoter (e.g. the −35 region on the transposon with the −10 region on the chromosome); or (4) it could inactivate a repressor or inhibitory region of the upstream DNA while still using the native flhDC promoter. All of these mechanisms have been documented in previous studies with various operons [23, 57–59]. We favour the last of these four mechanisms for flhDC activation, in agreement with previously published work [17, 28]. The fact that activation of the flhDC promoter occurs to a similar degree regardless of IS element direction (orientation) is in agreement with this conclusion. In this regard, it would be interesting to investigate whether the presence of these IS elements in the flhDC upstream region affects the binding and action of any of the many transcription factors that influence flhDC expression.
No apparent correlation between the insertion sites and swarming rates was seen for IS5, but a possible correlation could be seen for IS1 (Fig. 9). Although the detailed mechanisms by which IS elements influence motility are unknown, insertions at sites immediately upstream of the flhDC promoter yielded strains with higher expression based on increased motility in soft agar (see also [28]).
Fig. 9.
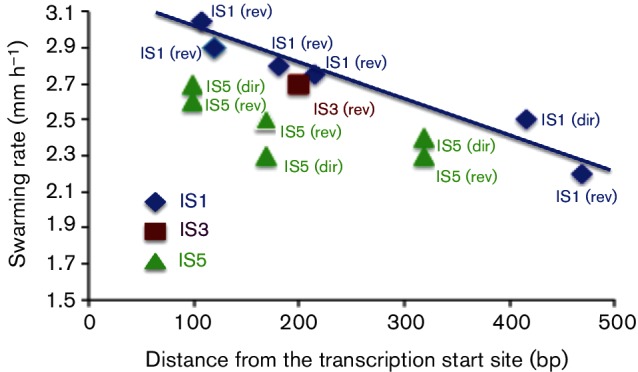
Insertion sites versus swarming rates. Colony expansion swarming rates in soft agar for the different insertion mutants were plotted as a function of distance from the flhD transcriptional start site. The data are from Table 1 (no glucose). The IS element type and its insertional orientation are indicated [direct (dir) or reverse (rev)]. Distance from the transcriptional start site is indicated on the x-axis. Errors of triplicate determinations were usually within 5 % of the measured values.
Glucose effect on motility
The IS insertional mutants showed greater rates of colony expansion (swarming) in the presence of glucose than in the absence of glucose, although glucose strongly inhibited motility of the wild-type strains, presumably by repressing flagellar synthesis (Table 1). The native flhDC operon is positively activated by the cyclic AMP receptor protein (Crp), which forms a complex with cAMP to bind to a specific site in the promoter [60]. Wild-type parental E. coli K12 cells lose their motility when glucose is present and cAMP levels are low [61], presumably because flhDC expression is dependent on the Crp-cAMP complex [60, 62]. By contrast, the increased rates of colony expansion of the mutants in the presence of glucose are paradoxical because the mutants also proved to be Crp-cAMP dependent. Whether a lower cAMP concentration is sufficient for motility in the mutants or whether the glucose-repressive effect observed in wild-type cells is partially or fully independent of Crp remains to be investigated. The increased swarming of the mutants in the presence of glucose might be due to an increased cellular energy level driving motility rather than an effect on gene expression. This suggestion will be examined in future experiments.
Role of DNA structural features in IS insertion and gene activation
Different transposable genetic elements display different target-site preferences. Even though there is a preference for short sequences (3–10 bp), this preference is not absolute for many IS elements, and genomic IS element distributions have been interpreted to suggest that transposition is more-or-less random, although with striking preferences for intergenic regions [63]. The question of what drives insertions to specific targets in experimental systems such as those discussed here is largely unexplored. Interestingly, the 370 bp region into which IS elements are inserted in the flhDC upstream region has an embedded SIDD element (for SIDD), and every inserted IS element occurs within this region (Fig. 5b). Examination of other hot spots of insertion elsewhere on the E. coli chromosome has revealed a strong preference for SIDD-overlapping regions. This last-mentioned work will be reported in a subsequent communication (M. Z. Humayun, Z. Zhang and M. H. Saier, manuscript in preparation).
In future studies, there are many additional aspects of IS-mediated flhDC mutational activation that need to be examined. For example, how does each IS element activate flhDC transcription? Which IS elements are transposed to which locations and why? How do the environmental cues trigger transposition to the upstream sites? These exciting questions will be of interest to many microbial physiologists.
Funding information
This work was supported by the National Institutes of Health (GM077402 and GM109895).
Acknowledgements
We thank Professor James Golden (University of California-San Diego) for valuable discussions, Professor Thomas Wood (Pennsylvania State University) and Dr Xiaoxue Wang (Chinese Academy of Sciences) for critically reading the manuscript prior to submission for publication and Anne Chu, Harry Zhou and Sabrina Phan of University of California-San Diego for assistance with the preparation of this manuscript.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Supplementary Data
Footnotes
One supplementary figure and two supplementary tables are available with the online Supplementary Material.
Edited by: A. J. Roe and M. Whiteley
References
- 1.Muñoz-Lopez M, Vilar-Astasio R, Tristan-Ramos P, Lopez-Ruiz C, Garcia-Pérez JL. Study of transposable elements and their genomic impact. Methods Mol Biol. 2016;1400:1–19. doi: 10.1007/978-1-4939-3372-3_1. [DOI] [PubMed] [Google Scholar]
- 2.Blot M. Transposable elements and adaptation of host bacteria. Genetica. 1994;93:5–12. doi: 10.1007/BF01435235. [DOI] [PubMed] [Google Scholar]
- 3.Bonchev G, Parisod C. Transposable elements and microevolutionary changes in natural populations. Mol Ecol Resour. 2013;13:765–775. doi: 10.1111/1755-0998.12133. [DOI] [PubMed] [Google Scholar]
- 4.Zhang Z, Saier MH., Jr Transposon-mediated adaptive and directed mutations and their potential evolutionary benefits. J Mol Microbiol Biotechnol. 2011;21:59–70. doi: 10.1159/000333108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derbyshire KM, Grindley ND. Replicative and conservative transposition in bacteria. Cell. 1986;47:325–327. doi: 10.1016/0092-8674(86)90586-6. [DOI] [PubMed] [Google Scholar]
- 6.Haniford DB, Ellis MJ. Transposons Tn10 and Tn5. Microbiol Spectr. 2015;3:MDNA3-0002-2014. doi: 10.1128/microbiolspec.MDNA3-0002-2014. [DOI] [PubMed] [Google Scholar]
- 7.Skipper KA, Andersen PR, Sharma N, Mikkelsen JG. DNA transposon-based gene vehicles – scenes from an evolutionary drive. J Biomed Sci. 2013;20:92. doi: 10.1186/1423-0127-20-92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chandler M, Fayet O, Rousseau P, Ton Hoang B, Duval-Valentin G. Copy-out–paste-in transposition of IS911: a major transposition pathway. Microbiol Spectr. 2015;3 doi: 10.1128/microbiolspec.MDNA3-0031-2014. [DOI] [PubMed] [Google Scholar]
- 9.Sousa A, Bourgard C, Wahl LM, Gordo I. Rates of transposition in Escherichia coli. Biol Lett. 2013;9:20130838. doi: 10.1098/rsbl.2013.0838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Yen MR, Saier MH., Jr Precise excision of IS5 from the intergenic region between the fucPIK and the fucAO operons and mutational control of fucPIK operon expression in Escherichia coli. J Bacteriol. 2010;192:2013–2019. doi: 10.1128/JB.01085-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziebuhr W, Krimmer V, Rachid S, Lössner I, Götz F, et al. A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol. 1999;32:345–356. doi: 10.1046/j.1365-2958.1999.01353.x. [DOI] [PubMed] [Google Scholar]
- 12.Siguier P, Gourbeyre E, Chandler M. Bacterial insertion sequences: their genomic impact and diversity. FEMS Microbiol Rev. 2014;38:865–891. doi: 10.1111/1574-6976.12067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaffé J, Mckenzie C, Maharjan RP, Coursange E, Ferenci T, et al. Insertion sequence-driven evolution of Escherichia coli in chemostats. J Mol Evol. 2011;72:398–412. doi: 10.1007/s00239-011-9439-2. [DOI] [PubMed] [Google Scholar]
- 14.Bonnefoy V, Fons M, Ratouchniak J, Pascal MC, Chippaux M. Aerobic expression of the nar operon of Escherichia coli in a fnr mutant. Mol Microbiol. 1988;2:419–425. doi: 10.1111/j.1365-2958.1988.tb00047.x. [DOI] [PubMed] [Google Scholar]
- 15.Sawers RG. Expression of fnr is constrained by an upstream IS5 insertion in certain Escherichia coli K-12 strains. J Bacteriol. 2005;187:2609–2617. doi: 10.1128/JB.187.8.2609-2617.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li B, Li N, Wang F, Guo L, Huang Y, et al. Structural insight of a concentration-dependent mechanism by which YdiV inhibits Escherichia coli flagellum biogenesis and motility. Nucleic Acids Res. 2012;40:11073–11085. doi: 10.1093/nar/gks869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee C, Park C. Mutations upregulating the flhDC operon of Escherichia coli K-12. J Microbiol. 2013;51:140–144. doi: 10.1007/s12275-013-2212-z. [DOI] [PubMed] [Google Scholar]
- 18.Chen YM, Lu Z, Lin EC. Constitutive activation of the fucAO operon and silencing of the divergently transcribed fucPIK operon by an IS5 element in Escherichia coli mutants selected for growth on l-1,2-propanediol. J Bacteriol. 1989;171:6097–6105. doi: 10.1128/jb.171.11.6097-6105.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hall BG. Activation of the bgl operon by adaptive mutation. Mol Biol Evol. 1998;15:1–5. doi: 10.1093/oxfordjournals.molbev.a025842. [DOI] [PubMed] [Google Scholar]
- 20.Barker CS, Prüss BM, Matsumura P. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J Bacteriol. 2004;186:7529–7537. doi: 10.1128/JB.186.22.7529-7537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Saier MH, Jr, Zhang Z. Transposon-mediated directed mutation controlled by DNA binding proteins in Escherichia coli. Front Microbiol. 2014;5:390. doi: 10.3389/fmicb.2014.00390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Saier MH, Jr, Zhang Z. Control of transposon-mediated directed mutation by the Escherichia coli phosphoenolpyruvate:sugar phosphotransferase system. J Mol Microbiol Biotechnol. 2015;25:226–233. doi: 10.1159/000375375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang Z, Saier MH., Jr A novel mechanism of transposon- mediated gene activation. PLoS Genet. 2009;5:e1000689. doi: 10.1371/journal.pgen.1000689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang Z, Saier MH., Jr A mechanism of transposon-mediated directed mutation. Mol Microbiol. 2009;74:29–43. doi: 10.1111/j.1365-2958.2009.06831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Z, Saier MH., Jr Transposon-mediated activation of the Escherichia coli glpFK operon is inhibited by specific DNA-binding proteins: Implications for stress-induced transposition events. Mutat Res. 2016;793-794:22–31. doi: 10.1016/j.mrfmmm.2016.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Soutourina OA, Bertin PN. Regulation cascade of flagellar expression in Gram-negative bacteria. FEMS Microbiol Rev. 2003;27:505–523. doi: 10.1016/S0168-6445(03)00064-0. [DOI] [PubMed] [Google Scholar]
- 27.Fitzgerald DM, Bonocora RP, Wade JT. Comprehensive mapping of the Escherichia coli flagellar regulatory network. PLoS Genet. 2014;10:e1004649. doi: 10.1371/journal.pgen.1004649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fahrner KA, Berg HC. Mutations that stimulate flhDC expression in Escherichia coli K-12. J Bacteriol. 2015;197:3087–3096. doi: 10.1128/JB.00455-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rahimpour M, Montero M, Almagro G, Viale AM, Sevilla Á, et al. GlgS, described previously as a glycogen synthesis control protein, negatively regulates motility and biofilm formation in Escherichia coli. Biochem J. 2013;452:559–573. doi: 10.1042/BJ20130154. [DOI] [PubMed] [Google Scholar]
- 30.Vikram A, Jayaprakasha GK, Uckoo RM, Patil BS. Inhibition of Escherichia coli O157:H7 motility and biofilm by β-sitosterol glucoside. Biochim Biophys Acta. 2013;1830:5219–5228. doi: 10.1016/j.bbagen.2013.07.022. [DOI] [PubMed] [Google Scholar]
- 31.Allison SE, Silphaduang U, Mascarenhas M, Konczy P, Quan Q, et al. Novel repressor of Escherichia coli O157:H7 motility encoded in the putative fimbrial cluster OI-1. J Bacteriol. 2012;194:5343–5352. doi: 10.1128/JB.01025-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kitagawa R, Takaya A, Yamamoto T. Dual regulatory pathways of flagellar gene expression by ClpXP protease in enterohaemorrhagic Escherichia coli. Microbiology. 2011;157:3094–3103. doi: 10.1099/mic.0.051151-0. [DOI] [PubMed] [Google Scholar]
- 33.Lehti TA, Bauchart P, Dobrindt U, Korhonen TK, Westerlund-Wikström B. The fimbriae activator MatA switches off motility in Escherichia coli by repression of the flagellar master operon flhDC. Microbiology. 2012;158:1444–1455. doi: 10.1099/mic.0.056499-0. [DOI] [PubMed] [Google Scholar]
- 34.Reiss DJ, Mobley HL. Determination of target sequence bound by PapX, repressor of bacterial motility, in flhD promoter using systematic evolution of ligands by exponential enrichment (SELEX) and high throughput sequencing. J Biol Chem. 2011;286:44726–44738. doi: 10.1074/jbc.M111.290684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Theodorou MC, Theodorou EC, Kyriakidis DA. Involvement of AtoSC two-component system in Escherichia coli flagellar regulon. Amino Acids. 2012;43:833–844. doi: 10.1007/s00726-011-1140-7. [DOI] [PubMed] [Google Scholar]
- 36.Wiebe H, Gürlebeck D, Groß J, Dreck K, Pannen D, et al. YjjQ represses transcription of flhDC and additional loci in Escherichia coli. J Bacteriol. 2015;197:2713–2720. doi: 10.1128/JB.00263-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wada T, Hatamoto Y, Kutsukake K. Functional and expressional analyses of the anti-FlhD4C2 factor gene ydiV in Escherichia coli. Microbiology. 2012;158:1533–1542. doi: 10.1099/mic.0.056036-0. [DOI] [PubMed] [Google Scholar]
- 38.De Lay N, Gottesman S. A complex network of small non-coding RNAs regulate motility in Escherichia coli. Mol Microbiol. 2012;86:524–538. doi: 10.1111/j.1365-2958.2012.08209.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomason MK, Fontaine F, De Lay N, Storz G. A small RNA that regulates motility and biofilm formation in response to changes in nutrient availability in Escherichia coli. Mol Microbiol. 2012;84:17–35. doi: 10.1111/j.1365-2958.2012.07965.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chilcott GS, Hughes KT. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar Typhimurium and Escherichia coli. Microbiol Mol Biol Rev. 2000;64:694–708. doi: 10.1128/MMBR.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Guttenplan SB, Kearns DB. Regulation of flagellar motility during biofilm formation. FEMS Microbiol Rev. 2013;37:849–871. doi: 10.1111/1574-6976.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sule P, Horne SM, Logue CM, Prüss BM. Regulation of cell division, biofilm formation, and virulence by FlhC in Escherichia coli O157:H7 grown on meat. Appl Environ Microbiol. 2011;77:3653–3662. doi: 10.1128/AEM.00069-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Copeland MF, Weibel DB. Bacterial swarming: a model system for studying dynamic self-assembly. Soft Matter. 2009;5:1174–1187. doi: 10.1039/B812146J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang X, Wood TK. IS5 inserts upstream of the master motility operon flhDC in a quasi-Lamarckian way. ISME J. 2011;5:1517–1525. doi: 10.1038/ismej.2011.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang H, Noordewier M, Benham CJ. Stress-induced DNA duplex destabilization (SIDD) in the E. coli genome: SIDD sites are closely associated with promoters. Genome Res. 2004;14:1575–1584. doi: 10.1101/gr.2080004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhabinskaya D, Madden S, Benham CJ. SIST: stress-induced structural transitions in superhelical DNA. Bioinformatics. 2015;31:421–422. doi: 10.1093/bioinformatics/btu657. [DOI] [PubMed] [Google Scholar]
- 48.Benham CJ. Duplex destabilization in superhelical DNA is predicted to occur at specific transcriptional regulatory regions. J Mol Biol. 1996;255:425–434. doi: 10.1006/jmbi.1996.0035. [DOI] [PubMed] [Google Scholar]
- 49.Bi C, Benham CJ. WebSIDD: server for predicting stress-induced duplex destabilized (SIDD) sites in superhelical DNA. Bioinformatics. 2004;20:1477–1479. doi: 10.1093/bioinformatics/bth304. [DOI] [PubMed] [Google Scholar]
- 50.Wang H, Benham CJ. Superhelical destabilization in regulatory regions of stress response genes. PLoS Comput Biol. 2008;4:e17. doi: 10.1371/journal.pcbi.0040017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zimm BH, Levene SD. Problems and prospects in the theory of gel electrophoresis of DNA. Q Rev Biophys. 1992;25:171–204. doi: 10.1017/S0033583500004662. [DOI] [PubMed] [Google Scholar]
- 52.Martinez-Vaz BM, Xie Y, Pan W, Khodursky AB. Genome-wide localization of mobile elements: experimental, statistical and biological considerations. BMC Genomics. 2005;6:81. doi: 10.1186/1471-2164-6-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Stoebel DM, Hokamp K, Last MS, Dorman CJ. Compensatory evolution of gene regulation in response to stress by Escherichia coli lacking RpoS. PLoS Genet. 2009;5:e1000671. doi: 10.1371/journal.pgen.1000671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gode-Potratz CJ, Kustusch RJ, Breheny PJ, Weiss DS, McCarter LL. Surface sensing in Vibrio parahaemolyticus triggers a programme of gene expression that promotes colonization and virulence. Mol Microbiol. 2011;79:240–263. doi: 10.1111/j.1365-2958.2010.07445.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kim YK, McCarter LL. Cross-regulation in Vibrio parahaemolyticus: compensatory activation of polar flagellar genes by the lateral flagellar regulator LafK. J Bacteriol. 2004;186:4014–4018. doi: 10.1128/JB.186.12.4014-4018.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.McCarter LL. Dual flagellar systems enable motility under different circumstances. J Mol Microbiol Biotechnol. 2004;7:18–29. doi: 10.1159/000077866. [DOI] [PubMed] [Google Scholar]
- 57.Vandecraen J, Monsieurs P, Mergeay M, Leys N, Aertsen A, et al. Zinc-induced transposition of insertion sequence elements contributes to increased adaptability of Cupriavidus metallidurans. Front Microbiol. 2016;7:359. doi: 10.3389/fmicb.2016.00359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Al-Natour S. Lipoid proteinosis: a report of 2 siblings and a brief review of the literature. Saudi Med J. 2008;29:1188–1191. [PubMed] [Google Scholar]
- 59.Teras R, Hõrak R, Kivisaar M. Transcription from fusion promoters generated during transposition of transposon Tn4652 is positively affected by integration host factor in Pseudomonas putida. J Bacteriol. 2000;182:589–598. doi: 10.1128/JB.182.3.589-598.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Soutourina O, Kolb A, Krin E, Laurent-Winter C, Rimsky S, et al. Multiple control of flagellum biosynthesis in Escherichia coli: role of H-NS protein and the cyclic AMP-catabolite activator protein complex in transcription of the flhDC master operon. J Bacteriol. 1999;181:7500–7508. doi: 10.1128/jb.181.24.7500-7508.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Saier MH., Jr Protein phosphorylation and allosteric control of inducer exclusion and catabolite repression by the bacterial phosphoenolpyruvate: sugar phosphotransferase system. Microbiol Rev. 1989;53:109–120. doi: 10.1128/mr.53.1.109-120.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stella NA, Kalivoda EJ, O'Dee DM, Nau GJ, Shanks RM. Catabolite repression control of flagellum production by Serratia marcescens. Res Microbiol. 2008;159:562–568. doi: 10.1016/j.resmic.2008.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lee H, Doak TG, Popodi E, Foster PL, Tang H. Insertion sequence-caused large-scale rearrangements in the genome of Escherichia coli. Nucl Acids Res. 2016;44:7109–7119. doi: 10.1093/nar/gkw647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.