Abstract
Emerging clinical trial data implicate progestins in the development of breast cancer. While the role for the progesterone receptor (PR) in this process remains controversial, it is clear that PR, a steroid-activated nuclear receptor, alters the transcriptional landscape of breast cancer. PR interacts with many different types of proteins, including transcriptional co-activators and co-repressors, transcription factors, nuclear receptors, and proteins that post-translationally modify PR (i.e., kinases and phosphatases). Herein, we identify a novel interaction between PR and O-GlcNAc transferase (OGT), the enzyme that catalyzes the addition of a single N-acetylglucosamine sugar, referred to as O-GlcNAc, to acceptor serines and threonines in target proteins. This interaction between PR and OGT leads to the post-translational modification of PR by O-GlcNAc. Moreover, we show that O-GlcNAcylated PR is more transcriptionally active on PR-target genes, despite the observation that PR messenger RNA and protein levels are decreased when O-GlcNAc levels are high. O-GlcNAcylation in breast cancer is clinically relevant, as we show that O-GlcNAc levels are higher in breast cancer as compared to matched normal tissues, and PR-positive breast cancers have higher levels of OGT. These data predict that under conditions where O-GlcNAc levels are high (breast cancer), PR, through an interaction with the modifying enzyme OGT, will exhibit increased O-GlcNAcylation and potentiated transcriptional activity. Therapeutic strategies aimed at altering cellular O-GlcNAc levels may have profound effects on PR transcriptional activity in breast cancer.
Electronic supplementary material
The online version of this article (10.1007/s12672-017-0310-9) contains supplementary material, which is available to authorized users.
Keywords: PR Function, Transcriptional Landscape, Acceptor Serine, Interferon-stimulated Genes (ISGs), Positive Breast Cancer Cell Lines
Introduction
Upon diagnosis, nearly 70% of breast cancers express progesterone receptor (PR) and the estrogen receptor (ER). ER action in breast cancer has been well studied and as a result, ER has proven to be an excellent target for current endocrine-based therapies. Controversial clinical trial data have implicated progestins in the development of invasive breast cancer; however, the role of progesterone/PR in breast cancer has been largely understudied [1, 2]. What is clear from the combined years of clinical and laboratory based data is that PR, either alone or through modulation of the ER transcriptome, affects the transcriptional landscape in breast cancer (reviewed in [3]). Cumulatively, these data suggest that deciphering PR transcriptional activity in breast cancer is of great clinical importance.
PR is significantly post-translationally modified, primarily through phosphorylation [4]. Mitogenic kinases (i.e., cdk2, ck2, MAPK) upregulated in breast cancer modify PR function through phosphorylation [5–8]. Under normal (non-cancerous) conditions, progesterone is required for these phosphorylation events. However, in high kinase activity environments, such as breast cancer, PR becomes inappropriately activated (phosphorylated) in the absence of ligand, driving PR-dependent transcriptional programs. Recently, our lab detailed the mechanism and biological significance of PR phosphorylation on Ser81 by the mitogenic kinase, ck2 [9], an ideal example of how post-translational modifications can dramatically alter PR function. In addition to phosphorylation, PR modifications by acetylation, ubiquitination, sumoylation, and methylation have been reported to modify receptor function, co-factor interactions, and target gene promoter specificity [10].
O-GlcNAc is a ubiquitous post-translational modification of a single N-acetylglucosamine sugar that cycles on and off serine or threonine residues in nuclear, cytoplasmic, and mitochondrial proteins. Two enzymes control O-GlcNAc cycling: O-GlcNAc transferase (OGT) adds the modification, while O-GlcNAcase (OGA) removes the modification [11]. O-GlcNAc is a regulator of protein function, specifically within transcription complexes [11]. O-GlcNAcylation of target proteins can alter protein stability, protein-protein interactions, and phosphorylation [12]. Data are mounting that implicate O-GlcNAcylation and its regulators (OGT and OGA) in multiple tumor types, including the lung, colon, leukemia, prostate, endometrial, and ovarian cancers [13]. Key oncogenes and tumor suppressor genes, such as p53 and c-myc, are modified by O-GlcNAcylation, potentially contributing to their roles in oncogenesis [14, 15]. Of note, the role for O-GlcNAcylation in breast cancer remains an area of intense debate; however, increasing data from multiple groups suggest that O-GlcNAcylation levels are high in primary and metastatic breast cancer, although these studies have primarily been conducted in breast cancer cell line models [16–21]. Herein, we show that PR is O-GlcNAcylated, and this modification promotes the transcriptional activity of PR. These data argue that modifying/blocking PR O-GlcNAcylation could have significant impact on PR transcriptional activity in breast cancer.
Methods
Cell Lines and Constructs
T47D-co, T47D-Y, T47D-YA, T47D-YB, and T47D-S79/81A PR cells have been previously described and were a generous gift of Dr. Carol Lange (Minnesota) [9, 22]. MCF7 cells were cultured in RPMI-1640 with 10% FBS, 1% Penn/Strep, 6 ng/ml insulin, and 1% non-essential amino acids.
Cells were treated with the following reagents (when applicable), as described in the figure legends: R5020 (10 nM; Sigma) and Thiamet-G (TMG; 10uM; S.D. Specialty Chemicals).
RIME
RIME experiments were performed as previously described [23–25] in R5020-treated (10 nM for 60 min) T47D-YB or T47D-Y cell lines that were cross-linked with 1% formaldehyde for 10 min. Immunoprecipitations were performed using a PR antibody (20μg, Santa Cruz Biotechnology, sc-7208). The peptide samples were analyzed on a Dionex Ultimate 3000 UHPLC system coupled with the LTQ Orbitrap velos mass spectrometer (Thermo Scientific). For the separation of the peptides, a multistep gradient elution was used: Mobile phase (A) was composed of 2% acetonitrile, 0.1% formic acid, and 5% DMSO, and mobile phase (B) was composed of 80% acetonitrile, 0.1% formic acid, and 5% DMSO. The gradient elution method at flow rate 300 nL/min was as follows: for 65 min gradient up to 45% (B), for 10 min gradient up to 95% (B), for 10 min isocratic 95% (B), for 5 min down to 5% (B), and for 10 min isocratic equilibration 5% (B) at 40 °C. The full scan was performed in the Orbitrap in the range of 400–1600 m/z at 60 K resolution. The MS2 scan was performed with CID collision energy 30% and exclusion duration 30s. The raw data were processed in Proteome Discoverer 1.4 using the SequestHT search engine. The node for SequestHT included the following parameters: precursor mass tolerance 20 ppm, fragment mass tolerance 0.5 Da, dynamic modifications were oxidation of M (+ 15.995 Da), and deamidation of N, Q (+ 0.984 Da). Significant peptides were filtered at FDR < 1%, and specific interactors were considered if they were identified in both RIME replicate experiments in T47D-YB cells, but not in the PR-null cell line.
Breast Cancer Tissue Microarray
Tissue microarrays (TMAs) were constructed from archival formalin-fixed, paraffin-embedded samples of invasive mammary carcinoma (43 patients) as well as matched benign breast tissue (35 patients). These samples were identified from the pathology departmental archives of the University of Kansas Medical Center from 1997 to 2011. Based on review of the original pathology reports, the invasive mammary carcinomas were typed as invasive ductal carcinoma (39 patients), metaplastic carcinoma (1 patient), invasive mammary carcinoma with ductal and lobular features (1 patient), invasive ductal carcinoma with a minor lobular component (1 patient), and invasive lobular carcinoma (1 patient). Using the semi-automated TMArrayer (Pathology Devices, Inc., Westminster, MD), TMA paraffin blocks were assembled with 2.0 mm cores.
Immunohistochemistry
O-GlcNAc and OGT antibodies (see Immunoblotting) were used for Immunohistochemical staining according to the following procedure: Four micron paraffin sections are mounted on Fisherbrand Superfrost slides and baked for 60 min at 60 °C then deparaffinized. Epitope retrieval was performed in Biocare Decloaking Chamber (pressure cooker), under pressure for 5 min, using pH 6.0 Citrate buffer followed by a 10-min cool down period. Endogenous peroxidase is blocked with 3% H2O2 for 10 min followed by incubation with O-GlcNAc (1:300) or OGT (1:100) primary antibody for 30 min, followed by Envision+ Anti-Mouse, (Dako; Carpinteria, CA) for 30 min and DAB+ chromogen (Dako) for 5 min. Immunohistochemical staining was performed using the IntelliPATH FLX Automated Stainer at room temperature. A light hematoxylin counterstain was performed, following which the slides were dehydrated, cleared, and mounted using permanent mounting media. A pathologist then scored the slides according to the intensity of staining (0 = no staining, 1 = mild intensity, 2 = moderate intensity, 3 = strong intensity), percentage of positive cells, and the subcellular localization of the staining. Statistical analysis: the difference in O-GlcNAc staining intensities between cancer and normal specimens was assessed using Wilcoxon signed rank test. The Wilcoxon signed rank test compares the number of times a score from one group is ranked higher than a score from another group pairwise. The Wilcoxon rank sum test was used to assess the differences in the OGT intensities between two groups of subjects (PR-positive vs PR-negative tumors).
Co-immunoprecipitations
For Co-IP experiments, cell lysates were collected in radioimmunoprecipitation assay (RIPA) buffer (supplemented with protease/phosphatase inhibitors) and incubated on ice for 30 min. Cell lysates containing equivalent protein concentrations (1000 μg) were incubated overnight at 4 °C with 2 μg appropriate antibody or control IgG. Protein G agarose (Roche Diagnostics, Indianapolis, IN) was added for the final 2 h of incubation time. Immune complexes were washed three times with supplemented RIPA buffer, resuspended in Laemmli sample buffer containing β-mercaptoethanol, boiled for 5 min, and subjected to Western blotting analysis.
Immunoblotting
Immunoblotting/Western blotting was performed as previously described [9, 22, 26]. Membranes were probed with primary antibodies recognizing total PR (Santa Cruz Biotechnology, sc-7208 or ThermoScientific, MS-298-P), OGT (a kind gift from Dr. Gerald Hart at the Johns Hopkins School of Medicine), and O-GlcNAc (Abcam, ab2739). All Western blotting experiments were performed in triplicate, and representative experiments are shown.
Real-Time qPCR
RNA isolation, cDNA creation, and qPCR were performed as previously described [9, 22, 26], with modifications noted here and in the figure legends. qPCR was performed using the Faststart Essential DNA Green Master (Roche) on a Roche LightCycler96. Relative concentrations were quantified using the LightCycler96 (Roche, Software 1.1, Absolute Quantification Analysis), using a six-point standard curve.
Subcellular Fractionation
Subcellular fractionation studies were performed as described previously [27], with modifications noted in the figure legend.
Statistics
Statistical significance for all qPCR experiments was determined using an unpaired Student’s t test, unless otherwise specified. A p value ≤ 0.05 is considered statistically significant.
Results
PR Interacts with OGT
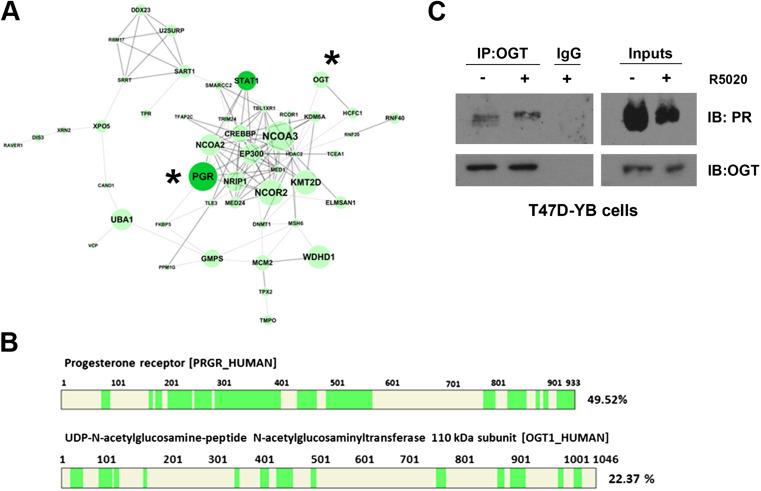
Our lab has a long-standing interest in defining PR protein-protein interaction complexes and defining how these protein interactions alter PR transcriptional activity in breast cancer. We used Rapid Immunoprecipitation Mass spectrometry of Endogenous proteins or RIME, a technique developed by the Carroll lab [23–25], as an unbiased approach to identify novel PR protein-protein interactions. This technique allows for the identification of protein interactors (using mass spectrometry) of nuclear proteins, making this technique ideal for studying protein interactions of transcriptional co-factors and chromatin-associated proteins. A brief note about our most frequently used model system: PR is an important target gene of ER and, as such, PR expression is regulated by estrogen in most tissues [28, 29]. In order to differentiate between the effects of ER/estrogen and PR/progesterone, our laboratory uses PR-positive (T47D-co) and PR-null (T47D-Y) variants of the ER/PR+ breast cancer cell line, T47D [30]. T47D-co cells endogenously express both isoforms of PR, PR-A and PR-B, without the need for exogenously added estrogen, allowing us to study the function of PR without the confounding effects of estrogen. T47D-Y (PR-null) cells can also be used to reintroduce single isoform variants of PR, such as PR-A (T47D-YA cells) and PR-B (T47D-YB cells). We have published extensively using these cell line models to define isoform- and phosphorylation-specific PR gene regulation and protein-protein interactions [4, 9, 22, 26, 31, 32], and this cell line model remains a powerful and well-established system for studying PR activity [3]. We used T47D-YB (stably expressing the full length PR-B isoform) and PR-null (T47D-Y) cells as a model system to study PR protein interactions using RIME. Briefly, T47D-YB and T47D-Y cells were cross-linked following treatment with a synthetic PR ligand (R5020) or vehicle (EtOH). PR was immunoprecipitated (IP’d) using a PR antibody from isolated nuclear lysates, and PR-interacting proteins were analyzed using mass spectrometry. Among the top 60 identified PR-interacting proteins was OGT, the enzyme that catalyzes the addition of O-GlcNAc to target proteins (Fig. 1a); peptide sequence coverage information for both PR and OGT is shown in Fig. 1b. We verified the interaction between PR and OGT in T47D-YB (Fig. 1c) using co-immunoprecipitation (co-IP); PR could be detected in a complex with IP’d OGT in vehicle and ligand (R5020)-treated cells. Both PR-A and PR-B can interact with OGT, as both isoforms were detected in OGT immunoprecipitates from T47D-co cells (endogenously expressing both PR isoforms; Supplementary Fig. 1).
Fig. 1.
PR interacts with OGT. a STRING network of the top 60 PR interactors identified in RIME experiments. The size of the node increases proportionally to the number of identified peptides, and thick edges denote high confidence STRING interactions (0.7–0.99). RIME was performed in T47D-YB cells on two biological replicates, and proteins identified in PR-null cells were not included in the PR-specific interactor list. b Sequence coverage of PR and OGT in both replicate RIME experiments. Green highlights high confidence peptides at FDR < 1%. PR and OGT have been identified by 27 and 12 unique peptides, respectively. c OGT was immunoprecipitated from T47D-YB cell lysates (+/− R5020), and the resulting associated protein complexes were analyzed by Western blotting. Right panels represent total input cell lysates (color figure online)
PR Is Modified by O-GlcNAc
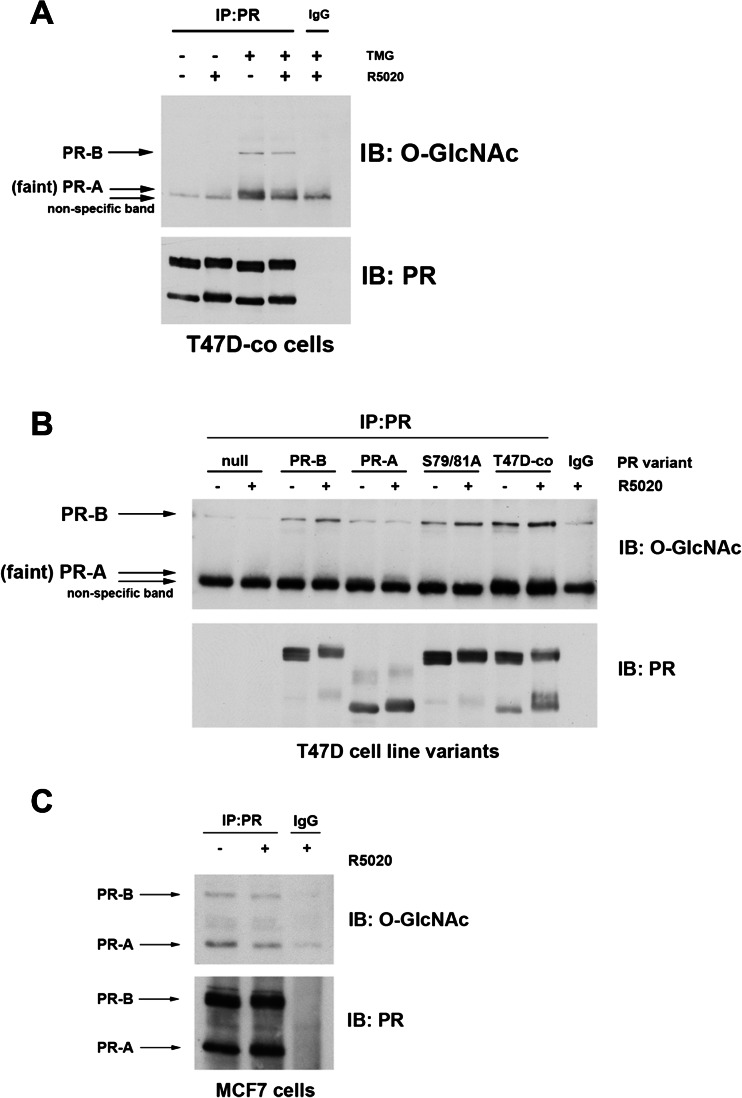
The interaction between PR and OGT suggested that PR may be modified by O-GlcNAc. Using an antibody that recognizes the O-GlcNAc modification, we detected O-GlcNAcylation on PR immunoprecipitated from T47D-co cells, both basally (ligand-independent) and in response to PR ligand (Fig. 2a). In order to detect O-GlcNAcylation, cells were pre-treated with Thiamet-G (TMG), a commonly used competitive inhibitor of OGA, which leads to decreased O-GlcNAc cycling (reduced turnover of the modification), thus allowing for an increase in detectable O-GlcNAcylated proteins [27, 33–35]. O-GlcNAcylated PR was undetectable in the non-TMG-treated cells, indicating that this modification is cycled on and off rapidly under normal conditions. Although the O-GlcNAcylated PR-A band is faint (and situated just above a non-specific band), it appears that both PR-B and PR-A are modified by O-GlcNAc. To further characterize PR O-GlcNAcylation, we measured levels of O-GlcNAc in TMG-treated T47D cells stably expressing only PR-B, PR-A, a phospho-mutant version of PR that we have previously characterized (S79/81A), parental T47D-co (expresses both PR isoforms), and PR-null (T47D-Y) cells serve as the negative control (Fig. 2b). As previously seen in Fig. 2a, we observed O-GlcNAcylation on both PR-B and PR-A (faint) isoforms. Phosphorylation and O-GlcNAcylation can often compete for the same target serines/threonine residues. As the S79/81A PR phospho-mutant remains O-GlcNAcylated, these sites do not appear to be critical for blocking/promoting PR O-GlcNAcylation. Finally, these data were repeated in two additional PR positive breast cancer cell lines: MCF7 (Fig. 2c) and BT474 (data not shown) cells. In both cell lines, PR-B and PR-A were modified by O-GlcNAc. Cumulatively, these data show that PR is post-translationally modified by O-GlcNAc.
Fig. 2.
PR is post-translationally modified by O-GlcNAc. a PR was immunoprecipitated from T47D-co cell lysates pre-treated with TMG for 18 h, followed by R5020 treatment for 60 min. The resulting associated protein complexes were analyzed for O-GlcNAcylation by Western blotting. b Experiments were performed as in a, but the following T47D cell line variants were used: T47D-YB, T47D-YA, T47D-S79/81A, T47D-co, and T47D-Y (PR-null). All cells were pre-treated with TMG for 18 h. c Experiments were performed as in b in MCF7 cells. All experiments were performed in triplicate, and representative experiments are shown here
PR Levels Are Lowered by O-GlcNAc
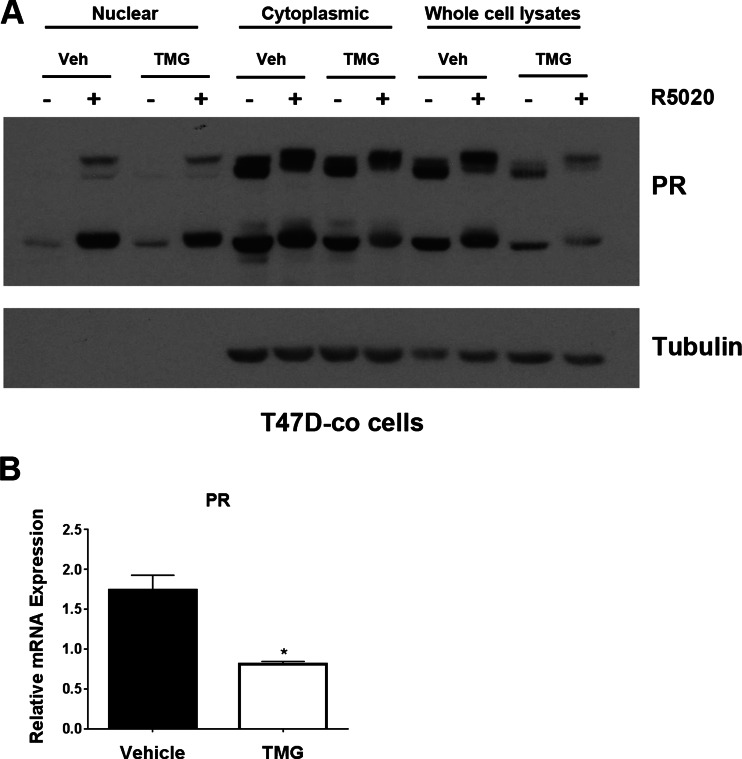
To determine how persistently elevated O-GlcNAc levels affect PR function, we treated cells with TMG (or vehicle control) for 3 weeks, thus stably elevating O-GlcNAc levels. Post-translational modifications on PR have been linked with differences in PR nucleo-cytoplasmic shuttling [4]. We assayed how O-GlcNAcylation on PR affected its transit into the nucleus in cells treated with TMG. Under normal conditions in response to progestins, PR translocates to the nucleus to bind DNA and activate target genes. We observed PR nuclear translocation in both vehicle and TMG-treated cells; however, there was less nuclear PR overall in the TMG-treated cells (Fig. 3a; left portion of the gel). Similarly, there was less PR in the cytoplasmic fraction of TMG-treated cells (middle portion). These data reflected what was seen in the whole cell lysates (right portion): cells treated with TMG had less total PR, and this was observed throughout the cell and not just in one particular compartment. qPCR revealed that PR messenger RNA (mRNA) was similarly decreased in TMG-treated cells, suggesting that the downregulation of PR occurs at the transcriptional/mRNA level (Fig. 3b). Together, these data suggest that high levels of O-GlcNAc lead to downregulation of PR.
Fig. 3.
PR levels are lower in TMG-treated cells. a T47D-co cells were continually passed in the presence of TMG (or vehicle) for 3 weeks, followed by R5020 or vehicle (EtOH) for 1 h. Nuclear, cytoplasmic, and whole cell lysates were separated and blotted with antibodies to PR and tubulin (cytoplasmic marker and loading control). b Cells were treated with TMG as in a, and RNA was isolated after 3 weeks of TMG treatment. Isolated RNA was analyzed for PR. Gene values were normalized to an internal control (β-actin). Error bars represent standard deviation between biological triplicates. Asterisks represent statistical significance between the vehicle and TMG-treated groups; p < 0.05, as determined using an unpaired Student’s t test. The experiments shown here were performed in triplicate, and a representative experiment is shown here
O-GlcNAcylated PR Has Altered Transcriptional Activity
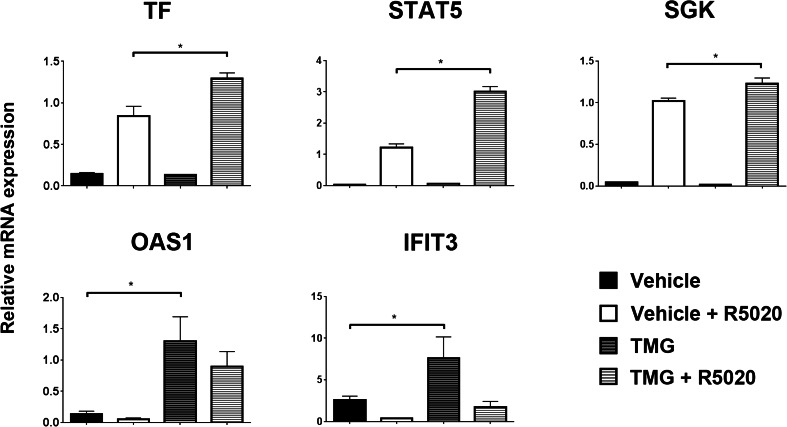
To determine how elevated O-GlcNAc affects the transcription factor activity of PR, we measured transcription of key PR-target genes in TMG-treated cells. Interestingly, and surprisingly given that PR protein levels were lower in TMG-treated cells, we saw that the transcriptional response following treatment with PR ligand (R5020) was potentiated in cells treated with TMG, as compared to the vehicle control (Fig. 4—top). These data suggest that O-GlcNAcylation on PR makes it a more potent transcription factor at activating gene expression in response to ligand. Moreover, these data indicate that although there is less PR protein under conditions where O-GlcNAc is high (see Fig. 3), the remaining PR is more active (as measured by transcriptional activation of PR-target genes). These data were replicated in another PR positive breast cancer cell line, BT474: PR RNA levels were decreased in TMG-treated cells; however, the remaining PR was more transcriptionally active on PR target genes (data not shown). Of note, our lab recently described the PR-dependent transcriptional repression of a subset of genes called interferon-stimulated genes (ISGs) [26]. In addition to ligand-dependent transcriptional repression, ISGs are also regulated by PR in the absence of ligand; in cells where PR levels have been knocked-down, basal levels of ISGs are high when compared to cells expressing PR. Interestingly, we observed the same phenotype of high basal ISG expression in TMG-treated cells, another condition where PR levels are reduced; levels of OAS1 and IFIT3 are shown as representative examples (Fig. 4—bottom).
Fig. 4.
PR-target genes are differentially regulated by O-GlcNAcylated PR. Starved T47D-co cells were continually passed in the presence of TMG (or vehicle) for 3 weeks, followed by R5020 or vehicle (EtOH) for 6 h. Isolated RNA was analyzed for select genes. Gene values were normalized to an internal control (β-actin). Error bars represent standard deviation between biological triplicates. Asterisks represent statistical significance; p < 0.05, as determined using an unpaired Student’s t test. This experiment was performed in triplicate, and a representative experiment is shown here
O-GlcNAc Levels Are Elevated in Breast Cancer
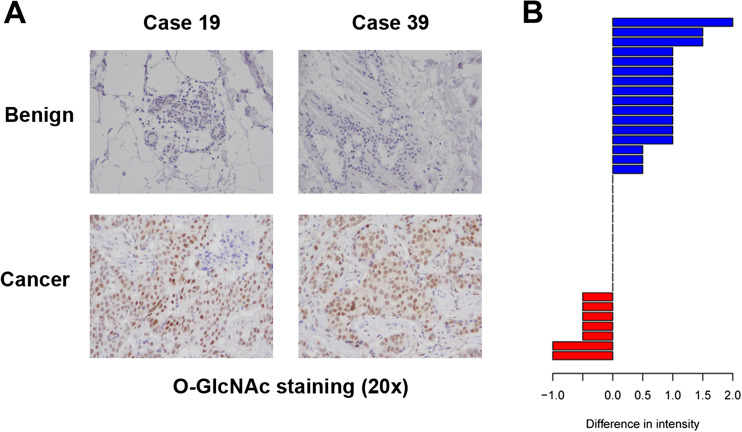
Previous studies looking at levels of O-GlcNAc in breast cancer have primarily been conducted in human breast tumor cell lines. To determine if O-GlcNAcylation levels are increased in breast cancer patient samples compared to matched benign tissue, we created a breast tissue microarray (TMA) composed of specimens collected from 43 breast cancer patients seen at the University of Kansas Medical Center; 22 of the breast cancers were PR-positive. De-identified clinical characteristics, including PR-positivity, are presented in Supplemental Table 1. Additionally, benign breast tissue was collected from 35 of these patients, thereby representing matched benign and tumor tissue obtained from the same patient. These 35 paired samples are a powerful tool to measure how protein expression changes in tumor tissues as compared to the matched benign controls, within the same patient. We stained this breast TMA with antibodies to recognize O-GlcNAc, and staining intensities were scored by a clinical pathologist. Interestingly, and in agreement with what has previously been reported in breast cancer cell lines [20], the average O-GlcNAc intensity staining levels were significantly higher in the cancer samples when compared to their matched benign controls (n = 35; p = 0.007); select examples are shown in Fig. 5a. There was no correlation between O-GlcNAc staining intensity and PR-positivity, perhaps due to the small number of PR-positive samples. When representing the staining intensity data as changes within each individual patient, when O-GlcNAc levels change between benign and cancer, significantly more patients show an increase in O-GlcNAc staining (blue bars) intensity in the cancer tissue (Fig. 5b). These data show that O-GlcNAc levels increase in breast cancer.
Fig. 5.
O-GlcNAc levels are high in breast cancer. a Tissue microarray (TMA) analysis was performed using immunohistochemical staining with an O-GlcNAc antibody. Select benign and normal cases from the same patient are shown here at × 20 magnification. b Staining intensity changes between normal and cancer are shown for each patient in the TMA for which matched benign and cancer tissue was collected (n = 35). Data is shown in bar graph format; each bar represents an individual patient. Blue bars (top) represent an increase in staining intensity when comparing cancer to benign; red bars represent a decrease in staining intensity when comparing cancer to benign. No bar indicates unchanged. The Wilcoxon signed rank test was used to compare O-GlcNAc levels between matched benign and cancer samples, as described in the “Methods” section (color figure online)
OGT Levels in Breast Cancer Correlate with PR Expression
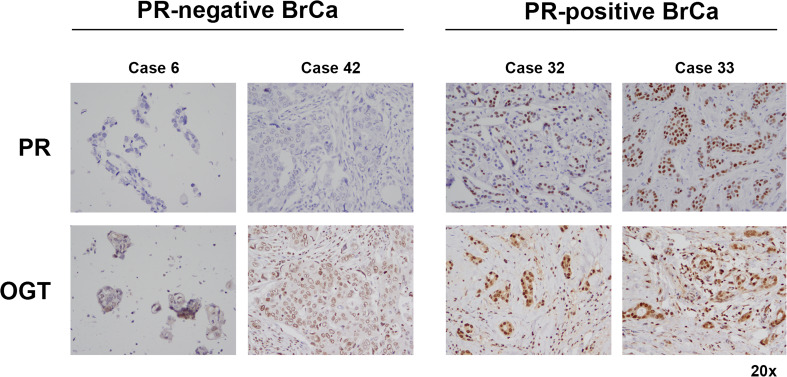
Given the interaction between PR and OGT, we used the breast TMA to determine if a correlation existed between PR and OGT expression in breast cancer patients. As above, the breast cancer TMA was stained with OGT and staining intensity was scored by a clinical pathologist. Interestingly, PR-positive breast cancers (22 out of 43 total breast cancers on the TMA) had significantly higher staining intensity when compared to PR-negative breast cancers (p = 0.02) for OGT cytoplasmic staining; OGT nuclear staining also trended towards significance as higher in PR-positive tumors (p = 0.08). Select images of PR-positive and PR-negative tumors with correlating OGT staining are shown in Fig. 6. These data indicate that there is a correlation between PR positivity and OGT expression in breast cancer.
Fig. 6.
OGT levels are higher in PR-positive breast cancers. Tissue microarray analysis was performed using immunohistochemical staining with PR and OGT antibodies. Select PR-negative (left) and PR-positive (right) breast cancer (BrCa) cases are shown here at × 20 magnification. The Wilcoxon rank sum test was used to compare OGT levels in PR-positive vs PR-negative breast cancer, as described in the “Methods” section
Discussion
Herein, we present data that O-GlcNAc levels are high in breast cancer and that PR-positive patients have significantly higher levels of OGT expression, the enzyme that catalyzes the addition of O-GlcNAc to target proteins. We show an interaction between PR and OGT and show that this interaction leads to O-GlcNAcylation on PR. Under conditions where O-GlcNAc levels are high and PR is O-GlcNAcylated, there is a reduced amount of PR; however, this remaining fraction of PR is more transcriptionally active, as key PR-target genes are more potently activated when PR is O-GlcNAcylated. Cumulatively, these data suggest that high O-GlcNAc levels seen in breast cancer may drive/alter PR transcriptional programs in breast cancer.
Our breast cancer TMA data showed a positive correlation between PR-positivity and OGT expression. As OGT is required to add O-GlcNAc to target proteins, these data imply that PR-positive breast cancers are not only positive for PR, but that a population of that PR is very likely O-GlcNAcylated. Site mapping of the O-GlcNAc site(s) on PR is underway and will allow for the creation of an O-GlcNAc-PR-specific antibody to further test this correlation. Moreover, the PR-target gene data presented herein suggest that O-GlcNAcylated PR is more transcriptionally active, promoting transcription of classic PR-target genes. Interestingly, a family of PR-target genes whose basal levels are increased under conditions where PR O-GlcNAcylation is high belong to a family of genes involved in interferon signaling, called interferon-stimulated genes (ISGs). We have recently described PR-dependent transcriptional repression of ISGs [26]. Of note clinically, high expression of these ISGs has been correlated with resistance to chemotherapy and radiation therapy in breast cancer patients [36, 37]. These data presented herein, where ISG levels are basally higher in conditions where PR is O-GlcNAcylated, could translate to negative therapeutic responses in breast cancer patients. Cumulatively, these data suggest that modulation of PR transcriptional activity by O-GlcNAc underscore the importance of understanding how this post-translational modification on PR may alter PR action in breast tumorigenesis.
O-GlcNAcylation has been shown on many different transcription factors (reviewed in [11, 38]). Most notably, and related to the O-GlcNAcylation of PR, is the reported modification of murine ER by O-GlcNAc [39, 40]. Further work showed that elevated O-GlcNAcylation lead to decreased RNA and protein levels of ER, due to a transcriptional repression mechanism possibly involving lack of function of key transcription factors (i.e., Sp1) needed to activate transcription of the ESR1 gene [41]. These data presented herein suggest a similar phenotype for O-GlcNAcylated PR, as we observe less PR RNA and protein levels under conditions where PR is O-GlcNAcylated. Experiments are currently underway to determine the mechanism through which PR mRNA is downregulated when O-GlcNAc levels are high. Finally, our data are in agreement with previous published reports of lower levels of PR in conditions where O-GlcNAc levels were high [42]. However, the work presented herein is the first reporting of direct modification of PR by O-GlcNAcylation. O-GlcNAcylation of other nuclear receptors known to be highly expressed in ER/PR-positive tumors, such as glucocorticoid receptor and androgen receptor, has not been published.
Interestingly, although there is less PR under conditions where O-GlcNAc levels are high, the remaining PR is more transcriptionally active on PR-target genes. This somewhat paradoxical situation suggests a complex regulation between PR RNA/protein levels and PR presence/recruitment to PR-target genes. Similar phenotypes have been observed with phosphorylated PR; PR target-gene programs linked to phosphorylation on PR Ser294 are active in human breast tumors that have been clinically designated as PR-null or PR-low [43]. These data suggest that a small minority of highly active PRs are driving phospho-PR-dependent transcriptional programs in these breast cancers. Moreover, phosphorylation on PR is linked to increased protein turnover of PR (reviewed in [44]). These same phenotypes (less protein, but more activity) may be true for O-GlcNAcylated PR, as both post-translational modifications may be regulating PR activity in a similar fashion. We find this result, less PR protein with more activity, very intriguing, and certainly warrants further investigation. The creation of a PR O-GlcNAc-specific antibody will aid in these studies.
In summary, our results show that O-GlcNAcylation is upregulated in breast cancer patients. The positive correlation between PR and OGT levels in PR-positive breast cancers creates a scenario where PR may be more heavily O-GlcNAcylated, and thus more transcriptionally active, in breast cancer. These data suggest that targeting PR O-GlcNAcylation may be a novel mechanism for altering PR action in breast tumors.
Electronic Supplementary Material
Both isoforms of PR interact with OGT. OGT was immunoprecipitated from T47D-co cell lysates (+/- R5020) and the resulting associated protein complexes were analyzed by Western blotting. Right panels represent total input cell lysates. (DOCX 1545 kb)
Deidentified patient data for tumors included on the breast cancer tissue microarray. (PDF 10 kb)
Acknowledgements
We acknowledge support from the University of Kansas (KU) Cancer Center’s Biospecimen Repository Core Facility staff, in particular Tara Meyer, for helping obtain human specimens and for performing histological work.
Funding Information
The authors also acknowledge support from the KU Cancer Center’s Cancer Center Support Grant (P30 CA168524). This work was supported by R01DK100595 (CS), R00CA166643 (CRH), DOD BCRP W81XWH-16-1-0320 (CRH), Susan G Komen Foundation CCR16376147 (CRH), V Foundation V2015-025 (CRH), and the NCI Cancer Center Support Grant P30 CA168524 (CRH).
Compliance with Ethical Standards
Conflict of Interest
The authors declare no conflict of interest.
References
- 1.Beral V. Breast cancer and hormone-replacement therapy in the Million Women Study. Lancet. 2003;362(9382):419–427. doi: 10.1016/S0140-6736(03)14065-2. [DOI] [PubMed] [Google Scholar]
- 2.Anderson GL, et al. Effects of conjugated equine estrogen in postmenopausal women with hysterectomy: the Women’s Health Initiative randomized controlled trial. JAMA. 2004;291(14):1701–1712. doi: 10.1001/jama.291.14.1775. [DOI] [PubMed] [Google Scholar]
- 3.Carroll JS, et al. Deciphering the divergent roles of progestogens in breast cancer. Nat Rev Cancer. 2017;17(1):54–64. doi: 10.1038/nrc.2016.116. [DOI] [PubMed] [Google Scholar]
- 4.Hagan CR, et al. Role of phosphorylation in progesterone receptor signaling and specificity. Mol Cell Endocrinol. 2012;357(1–2):43–49. doi: 10.1016/j.mce.2011.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gregory CW, et al. Epidermal growth factor increases coactivation of the androgen receptor in recurrent prostate cancer. J Biol Chem. 2004;279(8):7119–7130. doi: 10.1074/jbc.M307649200. [DOI] [PubMed] [Google Scholar]
- 6.Wilson GR, et al. Activated c-SRC in ductal carcinoma in situ correlates with high tumour grade, high proliferation and HER2 positivity. Br J Cancer. 2006;95(10):1410–1414. doi: 10.1038/sj.bjc.6603444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Steeg PS, Zhou Q. Cyclins and breast cancer. Breast Cancer Res Treat. 1998;52(1–3):17–28. doi: 10.1023/A:1006102916060. [DOI] [PubMed] [Google Scholar]
- 8.Tawfic S, et al. Protein kinase CK2 signal in neoplasia. Histol Histopathol. 2001;16(2):573–582. doi: 10.14670/HH-16.573. [DOI] [PubMed] [Google Scholar]
- 9.Hagan CR, Knutson TP, Lange CA. A common docking domain in progesterone receptor-B links DUSP6 and CK2 signaling to proliferative transcriptional programs in breast cancer cells. Nucleic Acids Res. 2013;41(19):8926–8942. doi: 10.1093/nar/gkt706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leehy KA, et al. Modifications to glucocorticoid and progesterone receptors alter cell fate in breast cancer. J Mol Endocrinol. 2016;56(3):R99–R114. doi: 10.1530/JME-15-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Slawson C, Hart GW. O-GlcNAc signalling: implications for cancer cell biology. Nat Rev Cancer. 2011;11(9):678–684. doi: 10.1038/nrc3114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hart GW, et al. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu Rev Biochem. 2011;80:825–858. doi: 10.1146/annurev-biochem-060608-102511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Queiroz RM, Carvalho E, Dias WB. O-GlcNAcylation: the sweet side of the cancer. Front Oncol. 2014;4:132. doi: 10.3389/fonc.2014.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shaw P, et al. Regulation of specific DNA binding by p53: evidence for a role for O-glycosylation and charged residues at the carboxy-terminus. Oncogene. 1996;12(4):921–930. [PubMed] [Google Scholar]
- 15.Chou TY, Hart GW, Dang CV. c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J Biol Chem. 1995;270(32):18961–18965. doi: 10.1074/jbc.270.32.18961. [DOI] [PubMed] [Google Scholar]
- 16.Gu Y, et al. GlcNAcylation plays an essential role in breast cancer metastasis. Cancer Res. 2010;70(15):6344–6351. doi: 10.1158/0008-5472.CAN-09-1887. [DOI] [PubMed] [Google Scholar]
- 17.Champattanachai V, et al. Proteomic analysis and abrogated expression of O-GlcNAcylated proteins associated with primary breast cancer. Proteomics. 2013;13(14):2088–2099. doi: 10.1002/pmic.201200126. [DOI] [PubMed] [Google Scholar]
- 18.Ferrer CM, et al. O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol Cell. 2014;54(5):820–831. doi: 10.1016/j.molcel.2014.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Krzeslak A, et al. Gene expression of O-GlcNAc cycling enzymes in human breast cancers. Clin Exp Med. 2012;12(1):61–65. doi: 10.1007/s10238-011-0138-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Caldwell SA, et al. Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene. 2010;29(19):2831–2842. doi: 10.1038/onc.2010.41. [DOI] [PubMed] [Google Scholar]
- 21.Sodi VL, et al. mTOR/MYC Axis regulates O-GlcNAc transferase expression and O-GlcNAcylation in breast cancer. Mol Cancer Res. 2015;13(5):923–933. doi: 10.1158/1541-7786.MCR-14-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagan CR, et al. ck2-Dependent phosphorylation of progesterone receptors (PR) on Ser81 regulates PR-B isoform-specific target gene expression in breast cancer cells. Mol Cell Biol. 2011;31(12):2439–2452. doi: 10.1128/MCB.01246-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mohammed H, et al. Endogenous purification reveals GREB1 as a key estrogen receptor regulatory factor. Cell Rep. 2013;3(2):342–349. doi: 10.1016/j.celrep.2013.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mohammed H, et al. Progesterone receptor modulates ERalpha action in breast cancer. Nature. 2015;523(7560):313–317. doi: 10.1038/nature14583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohammed H, et al. Rapid immunoprecipitation mass spectrometry of endogenous proteins (RIME) for analysis of chromatin complexes. Nat Protoc. 2016;11(2):316–326. doi: 10.1038/nprot.2016.020. [DOI] [PubMed] [Google Scholar]
- 26.Walter K, Goodman M, Singhal H, et al (2017) Interferon-stimulated genes are transcriptionally repressed by PR in breast cancer. Mol Cancer Res. 10.1158/1541-7786.MCR-17-0180. (in press) [DOI] [PMC free article] [PubMed]
- 27.de Queiroz RM, et al. Changes in O-linked N-acetylglucosamine (O-GlcNAc) homeostasis activate the p53 pathway in ovarian cancer cells. J Biol Chem. 2016;291(36):18897–18914. doi: 10.1074/jbc.M116.734533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horwitz KB, Koseki Y, McGuire WL. Estrogen control of progesterone receptor in human breast cancer: role of estradiol and antiestrogen. Endocrinology. 1978;103(5):1742–1751. doi: 10.1210/endo-103-5-1742. [DOI] [PubMed] [Google Scholar]
- 29.Horwitz KB, McGuire WL. Estrogen control of progesterone receptor induction in human breast cancer: role of nuclear estrogen receptor. Adv Exp Med Biol. 1979;117:95–110. doi: 10.1007/978-1-4757-6589-2_5. [DOI] [PubMed] [Google Scholar]
- 30.Sartorius CA, et al. New T47D breast cancer cell lines for the independent study of progesterone B- and A-receptors: only antiprogestin-occupied B-receptors are switched to transcriptional agonists by cAMP. Cancer Res. 1994;54(14):3868–3877. [PubMed] [Google Scholar]
- 31.Dressing GE, et al. Progesterone receptor-cyclin d1 complexes induce cell cycle-dependent transcriptional programs in breast cancer cells. Mol Endocrinol. 2014;28(4):442–457. doi: 10.1210/me.2013-1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hagan CR, Faivre EJ, Lange CA. Scaffolding actions of membrane-associated progesterone receptors. Steroids. 2009;74(7):568–572. doi: 10.1016/j.steroids.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang Z, et al. O-GlcNAcase expression is sensitive to changes in O-GlcNAc homeostasis. Front Endocrinol (Lausanne) 2014;5:206. doi: 10.3389/fendo.2014.00206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tan EP, et al. O-linked N-acetylglucosamine cycling regulates mitotic spindle organization. J Biol Chem. 2013;288(38):27085–27099. doi: 10.1074/jbc.M113.470187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Z, et al. O-linked N-acetylglucosamine (O-GlcNAc) transferase and O-GlcNAcase interact with Mi2beta protein at the Agamma-globin promoter. J Biol Chem. 2016;291(30):15628–15640. doi: 10.1074/jbc.M116.721928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Weichselbaum RR, et al. An interferon-related gene signature for DNA damage resistance is a predictive marker for chemotherapy and radiation for breast cancer. Proc Natl Acad Sci U S A. 2008;105(47):18490–18495. doi: 10.1073/pnas.0809242105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Minn AJ. Interferons and the immunogenic effects of cancer therapy. Trends Immunol. 2015;36(11):725–737. doi: 10.1016/j.it.2015.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ozcan S, Andrali SS, Cantrell JE. Modulation of transcription factor function by O-GlcNAc modification. Biochim Biophys Acta. 2010;1799(5–6):353–364. doi: 10.1016/j.bbagrm.2010.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cheng X, et al. Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor beta. Biochemistry. 2000;39(38):11609–11620. doi: 10.1021/bi000755i. [DOI] [PubMed] [Google Scholar]
- 40.Cheng X, Hart GW. Alternative O-glycosylation/O-phosphorylation of serine-16 in murine estrogen receptor beta: post-translational regulation of turnover and transactivation activity. J Biol Chem. 2001;276(13):10570–10575. doi: 10.1074/jbc.M010411200. [DOI] [PubMed] [Google Scholar]
- 41.Kanwal S, et al. O-GlcNAcylation-inducing treatments inhibit estrogen receptor alpha expression and confer resistance to 4-OH-tamoxifen in human breast cancer-derived MCF-7 cells. PLoS One. 2013;8(7):e69150. doi: 10.1371/journal.pone.0069150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bowe DB, et al. O-GlcNAc integrates the proteasome and transcriptome to regulate nuclear hormone receptors. Mol Cell Biol. 2006;26(22):8539–8550. doi: 10.1128/MCB.01053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Knutson TP, et al. Posttranslationally modified progesterone receptors direct ligand-specific expression of breast cancer stem cell-associated gene programs. J Hematol Oncol. 2017;10(1):89. doi: 10.1186/s13045-017-0462-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hagan CR, Lange CA. Molecular determinants of context-dependent progesterone receptor action in breast cancer. BMC Med. 2014;12:32. doi: 10.1186/1741-7015-12-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Both isoforms of PR interact with OGT. OGT was immunoprecipitated from T47D-co cell lysates (+/- R5020) and the resulting associated protein complexes were analyzed by Western blotting. Right panels represent total input cell lysates. (DOCX 1545 kb)
Deidentified patient data for tumors included on the breast cancer tissue microarray. (PDF 10 kb)