Summary
Protein-protein interactions mediate essential cellular processes, however the detection of native interactions is challenging since they are often low affinity and context dependent. Here, we develop a chemical proteomics approach in vivo CLASPI [iCLASPI] (in vivo crosslinking-assisted and stable isotope labeling by amino acids in cell culture [SILAC}-based protein identification) relying upon photo-crosslinking, amber suppression, and SILAC-based quantitative proteomics to profile context-dependent protein-protein interactions in living cells. First, we use iCLASPI to profile in vivo binding partners of the N-terminal tails of soluble histone H3 or H4. We identify known histone chaperones and modifying proteins, thereby validating our approach, and find an interaction between soluble histone H3 and UBR7, an E3 ubiquitin ligase, mediated by UBR7’s PHD domain. Furthermore, we apply iCLASPI to profile the context-dependent protein-protein interactions of chromatin-associated histone H3 at different cell cycle stages, and identify ANP32A as a mitosis-specific interactor. Our results demonstrate that the iCLASPI approach can provide a general strategy for identifying native, context-dependent direct protein-protein interactions using photo-crosslinking and quantitative proteomics.
eTOC

Protein-protein interactions mediate essential biological processes, but characterizing these interactions in cells presents a major challenge. Kleiner et al. describe a chemical proteomics approach combining photocrosslinking and SILAC-based proteomics to characterize context-dependent direct protein-protein interactions in live cells. They apply their approach to profile interactions involving core histones H3 and H4.
Introduction
The identification of protein interaction networks provides important insights into protein function and regulation (Aebersold and Mann, 2016). Notably, advances in mass spectrometry-based proteomics have enabled large-scale interactome analysis of many essential cellular proteins (Kim et al., 2014; Wilhelm et al., 2014); however, identifying direct protein-protein interactions by mass spectrometry (or other methods) typically requires stringent affinity purification of binding partners, which remains challenging since the underlying interactions can be low affinity and context dependent.
Low-affinity protein-protein interactions can be stabilized by chemical crosslinking with bifunctional crosslinkers; however, such approaches are often inefficient because they require precise orientation of compatible amino acid side chains and delivery of the reactive crosslinker to the interaction of interest (Leitner et al., 2010). An alternative approach to protein-protein crosslinking relies upon highly reactive intermediates that can be generated in situ by photoactivation (“photo-crosslinking”), such as a 1,2-diradical or carbene produced by UV irradiation of benzophenone or diazirine functionalities, respectively (Pham et al., 2013). These photo-excited species are more broadly reactive than chemical crosslinkers and have shorter lifetimes, potentially improving their efficiency and reducing off-target crosslinking. However, photo-crosslinking strategies require incorporation of an appropriate photo-cross-linker into the target molecule, which has primarily restricted photo-affinity labeling to small molecules or peptide reagents (MacKinnon et al., 2007; Vila-Perello et al., 2007) that can be made by total chemical synthesis, or non-specific incorporation of photo-crosslinkable amino acids throughout the proteome (Suchanek et al., 2005; Yang et al., 2016).
Genetic code expansion strategies (i.e. “amber suppression”) provide a powerful tool for the site-specific incorporation of photo-crosslinkable amino acids containing benzophenone or aliphatic diazirines into cellular proteins(Ai et al., 2011; Chin et al., 2002; Chou et al., 2011; Zhang et al., 2011). These approaches are enabled by orthogonal tRNA/aminoacyl tRNA-synthetase pairs that can insert non-canonical amino acids at an amber stop codon in the gene of interest, and have been shown to work in bacteria, yeast, insects, and cultured mammalian cells. Amber suppression-mediated photo-crosslinking can be used for the analysis of protein-protein interactions in living cells(Hino et al., 2005; Zhang et al., 2011), although low crosslinking efficiency poses a considerable challenge to the detection of native interactions and interactome profiling efforts.
Previously, we developed a photo-crosslinking and stable isotope labeling by amino acids in cell culture (SILAC)-based proteomics approach (crosslinking-assisted and SILAC-based protein identification [CLASPI]) to profile post-translational modification-dependent protein-protein interactions (Kleiner et al., 2015; Li et al., 2013; Li et al., 2012). This approach relied on short synthetic peptides modified with a photo-crosslinkable p-benzoylphenylalanine residue, and was therefore limited in its ability to reveal context-dependent interactions occurring in living cells. Here, we present a chemical proteomics approach (named iCLASPI, or in vivo CLASPI) that enables profiling of context-dependent protein-protein interactions in living cells. This approach relies upon site-specific photo-crosslinking in living cells enabled by amber suppression-mediated incorporation of a diazirine-containing amino acid, combined with quantitative SILAC-based (Ong et al., 2002) mass spectrometry to detect crosslinked proteins (Figure 1). We apply iCLASPI to characterize the interactomes of soluble and chromatin-bound histones during different stages of the cell cycle, and identify known histone chaperones and modifying proteins as well as cell-cycle-specific chromatin binders. Taken together, our study highlights the dynamic nature of chromatin and histone interactions and provides a general method for profiling protein-protein interactions in their native context.
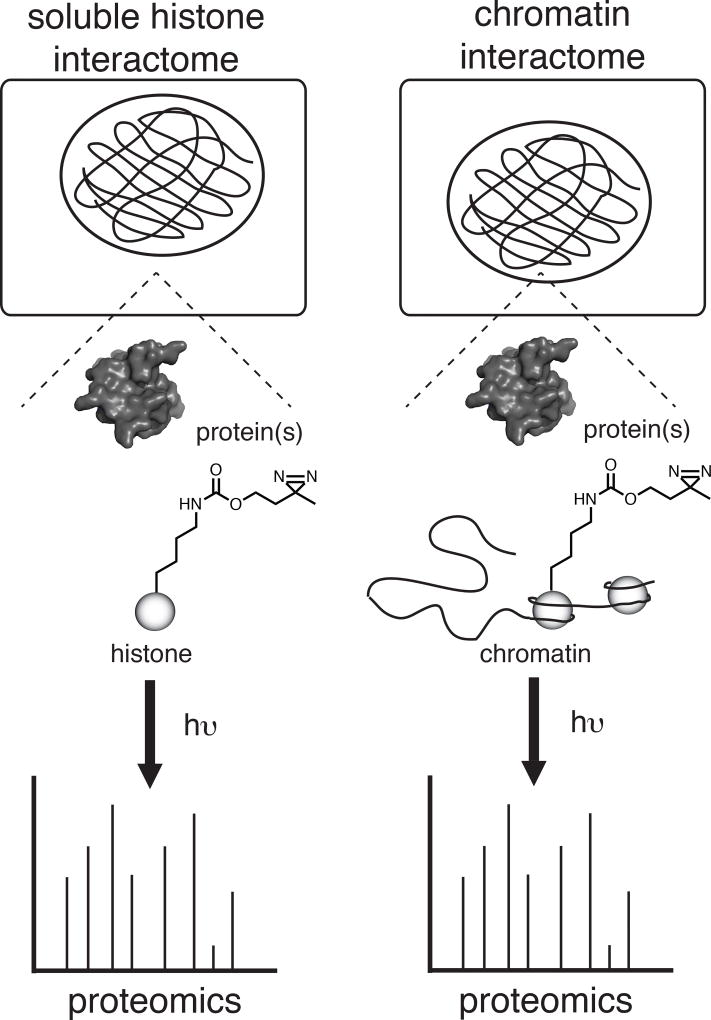
Figure 1.
The iCLASPI approach for profiling context-dependent direct protein-protein binding interactions in living cells. Amber suppression-mediated incorporation of a diazirine-containing amino acid enables live-cell photo-crosslinking and quantitative proteomics is used to identify protein-protein crosslinks.
Results
Amber suppression enables generation of photo-crosslinkable histone H3 and H4
To stabilize direct interactions between histones H3 and H4 and their associated proteins in living cells, we tested the feasibility of incorporating photo-crosslinkable amino acids into cellular histones using amber suppression (Figure 2a) (Liu and Schultz, 2010). We chose to modify the N-terminal tails of these two core histones since they are known “hot-spots” for protein-protein interactions and post-translational modifications. In breif, HEK293T cells were transfected with plasmids encoding orthogonal tRNA and aminoacyl-tRNA synthetase from Methanosarcina barkeri and an affinity-tagged histone H3 gene with an amber codon at position 7. In parallel, we generated an affinity-tagged histone H4 construct containing an amber codon at position 10. We then analyzed amber suppression efficiency in the presence of the non-canonical amino acids 3’-azibutyl-N-carbamoyl-lysine (AbK)(Ai et al., 2011), a diazirine-modified lysine derivative that facilitates efficient protein-protein photo-crosslinking, and Ne-Boc-lysine (BocK), which can be used as a negative control in photo-crosslinking experiments. Western blots of soluble extracts from cells grown in the presence of AbK or BocK indicated successful amber suppression of the site-specifically modified histone H3 and H4 constructs, with minimal translation of affinity-tagged histone in the absence of these amino acids (Figures 2b and 2c). We quantified relative levels of the modified H3 and H4 proteins by western blot with antibodies for the endogenous histones. Our results show that AbK-modified and affinity-tagged H3 protein comprises 27% of total soluble H3 in the cell. We were unable to robustly detect AbK-modified and affinity-tagged H4 protein using an anti-histone H4 antibody, suggesting that its relative expression levels are lower than what can be reliably measured using this approach. In addition, we assayed for potential in vivo protein photo-crosslinking to soluble histones by subjecting cells expressing AbK- or BocK-modified histones to 365 nM UV light and analyzing the soluble fraction by anti-FLAG western blot. We observed a number of slower migrating bands consistent with photo-crosslinked complexes specific to the cells expressing AbK-modified H3 or H4 (Figures 2d and 2e). Taken together, our results show that amber suppression enables the expression of site-specifically modified and photo-cross-linkable soluble histone H3 and H4, which can be used to capture direct interaction partners.
Figure 2.
Amber suppression enables the generation of photo-crosslinkable soluble histone H3 and H4. (a) HEK293T cells expressing amber suppression constructs and grown in the presence of 3’-azibutyl-N-carbamoyl-lysine (AbK) can be used to produce photo-crosslinkable histones. (b) Anti-H3 and anti-FLAG western blot analyses of HEK293T cells expressing affinity-tagged and amber codon containing histone H3 (with the amber codon at position 7) (H3-A7TAG) transgene and orthogonal aminoacyl-tRNA synthetase and tRNA. (c) Anti-FLAG and anti-tubulin western blot of HEK293T cells expressing affinity-tagged and amber codon containing histone H4 (with the amber codon at position 10) (H4-L10TAG) transgene and orthogonal aminoacyl-tRNA synthetase and tRNA. (d) Cell prepared as in (b) were grown on AbK or BocK, irradiated with 365 nm light, and analyzed by anti-FLAG western blot. Red arrowheads indicate putative photo-cross-linked protein complexes. (e) Cells prepared as in (c) were grown on AbK or BocK, irradiated with 365 nm light, and analyzed by anti-FLAG Western blot. Red arrowheads indicate putative phot-cross-linked protein complexes.
Soluble AbK-modified H3 and H4 photo-crosslink to known histone chaperones and modifying proteins
Having validated expression and photo-crosslinking of soluble AbK-modified H3 and H4, we next used affinity purification and SILAC-based proteomics (Ong et al., 2002) to profile their respective interactomes. For each histone construct, we set up a comparative proteomics experiment consisting of isotopically labeled HEK293T cells subjected to two conditions: 1) a photo-crosslinking sample expressing the AbK-modified histone protein and treated with UV, and 2) a negative control sample consisting of cells expressing AbK-modified histone but not subjected to UV, or cells expressing BocK-modified histone and treated with UV (Figure 3a). After photo-cross-linking, covalent protein complexes were isolated from the soluble histone pool, affinity enriched under denaturing conditions to favor isolation of direct binders, and analyzed by liquid chromatography-tandem mass spectrometry. We performed two proteomics experiments (termed “forward” and “reverse” SILAC experiments) for histone H3 and H4 while switching the SILAC labels and treatment conditions to increase our confidence in putative “hits” and account for differences in protein abundance that is a result of isotopic labeling.
Figure 3.
iCLASPI analysis of soluble histone H3 and H4 interactors. (a) Scheme for iCLASPI analysis of direct interaction partners of soluble histones H3 and H4. (b) SILAC ratios for forward and reverse iCLASPI experiments with soluble histone H3 (AbK at position 7). The dashed red box indicates SILAC ratios > 1. (c) SILAC ratios for forward and reverse iCLASPI experiments with soluble histone H4 (AbK at position 10). The dashed red box indicates SILAC ratios > 1.
Our analysis of forward and reverse experiments identified several proteins photo-crosslinked to soluble histone H3 and soluble histone H4 (Figures 3b and 3c, top right quadrants). Notably, we find NASP, a histone chaperone protein that regulates levels of soluble H3 and H4 (Cook et al., 2011), enriched between 5- and 14-fold in both soluble histone interactome datasets (Figures 3b and 3c, Table S1). In addition, RBBP7, a known H3 and H4 binding protein involved in chromatin assembly and remodeling complexes (Lai and Wade, 2011; Murzina et al., 2008), was identified in both of our datasets with an enrichment value of ~12. These datasets also revealed differences between the soluble histone H3 and H4 interactome. Specific H4 interactors enriched between 3- and 12-fold included RBBP4, HAT1, NPM1 and ZNF727. HAT1 is a major histone acetyltransferase that targets sites on the N terminus of soluble histone H4 (Benson et al., 2007), and NPM1 (Lindstrom, 2011), and RBBP4 (Verreault et al., 1996) are known histone chaperones; none of these proteins was identified in the soluble H3 dataset. Instead, we identified interactions between soluble H3 and SET, UBR7, and ANP32A, all reported histone binding proteins (Li et al., 2012; Seo et al., 2001), and enriched 7- to 12-fold in our proteomics experiment. In addition, the protein disulfide isomerases PDIA3 and PDIA1, as well as G3BP1, ATXN2L, and NFXL1 were identified as soluble H3 interactors. While PDIA3 was highly enriched, we observed a large discrepancy (26-fold) between forward and reverse SILAC ratios and therefore did not pursue this potential interaction further (Figure 3b). We did validate the formation of a subset of the photo-crosslinked complexes identified in the proteomic data by western blot with anti-FLAG (to detect the modified histone) and gene-specific antibodies. Consistent with our proteomic dataset, we were able to observe photo-crosslinking between histone H4 and HAT1, and histone H4 and RBBP7 (Figures 4a and 4b). We were also able to validate the interaction between histone H3 and UBR7 (Figure 4c).
Figure 4.
Validation of H3 and H4 interaction partners. (a) HEK293T cells expressing AbK/BocK-modified H4 were UV irradiated, and covalent H4 complexes were affinity purified and subjected to anti-FLAG and anti-HAT1 western blot analysis. The red arrow indicates the H4-HAT1 crosslinked complex. (b) Samples were treated as in (a) but were analyzed by anti-RBBP7 western blot. The red arrows indicate H4-RBBP7 crosslinks. (c) HEK293T cells expressing AbK/BocK-modified H3 and GFP-UBR7 were UV irradiated, and subjected to anti-FLAG and anti-GFP western blot analysis. The red arrow indicates the H3-UBR7 crosslinked complex. (d) HEK293T cells expressing AbK-modified H3 and the indicated GFP-UBR7 constructs were treated as in (c). (e) Equilibrium binding between H3 N-terminal peptides and recombinant, purified UBR7-PHD domain. Binding of UBR7-PHD to fluorescein-labeled H3 peptides was quantified by fluorescence anisotrophy. Values represent mean +/− s. d. (n = 3).
UBR7 PHD finger is critical for the UBR7-H3 interaction
We chose to further pursue the direct interaction between UBR7 and soluble histone H3 since it had not previously been characterized. Having successfully detected photo-crosslinking between AbK-modified histone H3 and a UBR7 transgene by western blot, we employed this assay to identify the region of UBR7 that mediated binding in cells. UBR7 contains a PHD finger domain, which is a well-established “reader” domain for methylated lysine residues in histone proteins (Sanchez and Zhou, 2011). Indeed, alignment of the UBR7 PHD finger with the structurally characterized PHD finger from ING2 shows conservation of residues that form the “aromatic cage” that is important for recognition (Figure S1). Therefore, we generated UBR7 expression constructs containing single point mutations in several of these aromatic residues and compared their photo-crosslinking behavior to the wild-type protein. Mutation of the aromatic residues Tyr134, Tyr141 or Trp161 to Ala reduced the amount of photo-crosslinked H3-UBR7 product (Figure 4d), indicating that these residues are important for binding to H3. In contrast, mutation of Glu151 to Ala did not substantially alter photo-crosslinking efficiency (Figure 4d). To further characterize the interaction between UBR7 PHD finger and the N terminus of histone H3, we generated a recombinant UBR7 PHD finger in Escherichia coli and measured its binding to fluorescein-labeled histone H3 N-terminal peptides (residues 1–21) by fluorescence anisotropy (Figure 4e). We observed a dose-dependent increase in millipolarization that did not saturate at 80 µM UBR7 (the highest concentration that we could assay), indicating a low affinity interaction between UBR7 and H3 N terminus (estimated Kd ~ 50–100 µM). Since PHD finger domains often bind most tightly to methylated lysine residues, we also performed the same assay using an H3 peptide containing methyl-lysine at position 4, and observed similar affinity binding, indicating that methylation at lysine-4 does not have a strong effect on the interaction (Figure 4e). Taken together, our results show that the PHD domain is needed for UBR7 binding to the N-terminal tail of histone H3, suggest the involvement of several aromatic residues in histone recognition, and establish that post-translational methylation is not required for UBR7-H3 recognition.
Analysis of chromatin-bound AbK-modified histone H3
As we found that iCLASPI could identify binding partners of soluble histones, we reasoned that this approach might also enable profiling of direct and cell-cycle-specific interactions between chromatin-bound histones and their associated proteins in living cells. To test this, we isolated chromatin fractions from HEK293T cells expressing an amber-codon containing H3 construct and grown separately in the presence of AbK, BocK, or no non-canonical amino acid, and analyzed them by anti-FLAG and anti-histone H3 western blot to detect the presence of modified H3 proteins. As expected, we observed expression of the modified chromatin-bound H3 protein predominantly in cells grown with AbK or BocK (Figures 5a and 5b). In these cells, we estimate that these site-specifically modified histones constitute ~33% of total chromatin-bound H3 (Figure 5b), a level that we expected would minimally perturb cell physiology but still allow for detectable amounts of protein-protein crosslinking. We also assayed whether Abk-modified H3 was a substrate for endogenous histone-modifying enzymes, particularly those that modify residues proximal to our AbK residue at position 7. Towards this end, we expressed AbK-modified H3 as described above, and induced histone phosphorylation (and mitotic arrest) by treatment with anti-mitotic compounds nocodazole or S-trityl-L-cysteine (STLC). We then used western blot to detect phosphorylation of endogenous and AbK-modified H3 on Ser10, a well established histone “mark” associated with mitosis (Garcia et al., 2005). After treatment with STLC or nocodazole, we were able to detect H3 S10-phosphorylation on both endogenous and AbK-modified H3 (Figure 5c), indicating that the modified histone did accumulate endogenous post-translational modifications at sites near AbK. We confirmed the specificity of the anti-H3S10phos antibody by performing an analogous experiment with an H3 transgene containing an S10A mutation (Figure 5c). Finally, we assayed whether we could detect the occurrence of in vivo photo-crosslinking events between AbK-modified H3 and other cellular proteins. We subjected cells expressing AbK- or BocK-modified H3 to 365 nm UV light, and analyzed the chromatin fraction by anti-FLAG western blot. We were able to detect slower migrating species specific to the AbK-containing sample (Figure 5d), consistent with the formation of protein-protein crosslinks between AbK-modified H3 and interacting proteins. Taken together, our results demonstrate that AbK-modified H3 displays comparable chromatin incorporation and post-translational modification state as the endogenous histone, and forms UV-dependent cross-links with cellular proteins.
Figure 5.
Amber suppression generates photo-crosslinkable histone H3 in human cells. (a) HEK293T cells expressing amber suppression constructs and grown in the presence of 3’-azibutyl-N-carbamoyl-lysine (AbK) can be used to produce photo-crosslinkable histones. (b) Anti-H3 and anti-FLAG western blot analyses of the chromatin fraction of HEK293T cells expressing affinity-tagged and amber-containing histone H3 transgene (with an amber codon at position 7) and orthogonal aminoacyl-tRNA synthetase and tRNA. (c) Anti-FLAG and anti-H3 S10phos western blot analysis of HEK293T cells expressing an amber codon containing H3 (H3A7TAG) or an amber codon containing H3 with the S10A mutation (H3A7TAG S10A). Cells were grown in medium containing BocK and arrested with nocodazole or STLC. (d) Cells prepared as in (b) were grown on AbK or BocK, irradiated with 365 nm light, and analyzed by anti-FLAG western blot. Black unlabeled arrowheads indicate putative photo-cross-linked protein complexes.
Profiling chromatin-associated H3 interactome in asynchronous cells
Next we used iCLASPI to profile the chromatin-associated H3 interactome in asynchronous cells (Figure 6a). Similar to the iCLASPI experiment probing the interactome of soluble histones, pairs of experiments in which the SILAC labels and UV treatment conditions were switched (termed “forward” and “reverse” SILAC experiments) were performed to account for potential differences in protein abundance due to isotopic labeling. Our analysis of the forward and reverse experiments revealed a number of proteins enriched upon UV treatment (Figure 6b, top right quadrant). These included all of the core histones (i.e. H2A, H2B, H3 and H4) and the linker histone H1, which displayed enrichment values ranging from 3.4 to 11 (Figure 6b, Table S2). In addition, we identified the histone chaperones NPM1, NPM3, NAP1L1, NCL, ANP32B, ANP32E and SSRP1 (a member of the facilities chromatin transcription [FACT] complex)(Hammond et al., 2017), and the proteins SET, RBBP7 and RBBP4, part of complexes regulating chromatin acetylation and remodeling (Lai and Wade, 2011; Seo et al., 2001), which all displayed enrichment values ≥5.0 (Figure 6b, Table S2). Finally, SPIN1, a “reader” of H3 K4Me3 (Li et al., 2012; Yang et al., 2012), CENP-B, a DNA-binding protein found in the mammalian centromere (Cooke et al., 1990), and UBF1, a transcription factor, were enriched 30-, 6.8-, and 3.3-fold, respectively (Figure 6b, Table S2). We validated photo-cross-linking between H3 and SPIN1 via western blotting by analyzing UV-treated cells expressing AbK-modified H3 and GFP-SPIN1 (Figure S2), and observed efficient crosslinking, consistent with our proteomic dataset.
Figure 6.
iCLASPI analysis of chromatin-associated histone H3 during interphase and mitosis. (a) Scheme for iCLASPI analysis of direct interaction partners of chromatin-associated histone H3. (b) SILAC ratios for forward and reverse iCLASPI experiments with chromatin-associated histone H3 (AbK at position 7) during interphase. The dashed red box indicates SILAC ratios > 1. (c) SILAC ratios for forward and reverse iCLASPI experiments with chromatin-associated histone H3 (AbK at position 7) during mitosis. The dashed red box indicates SILAC ratios > 1.
Profiling mitotis-specific chromatin-associated H3 interactome
We next profiled the chromatin-associated H3 interactome during M phase. Chromatin exists in a condensed and mainly transcriptionally silent state during mitosis (Gottesfeld and Forbes, 1997) and contains distinct post-translational modifications, including phosphorylation of Thr3 and Ser10 on the N terminus of H3 (Garcia et al., 2005). These changes in post-translational modifications can regulate interactions with binding partners(Fischle et al., 2005; Kelly et al., 2010), however, few systematic studies of mitosis-specific chromatin interacting proteins have been performed. To probe for mitosis-specific binders of chromatin-bound H3, we performed essentially the same experiment as described above for asynchronous cells, except that, prior to UV photo-crosslinking, cells were arrested in mitosis by treatment with nocodazole. Similar to our results in asynchronous cells, we found a number of proteins enriched upon UV treatment (Figure 6c, top right quadrant). Again, all of the core histones and the linker histone were enriched, albeit with ~2-fold lower enrichment values than those measured in interphase cells. In addition, we found the histone chaperone/chromatin remodeling proteins SET, NPM1, NPM3, ANP32E, NAP1L1, ANP32B, NCL, RBBP4 and SSRP1, as well as the centromere-specific protein CENP-B and the transcription factor UBF1, which were all identified in the interphase dataset. Interestingly, our results identified several proteins that displayed mitosis specific interactions with the N terminus of H3 including ANP32A, HTATSF1, NAP1L4, H2AX, HSPA1, HNRNPH, and HSC70, enriched between 3.7- and 13-fold (Figure 6c, Table S2). None of these proteins were enriched in the corresponding interphase dataset. Conversely, several proteins identified as interphase binders were absent from the mitotic dataset. Our data predicts that SPIN1, RBBP7, and HNRNPF are excluded from interacting with chromatin-bound H3 during mitosis (Figure 6b, Figure 6c). Our finding that RBBP7 does not bind to H3 during mitosis is consistent with the report that this protein binds more tightly to an unmodified N-terminal H3 peptide, compared with the same peptide containing the mitotic phosphorylation “mark” at Ser10 (Klingberg et al., 2015).
We chose to characterize the interaction of ANP32A with chromatin, since our analysis predicts that this interaction is direct, and specific to nocodazole-arrested cells. We generated recombinant full-length ANP32A and a deletion construct lacking the acidic C-terminal tail (residues 1–149) (Figure 7a), and assayed them for binding to an H3 N-terminal peptide (residues 1–21) by affinity pull-down. Consistent with literature precedent (Schneider et al., 2004), and our live-cell photo-crosslinking data, our results show that full-length ANP32A interacts with the N terminus of H3. Interestingly, we did not observe an interaction between H3 and ANP32A lacking its acidic C-terminal region, suggesting that this region is required for H3 recognition (Figure 7a). To further characterize the mitosis-specific behavior of ANP32A, we generated RPE-1 cells expressing a GFP-ANP32A transgene and used live-cell confocal microscopy to track the localization of ANP32A as the cells progressed through mitosis. During interphase, we found that ANP32A was localized exclusively to the nucleus (Figure 7b). As the nuclear membrane breaks down in early mitosis, the protein exhibits diffuse staining throughout the cell. This distribution persists through metaphase. In the final stages of cell division, we observed the accumulation of ANP32A on chromosomes. Taken together, our findings show that ANP32A localizes to chromatin in dividing cells and suggests that it may be involved in cell signaling in this context.
Figure 7.
ANP32A is a mitosis-specific interactor of chromatin-bound H3. (a) ANP32A interacts directly with the N terminus of histone H3, most likely through its acidic C-terminal region. Dynabeads displaying a histone H3 peptide (residues 1–21) or no peptide were incubated with either full-length recombinant ANP32A protein or a truncated construct (residues 1–149). Input and elution were separated by SDS-PAGE and imaged with Coomassie Blue stain. (b) Live-cell confocal imaging of GFP-ANP32A in RPE-1 cells. Cells were imaged by fluorescence and differential interference contrast (DIC) confocal microscopy as they progressed through the cell cycle.
Discussion
Here we have established a photo-crosslinking, amber suppression, and SILAC-based proteomics approach for detecting direct protein-protein interactions in living cells, which we have termed iCLASPI. In contrast to other affinity pull-down approaches that have been combined with quantitative proteomics, our method is designed to be specific for direct interactions since we isolate photo-crosslinked binding partners under denaturing conditions. Chemical proteomics has emerged as a powerful method for capturing small molecule-protein interactions; however, it has not been broadly applied to investigate protein-protein interaction networks, largely due to the challenge of incorporating reactive handles into full-length proteins expressed in living cells. Our work shows how the specific incorporation of a diazirine-containing amino acid into a cellular protein, enabled by amber suppression, facilitates light-mediated chemical crosslinking of interacting proteins. This approach has the potential to stabilize both high- and low-affinity interactions, and greatly simplifies the extraction and affinity purification of protein complexes from the cell. Since photo-crosslinked products are often present in low stoichiometry, we use quantitative SILAC-based proteomics as a general method for detecting covalent complexes. Due to the site-specific incorporation of the photo-crosslinker, our approach provides region-specific interactomes and therefore proper selection of protein-protein interaction hotspots is critical to its success.
We apply iCLASPI to profile the interactome of two different soluble histone proteins, H3 and H4, providing additional insight into the specificity of native histone-mediated interactions. While two proteins, histone chaperones NASP and RBBP7, were identified in both soluble H3 and H4 datasets, we also found several direct binding partners specific to each protein, including HAT1, RBBP4, and NPM1 for H4, and SET, G3BP1, and UBR7 for H3. We further characterized the H3 interaction with UBR7, a member of the N-recognin family of E3 ubiquitin ligases that facilitate degradation of proteins containing N-degron sequences (Tasaki et al., 2005). UBR7 contains the UBR box motif; however, in vitro binding studies have shown that it does not recognize the canonical N-degron sequence (Tasaki et al., 2009). Notably, and in contrast to the other six UBR proteins, UBR7 contains a PHD domain, a known histone recognition motif. Importantly, our data shows that this interaction predominantly occurs in the soluble histone pool. While UBR7 has not been shown to be a competent E3 ubiquitin ligase, it is tempting to speculate that it may be involved in ubiquitin-dependent turnover of soluble histone H3.
Our study also demonstrates that iCLASPI can be used to profile context-dependent interactions, as we identified interaction partners of chromatin-bound histone H3 in asynchronous and nocodazole-arrested cells. In asynchronous cells, we identified interactions with a number of histone chaperone and nucleosome assembly proteins including members of the nucleophosmin family, NAP1-like proteins, nucleolin, FACT complex, and acidic nuclear phosphoproteins, as well as proteins involved in chromatin remodeling and acetylation. While many of these proteins are known to be associated with chromatin, we do not know how they interact in vivo with the nucleosome particle. Our data suggests that these binding events involve a direct interaction with the N-terminus of histone H3. We also identified CENP-B, a centromeric protein that binds to DNA in a sequence-specific manner. CENP-B binds preferentially to nucleosomes containing the H3 variant CENP-A, but can also recognize canonical H3-containing nucleosomes (Fujita et al., 2015). Since the stoichiometry of CENP-A nucleosomes at the centromere is low (Bodor et al., 2014), it is likely that CENP-B retention at centromeres requires auxiliary interactions, and our data show that these interactions include binding to the N-terminus of H3.
During mitosis, chromatin exists in a condensed state and fundamental cellular processes that are active during interphase, such as transcription and DNA repair, are largely silenced or inhibited (Gottesfeld and Forbes, 1997; Orthwein et al., 2014). Interestingly, for AbK-modified H3 isolated from mitotic cells, we find that most chromatin-associated proteins present during interphase remain bound to chromatin during mitosis. This is somewhat unexpected since these proteins are largely histone chaperones and chromatin remodeling proteins that facilitate dynamic processes (e.g., the FACT complex mediates transcriptional elongation (Orphanides et al., 1998)) that are predominantly repressed during mitosis. One possibility is that these proteins remain bound (but functionally inactive) throughout mitosis as a form of “bookmarking” (Kadauke and Blobel, 2013). Notably, analysis of the interphase and mitosis datasets did identify a subset of proteins that bind during only one of these cell-cycle stages, including ANP32A, HTATSF1, H2AX, SPIN1 and RBBP7. Several of these proteins, including ANP32A and H2AX, have been implicated in mitosis-specific processes (McManus and Hendzel, 2005; Nalepa et al., 2013). We imaged ANP32A during mitosis and found that it localizes to chromosomes during the final stages of cell division. ANP32A has been implicated as a regulator of PP2A phosphatase activity(Li et al., 1996), and therefore it may be involved in mitotic progression. Further investigation will be needed to decipher the functional significance of these cell-cycle-specific interactions.
In conclusion, our study demonstrates the generality of the iCLASPI approach, as we have successfully used it to profile native protein-protein interactions involving two different histone proteins, different cell-cycle stages, and different cellular localization. Extending this methodology to other protein families, and expanding its scope to study post-translational modification-dependent interactions, should provide a powerful means of illuminating important biological processes.
Significance
Methods for identifying low-affinity protein-protein interactions are critical for advancing our understanding of important biological processes. Development of a chemical proteomics-based approach that combines photo-crosslinking, amber suppression, and SILAC, which we term iCLASPI (in vivo crosslinking-assisted and SILAC-based protein identification), enables us to profile the interactome of target proteins in living cells. Our approach allows the elucidation of context-specific as well as site-specific direct interactions. We envision that iCLASPI methodology can be broadly applied to various protein families in live cells.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Tarun Kapoor (kapoor@rockefeller.edu).
Experimental Model and Subject Details
Cell culture
HEK293T cells were grown at 37°C in a humidified atmosphere with 5% CO2 in Dulbecco's Modified Eagle Medium (DMEM) medium containing 10% bovine calf serum (HyClone), penicillin-streptomycin, and 2mM L-glutamine. RPE1 cells were grown at 37°C in a humidified atmosphere with 5% CO2 in Dulbecco's Modified Eagle Medium (DMEM) medium containing 10% fetal bovine serum (HyClone), penicillin-streptomycin, and 2mM L-glutamine. RPE1 cells stably expressing GFP-ANP32A were generated by retroviral transduction using retrovirus generated in 293-ampho cells.
Method Details
Plasmid Construction
Amber suppression plasmid pCMV-AbK (Ai et al., 2011) was a gift from Peter Schultz. Mammalian expression plasmids encoding amber-containing histone H3.2 or histone H4 with tandem C-terminal 12X-His and 3X-FLAG tags were generated using pCDNA3.1-Myc-HisA (Life Technologies). Expression plasmids for GFP-UBR7 constructs were generated by restriction cloning of a GFP-UBR7 PCR cassette into pCDNA3.1-Myc-HisA. An expression plasmid for GFP-SPIN1 was generated by restriction cloning of a GFP-SPIN1 PCR cassette into pCDNA3.1-Myc-HisA. The bacterial expression plasmid for full-length ANP32A (pET30a-PP32) was a gift from Judy Lieberman (Addgene plasmid # 8824) (Beresford et al., 2001). The bacterial expression plasmid for truncated ANP32A (residues 1–149) was generated by restriction cloning into the same vector. UBR7-PHD was cloned into pGEX-6P-1 (GE Healthcare) for bacterial expression. For generating GFP-ANP32A retrovirus, ANP32A was introduced into a modified pMSCV-based vector (pEA22-ANP32A).
Amino Acids
3’-azibutyl-N-carbamoyl-lysine (AbK) was synthesized as described previously (Ai et al., 2011). Nε-Boc-L-lysine (BocK) was purchased from Chem-Impex International.
Transfections and Photocrosslinking
For histone H3 experiments, cells were transfected with the histone H3 plasmid and pCMV-AbK using the CaPO4 method in medium supplemented with 0.5 mM AbK or 1 mM BocK. For histone H4 experiments, cells were transfected with histone H4 and pCMV-AbK using Lipofectamine 2000 (Life Technologies) and were grown in medium supplemented with 0.4mM AbK or 1.2mM BocK. 24 hours later, medium was replaced with cold PBS and cells were exposed to 365 nm UV (Spectroline ML-3500S UV lamp) for 15 minutes on ice. Cells were harvested and stored at −80°C prior to lysis and affinity purification. For nocodazole arrest, 330 nM nocodazole (Sigma) was added to the medium 24 hrs after transfection, and cells were incubated for 20 hr prior to UV treatment. For SILAC experiments, cells were cultured in DMEM –Arg –Lys (ThermoFisher Scientific) containing 10% dialyzed fetal bovine serum (ThermoFisher), penicillin–streptomycin, and 2 mM l-glutamine supplemented with either 22 mg/L 13C6 15N4-L-arginine (ThermoFisher) and 50 mg/L 13C6 15N2-L-lysine (Sigma) to make “heavy” medium, or the corresponding non-labeled amino acids (Sigma) to make the “light” medium.
Preparation of Soluble Extracts and Enrichment of Photo-Crosslinked Proteins
To prepare soluble extracts, cell pellets were resuspended in 20 mM HEPES, pH 7.5, 150mM NaCl, 0.2% Triton-X 100, 10 mM beta-mercaptoethanol, 20 mM Imidazole, 1 mM phenylmethylsulfonyl fluoride (PMSF), and Complete EDTA-free protease inhibitors (Roche), and incubated for 10 min at 4°C. The suspension was centrifuged at 20,000g for 15 min at 4°C and the supernatant was collected. The soluble extract was then diluted with 8 M urea, and incubated with His-tag Isolation and Pulldown Dynabeads (Life Technologies) at room temperature for 2 hr. Beads were washed 6X with 20 mM HEPES pH 7.5, 1 M NaCl, 20 mM Imidazole, 10 mM beta-mercaptoethanol, 0.1% Triton X-100 and 8 M Urea. Proteins were eluted in 20 mM HEPES, pH 7.5, 150 mM NaCl, 500 mM Imidazole, 10 mM beta-mercaptoethanol and 8 M Urea.
Preparation of Chromatin Extracts and Enrichment of Photo-Crosslinked Proteins
To prepare chromatin extracts, cell pellets were first resuspended in 20 mM HEPES, pH 7.5, 150mM NaCl, 0.2% Triton-X 100, 10 mM beta-mercaptoethanol, 20 mM Imidazole, 1 mM phenylmethylsulfonyl fluoride (PMSF), and Complete EDTA-free protease inhibitors (Roche), and incubated for 10 min at 4°C. The suspension was centrifuged at 20,000g for 15 min at 4°C and the supernatant was removed. The chromatin pellet was then digested with Micrococcal Nuclease (Worthington Biochemical Corporation) at room temperature, and subsequently extracted with buffer containing 20 mM HEPES pH 7.5, 8 M urea, and 500 mM NaCl. The clarified chromatin extract was then incubated with His-tag Isolation and Pulldown Dynabeads (Life Technologies) at room temperature for 2 hr. Beads were washed 6X with 20 mM HEPES pH 7.5, 1 M NaCl, 20 mM Imidazole, 10 mM beta-mercaptoethanol, 0.1% Triton X-100 and 8 M Urea. Proteins were eluted in 20 mM HEPES, pH 7.5, 150 mM NaCl, 500 mM Imidazole, 10 mM beta-mercaptoethanol and 8 M Urea.
Mass Spectrometry
Mass spectrometry was performed essentially as described (Kleiner et al., 2015). Protein samples were reduced and alkylated, and then separated on a 4–12% Bis-Tris gradient gel (Life Technologies), followed by in-gel trypsin digestion. Tryptic peptides were purified and analyzed on a Q Exactive mass spectrometer (Thermo Scientific). Protein identification and quantitation of SILAC peptide ratios was performed using MaxQuant version 1.2.2.5.
Protein Purification and Peptide Pulldown
ANP32A proteins were expressed in Escherichia coli BL21 (Rosetta) by induction overnight with IPTG (0.5 mM) at 18 °C. After cell lysis, His6-tagged proteins were captured on Ni-NTA agarose (Qiagen) and subsequently purified by anion-exchange (MonoQ) and size-exclusion (Superdex 200) chromatography. Peptide pulldowns were performed using Streptavidin Dynabeads (Life Technologies) displaying a biotinylated H3 peptide (residues 1–21) (Anaspec). UBR7-PHD was purified using glutathione sepharose (GE Healthcare), and purified by by size-exclusion (Superdex 75) chromatography after cleavage of the GST tag.
Western Blot
For Western blot analysis, the following antibodies were used: anti-GFP, anti-H3 (Cell Signaling Technology #4499), anti-H3 S10phos (Cell Signaling Technology #3377), anti-alpha tubulin (Serotec MCA77G), anti-RBBP7 (Cell Signalling #6882), anti-Hat1 (Bethyl A305-360A), and anti-FLAG M2 (Sigma). IRDye-conjugated secondary antibodies raised in goat were purchased from LI-COR Biosciences and used according to the manufacturer’s instructions. Estimation of relative modified soluble and chromatin-bound H3 proteins was performed by densitometry analysis.
Fluorescence Anisotropy
Binding assays with UBR7-PHD and fluorescein-labeled H3 peptides (Anaspec) were performed by combining peptide (50 nM) with varying concentration of purified protein in buffer (50 mM HEPES pH 7.5, 150 mM NaCl, 1 mM DTT). Fluorescence anisotropy was measured using a BioTek Synergy Neo microplate reader.
Microscopy
Live-cell imaging was performed according to methods we have previously reported (Foley et al., 2011).
Quantification and Statistical Analysis
SILAC ratios for mass spectrometry experiments were calculated using MaxQuant version 1.2.2.5. Dissociation constants for fluorescence anisotropy experiments were determined by fitting the data to a four-parameter sigmoidal dose-response curve using DeltaGraph. Data are shown as mean +/− s.d.
Supplementary Material
Document S1. Tables S1–S2, Figures S1–S2.
Table S4. SILAC ratios for soluble H4 interactome in asynchronous cells, related to Figure 3.
Table S5. SILAC ratios for H3 interactome in nocodazole-arrested cells, related to Figure 6.
Highlights.
Photocrosslinkable histones capture direct protein-protein interactions in cells
SILAC-based proteomics profiles the live-cell interactome of histone H3 and H4
UBR7 binds to soluble histone H3; the interaction is mediated by its PHD domain
ANP32A is a mitosis-specific interactor of chromatin-bound histone H3
Acknowledgments
We are grateful to Peter Schultz for the generous gift of amber suppression plasmid pCMV-AbK. R.E.K. acknowledges support from the Charles H. Revson Foundation through a Revson Senior Fellowship in Biomedical Science. L.E.H. was supported by NRSA Training Grant #CA009673-37. The authors acknowledge support from the US National Institutes of Health (T.M.K- GM98579 and B.T.C.- PHS GM103314 and PHS GM109824).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Author Contributions
T. M. K. and R. E. K. designed the study. R. E. K., L. E. H., and K. R. M. conducted experiments. B. T. C. directed K. R. M. and T. M. K. directed R. E. K. and L. E. H. R. E. K., L. E. H. and T. M. K. wrote the manuscript.
References
- Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. 2016;537:347–355. doi: 10.1038/nature19949. [DOI] [PubMed] [Google Scholar]
- Ai HW, Shen W, Sagi A, Chen PR, Schultz PG. Probing protein-protein interactions with a genetically encoded photo-crosslinking amino acid. Chembiochem. 2011;12:1854–1857. doi: 10.1002/cbic.201100194. [DOI] [PubMed] [Google Scholar]
- Benson LJ, Phillips JA, Gu Y, Parthun MR, Hoffman CS, Annunziato AT. Properties of the type B histone acetyltransferase Hat1: H4 tail interaction, site preference, and involvement in DNA repair. J Biol Chem. 2007;282:836–842. doi: 10.1074/jbc.M607464200. [DOI] [PubMed] [Google Scholar]
- Beresford PJ, Zhang D, Oh DY, Fan Z, Greer EL, Russo ML, Jaju M, Lieberman J. Granzyme A activates an endoplasmic reticulum-associated caspase-independent nuclease to induce single-stranded DNA nicks. J Biol Chem. 2001;276:43285–43293. doi: 10.1074/jbc.M108137200. [DOI] [PubMed] [Google Scholar]
- Bodor DL, Mata JF, Sergeev M, David AF, Salimian KJ, Panchenko T, Cleveland DW, Black BE, Shah JV, Jansen LE. The quantitative architecture of centromeric chromatin. Elife. 2014;3:e02137. doi: 10.7554/eLife.02137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW, Martin AB, King DS, Wang L, Schultz PG. Addition of a photocrosslinking amino acid to the genetic code of Escherichiacoli. Proc Natl Acad Sci U S A. 2002;99:11020–11024. doi: 10.1073/pnas.172226299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou CJ, Uprety R, Davis L, Chin JW, Deiters A. Genetically encoding an aliphatic diazirine for protein photocrosslinking. Chem Sci. 2011;2:480–483. [Google Scholar]
- Cook AJ, Gurard-Levin ZA, Vassias I, Almouzni G. A specific function for the histone chaperone NASP to fine-tune a reservoir of soluble H3-H4 in the histone supply chain. Mol Cell. 2011;44:918–927. doi: 10.1016/j.molcel.2011.11.021. [DOI] [PubMed] [Google Scholar]
- Cooke CA, Bernat RL, Earnshaw WC. CENP-B: a major human centromere protein located beneath the kinetochore. J Cell Biol. 1990;110:1475–1488. doi: 10.1083/jcb.110.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischle W, Tseng BS, Dormann HL, Ueberheide BM, Garcia BA, Shabanowitz J, Hunt DF, Funabiki H, Allis CD. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- Foley EA, Maldonado M, Kapoor TM. Formation of stable attachments between kinetochores and microtubules depends on the B56-PP2A phosphatase. Nat Cell Biol. 2011;13:1265–1271. doi: 10.1038/ncb2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita R, Otake K, Arimura Y, Horikoshi N, Miya Y, Shiga T, Osakabe A, Tachiwana H, Ohzeki J, Larionov V, et al. Stable complex formation of CENP-B with the CENP-A nucleosome. Nucleic Acids Res. 2015;43:4909–4922. doi: 10.1093/nar/gkv405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia BA, Barber CM, Hake SB, Ptak C, Turner FB, Busby SA, Shabanowitz J, Moran RG, Allis CD, Hunt DF. Modifications of human histone H3 variants during mitosis. Biochemistry. 2005;44:13202–13213. doi: 10.1021/bi050906n. [DOI] [PubMed] [Google Scholar]
- Gottesfeld JM, Forbes DJ. Mitotic repression of the transcriptional machinery. Trends Biochem Sci. 1997;22:197–202. doi: 10.1016/s0968-0004(97)01045-1. [DOI] [PubMed] [Google Scholar]
- Hammond CM, Stromme CB, Huang H, Patel DJ, Groth A. Histone chaperone networks shaping chromatin function. Nat Rev Mol Cell Biol. 2017;18:141–158. doi: 10.1038/nrm.2016.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hino N, Okazaki Y, Kobayashi T, Hayashi A, Sakamoto K, Yokoyama S. Protein photo-cross-linking in mammalian cells by site-specific incorporation of a photoreactive amino acid. Nat Methods. 2005;2:201–206. doi: 10.1038/nmeth739. [DOI] [PubMed] [Google Scholar]
- Kadauke S, Blobel GA. Mitotic bookmarking by transcription factors. Epigenetics Chromatin. 2013;6:6. doi: 10.1186/1756-8935-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly AE, Ghenoiu C, Xue JZ, Zierhut C, Kimura H, Funabiki H. Survivin reads phosphorylated histone H3 threonine 3 to activate the mitotic kinase Aurora B. Science. 2010;330:235–239. doi: 10.1126/science.1189505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim MS, Pinto SM, Getnet D, Nirujogi RS, Manda SS, Chaerkady R, Madugundu AK, Kelkar DS, Isserlin R, Jain S, et al. A draft map of the human proteome. Nature. 2014;509:575–581. doi: 10.1038/nature13302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner RE, Verma P, Molloy KR, Chait BT, Kapoor TM. Chemical proteomics reveals a gammaH2AX-53BP1 interaction in the DNA damage response. Nat Chem Biol. 2015;11:807–814. doi: 10.1038/nchembio.1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klingberg R, Jost JO, Schumann M, Gelato KA, Fischle W, Krause E, Schwarzer D. Analysis of phosphorylation-dependent protein-protein interactions of histone h3. ACS Chem Biol. 2015;10:138–145. doi: 10.1021/cb500563n. [DOI] [PubMed] [Google Scholar]
- Lai AY, Wade PA. Cancer biology and NuRD: a multifaceted chromatin remodelling complex. Nat Rev Cancer. 2011;11:588–596. doi: 10.1038/nrc3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Probing native protein structures by chemical cross-linking, mass spectrometry, and bioinformatics. Mol Cell Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Makkinje A, Damuni Z. Molecular identification of I1PP2A, a novel potent heat-stable inhibitor protein of protein phosphatase 2A. Biochemistry. 1996;35:6998–7002. doi: 10.1021/bi960581y. [DOI] [PubMed] [Google Scholar]
- Li X, Foley EA, Kawashima SA, Molloy KR, Li Y, Chait BT, Kapoor TM. Examining post-translational modification-mediated protein-protein interactions using a chemical proteomics approach. Protein Sci. 2013;22:287–295. doi: 10.1002/pro.2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Foley EA, Molloy KR, Li Y, Chait BT, Kapoor TM. Quantitative chemical proteomics approach to identify post-translational modification-mediated protein-protein interactions. J Am Chem Soc. 2012;134:1982–1985. doi: 10.1021/ja210528v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstrom MS. NPM1/B23: A Multifunctional Chaperone in Ribosome Biogenesis and Chromatin Remodeling. Biochem Res Int. 2011;2011:195209. doi: 10.1155/2011/195209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CC, Schultz PG. Adding new chemistries to the genetic code. Annu Rev Biochem. 2010;79:413–444. doi: 10.1146/annurev.biochem.052308.105824. [DOI] [PubMed] [Google Scholar]
- MacKinnon AL, Garrison JL, Hegde RS, Taunton J. Photo-leucine incorporation reveals the target of a cyclodepsipeptide inhibitor of cotranslational translocation. J Am Chem Soc. 2007;129:14560–14561. doi: 10.1021/ja076250y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McManus KJ, Hendzel MJ. ATM-dependent DNA damage-independent mitotic phosphorylation of H2AX in normally growing mammalian cells. Mol Biol Cell. 2005;16:5013–5025. doi: 10.1091/mbc.E05-01-0065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murzina NV, Pei XY, Zhang W, Sparkes M, Vicente-Garcia J, Pratap JV, McLaughlin SH, Ben-Shahar TR, Verreault A, Luisi BF, et al. Structural basis for the recognition of histone H4 by the histone-chaperone RbAp46. Structure. 2008;16:1077–1085. doi: 10.1016/j.str.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nalepa G, Barnholtz-Sloan J, Enzor R, Dey D, He Y, Gehlhausen JR, Lehmann AS, Park SJ, Yang Y, Yang X, et al. The tumor suppressor CDKN3 controls mitosis. J Cell Biol. 2013;201:997–1012. doi: 10.1083/jcb.201205125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong SE, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- Orphanides G, LeRoy G, Chang CH, Luse DS, Reinberg D. FACT, a factor that facilitates transcript elongation through nucleosomes. Cell. 1998;92:105–116. doi: 10.1016/s0092-8674(00)80903-4. [DOI] [PubMed] [Google Scholar]
- Orthwein A, Fradet-Turcotte A, Noordermeer SM, Canny MD, Brun CM, Strecker J, Escribano-Diaz C, Durocher D. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science. 2014;344:189–193. doi: 10.1126/science.1248024. [DOI] [PubMed] [Google Scholar]
- Pham ND, Parker RB, Kohler JJ. Photocrosslinking approaches to interactome mapping. Curr Opin Chem Biol. 2013;17:90–101. doi: 10.1016/j.cbpa.2012.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez R, Zhou MM. The PHD finger: a versatile epigenome reader. Trends Biochem Sci. 2011;36:364–372. doi: 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider R, Bannister AJ, Weise C, Kouzarides T. Direct binding of INHAT to H3 tails disrupted by modifications. J Biol Chem. 2004;279:23859–23862. doi: 10.1074/jbc.C400151200. [DOI] [PubMed] [Google Scholar]
- Seo SB, McNamara P, Heo S, Turner A, Lane WS, Chakravarti D. Regulation of histone acetylation and transcription by INHAT, a human cellular complex containing the set oncoprotein. Cell. 2001;104:119–130. doi: 10.1016/s0092-8674(01)00196-9. [DOI] [PubMed] [Google Scholar]
- Suchanek M, Radzikowska A, Thiele C. Photo-leucine and photo-methionine allow identification of protein-protein interactions in living cells. Nat Methods. 2005;2:261–267. doi: 10.1038/nmeth752. [DOI] [PubMed] [Google Scholar]
- Tasaki T, Mulder LC, Iwamatsu A, Lee MJ, Davydov IV, Varshavsky A, Muesing M, Kwon YT. A family of mammalian E3 ubiquitin ligases that contain the UBR box motif and recognize N-degrons. Mol Cell Biol. 2005;25:7120–7136. doi: 10.1128/MCB.25.16.7120-7136.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasaki T, Zakrzewska A, Dudgeon DD, Jiang Y, Lazo JS, Kwon YT. The substrate recognition domains of the N-end rule pathway. J Biol Chem. 2009;284:1884–1895. doi: 10.1074/jbc.M803641200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verreault A, Kaufman PD, Kobayashi R, Stillman B. Nucleosome assembly by a complex of CAF-1 and acetylated histones H3/H4. Cell. 1996;87:95–104. doi: 10.1016/s0092-8674(00)81326-4. [DOI] [PubMed] [Google Scholar]
- Vila-Perello M, Pratt MR, Tulin F, Muir TW. Covalent capture of phospho-dependent protein oligomerization by site-specific incorporation of a diazirine photo-cross-linker. J Am Chem Soc. 2007;129:8068–8069. doi: 10.1021/ja072013j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilhelm M, Schlegl J, Hahne H, Gholami AM, Lieberenz M, Savitski MM, Ziegler E, Butzmann L, Gessulat S, Marx H, et al. Mass-spectrometry-based draft of the human proteome. Nature. 2014;509:582–587. doi: 10.1038/nature13319. [DOI] [PubMed] [Google Scholar]
- Yang N, Wang W, Wang Y, Wang M, Zhao Q, Rao Z, Zhu B, Xu RM. Distinct mode of methylated lysine-4 of histone H3 recognition by tandem tudor-like domains of Spindlin1. Proc Natl Acad Sci U S A. 2012;109:17954–17959. doi: 10.1073/pnas.1208517109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Li XM, Bao X, Fung YM, Li XD. Photo-lysine captures proteins that bind lysine post-translational modifications. Nat Chem Biol. 2016;12:70–72. doi: 10.1038/nchembio.1990. [DOI] [PubMed] [Google Scholar]
- Zhang M, Lin S, Song X, Liu J, Fu Y, Ge X, Fu X, Chang Z, Chen PR. A genetically incorporated crosslinker reveals chaperone cooperation in acid resistance. Nat Chem Biol. 2011;7:671–677. doi: 10.1038/nchembio.644. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Document S1. Tables S1–S2, Figures S1–S2.
Table S4. SILAC ratios for soluble H4 interactome in asynchronous cells, related to Figure 3.
Table S5. SILAC ratios for H3 interactome in nocodazole-arrested cells, related to Figure 6.