Abstract
MicroRNAs (miRNAs) are small RNAs that bind to mRNA targets and regulate their translation. A functional study of miRNAs and exploration of their utility as disease markers require miRNA extraction from biological samples, which contain large amounts of interfering compounds for downstream RNA identification and quantification. The most common extraction methods employ silica columns or the TRIzol reagent but give out low recovery for small RNAs probably due to their short strand lengths. Herein, we fabricated the titanium dioxide nanofibers using electrospinning to facilitate miRNA extraction and developed the optimal buffer conditions to improve miRNA recovery from biological matrices of cell lysate and serum. We found that our TiO2 fibers could obtain a recovery of 18.0 ± 3.6% for miRNA fibers while carrying out the extraction in the more complex medium of cell lysate, much higher than the 0.02 ± 0.0001% recovery from the commercial kit. The much improved extraction of miRNAs from our fibers could be originated from the strong coordination between TiO2 and RNA’s phosphate backbone. In addition, the binding, washing, and elution buffers judiciously developed in the present study can achieve selective extraction of small RNA shorter than 500 nucleotides in length. Our results demonstrate that TiO2 nanofibers can work as a valuable tool for extraction of miRNAs from biological samples with high recovery.
Keywords: Electrospinning, Nanofibers, Titanium oxide, MicroRNA, Solid phase extraction, Recovery
Introduction
Small RNAs are noncoding RNAs shorter than 100 nucleotides. One of the most commonly studied small RNA families are microRNAs (miRNA) that can bind to target mRNAs and inhibit their translation or induce degradation [1, 2]. Expression of miRNA often varies during pathological processes [3, 4], making them promising biomarkers for disease diagnosis and prognosis [5–7]. Discovery or detection of the miRNA-based biomarkers requires extraction and purification of miRNAs from biological samples, because the interfering molecules, like the abundant proteins, proteinases, nucleases, salts, etc., present in the matrix can inhibit the downstream analytical steps, such as PCR or next-generation sequencing, preventing sensitive and accurate miRNA identification and quantification [8, 9]. The widely implemented methods for miRNA extraction employ either organic solvents or columns packed with solid sorbents. The most common solvent used is the TRIzol reagent, an improved and simplified format of liquid-liquid extraction (LLE) with phenol and chloroform. The TRIzol reagent extracts the nucleic acids to an aqueous phase, which are then precipitated by alcohol, and leaves the proteins to the organic phase, including enzymes that degrade the nucleic acids [10]. TRIzol offers the advantages of providing both the nucleic acid and protein partitions from the same sample and offering good consistency in recovery. However, TRIzol contains phenol, a volatile and corrosive chemical and can take more than 12 h to obtain optimal yield. In addition, LLE is labor-intensive and with low throughput [11]. Solid phase extraction (SPE) columns can replace organic solvents to improve sample throughput and extraction efficiency. The most representative sorbent is silicone dioxide (SiO2) that is prepared in the form of columns or membranes for nucleic acid binding. RNA and DNA can be adsorbed onto SiO2 with the aid of chaotropic reagents like guanidine salts that can denature nucleic acids and serve as salt bridges to enhance the binding of the negatively charged nucleic acids to silica [12]. However, SPE columns cannot yield high recovery to small RNAs due to insufficient adsorption on the silica-based column [13]. New approaches have been developed mainly for DNA using various nanomaterials that include graphene oxide, carbon nanotubes, zinc oxide, and metal nanoparticles [14–19], and some DNA isolation approaches were directly coupled with detection assays [14–16, 18, 20–25]. But applications of nanomaterials in extraction of small RNAs are very limited.
The regulating roles of miRNAs determine that miRNA abundance is not high and always changing in biological samples. Thus, speedy extraction with high recovery and purity is very critical for identification of potential miRNA markers, in particular for the downregulated ones present at trace levels. In addition, biomarker discovery and validation should be carried out in cells, animal tissues, and clinical specimens, the availability of which could be highly limited with only small aliquots attainable. It has been demonstrated that with the conventional TRIzol-based LLE or SiO2-based SPE, a decrease in sample amount or volume leads to lower recovery of miRNAs [26]. All these challenges demand the development of new extraction techniques for rapid and simple miRNA extraction with high recovery.
Titanium dioxide (TiO2)-based materials have been widely employed for enrichment of phosphorylated peptides and proteins because TiO2 can interact strongly with the phosphate groups [27–31]. Adsorption of DNA onto the TiO2 nanoparticles has also been observed [32]. These pioneering works point out the possibility of employing the TiO2-based nanomaterials for RNA extraction by adjusting the binding conditions. However, the strong interaction between TiO2 and the nucleic acid phosphate backbone enhances the difficulty in RNA elution for downstream analysis. Development of the suitable binding and elution conditions becomes very critical in order to achieve high RNA recovery. In addition, the TiO2-based material to be employed should be easily fabricated and handled for simple operation, as well as provide large specific surface area for RNA adsorption. TiO2 nanofibers can be fabricated by the well-established electrospinning method [19, 33, 34]. Compared to the zero-dimensional nanoparticles, the 1-dimensional (1D) fibers possess higher surface area-to-volume ratios for adsorption of target molecules. The long fibers can also be easily separated and purified from solutions using filtration, simplifying the procedure of removing the fibers from a series of solutions needed for RNA binding, washing, and elution. Hence, in the present work, we synthesized TiO2 nanofibers and applied them for specific extraction of RNAs shorter than 500 nt, in which size region miRNAs locate. To improve miRNA recovery from biological samples, three solutions required for all SPE-based nucleic acid extraction were optimized: the binding, washing, and elution buffers [32, 35]. Spiked or endogenous miRNAs in serum or cell lysate were quantified by RT-quantitative polymerase chain reaction (qPCR), and much higher recovery was obtained with our fibers, showing high promise of this material in extraction of miRNAs for functional study and biomarker discovery.
Materials and methods
Chemical and biochemical
Glacial acetic acid, hydrochloric acid, DMSO, titanium (IV) isopropoxide, guanidine HCl (Gu-HCl), and potassium chloride (KCl) were obtained from Fisher Scientific (Waltham, MA, USA). Polyvinylpyrrolidone (PVP), polyvinyl alcohol (PVA), tetraethyl orthosilicate (TEOS), and guanidine thiocyanate (GuSCN) were acquired from Sigma-Aldrich (St. Louis, MO, USA). Tris base, EDTA, and ethanol were obtained from Acros Organics (part of Thermo Fisher), Promega (Madison, WI, USA), and Decon Labs (King of Prussia, PA, USA), respectively. The Multiscribe Reverse Transcription kit, TaqMan probes, PureLink™ DNA, and miRNA isolation kits were from Life Technologies (Carlsbad, CA, USA). The 5× Taq polymerase and the 25-mM magnesium chloride (MgCl2) solution were from New England Biolabs (Ipswich, MA). All nucleic acids used in the work were purchased from Integrated DNA Technologies, Inc. (IA, USA) with their sequences listed in Table S1 (see Electronic Supplementary Material, ESM).
Fiber fabrication
We fabricated the TiO2 fibers following the method published by Li and Xia [36]. A mixture of 0.9 g of PVP with a molecular weight of ~ 1,300,000 in 7.5 mL of 200 proof ethanol was prepared and mixed in a vial. In a separate vial, 3 mL of ethanol, 3 mL of glacial acetic acid, and 1.5 mL of titanium isopropoxide were mixed and stirred on a stir plate for 20 min. The two solutions were then combined into one vial and stirred for 20 to 30 min at room temperature.
Once the mixture was fully dissolved, the solution was added into a 10-mL syringe with a 1-in, 21-gauge needle (Zephytronics, Pomona, CA, USA). The syringe was then placed on a syringe pump, set to dispense the titanium isopropoxide-PVP solution at 3.6 mL/h. The solution then underwent electrospinning at 20 kV, and the product was collected on a cylindrical drum with an aluminum surface at a distance of 10 cm. After electrospinning, the fibers were calcinated in a furnace, with the temperature increasing at the rate of 17 °C/min until 600 °C, where the temperature was held constant for 3 h before decreasing to room temperature at the rate of 1 °C/min. This process should remove the polymer, leaving behind the titanium dioxide fiber. Fibers were stored in tubes as dry solid and suspended in solution prior to use. For extraction, the fiber batch was added to a 2-mL microcentrifuge tube and diluted with water down to 25 mg/mL. The fiber suspension was vortexed for 30 s to 1 min at high speed prior to use to break down the long fibers into shorter ones.
SiO2 fibers were prepared by dissolving 0.24 g of PVA in 2.76 mL H2O to produce an 8% PVA solution at 60 °C until fully dissolved, as reported in Pirzada et al. [37]. Separately, we mixed a solution of 2.23 mL of TEOS and 1.82 mL of ethanol and added 1.44 mL of H2O. While this solution was mixing, 33.2 μL of 6 N HCl was added dropwise. This second solution was also heated to 60 °C and stirred for 1 h. After the solutions were mixed, 1.38 mL of the 8% PVA solution was added to the TEOS solution and mixed and incubated for another hour at 60 °C. This mixture was then electrospun with a tip flow rate of 1.2 mL/h at 15 cm from the aluminum collection drum and 20 kV.
MiRNA extraction and quantitation
Solutions for binding and elution with the TiO2 fibers were optimized for the extraction process. The binding buffer was mainly composed of 3 M guanidine isothiocyanate (GuSCN), 2 M guanidine hydrochloride (Gu-HCl), and 0.1% Tween-20. The elution buffer was the common TE buffer (20 mM Tris-EDTA) at pH 8.4. These conditions were applied directly for miRNA extraction from water. Fifty microliters of water or human serum spiked with 1 pmol of cel-miR-54 was mixed with 150 μL of binding buffer and 250 μg of fibers (in 25 mg/mL suspension). Additional 20 μL absolute ethanol was added to serum for denaturation of proteins. The mixture was incubated for 5 min on a rotator at room temperature and then transferred into a filter tube. The filter set was centrifuged for 5 min at 5000×g, and the filtrate was discarded. Five hundred microliters of 60% ethanol in 1 M GuSCN was added to the filter top sequentially and pipetted several times for thorough mixing to complete the first washing step. Two more rounds of wash were carried out, each with 500 μL of 80 and 90% ethanol, respectively. At last, a single centrifugation round of 2 min at 16,000×g was used to remove the residual washing solution on the filter top, and 50 μL of elution buffer was added to the fibers to elute the miRNAs by 5 min centrifugation at 5000×g. The eluent was either used immediately or stored at −20 °C. The initial SiO2 and TiO2 fiber extractions prior to optimization utilized a binding buffer primarily composed of 2.5 M GuSCN and 2 M Gu-HCl. The fibers were then washed twice with 90% ethanol and eluted in 20 mM phosphate buffer (PB).
Reverse transcription (RT) of the recovered RNAwas done using the TaqMan Probe primers and TaqMan MicroRNA Reverse Transcription Kit. The reverse transcription mix was composed of 1.1 μL of RNase-free water, 1.0 μL 10× reverse transcriptase buffer, 0.13 μL RNase inhibitor (20 U/μL), 0.1 μL dNTP mix (100 mM), 0.67 μL Multiscribe RT enzyme (50 U/μL), and 2.0 μL cel-miR-54 RT primer. Five microliters of the RT mix and 5 μL of the sample were added to individual tubes and underwent RTon a Bio-Rad CFX thermocycler. The reaction protocol was as follows: 16 °C for 30 min, 42 °C for 32 min, and 85 °C for 5 min. Following RT, qPCR was carried out. The reaction mixture contained 2 μL of the RT product, 0.1 μL DMSO, 1.0 μL ethylene glycol (> 99%), 0.5 μL magnesium chloride (25 mM), 3.9 μL RNase-free water, 2 μL 5× Taq polymerase, and 0.5 μL cel-miR-54 or endogenous miRNA TaqMan probes. The reaction protocol was as follows: 95 °C for 1.5 min, 59 °C for 50 s, followed by a denaturing step at 95 °C for 35 s and a combined annealing and extension step at 53 °C for 1 min and 10 s that cycled 45 times.
Following qPCR with the Bio-Rad CFX connect amplification and detection, recovery was calculated using the spiked cel-miR-54. A standard curve of cel-miR-54 was created and run in the qPCR to quantify the RNA recovered. Along with the extracted cel-miR-54, spiked samples were used to calculate recovery. All Cq (cycle of quantitation) values obtained from the qPCR were converted to copy number and the following equation was then used to calculate recovery:
Qubit protocol for the RNA HS Assay Kit was followed for quantifying recovered RNA, with 5 μL of the sample solution added to the working solution prior to detection.
MDA-MB-231 cells were used to test small RNA recovery efficiency of both fibers and commercial columns. The cells were lysed with RIPA lysis buffer (Santa Cruz Biotechnology) prior to proceeding with each extraction protocol, and 50 μL cell lysate, containing about 105 cells, was used per extraction. The extraction protocol followed for the fiber extraction was the optimized extraction stated above. The commercial column extraction protocol that was followed was the optimized protocol provided by Life Technologies for cell lysate with the provided buffers. An Agilent small RNA Chip was used to determine the quality of the small RNA collected by the fibers from cell lysate. The chip was run on an Agilent 2100 Bioanalyzer Instrument using the Small RNA Analysis Kit with 1 μL of extraction sample following the protocol provided by Agilent.
Results and discussion
Titanium dioxide fibers
To confirm the utility of titania fibers in nucleic acid enrichment, we initially evaluated their performance in extraction of short single-stranded nucleotides (ssDNA), in comparison with the silica-based materials, including the electrospun silica fibers, silica nanoparticles (Bioclone Inc., 1 μm diameter), and the silica-based columns. The postcalcinated titania fibers were observably more brittle and were easier to break down than the silica fibers, making it more consistent and simpler to prepare the fiber suspension by just breaking down the fibers with vortexing force. On the contrary, silica (SiO2) fibers produced with the similar electrospinning procedure needed to be ground to obtain the short fibers for suspension, and the residual polymer was harder to be cleaned after calcination. These difficulties reduced the consistency in applying the same amount of fibers during extraction. Scanning electron microscopy demonstrated that the silica fibers produced were larger in diameter, ranging from 500 to 800 nm, while the titania fibers were smaller and more uniform in size, with the average diameter found at 126.5 ± 13.4 nm (see ESM, Fig. S1). Using the Brunauer-Emmett-Teller (BET) analysis, the surface area was found to be 16.5 m2/g for the TiO2 fibers. The small diameter, large specific surface area, and simplicity in generating reproducible fiber suspension will lead to high recovery and high consistency in extraction, two important criteria for materials used in target enrichment (Fig. 1).
Fig. 1.
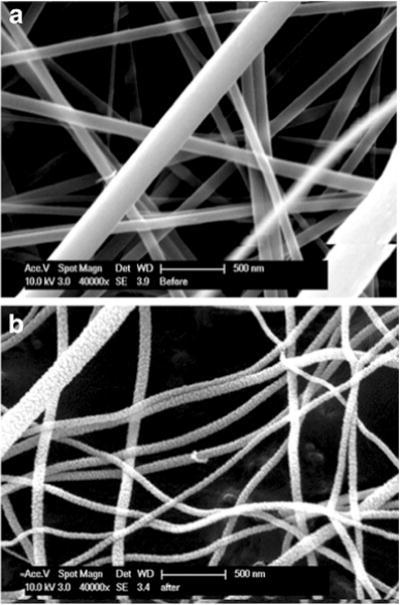
Transmission election microscope (TEM) image of the TiO2 fibers a pre- and b postcalcination. The precalcinated fibers had a smoother appearance compared to the rougher surface on the postcalcinated fibers
Commercial buffer sets included in the silica SPE column (PureLink™, Life Technologies) were used to recover the 80-nt ssDNA (WA DNA in ESM Table S1) spiked in water at 1 pmol using the aforementioned materials (for the fibers and beads, the same mass of the nanomaterial was employed) (Fig. 2). The lowest recoveries were found in the SiO2 columns (following the protocol and solutions provided by the manufacturer) and the 1-μm beads (following the optimized protocol we developed in the lab, the same as employed for the extraction with the silica fibers described in “Materials and methods”) at 6.73 ± 1.22 and 4.95 ± 4.95%, respectively. The SiO2 fibers that we fabricated yielded a recovery of 20.9 ± 2.2%, higher than the commercial SiO2 products, but still much lower than the TiO2 fibers, which attained a recovery of 87.6 ± 4.4%. Therefore, we continued with the TiO2 fibers for optimization of the extraction conditions to recover small RNAs.
Fig. 2.
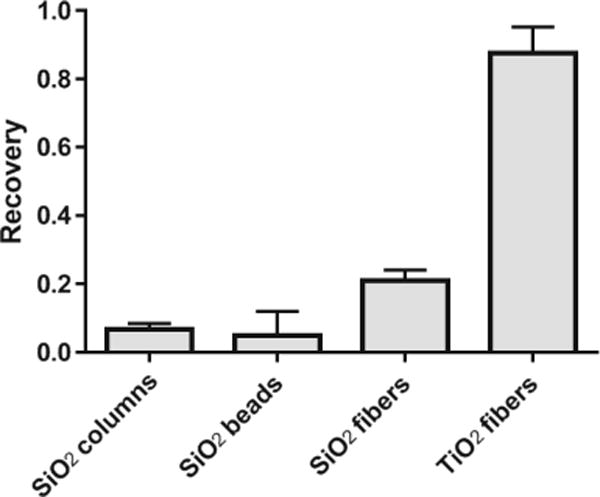
Comparison of extraction with the TiO2 fibers (n = 3) and the SiO2 (n = 2)-based methods tested with 1 pmol of ssDNA spiked in water using the commercially available buffers
Binding buffer optimization for RNA adsorption
Three conditions were considered in our development: pH, denaturant, and additive. DNA adsorption to SiO2 is most efficient when pH is above the pKa of silanol, allowing for a salt bridge to form and bind a negatively charged phosphate backbone on nucleic acids. We reason that TiO2, at the correct pH, would interact with the nucleic acids directly via formation of coordination between Ti(IV) and phosphate, or through a salt bridge, similar to SiO2. A strong chaotropic salt is definitely required to promote RNA denaturation and efficient fiber binding. We chose GuSCN in the present work, because it is also strong enough to assist in lysing cells and denaturing nucleases that affect RNA stability. Addition of ethanol (EtOH) or ethylene glycol (EG) was considered because they both could reduce the relative permittivity of the solution, thus lowering the Coulomb’s constant and allowing for the salt and phosphate in nucleic acids to interact more easily. Charge neutralization then allows nucleic acids to become more hydrophobic and easier to precipitate out of solution to bind to the fibers.
Initial miRNA extractions were performed in water, using GuSCN, which allowed easy dissolution of the salt and a good extraction efficiency at 79.8 ± 0.05% (Fig. S2, see ESM). Effects from addition of ethanol or ethylene glycol as well as varied pH values were investigated (Fig. 3a). Addition of EtOH or EG enhanced the recovery of miRNA from water by at least 17-folds. Reducing the pH to 5.0 increased the recovery by nearly 50-folds. The final optimal pH was found to be around 4.0 to 4.1, yielding a recovery of 61.1 ± 16.8% at pH 4, at least 2000-folds higher compared to that attained at pH 6 (0.03 ± 0.02%) (Fig. 3b).
Fig. 3.
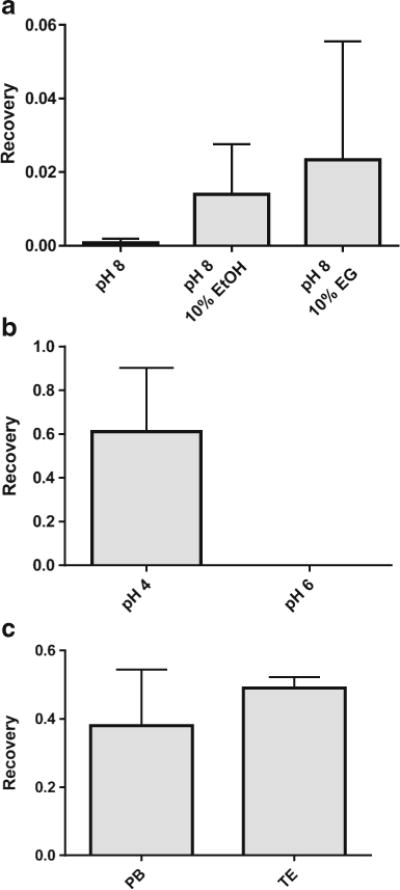
Binding buffer optimization of fiber-miRNA extractions from human serum. a Ethanol and ethylene glycol increase recovery of miRNA (n = 2). b A pH of 4 (n = 3) is important to keeping a recovery as high as 60% compared to near 0% recovery at pH 6 (n = 3). c Tris-EDTA elution buffer (n = 3) worked just as well as phosphate buffer (n = 3)
Washing and elution conditions
Following binding, wash buffers have an important role in removing any excess salts that cause interference in downstream applications, such as qPCR amplification. They should also keep miRNAs bound on the fibers so that high recovery with high purity can be obtained simultaneously. Thus, a high content of denaturants like guanidine salts needs to be included in the washing buffer. However, residual denaturants from the washing buffer could denature the enzymes used in downstream processing and need to be removed completely at the end of washing. We implemented the three-step gradient washing to gradually reduce the content of the guanidine salts and increase EtOH volume fraction, which could be thoroughly removed by evaporation before elution. Guanidine contamination was monitored by UV absorption (Nanodrop) at λ = 230 nm. We found that when the absorption at this wavelength was below 0.1, impact to the amplification step was observed (Table S2, see ESM). This was the criterion we used to judge adequate removal of the denaturants from our samples. We employed EtOH in washing because it can keep RNA bound to the fibers and be removed after washing by evaporation, and three rounds of washing with increasing EtOH concentrations were found adequate to remove almost all guanidine off the fibers and provide sufficient purity of the eluted RNA after evaporation.
Initially, we eluted the extracted miRNA with 20 mM phosphate buffer at pH 8.5, in consideration that phosphate ions can displace DNA/RNAs off the fibers by competitive binding to Ti(IV) and the more basic pH also makes the fiber surface more negative to repel the nucleic acids. The concentration of 20 mM phosphate in the elution was chosen because it showed no negative impact to PCR and led to high RNA recovery (Fig. S3, see ESM). However, other downstream applications like Bioanalyzer analysis require or prefer purified RNA to be in RNase-free water or in TE buffer, with the concerns that phosphate ions will affect enzyme performance, nucleic acid separation, or other processing. Both RNase-free water and TE buffer provide a neutral to basic pH and lack salt that facilitates RNA binding to fibers, and should work for the elution purpose. Thus, we tested the common TE buffer (20 mM Tris-base and 1 mM EDTA, pH 8.0), in which Tris has a primary role of maintaining a stable pH, and EDTA binds to cations and helps maintain the integrity of enzymes and lipids. TE provided a recovery of 49.0 ± 1.9%, slightly higher but more reproducible than the 38.0 ± 9.5% recovery from PB (Fig. 3c). Thus, the TE buffer was chosen in the subsequent analysis.
RNA size selectivity of the fibers
Following optimization of the extraction, washing, and elution buffers, we tested RNA size selectivity of our extraction system. Size selectivity can improve specificity and/or lower the background in miRNA detection by removing their long precursors or other long RNAs that may contain similar sequences as the short miRNAs. A ssRNA ladder was chosen that covered the range of small RNA (25 nt) to shorter mRNA (1000 nt). Extraction was performed in water with the optimized extraction and elution conditions found above. The PAGE result clearly showed that all ssRNA bands below 500 nt were extracted efficiently while the 1000-nt band was not visible (Fig. 4). Such a size selectivity makes our fiber and solution combination valuable in extraction of small RNA by eliminating the contamination from mRNA or other long nucleic acids. It is not yet clear about the mechanism of such a size selectivity. We suspect that the long RNA could be harder to be denatured due to their entangled/folded structures, and once bound, they could be more difficult to be eluted off because of their large molecular weights, compared to the short RNAs. Since we optimized our binding and elution solutions using a miRNA standard, these conditions may not be harsh enough to recover the long RNA, bringing in the unique feature of our method to enrich only the short RNA population. On the other hand, the silica-based materials or kits cannot provide a strong enough interaction with short RNA, and thus, they do not work well in enriching small RNA.
Fig. 4.
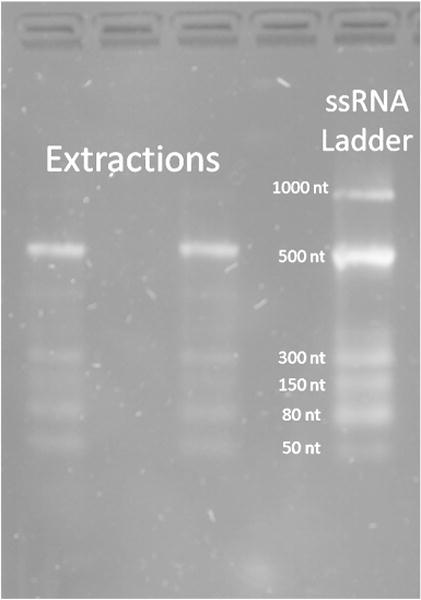
ssRNA ladder was extracted from water to test recovery of various RNA lengths. ssRNA with 500 nucleotide length and below is efficiently extracted, while the 1000-nt long fragment was not detected on the gel
MiRNA fiber application in biological samples
Optimization was continued with the fiber-RNA extraction from blood serum, to demonstrate the effectiveness of our method in a more complex biological matrix. Direct implementation of the fibers for serum extraction did not initially lead to a good recovery (< 0.01%). After testing the pH of the serum sample mixed with the binding solution, we found that addition of the binding buffer increased the pH of the serum sample from 7.4 to > 8.0, which is too high compared to the optimal pH of 4.1. At such a high pH, TiO2 has fewer negatively charged groups and is more dependent on the formation of salt bridge for nucleic acid extraction. But the concentration of salt in our binding buffer may not be high enough to form enough salt bridges for RNA binding.
The pH of the serum sample was then adjusted using hydrochloric acid to around pH 4.1 prior to spiking in the internal standard of cel-miR-54 when carrying out extraction of miRNAs from complex biological matrices. Reduction of pH led to the much improved and more consistent extraction compared to no adjustment. Triplicate extractions were performed from the lysate of MDA-MB-231 cells spiked with 1 pmol of cel-miR-54. For the PureLink kit, the manufacturer suggested that the extraction protocol is followed. Extraction with the TiO2 fibers yielded a recovery of 18.0 ± 3.6%, while the columns gave out a recovery of 0.02 ± 0.0001% (Fig. 5a). Such a big difference could be attributed to the stronger interaction of miRNAs to TiO2 than to SiO2 surface. In addition, direct interaction between the positively charged TiO2 and the negatively charged RNA at acidic pH makes the binding of miRNAs to TiO2 surface less dependent on the formation of salt bridges, like in the case of SiO2-RNA binding. Both make miRNAs the main occupants to the binding sites on the TiO2 fibers, instead of the interfering molecules in the sample matrix. In contrast, interaction between miRNAs and SiO2 relies significantly on salt bridge formation, which is easier to be outcompeted by the highly abundant matrix components, leading to low binding efficiency to the silica column surface. Figure 5b, c demonstrates the recovery of two endogenous miRNAs, hsa-miR-21 and hsa-miR-191. These two strands were quantified with qPCR. The amount of endogenous miRNA recovered from the cell lysate was about 200-folds higher with the fibers than with the column.
Fig. 5.
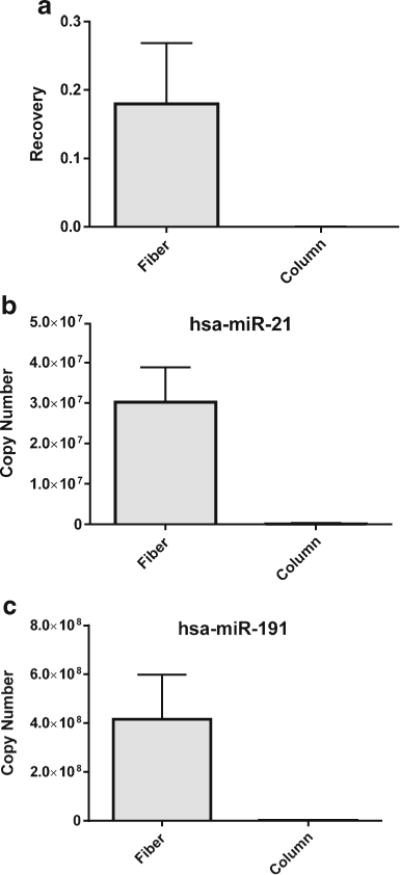
Extracting miRNA from MDA-MB-231 cells with TiO2 fibers and PureLink miRNA isolation columns. a Recovery of cel-miR-54 spiked into cell lysate was 18.0% with the fibers (n = 8) and 0.02% with columns (n = 4). Higher recoveries of endogenous b hsa-miR-21 and c hsa-miR-191 were found with TiO2 fibers
To demonstrate that our extraction method is compatible with Bioanalyzer, the common analysis used in next-generation sequencing for determination of RNA quality, the RNA extracted from 50-μL cell lysate (~ 105 cells) was run on an Agilent Bioanalyzer 2100, and the result was compared to those obtained using the commercial columns, the PureLink miRNA isolation columns. The RNA Pico chip was first used to see the full size range of the recovered RNA, and the RNA collected was all below 1000 nt (Fig. S4, see ESM). The more confined size range was proved to be from 20 to 150 nt using the small RNA chip, with the largest amount of RNA found to be 60 nt in length, which should be the highly abundant tRNA in cell lysate (Fig. 6) [38, 39]. The Bioanalyzer also quantified that the total small RNA concentration obtained with the TiO2 fibers was 985.2 pg/μL, much higher than that with the commercial column (10.2 pg/μL).
Fig. 6.
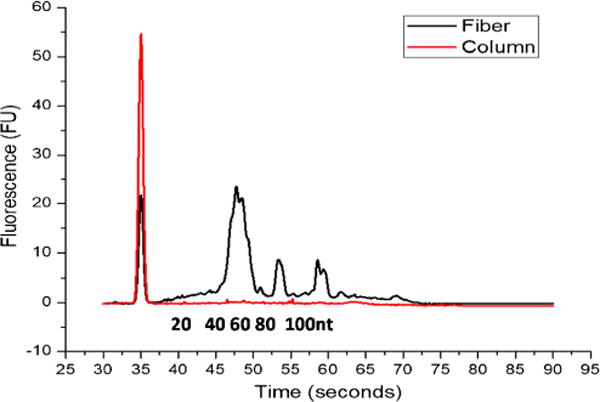
Comparison of small RNA extraction from MDA-MB-231 cells with fibers and columns analyzed by an Agilent 2100 Bioanalyzer. Small RNA recovery was as high as 985 pg/μL with the fibers and 10.2 pg/μL with the columns from as little as 134,000 cells
Conclusion
This work has demonstrated that the electrospun titanium dioxide nanofibers have the capability to consistently recover miRNA with higher recovery within comparable or shorter duration than the commercially available methods. The binding, washing, and elution conditions were optimized to improve recovery in complex biological matrices, and elution in the TE buffer permits good compatibility with diverse downstream RNA processing techniques. Quantitation by RTqPCR and quality evaluation in the Bioanalyzer of the miRNAs recovered from cell lysate and serum support the applicability of our method with complex samples. The high recovery of miRNA will allow consumption of small sample volumes and accurate quantitation of the low abundant miRNAs and other small RNAs, beneficial for miRNA functional study and biomarker discovery for disease diagnosis and prognosis.
Supplementary Material
Acknowledgments
We are thankful for the kind assistance from Mr. Joshua Belardes on the early stage of buffer optimization for DNA extraction with SiO2 fibers.
Funding information: The authors acknowledge the support by the National Cancer Institute of the National Institutes of Health under Award Number R01CA188991 to WZ. This award also provided support (3R01CA188991-02S1) under the Research Supplements to Promote Diversity in Health-Related Research Program for LAJ. MAG was supported by MARCU-STAR training grant from NIH. SS and YM were supported by the Research in Science and Engineering (RISE) undergraduate summer research program at UC Riverside. JGC was supported by UC Riverside’s office of Undergraduate Education (UE).
Footnotes
Published in the topical collection celebrating ABCs 16th Anniversary.
Electronic supplementary material: The online version of this article (https://doi.org/10.1007/s00216-017-0649-3) contains supplementary material, which is available to authorized users.
Compliance with ethical standards
Conflict of interest: The authors declare that they have no competing interests.
Informed consent: The serum samples were obtained from commercial sources with the individual information unknown to the researchers.
References
- 1.Djuranovic S, Nahvi A, Green R. miRNA-mediated gene silencing by translational repression followed by mRNA deadenylation and decay. Science. 2012;336:237–40. doi: 10.1126/science.1215691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Storz G. An expanding universe of noncoding RNAs. Science. 2002;296:1260–3. doi: 10.1126/science.1072249. [DOI] [PubMed] [Google Scholar]
- 3.Sassen S, Miska EA, Caldas C. MicroRNA: implications for cancer. Virchows Arch. 2008;452:1–10. doi: 10.1007/s00428-007-0532-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winter J, Jung S, Keller S, Gregory RI, Diederichs S. Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol. 2009;11:228–34. doi: 10.1038/ncb0309-228. [DOI] [PubMed] [Google Scholar]
- 5.Bekris LM, Leverenz JB. The biomarker and therapeutic potential of miRNA in Alzheimer’s disease. Neurodegener Dis Manag. 2015;5:61–74. doi: 10.2217/nmt.14.52. [DOI] [PubMed] [Google Scholar]
- 6.Alipoor SD, Adcock IM, Garssen J, Mortaz E, Varahram M, Mirsaeidi M, et al. The roles of miRNAs as potential biomarkers in lung diseases. Eur J Pharmacol. 2016;791:395–404. doi: 10.1016/j.ejphar.2016.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhao Z, Moley KH, Gronowski AM. Diagnostic potential for miRNAs as biomarkers for pregnancy-specific diseases. Clin Biochem. 2013;46:953–60. doi: 10.1016/j.clinbiochem.2013.01.026. [DOI] [PubMed] [Google Scholar]
- 8.Cao Y, Griffith JF, Weisberg SB. The next-generation PCR-based quantification method for ambient waters: digital PCR. Methods Mol Biol. 2016;1452:113–30. doi: 10.1007/978-1-4939-3774-5_7. [DOI] [PubMed] [Google Scholar]
- 9.Al-Soud WA, Rådström P. Purification and characterization of PCR-inhibitory components in blood cells. J Clin Microbiol. 2001;39:485–93. doi: 10.1128/JCM.39.2.485-493.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duy J, Koehler JW, Honko AN, Minogue TD. Optimized microRNA purification from TRIzol-treated plasma. BMC Genomics. 2015;16:95. doi: 10.1186/s12864-015-1299-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rio DC, Ares M, Hannon GJ, Nilsen TW. Purification of RNA using TRIzol (TRI reagent) Cold Spring Harb Protoc. 2010;2010 doi: 10.1101/pdb.prot5439. pdb.prot5439. [DOI] [PubMed] [Google Scholar]
- 12.Auffinger P, Bielecki L, Westhof E. Anion binding to nucleic acids. Structure. 2004;12:379–88. doi: 10.1016/j.str.2004.02.015. [DOI] [PubMed] [Google Scholar]
- 13.Simões AES, Pereira DM, Amaral JD, Nunes AF, Gomes SE, Rodrigues PM, et al. Efficient recovery of proteins from multiple source samples after TRIzol(®) or TRIzol(®)LS RNA extraction and long-term storage. BMC Genomics. 2013;14:181. doi: 10.1186/1471-2164-14-181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao X, Johnson JK. Simulation of adsorption of DNA on carbon nanotubes. J Am Chem Soc. 2007;129:10438–45. doi: 10.1021/ja071844m. [DOI] [PubMed] [Google Scholar]
- 15.Saha S, Sarkar P. Understanding the interaction of DNA-RNA nucleobases with different ZnO nanomaterials. Phys Chem Chem Phys. 2014;16:15355–66. doi: 10.1039/c4cp01041h. [DOI] [PubMed] [Google Scholar]
- 16.Park JS, Goo N-I, Kim D-E. Mechanism of DNA adsorption and desorption on graphene oxide. Langmuir. 2014;30:12587–95. doi: 10.1021/la503401d. [DOI] [PubMed] [Google Scholar]
- 17.Hashemi E, Akhavan O, Shamsara M, Valimehr S, Rahighi R. DNA and RNA extractions from eukaryotic and prokaryotic cells by graphene nanoplatelets. RSC Adv. 2014;4:60720–8. [Google Scholar]
- 18.Wang F, Liu B, Huang P-JJ, Liu J. Rationally designed nucleobase and nucleotide coordinated nanoparticles for selective DNA adsorption and detection. Anal Chem. 2013;85:12144–51. doi: 10.1021/ac4033627. [DOI] [PubMed] [Google Scholar]
- 19.Madhugiri S, Sun B, Smirniotis PG, Ferraris JP, Balkus KJ. Electrospun mesoporous titanium dioxide fibers. Microporous Mesoporous Mater. 2004;69:77–83. [Google Scholar]
- 20.Lu C, Liu Y, Ying Y, Liu J. Comparison of mos2, WS2, and graphene oxide for DNA adsorption and sensing. Langmuir. 2017;33:630–7. doi: 10.1021/acs.langmuir.6b04502. [DOI] [PubMed] [Google Scholar]
- 21.Kim J, Park S-J, Min D-H. Emerging approaches for graphene oxide biosensor. Anal Chem. 2017;89:232–48. doi: 10.1021/acs.analchem.6b04248. [DOI] [PubMed] [Google Scholar]
- 22.Pihlasalo S, Mariani L, Härmä H. Quantitative and discriminative analysis of nucleic acid samples using luminometric nonspecific nanoparticle methods. Nano. 2016;8:5902–11. doi: 10.1039/c5nr09252c. [DOI] [PubMed] [Google Scholar]
- 23.Jena PV, Safaee MM, Heller DA, Roxbury D. DNA-carbon nanotube complexation affinity and photoluminescence modulation are independent. ACS Appl Mater Interfaces. 2017;9:21397–405. doi: 10.1021/acsami.7b05678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vilela P, El-Sagheer A, Millar TM, Brown T, Muskens OL, Kanaras AG. Graphene oxide-upconversion nanoparticle based optical sensors for targeted detection of mRNA biomarkers present in Alzheimer’s disease and prostate cancer. ACS Sens. 2017;2:52–6. doi: 10.1021/acssensors.6b00651. [DOI] [PubMed] [Google Scholar]
- 25.Li F, Liu X, Zhao B, Yan J, Li Q, Aldalbahi A, et al. Graphene nanoprobes for real-time monitoring of isothermal nucleic acid amplification. ACS Appl Mater Interfaces. 2017;9:15245–53. doi: 10.1021/acsami.7b01134. [DOI] [PubMed] [Google Scholar]
- 26.Bielicka-Daszkiewicz K, Voelkel A. Theoretical and experimental methods of determination of the breakthrough volume of SPE sorbents. Talanta. 2009;80:614–21. doi: 10.1016/j.talanta.2009.07.037. [DOI] [PubMed] [Google Scholar]
- 27.Thingholm TE, Larsen MR. The use of titanium dioxide for selective enrichment of phosphorylated peptides. Methods Mol Biol. 2016;1355:135–46. doi: 10.1007/978-1-4939-3049-4_9. [DOI] [PubMed] [Google Scholar]
- 28.Eriksson AIK, Bartsch M, Bergquist J, Edwards K, Lind SB, Agmo Hernández V. On-target titanium dioxide-based enrichment for characterization of phosphorylations in the Adenovirus pIIIa protein. J Chromatogr A. 2013;1317:105–9. doi: 10.1016/j.chroma.2013.08.096. [DOI] [PubMed] [Google Scholar]
- 29.Wakabayashi M, Kyono Y, Sugiyama N, Ishihama Y. Extended coverage of singly and multiply phosphorylated peptides from a single titanium dioxide microcolumn. Anal Chem. 2015;87:10213–21. doi: 10.1021/acs.analchem.5b01216. [DOI] [PubMed] [Google Scholar]
- 30.Piovesana S, Capriotti AL, Cavaliere C, Ferraris F, Iglesias D, Marchesan S, et al. New magnetic graphitized carbon black TiO2 composite for phosphopeptide selective enrichment in shotgun phosphoproteomics. Anal Chem. 2016;88:12043–50. doi: 10.1021/acs.analchem.6b02345. [DOI] [PubMed] [Google Scholar]
- 31.Li Q, Ning Z, Tang J, Nie S, Zeng R. Effect of peptide-to-TiO2 beads ratio on phosphopeptide enrichment selectivity. J Proteome Res. 2009;8:5375–81. doi: 10.1021/pr900659n. [DOI] [PubMed] [Google Scholar]
- 32.Zhang X, Wang F, Liu B, Kelly EY, Servos MR, Liu J. Adsorption of DNA oligonucleotides by titanium dioxide nanoparticles. Langmuir. 2014;30:839–45. doi: 10.1021/la404633p. [DOI] [PubMed] [Google Scholar]
- 33.Mondal K, Ali MA, Agrawal VV, Malhotra BD, Sharma A. Highly sensitive biofunctionalized mesoporous electrospun TiO(2) nanofiber based interface for biosensing. ACS Appl Mater Interfaces. 2014;6:2516–27. doi: 10.1021/am404931f. [DOI] [PubMed] [Google Scholar]
- 34.Chen X, Mao SS. Titanium dioxide nanomaterials: synthesis, properties, modifications, and applications. Chem Rev. 2007;107:2891–959. doi: 10.1021/cr0500535. [DOI] [PubMed] [Google Scholar]
- 35.Vandeventer PE, Mejia J, Nadim A, Johal MS, Niemz A. DNA adsorption to and elution from silica surfaces: influence of amino acid buffers. J Phys Chem B. 2013;117:10742–9. doi: 10.1021/jp405753m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li D, Xia Y. Fabrication of titania nanofibers by electrospinning. Nano Lett. 2003;3:555–60. [Google Scholar]
- 37.Pirzada T, Arvidson SA, Saquing CD, Shah SS, Khan SA. Hybrid silica-PVA nanofibers via sol-gel electrospinning. Langmuir. 2012;28:5834–44. doi: 10.1021/la300049j. [DOI] [PubMed] [Google Scholar]
- 38.Baglio SR, Rooijers K, Koppers-Lalic D, Verweij FJ, Pérez Lanzón M, Zini N, et al. Human bone marrow- and adipose-mesenchymal stem cells secrete exosomes enriched in distinctive miRNA and tRNA species. Stem Cell Res Ther. 2015;6:127. doi: 10.1186/s13287-015-0116-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masotti A, Preckel T. Analysis of small RNAs with the Agilent 2100 Bioanalyzer. Nat Methods. 2006;3:iii–v. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


