Abstract
Purpose
To automate dynamic contrast-enhanced magnetic resonance imaging (DCE-MRI) data analysis by unsupervised pattern recognition (PR) to enable spatial mapping of intra-tumoral vascular heterogeneity.
Methods
Three steps were automated: (i) Determination of contrast agent arrival time at tumor, including calculation of pre-contrast signal. (ii) Four criteria-based algorithms for the slice-specific selection of number of patterns (NPs) were validated using 109 tumor slices from subcutaneous flank tumors of 5 different tumor models. The criteria were: half area under the curve, standard deviation thresholding, % signal enhancement, and signal-to-noise ratio (SNR). The performance of these criteria was assessed by comparing the calculated NPs to visually determined NPs. (iii) Spatial assignment of single patterns and/or pattern mixtures, obtained by constrained non-negative matrix factorization (cNMF).
Results
The determination of the contrast agent arrival time at the tumor slice was successfully automated. For the determination of NPs, the SNR-based approach outperformed other selection criteria by agreeing >97% with visual assessment. The spatial localization of single patterns and pattern mixtures, the latter inferring tumor vascular heterogeneity at sub-pixel spatial resolution, was established successfully by automated assignment from DCE-MRI signal-versus-time curves.
Conclusion
The PR-based DCE-MRI analysis was successfully automated to spatially map intra-tumoral vascular heterogeneity.
Keywords: DCE-MRI, pattern recognition analysis, principal component analysis, automation, intra-tumoral vascular heterogeneity
Introduction
The tumor microenvironment is heterogeneous, exhibiting severe functional vascular abnormalities (1–3). Dynamic contrast-enhanced (DCE)-MRI is used to assess tumor blood flow and permeability clinically and pre-clinically, after the administration of the contrast agent (CA) gadopentetate dimeglumine (Gd-DTPA), with <30 min (clinically typically 5–10 min) scan times and high spatial resolution (<200 µm pre-clinically and 1–2 mm clinically) (4–8). Parameters from tracer-kinetic modeling of signal-versus-time DCE-MRI curves (4,9,10) have been used to differentiate tumor microenvironments (5,6,11,12) and to longitudinally monitor vascular changes in response to treatments (6,13–15). Various pattern analysis approaches, including machine learning, have been used to extract features to improve tumor classification and, to a lesser extent, assess intra-tumoral heterogeneity to guide treatment or gauge prognosis (16–23).
Using preclinical in vivo imaging modalities coregistered with pathology, we have shown previously that well-vascularized (well-perfused) tumor areas are characterized by rapid Gd-DTPA uptake/washout, that hypoxic areas exhibit reduced vascular function associated with delayed Gd-DTPA uptake/washout, while necrotic areas exhibit slow or no CA uptake and no discernible washout over the experimental observation (12,24,25). We categorized these tumor microenvironments based on their representative DCE-MRI signal-versus-time curves by pattern recognition (PR), using the Gaussian mixture model or constrained non-negative matrix factorization (cNMF) (24,25). The semi-automatic PR approach required manual input of the number of patterns (NPs) in the DCE-MRI data. The variable (subjective) application of a fixed NPs for all tumor slices may lead to overfitting (or underfitting) in tumors or tumor slices that are characterized by more (or fewer) patterns than predefined, and thus, disregards intra-tumoral heterogeneity represented by disparate DCE-MRI curves and physiological environments across tumor slices (Figure 1).
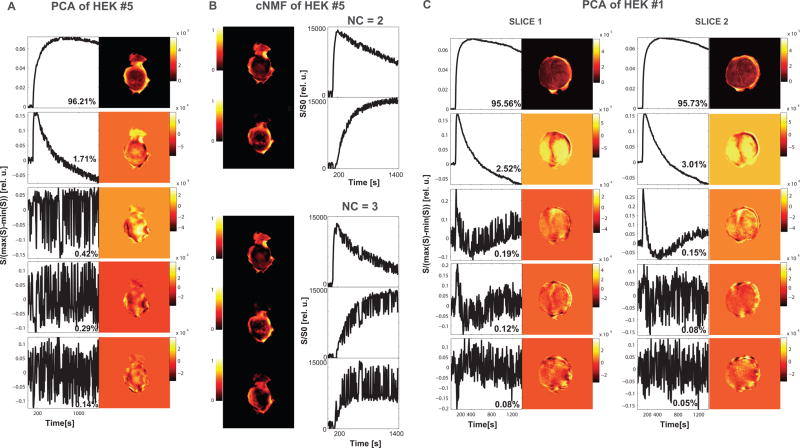
Figure 1. Significance of selecting number of patterns (NPs).
(A) As shown for a representative HEK tumor slice (HEK #5), PCA produced 2 distinct PCs followed by higher-order PCs depicting noise. The first 2 PCs explain 97.92% of signal-versus-time curves behavior in this tumor slice. (B) To the tumor slice depicted in (A) corresponding cNMF maps (left) with associated cNMF curves (right) for NPs set to 2 (top) and 3 (bottom), respectively. For NP = 2, two distinct cNMF patterns are identified; for NPs = 3 however, the second and third pattern are very noisy and show overlap due to data over-fitting resulting in the inability to reproducibly/accurately assign a pixel to pattern 2 or 3. (C) In a second example, PCA on two slices of a heterogeneous HEK flank tumor (HEK #1), the first five PCs and their corresponding weight maps are displayed. The % variability of each PC to explain the contrast agent (CA) uptake behavior in their respective slice is shown below each PC. Despite the % contribution to the overall signal of the 3rd PC in slice 1 and 2 being similar (0.19% and 0.15% respectively), the numbers of significant signal-related PCs identified visually for slice 1 and slice 2 respectively are 2 and 3, as the signal in the 3rd PC in slice 1 is within twice the size of the noise and similar to higher-order PCs. The overall signal explained by the number of significant signal-related PCs is 98.08% in slice 1 and 98.89% in slice 2. The varying NPs between slices attest to the vascular heterogeneity of this tumor across the thickness of the tumor.
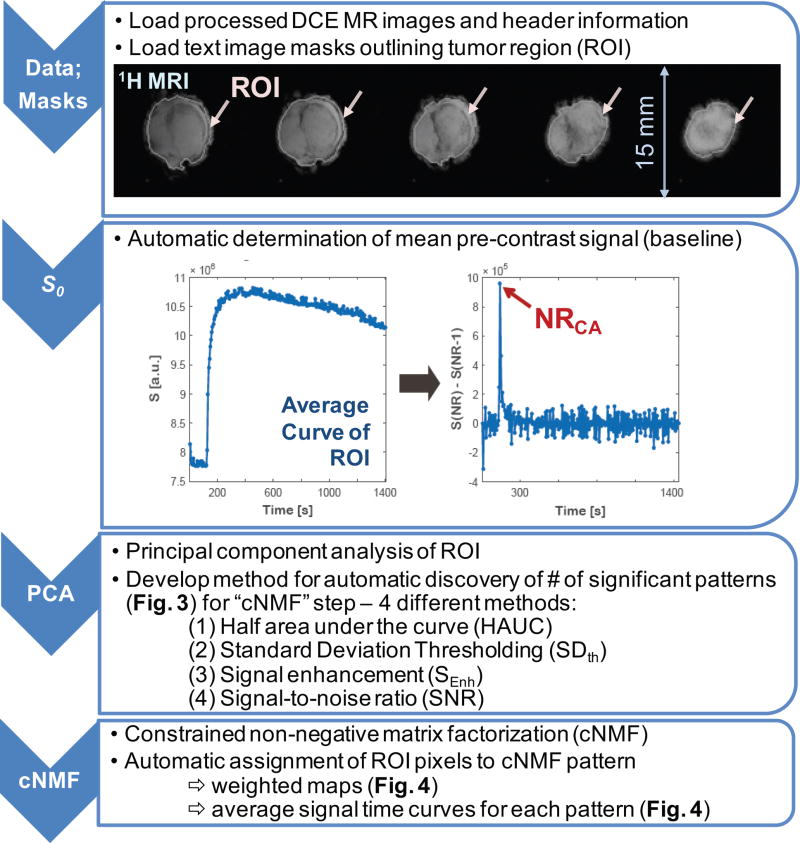
The goal of this study is to optimize and automate DCE-MRI data analysis by our previously described unsupervised PR approach (24) to accurately and fully-automated identify vascularity-driven intra-tumoral heterogeneity using cNMF. This involves novel automatic approaches to determine NPs for each DCE-MRI slice, to spatially map intra-tumoral heterogeneities and incorporates the computerized determination of the pre-contrast signal. A step-wise scheme of the analysis process is shown in Figure 2. All analysis steps were coded in MATLAB (The MathWorks, Inc. Natick, MA).
Figure 2. Processing steps for DCE-MRI data analysis. (Data; Masks).
Processed DCE-MRI images and manually contoured tumor region of interest (ROI) are up-loaded to the program. A representative example (HEK #1) showing the 1H MR images of the 5 tumor slices overlaid (white line) with the manually outlined tumor region (ROI) in each slice (arrows); (S0) Automatic determination of the time point of contrast agent arrival at tumor from an average signal-versus-time curve over tumor slice ROI; (PCA) Automatic determination of the number of patterns (NPs) using Principal Component Analysis (PCA); (cNMF) Automatic assignment of ROI pixels to constrained Non-negative Matrix Factorization (cNMF) pattern or pattern mixture. Note that three out of the four steps underwent automation.
Methods
Data Sets: Tumor Models, In Vivo DCE-MRI
We evaluated our approaches using 109 DCE-MR image slices of tumors from a tumorigenic, human embryonic kidney cell line (HEK, n = 6) and four prostate cancer cell lines: LAPC-4 (kindly provided by Dr. Sawyer (26), n = 7), Myc-CaP ((27), n = 2), PC-3 ((28), n = 2), and RM-1 ((29), n = 4).
In vivo DCE-MRI was performed using a custom-built, solenoid 1H MR coil on a horizontal-bore Bruker 7T magnet (Bruker Biospin, Germany). A bolus of 0.2 mmol/kg Gd-DTPA (Magnevist, Berlex Laboratories, Inc., Wayne, NJ) was administered i.v. via tail-vein catheter. During the MR experiment, mice were anesthetized with <2% isoflurane in oxygen. The breathing rate was kept at 50–90 breaths/min by adjusting the isoflurane level. The rodent core temperature was maintained at 34–37°C. After tumor positioning, 1H MR coil tuning and matching, the water line width was optimized to ~30–70 Hz full-width-half-maximum by field map-based shimming. To assess tumor vascularity, DCE-MRI data were acquired using a T1-weighted fast low-angle shot (FLASH (30)) sequence with 3.2 ms echo time, minimum repetition time (TRmin), 256 time points (NR256, number of frames/repetitions (NR) per image slice set of 5, 6, or 7 slices), 1 average, 15×15 mm2 field-of-view, 128×128 matrix, 1 mm slice thickness and 5–7 slices to cover the entire tumor. For 5, 6 and 7 slices respectively, TRmin were 42.875 ms, 51.450 ms, and 60.025 ms with a corresponding temporal resolution of 5.487 s, 6.585 s, and 7.683 s. All animal studies complied with protocols approved by the Institutional Animal Care and Use Committee of Memorial Sloan Kettering Cancer Center.
Data Loading and Determination of Mean Pre-contrast Signal S0
For each image slice, text image masks, outlining the entire tumor area (ROI), are created using ImageJ (http://imagej.nih.gov/ij/, NIH, Bethesda, MD) from ROIs drawn manually on processed (Fourier-transformed, magnitude calculated) MR images acquired with 1 NR and 500 ms TR (other parameters equal those for DCE-MRI) (Figure 2, 1st step). Sequence parameters, DCE-MR images, and ROI masks in each slice are loaded via a graphic user interface (Figure 2, 1st step). To automatically identify the time point of the CA arriving at the tumor tissue (NRCA), the derivative of the average signal-versus-time curve is calculated for each ROI. An example curve is depicted in Figure 2 (2nd step). The largest signal difference (red arrow, Figure 2, 2nd step) corresponds to NRCA. The lowest NR of the time points with the top 10 highest signal changes is selected to reduce the chance of erroneously identifying a later time point as NRCA. The pre-contrast signal S0 is obtained by averaging the signal between NR1 and NRCA with the first and last 5 NRs excluded to minimize errors due to signal distortions at the start of the DCE-MRI scan and due to potentially missing points that may already show enhancement but not the largest change (Figure 2, 2nd step). It is used to calculate baseline-corrected signal (signal-S0) and normalized signal (signal/S0) -versus-time curves.
Principal Component Analysis: Approaches for Automatic Determination of NPs
Principal component analysis (PCA, (31,32)) which identifies the sources of largest variations (principal components, PCs) is applied to the baseline-corrected signal (Figure 2, 3rd step). It was conducted through singular value decomposition of the data covariance matrix (24). For each pixel, signal-versus-time curves (S(x, t), with spatial location x and dynamic time frame t=NR1, …,NR256) extracted from DCE-MRI data were resolved as the weighted sum of the PCs. Orthonormal PCs were ordered by decreasing amounts of variability.
Comparing the NPs, determined by 1–3 readers (SH, EA, RS) based on PC curve characteristics (signal above noise, see also Figure 1) through visual inspection, we evaluated four criteria (Eq. 1 – Eq. 4) for their ability to automatically determine the number of signal-related PCs (equivalent to NPs) in each tumor-slice ROI. All criteria were calculated from PC curves. Let PC(tN, k) be the Nth point (N = 1, …, 256) in the kth PC (k = 1, …, 256).
The first criterion, half area under the curve (HAUC(k)), hypothesizes that the area of the first half of the points post-contrast in each PC is signal related if HAUC is above an empirically defined threshold of 0.5×HAUC(1).
| (Eq. 1) |
where k refers to the kth PC.
The second criterion, standard deviation thresholding (SDTh) hypothesizes that an empirically set cutoff value of 4× the standard deviation of , i.e. the sum of the pre-contrast time points of all PC curves (t=5 to t=(NRCA−5NR)) with kth PC weighted by its % contribution Fk to the overall signal, leads to the selection of only significant patterns (NPs), and is given by:
| (Eq. 2) |
The maximum i for which SDTh(i), i.e. the sum over all time points of all weighted PC curves (k=1 to NR256) minus the sum of all time points of k=1 to the ith weighted PC curves reaches the cutoff value, defines the NPs.
The third criterion, % signal enhancement (SEnh), is defined as:
| (Eq. 3) |
where max refers to the maximum ‘signal’ height, tEnh to the time frame from CA arrival until the end of the DCE-MRI acquisition (NRCA to NR256), mean to the average ‘signal’ height, and tBL to the time frame covering the pre-contrast acquisition from which S0 is calculated. A threshold of SEnh = 6000 was set empirically to select the NPs contributing significantly to the portrayal of the signal-versus-time curves, by optimizing agreement with visually determined NPs as more tumor slices were added to the analysis.
And the fourth criterion, signal-to-noise ratio (SNR), is defined as:
| (Eq. 4) |
with the noise (above which the ‘signal’ has to rise) defined by SD that is 4 times the standard deviation of the mean of PC(tBL,k). Guided by the Rose Criterion (33,34), a SNR threshold of 5 was set to assure 100% certainty in distinguishing the PC ‘signal’ from the noise (35). In cases of low or no contrast enhancement (e.g. necrotic tumors) with an SNR of the first PC between 2 and 5, the number of significant PCs was set to 1.
To assess the performance of these criteria, calculated NPs were compared to NPs determined visually.
Constrained Non-negative Matrix Factorization (cNMF) and Pattern Assignments
The orthonormal PCs are not able to represent signal-versus-time curves directly, as the latter are not commonly orthonormal (24). However, PCs are useful to characterize the number of uncorrelated, significant signal-related patterns (NP) underlying the signal-versus-time curves of DCE-MRI data. Therefore as described previously (24), constrained non-negative matrix factorization (cNMF, (36–38)), an unsupervised PR approach, is used to describe each pixel’s normalized signal-versus-time curve by the NP patterns (cNMF curves, which are not orthonormal unlike the PCs) and their corresponding weights without significant loss of information (Figure 1, 4th step). The weights determine the contribution of each representative cNMF curve to a given signal-versus-time curve, and thus, allows one to separate pixels dominated by one of the NP patterns of CA uptake/washout behavior from pixels that are characterized by a mixture of several cNMF patterns.
To generate cNMF curve pattern maps visualizing the contributions of the NP different patterns to each pixel in the ROI, the weights of each cNMF curve in a pixel are expressed as the fraction of sum of the weights in that pixel. Applying an encoder with NP binary cells which has 2 NP states, two different approaches were used to create pattern masks: (i) each pixel is assigned to the pattern with the maximum normalized weight (Decision Map 1), as done previously (24); (ii) each pixel is assigned either to a single pattern or a mixture out of 2 to NP patterns (Decision Map 2), that is if the normalized weight difference of the pattern with the maximum weight to one or more of the other pattern weights is less than 25%, the pixel is assigned to a pattern mixture, otherwise, it is assigned to the dominant pattern.
Pixels with a maximum signal enhancement (SEnh(k), Eq. 3) of less than 4 standard deviations of the pre-contrast signal (mean±SD) were assigned as no contrast regions, thus, unlike before (24), low contrast regions are included in the analysis and spatial mapping.
Results
Significance of Selection of NPs
The significance of choosing the NPs based on tumor characteristics is illustrated in Figure 1. As shown for a representative tumor slice in Figure 1A, PCA produced 2 distinct PCs followed by higher-order PCs depicting noise. The corresponding cNMF maps of this tumor slice with associated cNMF curves for NPs set to 2 and 3, respectively, are shown in Figure 1B. For 2 NPs, two distinct cNMF patterns are identified; for 3 NPs however, the second and third pattern are very noisy with visually overlapping pattern curves due to data over-fitting and the inability to reproducibly/accurately assign a pixel to pattern 2 or 3. In a 2nd example (Figure 1C), PCA on two different slices in a heterogeneous flank tumor demonstrates that the number of PCs, with a signal level significantly different from the noise on the PC curve, may vary between slices in a single tumor. Thus, to avoid over- or under-fitting to characterize the patterns present across a tumor, it is essential to adjust the NPs to reflect the number of physiological relevant patterns describing the tumor microenvironments present in each tumor slice.
Automation of DCE-MRI Analysis
Three out of four steps, involved in the proposed DCE-MRI data analysis (Figure 2) have been automated:
Automatic Determination of S0
The automated selection of the pre-contrast signal, as detailed in the method section, accounts for variable injection time points due to manual injection of the CA and specifies the actual arrival time of the CA at the tumor-healthy tissue interface (Figure 2, 2nd step; Supporting Figure S1 discusses ROI versus pixel-based calculation of the S0).
Automatic Selection of Significant NPs
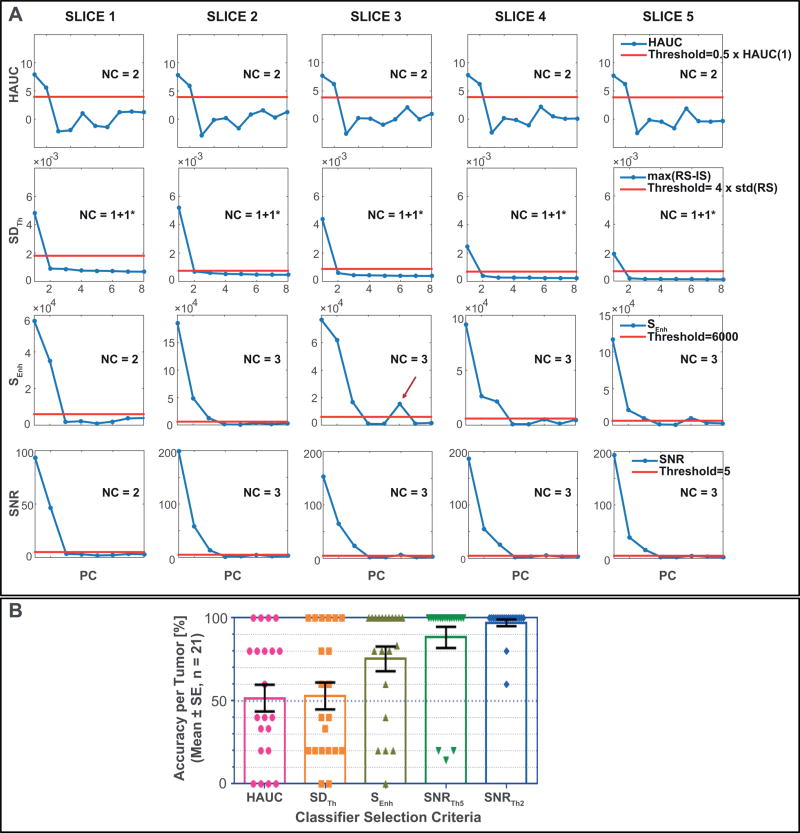
The automatic selection criteria of NPs were compared to NPs determined from visual inspection of PC curves (Figure 1) by up-to 3 readers (SH, EA, RS). Representative examples of the 4 methods for automatic selection of NPs are presented from tumor slices of HEK tumor #1 in Figure 3A. For the determination of NPs, only consecutive PCs above a pre-defined threshold were selected because higher PCs above the defined threshold, but following one or more PCs below the threshold (Figure 3A, red arrow), contribute typically less than 0.05% to the overall signal. As the SDTh method by definition assumes that the 1st PC (subtracted from the overall signal (Eq. 2)) is significant, NPs were calculated by adding one (depicted as +1* in Figure 3A) to the NPs determined from thresholding.
Figure 3. Automatic determination of number of patterns (NPs).
(A) Representative examples of each tested method for the automatic determination of NPs: HAUC (Eq. 1), SDTh (Eq. 2), SEnh (Eq. 3), and SNR (Eq. 4). The red horizontal lines represent the respective thresholds – 6000 for SEnh, 0.5×HAUC(1st PC) for HAUC, 4× standard deviation of the pre-contrast signal for SDTh, and either 5 or 2 (not shown) for SNR – as explained in the method section. The PCs are plotted along the x axis by decreasing amount of contribution to the total signal up-to the 8th PC. The red arrow denotes the occasional occurring PC above the defined threshold following one or more PCs below the defined threshold; these high-order PC contribute generally little (< 0.05%) to the total signal. (B) Accuracy per tumor is displayed for each selection criterion and each tumor by the closed signs, with the open bars displaying the corresponding mean±standard error (±SE) averaged over the 21 tumors. Accuracy per tumor is calculated as the percentage of total slices for which the NPs determined by each selection criterion matches the manually determined NPs. The method SNR had the highest accuracy, which could be further improved for tumors with low CA uptake by lowering the threshold from 5 to 2. The accuracy for the selection criteria HAUC, SDTh and SEnh deviated significantly from the desired 100%, contrary to SNR for both thresholds (P < 0.0001 for HAUC and SDTh; P = 0.0033 for SEnh, P = 0.0830 for SNRTh5, and P = 0.1864 for SNRTh2 – one sample t test with a theoretical mean of 100%, two-tailed P values).
Table 1 lists the accuracy for each tumor as the fraction of tumor slices where NPs from the four selection criteria and visual inspections agreed. The selection criterion SNR was applied with thresholds 5 and 2, whereby the latter improved the accuracy for tumors with low contrast-to-noise ratio (CNR, red numbers in Table 1). The corresponding overall accuracy for each method was 50%, 53%, 76%, 87%, and 97% for HAUC, SDth, SEnh, SNRTh5, and SNRTh2, respectively (Table 1, Total). Figure 3B shows the accuracy per tumor averaged over 21 tumors for each of the pattern selection criteria. The most accurate criterion to select NPs, SNR, is the only method (for both threshold levels) that does not significantly deviate from the desired 100% (P>0.19).
Table 1.
Accuracy of the determination of NPs from PC curves for diverse tumor models (HEK, LAPC-4, MycCaP, PC-3, RM-1) by (i) half area under the curve (HAUC), (ii) standard deviation thresholding (SDTh), (iii) % signal enhancement (SEnh), and (iv) signal-to-noise ratio with 2 different thresholds (SNRTh5 and and SNRTh2). The number of slices per total tumor slices in agreement with the manual NP determination is displayed for each tumor; the numbers in red show the improvement in NP determination by changing the SNR threshold in cases of overall low contrast-to-noise ratio (CNR) for the raw data – often in tumors with extensive necrosis. Further, the tumor volume covered by the DCE-MRI (VDCE) and the time post tumor cell injection1 (PTI) that the DCE-MRI experiments were performed are listed.
| Tumor type |
# | VDCE [mm3] |
PTI [days] |
HAUC | SDTh | SEnh | SNRTh5 | SNRTh2 |
|---|---|---|---|---|---|---|---|---|
| HEK | 1 | 182 | 13 | 1/5 | 1/5 | 5/5 | 5/5 | 5/5 |
| 2 | 241 | 18 | 2/6 | 2/6 | 6/6 | 6/6 | 6/6 | |
| 3 | 408 | 13 | 0/5 | 1/5 | 4/5 | 5/5 | 5/5 | |
| 4* | 400 | 19 | 4/5 | 1/5 | 1/5 | 1/5 | 3/5 | |
| 5 | 158 | 19 | 5/5 | 5/5 | 5/5 | 5/5 | 5/5 | |
| 6 | 239 | 20 | 4/5 | 4/5 | 5/5 | 5/5 | 5/5 | |
| LAPC-4 | 1* | 108 | 21 | 0/7 | 7/7 | 7/7 | 1/7 | 7/7 |
| 2 | 131 | 56 | 2/5 | 2/5 | 4/5 | 5/5 | 5/5 | |
| 3 | 112 | 57 | 2/5 | 1/5 | 4/5 | 5/5 | 5/5 | |
| 4 | 253 | 12 | 1/5 | 3/5 | 1/5 | 5/5 | 5/5 | |
| 5 | 331 | 13 | 4/5 | 5/5 | 1/5 | 5/5 | 5/5 | |
| 6 | 152 | 39 | 4/5 | 5/5 | 5/5 | 5/5 | 5/5 | |
| 7 | 136 | 40 | 4/5 | 5/5 | 5/5 | 5/5 | 5/5 | |
| MycCaP | 1 | 274 | 21 | 5/5 | 4/5 | 5/5 | 5/5 | 5/5 |
| 2 | 234 | 8 | 0/5 | 1/5 | 2/5 | 5/5 | 5/5 | |
| PC-3 | 1 | 179 | 27 | 3/5 | 1/5 | 5/5 | 5/5 | 5/5 |
| 2 | 182 | 22 | 5/5 | 0/5 | 3/5 | 5/5 | 5/5 | |
| RM-1 | 1 | 201 | 7 | 2/6 | 0/6 | 5/6 | 6/6 | 6/6 |
| 2 | 182 | 8 | 0/5 | 3/5 | 5/5 | 5/5 | 5/5 | |
| 3 | 300 | 6 | 2/5 | 2/5 | 5/5 | 5/5 | 5/5 | |
| 4 | 175 | 4 | 5/5 | 5/5 | 0/5 | 1/5 | 4/5 | |
| Total | 55/109 (50.46%) | 58/109 (53.21%) | 83/109 (76.15%) | 95/109 (87.16%) | 106/109 (97.25%) |
Tumors with very low contrast-to-noise ratio.
All cell lines were grown under sterile conditions in Dulbecco’s Modified Essential Medium, supplemented with 10% fetal bovine serum, 100 U/ml Penicillin and 100 µg/ml Streptomycin at 37°C in 5% CO2. Cancer cells were injected subcutaneously in the right flank of Nod/SCID mice (The Jackson Laboratory, Bar Harbor, ME).
Constrained Non-negative Matrix Factorization (cNMF) and Pattern Assignments
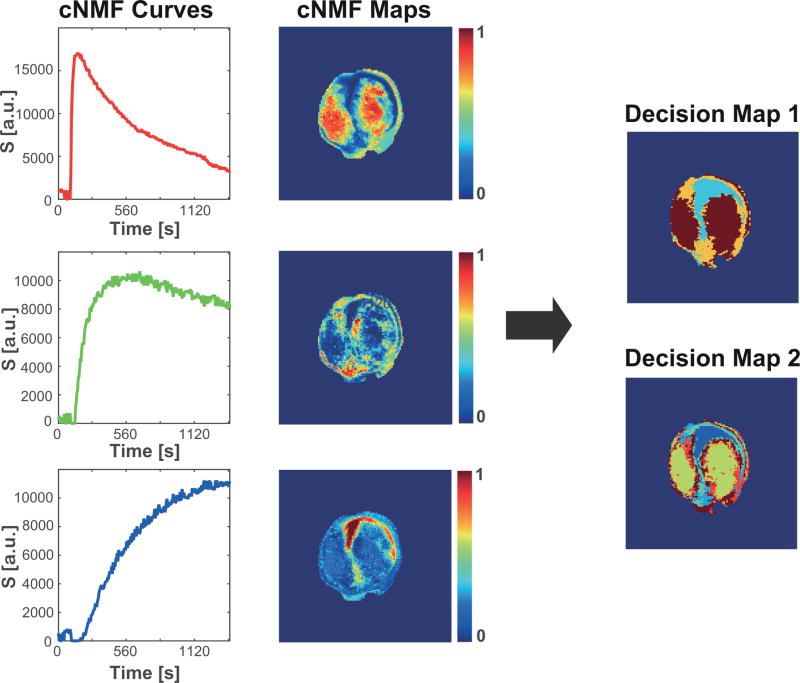
A representative example of cNMF curves and corresponding weight maps are shown in Figure 4 (left, center). Based on pattern shape and weight, single patterns or pattern mixtures were assigned automatically to each pixel and pattern masks created (Figure 4, right). Decision Map 1 (Figure 4) shows the spatial distribution of the dominant pattern in each pixel, while the Decision Map 2 (Figure 4) visualizes the spatial localization of single patterns and pattern mixtures, the latter inferring intra-tumoral heterogeneity at sub-pixel resolution. The in vivo DCE-MRI tumor data analyzed here do not have aligned ex vivo data. Thus, we validated the automated and optimized PR analysis by reanalyzing DCE-MRI data from experiments with aligned ex vivo data (Figure S2). While improving spatial mapping across tumor slices by the slice-wise analysis, we detect the same CA uptake behavior related to the tumor microenvironment as before (Figure S2).
Figure 4. Visualization of tumor environmental heterogeneity.
The red, green, and blue cNMF curves (left) from a representative tumor slice and their corresponding weight maps (center) show the spatial distribution and, as previously established (12,24) using similar experimental DCE-MRI acquisitions, represent the CA uptake behavior indicative of well-vascularized (Pattern 1), hypoxic (Pattern 2), and necrotic (Pattern 3) tumor areas, respectively. Decision Map 1 and 2 were determined as explained in the method section. In Decision Map 1, brown, orange, cyan reflect Pattern1 (indicative of well-vascularized tumor), Pattern 2 (indicative of hypoxia), and Pattern 3 (indicative of necrosis), respectively, while in Decision Map 2, Pattern 3 is displayed as dark blue, Pattern 1 as yellow, Pattern 2 as cyan, and corresponding mixtures as mixture colors. While Decision Map 1 characterizes the spatial distribution of the dominant pattern across the tumor slice, Decision Map 2 improves upon this by characterizing tumor heterogeneity at sub-pixel resolution and separating out pixels following a single pattern from pixels composed of pattern mixtures. The latter is especially of importance for quantifying pharmacokinetic parameters of a single (pure) pattern, as regional signal-versus-time curves of areas assigned to single patterns in Decision Map 1 may contain a noticeable contribution of pixels with pattern mixtures, affecting the signal-versus-time curve characteristic, and thus, the calculated pharmacokinetic parameter.
Discussion and Conclusions
As shown previously, an unsupervised PR approach, using PCA followed by cNMF, can visualize intra-tumoral microenvironmental heterogeneity based on tumor vascular features (24). Here, we successfully decreased user intervention and processing time by automating several analysis steps: (i) identification of the time period prior to CA arrival at tumor, resulting in an automated determination of the mean pre-contrast signal for signal-versus-time curve normalization; (ii) determination of NPs, previously obtained via visual inspection and required for cNMF analysis; and (iii) pattern assignments to visualize their spatial distribution across the tumor. Of the four developed and tested NP selection criteria, SNR showed the most promise with over 87% (threshold 5) or 97% (threshold 2) accuracy when compared to visual assessment. One limitation of this study is that the thresholds for the SNR criterion were determined empirically using solely preclinical tumor models, though over a wide range of tumor types. A second limitation is that the 25% threshold for the weight difference for assigning patterns to mixtures was also defined empirically.
The wider applicability of these settings to DCE-MRI data from other tumor sites (preclinical) and clinical tumors, including the impact of CNR, spatial resolution, temporal resolution and total acquisition time on the successful deconvolution of underlying patterns (Figure S3) and their interpretation and biological/physiological relevance will be the purpose of future research. Alone or in conjunction with other modalities assessing intra-tumor heterogeneity (18,19,22), the visualization of intra-tumoral vascular heterogeneity with fully-automated, combined PCA/cNMF analysis may provide in preclinical models (24,39), and after successful clinical translation (40), prognostic information, and useful information for monitoring, therapy planning, and follow up in longitudinal studies without the need for extensive tracer-kinetic modeling, while potentially improving and reducing computation time of tracer-kinetic modeling by using average signal-versus-time curves of assigned pattern areas.
Supplementary Material
Supporting Figure S1: Automatic determination of CA arrival time NRCA – ROI (Figure 2, 2nd step) versus pixel-based calculation. Example for the pixel-based CA arrival time, determined for each field-of-view pixel separately as outlined in the methods, mapped onto a representative tumor slice from tumor HEK #1. (Left) Example signal-versus-time-curve of a single pixel with high CNR; (Center) Example of the spatial localization and range of NRCA values across a tumor slice, and (Right) corresponding NRCA histogram, depicting the distribution of NRCA values.
While the distribution of NRCA reflects the arrival time of CA at different locations in the tumor, and thus, is reflective of plasma flow, the compared to an average tumor slice ROI curve much noisier signal-versus-time-curves may impact the robustness of this calculation significantly, as is evident from the background “NRCA” outside of the tumor area. Hence, the automatic determination of CA arrival time NRCA has been chosen based on the average tumor slice ROI curve, as shown in Figure 2 (2nd step).
To reduce the impact of signal disturbances at the beginning of the DCE-MRI acquisition and signal variations as a result of a significant number of pixels with a faster contrast agent arrival than the majority of pixels, potentially visible in a signal difference above noise just prior NRCA (Figure 1), the first 5 and last 5 frames from NR1 to NRCA are not included in the calculation of the average pre-contrast signal, resulting in a mean pre-contrast signal (±SD) of 780.5•104 (±4.1•104) for the tumor shown here.
Supporting Figure S2: Validation of the automation approaches of unsupervised PR DCE-MRI analysis. Previous experimental DCE-MRI data with existing aligned ex vivo data (12) were reanalyzed using the new code. (A) Proton MR reference images of the subcutaneous hind leg tumor (1230 mm3 by sliding-jaw caliper measurement). (B) The left panel shows the signal-versus-time curves for each of the three distinct patterns. On the right the corresponding weighted pattern maps are shown for each of the 5 DCE-MRI slices. (C) Top row: For the 4th slice from the left of this tumor, the three weighted pattern maps (cNMF maps 1–3) are shown enlarged with the merged pattern map shown to the right. Bottom row: To the in vivo slice shown in the top row corresponding ex vivo data (8 µm slice thickness) are shown. The hypoxic region shown in the 2nd panel was derived from the staining with the hypoxia marker pimonidazole (left panel). The autoradiography (3rd panel) of this tumor tissue slice depicts the accumulation of the hypoxia marker 18F-Fmisonidazole, which was administered prior to tumor excision for in vivo hypoxia imaging using positron emission tomography (data not shown). The 4th panel shows the corresponding H&E stain, visualizing viable and necrotic tissue. The holes in the tissue correspond to the fiduciary markers used to align the ex vivo data with the in vivo data.
The automation successfully detected and mapped the three distinct CA uptake behaviors seen previously (12,24), in this case by slice-based analysis. As before a delayed wash-in/wash-out behavior of the CA (green curve in B) was associated with tumor hypoxia (C). Pixels with low to no CA uptake with no CA washout (blue curve in B) tend to be associated with tumor necrosis. Of note is that the merged map demonstrates the appearance of pixels with mixtures of more than one pattern, indicating that a representative 8 µm-thick ex vivo slice may not capture fully the intra-pixel heterogeneity in a corresponding 0.79 mm-thick in vivo slice.
Supporting Figure S3: Effect of total acquisition time on pattern recognition in DCE-MRI analysis. (A, B) Automatic identification of NPs, as described in Methods and Figure 2, if only the first 5 min, 10 min, or 20 min of acquisition are used respectively. Representative results with the SEnh and SNRTh5 approaches for HEK tumor #1 are shown in (A). The table (B) lists the number of slices per total tumor slices in agreement with the manual NP determination for the six HEK tumors. The agreement between manual NP determination and NP determination by HAUC and SDTh respectively was affected by shortening the total acquisition time. However, the NPs determined manually and by SEnh, and SNRTh5 were unaffected by using only the first 5 min or 10 min of acquisition time, while agreement between manually and by SNRTh2 determined NPs increased using only the signal-versus-time-curves for the first 5 or 10 min acquisition time.
(C) Compared to a 20 min acquisition, visualization of intra-tumoral vascular heterogeneity by PR DCE-MRI analysis using the first 5 or 10 min acquisition time demonstrates that, at a respective temporal resolution of 5.487 s and 6.585 s, patterns can be deconvolved as well using the shorter acquisitions than using the longer acquisitions employed to develop the methods. As in Figure 4, the red, green, and blue cNMF curves (left) from a representative tumor slice (same as in Figure 4) and their corresponding weight maps (right) show the spatial distribution and CA uptake behavior of 3 distinct patterns, indicative of well-vascularized (Pattern 1), hypoxic (Pattern 2), and necrotic (Pattern 3) tumor areas, respectively, as previously established (12,24). Decision Maps 1 and 2 were determined as explained in the method section. In Decision Map 1, brown, orange, cyan reflect Pattern1 (indicative of well-vascularized tumor), Pattern 2 (indicative of hypoxia), and Pattern 3 (indicative of necrosis), respectively, while in Decision Map 2, Pattern 3 is displayed as dark blue, Pattern 1 as yellow, Pattern 2 as cyan, and corresponding mixtures as mixture colors.
Shorter acquisition times may affect the overall identifiable shape of the signal-versus-time curves, and thus, may affect their criteria for interpretation, e.g. if the acquisition time is so short as to measure no CA washout in well-vascularized areas. Further, tracer-kinetic modeling or semi-quantitative parameters may be affected as well by shortening of acquisition times, depending on model used. Association of patterns with clinically relevant tumor characteristics may also differ from their preclinical interpretation and requiring future research beyond the scope of this study.
Acknowledgments
We acknowledge support by NIH grants R01-CA163980, P50-CA092629 (Prostate SPORE (Specialized Programs of Research Excellence)), P30-CA008748 (Memorial Sloan Kettering Cancer Center Support Grant), and Future Research Funds 2013: Project No. 1.130030.01 and 2016: Project No. 1.160047.01 of Ulsan National Institute of Science and Technology (UNIST).
We gratefully acknowledge permission to use as test data DCE-MRI data acquired in collaboration between Dr. Ronald G. Blasberg’s (Dr. Ekaterina Moroz, Dr. Inna S. Serganova, technical support: Mr. Nisargbhai S. Shah) and Dr. Jason A. Koutcher’s (Dr. Ellen Ackerstaff, technical assistance: Ms. Natalia Kruchevsky) laboratory.
Footnotes
This research has been in part presented as Power Pitch #0191 in 2016 at the 24th Annual Meeting of the International Society for Magnetic Resonance in Medicine, Singapore.
References
- 1.Vaupel P, Kallinowski F, Okunieff P. Blood flow, oxygen and nutrient supply, and metabolic microenvironment of human tumors: a review. Cancer research. 1989;49(23):6449–6465. [PubMed] [Google Scholar]
- 2.Vaupel P. Tumor microenvironmental physiology and its implications for radiation oncology. Semin Radiat Oncol. 2004;14(3):198–206. doi: 10.1016/j.semradonc.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Carmeliet P, Jain RK. Molecular mechanisms and clinical applications of angiogenesis. Nature. 2011;473(7347):298–307. doi: 10.1038/nature10144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leach MO, Brindle KM, Evelhoch JL, Griffiths JR, Horsman MR, Jackson A, Jayson GC, Judson IR, Knopp MV, Maxwell RJ, McIntyre D, Padhani AR, Price P, Rathbone R, Rustin GJ, Tofts PS, Tozer GM, Vennart W, Waterton JC, Williams SR, Workman P. The assessment of antiangiogenic and antivascular therapies in early-stage clinical trials using magnetic resonance imaging: issues and recommendations. Br J Cancer. 2005;92(9):1599–1610. doi: 10.1038/sj.bjc.6602550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Egeland TA, Gaustad JV, Vestvik IK, Benjaminsen IC, Mathiesen B, Rofstad EK. Assessment of fraction of radiobiologically hypoxic cells in human melanoma xenografts by dynamic contrast-enhanced MRI. Magn Reson Med. 2006;55(4):874–882. doi: 10.1002/mrm.20852. [DOI] [PubMed] [Google Scholar]
- 6.Padhani AR, Husband JE. Dynamic contrast-enhanced MRI studies in oncology with an emphasis on quantification, validation and human studies. Clin Radiol. 2001;56(8):607–620. doi: 10.1053/crad.2001.0762. [DOI] [PubMed] [Google Scholar]
- 7.Salem A, O'Connor JP. Assessment of Tumor Angiogenesis: Dynamic Contrast-enhanced MR Imaging and Beyond. Magnetic resonance imaging clinics of North America. 2016;24(1):45–56. doi: 10.1016/j.mric.2015.08.010. [DOI] [PubMed] [Google Scholar]
- 8.Bhujwalla ZM, Artemov D, Glockner J. Tumor angiogenesis, vascularization, and contrast-enhanced magnetic resonance imaging. Topics in magnetic resonance imaging : TMRI. 1999;10(2):92–103. doi: 10.1097/00002142-199904000-00002. [DOI] [PubMed] [Google Scholar]
- 9.Sourbron SP, Buckley DL. Classic models for dynamic contrast-enhanced MRI. NMR in biomedicine. 2013;26(8):1004–1027. doi: 10.1002/nbm.2940. [DOI] [PubMed] [Google Scholar]
- 10.Hylton N. Dynamic contrast-enhanced magnetic resonance imaging as an imaging biomarker. J Clin Oncol. 2006;24(20):3293–3298. doi: 10.1200/JCO.2006.06.8080. [DOI] [PubMed] [Google Scholar]
- 11.Egeland TA, Gaustad JV, Galappathi K, Rofstad EK. Magnetic resonance imaging of tumor necrosis. Acta Oncol. 2011;50(3):427–434. doi: 10.3109/0284186X.2010.526633. [DOI] [PubMed] [Google Scholar]
- 12.Cho H, Ackerstaff E, Carlin S, Lupu ME, Wang Y, Rizwan A, O'Donoghue J, Ling CC, Humm JL, Zanzonico PB, Koutcher JA. Noninvasive multimodality imaging of the tumor microenvironment: registered dynamic magnetic resonance imaging and positron emission tomography studies of a preclinical tumor model of tumor hypoxia. Neoplasia (New York, NY. 2009;11(3):247–259. doi: 10.1593/neo.81360. 242p following 259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leach MO, Morgan B, Tofts PS, Buckley DL, Huang W, Horsfield MA, Chenevert TL, Collins DJ, Jackson A, Lomas D, Whitcher B, Clarke L, Plummer R, Judson I, Jones R, Alonzi R, Brunner T, Koh DM, Murphy P, Waterton JC, Parker G, Graves MJ, Scheenen TW, Redpath TW, Orton M, Karczmar G, Huisman H, Barentsz J, Padhani A Experimental Cancer Medicine Centres Imaging Network Steering C. Imaging vascular function for early stage clinical trials using dynamic contrast-enhanced magnetic resonance imaging. Eur Radiol. 2012;22(7):1451–1464. doi: 10.1007/s00330-012-2446-x. [DOI] [PubMed] [Google Scholar]
- 14.Ovrebo KM, Gulliksrud K, Mathiesen B, Rofstad EK. Assessment of tumor radioresponsiveness and metastatic potential by dynamic contrast-enhanced magnetic resonance imaging. Int J Radiat Oncol Biol Phys. 2011;81(1):255–261. doi: 10.1016/j.ijrobp.2011.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Ovrebo KM, Hompland T, Mathiesen B, Rofstad EK. Assessment of hypoxia and radiation response in intramuscular experimental tumors by dynamic contrast-enhanced magnetic resonance imaging. Radiother Oncol. 2012;102(3):429–435. doi: 10.1016/j.radonc.2011.11.013. [DOI] [PubMed] [Google Scholar]
- 16.Fusco R, Sansone M, Filice S, Carone G, Amato DM, Sansone C, Petrillo A. Pattern Recognition Approaches for Breast Cancer DCE-MRI Classification: A Systematic Review. Journal of medical and biological engineering. 2016;36(4):449–459. doi: 10.1007/s40846-016-0163-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim SG, Freed M, Leite APK, Zhang J, Seuss C, Moy L. Separation of benign and malignant breast lesions using dynamic contrast enhanced MRI in a biopsy cohort. J Magn Reson Imaging. 2017;45(5):1385–1393. doi: 10.1002/jmri.25501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stoyanova R, Pollack A, Takhar M, Lynne C, Parra N, Lam LL, Alshalalfa M, Buerki C, Castillo R, Jorda M, Ashab HA, Kryvenko ON, Punnen S, Parekh DJ, Abramowitz MC, Gillies RJ, Davicioni E, Erho N, Ishkanian A. Association of multiparametric MRI quantitative imaging features with prostate cancer gene expression in MRI-targeted prostate biopsies. Oncotarget. 2016;7(33):53362–53376. doi: 10.18632/oncotarget.10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Stoyanova R, Takhar M, Tschudi Y, Ford JC, Solorzano G, Erho N, Balagurunathan Y, Punnen S, Davicioni E, Gillies RJ, Pollack A. Prostate cancer radiomics and the promise of radiogenomics. Transl Cancer Res. 2016;5(4):432–447. doi: 10.21037/tcr.2016.06.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grimm LJ, Zhang J, Mazurowski MA. Computational approach to radiogenomics of breast cancer: Luminal A and luminal B molecular subtypes are associated with imaging features on routine breast MRI extracted using computer vision algorithms. J Magn Reson Imaging. 2015;42(4):902–907. doi: 10.1002/jmri.24879. [DOI] [PubMed] [Google Scholar]
- 21.Akbari H, Macyszyn L, Da X, Wolf RL, Bilello M, Verma R, O'Rourke DM, Davatzikos C. Pattern analysis of dynamic susceptibility contrast-enhanced MR imaging demonstrates peritumoral tissue heterogeneity. Radiology. 2014;273(2):502–510. doi: 10.1148/radiol.14132458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.O'Connor JP, Rose CJ, Waterton JC, Carano RA, Parker GJ, Jackson A. Imaging intratumor heterogeneity: role in therapy response, resistance, and clinical outcome. Clin Cancer Res. 2015;21(2):249–257. doi: 10.1158/1078-0432.CCR-14-0990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Eyal E, Degani H. Model-based and model-free parametric analysis of breast dynamic-contrast-enhanced MRI. NMR in biomedicine. 2009;22(1):40–53. doi: 10.1002/nbm.1221. [DOI] [PubMed] [Google Scholar]
- 24.Stoyanova R, Huang K, Sandler K, Cho H, Carlin S, Zanzonico PB, Koutcher JA, Ackerstaff E. Mapping Tumor Hypoxia In Vivo Using Pattern Recognition of Dynamic Contrast-enhanced MRI Data. Translational oncology. 2012;5(6):437–447. doi: 10.1593/tlo.12319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Han SH, Ackerstaff E, Stoyanova R, Carlin S, Huang W, Koutcher JA, Kim JK, Cho G, Jang G, Cho H. Gaussian mixture model-based classification of dynamic contrast enhanced MRI data for identifying diverse tumor microenvironments: preliminary results. NMR in biomedicine. 2013;26(5):519–532. doi: 10.1002/nbm.2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Craft N, Shostak Y, Carey M, Sawyers CL. A mechanism for hormone-independent prostate cancer through modulation of androgen receptor signaling by the HER-2/neu tyrosine kinase. Nature medicine. 1999;5(3):280–285. doi: 10.1038/6495. [DOI] [PubMed] [Google Scholar]
- 27.Watson PA, Ellwood-Yen K, King JC, Wongvipat J, Lebeau MM, Sawyers CL. Context-dependent hormone-refractory progression revealed through characterization of a novel murine prostate cancer cell line. Cancer research. 2005;65(24):11565–11571. doi: 10.1158/0008-5472.CAN-05-3441. [DOI] [PubMed] [Google Scholar]
- 28.Webber MM, Bello D, Quader S. Immortalized and tumorigenic adult human prostatic epithelial cell lines: characteristics and applications. Part I. Cell markers and immortalized nontumorigenic cell lines. The Prostate. 1996;29(6):386–394. doi: 10.1002/(SICI)1097-0045(199612)29:6<386::AID-PROS7>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 29.Baley PA, Yoshida K, Qian W, Sehgal I, Thompson TC. Progression to androgen insensitivity in a novel in vitro mouse model for prostate cancer. J Steroid Biochem Mol Biol. 1995;52(5):403–413. doi: 10.1016/0960-0760(95)00001-g. [DOI] [PubMed] [Google Scholar]
- 30.Haase A. Principles and applications of FLASH NMR imaging. Magma (New York, NY. 1994;2(3):157–160. [Google Scholar]
- 31.Stoyanova R, Brown TR. NMR spectral quantitation by principal component analysis. NMR in biomedicine. 2001;14(4):271–277. doi: 10.1002/nbm.700. [DOI] [PubMed] [Google Scholar]
- 32.Stoyanova R, Brown TR. NMR spectral quantitation by principal component analysis. III. A generalized procedure for determination of lineshape variations. J Magn Reson. 2002;154(2):163–175. doi: 10.1006/jmre.2001.2486. [DOI] [PubMed] [Google Scholar]
- 33.Burgess AE. The Rose model, revisited. Journal of the Optical Society of America A, Optics, image science, and vision. 1999;16(3):633–646. doi: 10.1364/josaa.16.000633. [DOI] [PubMed] [Google Scholar]
- 34.Haacke EM, Brown RW, Thompson MR, Venkatesan R. Magnetic Resonance Imaging: Physical Principles and Sequence Design. New York: Wiley; 1999. Object Visibility and the Rose Criterion; pp. 350–353. [Google Scholar]
- 35.Bushberg JT. The essential physics of medical imaging. Philadelphia: Lippincott Williams & Wilkins; 2002. p. xvi.p. 933. [Google Scholar]
- 36.Sajda P, Du S, Brown TR, Stoyanova R, Shungu DC, Mao X, Parra LC. Nonnegative matrix factorization for rapid recovery of constituent spectra in magnetic resonance chemical shift imaging of the brain. IEEE Trans Med Imaging. 2004;23(12):1453–1465. doi: 10.1109/TMI.2004.834626. [DOI] [PubMed] [Google Scholar]
- 37.Du S, Sajda P, Brown T, Stoyanova R. Recovery of Metabolomic Spectral Sources using Non-negative Matrix Factorization. Conf Proc IEEE Eng Med Biol Soc. 2005;2:1095–1098. doi: 10.1109/IEMBS.2005.1616610. [DOI] [PubMed] [Google Scholar]
- 38.Du S, Mao X, Sajda P, Shungu DC. Automated tissue segmentation and blind recovery of (1)H MRS imaging spectral patterns of normal and diseased human brain. NMR in biomedicine. 2008;21(1):33–41. doi: 10.1002/nbm.1151. [DOI] [PubMed] [Google Scholar]
- 39.Ackerstaff E, Kruchevsky N, Stoyanova R, Carlin S, Viola-Villegas NT, Sevak KK, Lewis JS, A KJ. Tumor pH and Vascularity in Human Prostate Cancer Models. Salt Lake City, Utah, USA: [2013 April 20–26]. 2013. [Google Scholar]
- 40.Stoyanova R, Ackerstaff E, Cho H, Koutcher JA, Pollack A. DCE-MRI for Delineation of Hypoxic Regions in Prostate Tumors. International Journal of Radiation Oncology Biology Physics. 2010;78(3):S337–S337. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure S1: Automatic determination of CA arrival time NRCA – ROI (Figure 2, 2nd step) versus pixel-based calculation. Example for the pixel-based CA arrival time, determined for each field-of-view pixel separately as outlined in the methods, mapped onto a representative tumor slice from tumor HEK #1. (Left) Example signal-versus-time-curve of a single pixel with high CNR; (Center) Example of the spatial localization and range of NRCA values across a tumor slice, and (Right) corresponding NRCA histogram, depicting the distribution of NRCA values.
While the distribution of NRCA reflects the arrival time of CA at different locations in the tumor, and thus, is reflective of plasma flow, the compared to an average tumor slice ROI curve much noisier signal-versus-time-curves may impact the robustness of this calculation significantly, as is evident from the background “NRCA” outside of the tumor area. Hence, the automatic determination of CA arrival time NRCA has been chosen based on the average tumor slice ROI curve, as shown in Figure 2 (2nd step).
To reduce the impact of signal disturbances at the beginning of the DCE-MRI acquisition and signal variations as a result of a significant number of pixels with a faster contrast agent arrival than the majority of pixels, potentially visible in a signal difference above noise just prior NRCA (Figure 1), the first 5 and last 5 frames from NR1 to NRCA are not included in the calculation of the average pre-contrast signal, resulting in a mean pre-contrast signal (±SD) of 780.5•104 (±4.1•104) for the tumor shown here.
Supporting Figure S2: Validation of the automation approaches of unsupervised PR DCE-MRI analysis. Previous experimental DCE-MRI data with existing aligned ex vivo data (12) were reanalyzed using the new code. (A) Proton MR reference images of the subcutaneous hind leg tumor (1230 mm3 by sliding-jaw caliper measurement). (B) The left panel shows the signal-versus-time curves for each of the three distinct patterns. On the right the corresponding weighted pattern maps are shown for each of the 5 DCE-MRI slices. (C) Top row: For the 4th slice from the left of this tumor, the three weighted pattern maps (cNMF maps 1–3) are shown enlarged with the merged pattern map shown to the right. Bottom row: To the in vivo slice shown in the top row corresponding ex vivo data (8 µm slice thickness) are shown. The hypoxic region shown in the 2nd panel was derived from the staining with the hypoxia marker pimonidazole (left panel). The autoradiography (3rd panel) of this tumor tissue slice depicts the accumulation of the hypoxia marker 18F-Fmisonidazole, which was administered prior to tumor excision for in vivo hypoxia imaging using positron emission tomography (data not shown). The 4th panel shows the corresponding H&E stain, visualizing viable and necrotic tissue. The holes in the tissue correspond to the fiduciary markers used to align the ex vivo data with the in vivo data.
The automation successfully detected and mapped the three distinct CA uptake behaviors seen previously (12,24), in this case by slice-based analysis. As before a delayed wash-in/wash-out behavior of the CA (green curve in B) was associated with tumor hypoxia (C). Pixels with low to no CA uptake with no CA washout (blue curve in B) tend to be associated with tumor necrosis. Of note is that the merged map demonstrates the appearance of pixels with mixtures of more than one pattern, indicating that a representative 8 µm-thick ex vivo slice may not capture fully the intra-pixel heterogeneity in a corresponding 0.79 mm-thick in vivo slice.
Supporting Figure S3: Effect of total acquisition time on pattern recognition in DCE-MRI analysis. (A, B) Automatic identification of NPs, as described in Methods and Figure 2, if only the first 5 min, 10 min, or 20 min of acquisition are used respectively. Representative results with the SEnh and SNRTh5 approaches for HEK tumor #1 are shown in (A). The table (B) lists the number of slices per total tumor slices in agreement with the manual NP determination for the six HEK tumors. The agreement between manual NP determination and NP determination by HAUC and SDTh respectively was affected by shortening the total acquisition time. However, the NPs determined manually and by SEnh, and SNRTh5 were unaffected by using only the first 5 min or 10 min of acquisition time, while agreement between manually and by SNRTh2 determined NPs increased using only the signal-versus-time-curves for the first 5 or 10 min acquisition time.
(C) Compared to a 20 min acquisition, visualization of intra-tumoral vascular heterogeneity by PR DCE-MRI analysis using the first 5 or 10 min acquisition time demonstrates that, at a respective temporal resolution of 5.487 s and 6.585 s, patterns can be deconvolved as well using the shorter acquisitions than using the longer acquisitions employed to develop the methods. As in Figure 4, the red, green, and blue cNMF curves (left) from a representative tumor slice (same as in Figure 4) and their corresponding weight maps (right) show the spatial distribution and CA uptake behavior of 3 distinct patterns, indicative of well-vascularized (Pattern 1), hypoxic (Pattern 2), and necrotic (Pattern 3) tumor areas, respectively, as previously established (12,24). Decision Maps 1 and 2 were determined as explained in the method section. In Decision Map 1, brown, orange, cyan reflect Pattern1 (indicative of well-vascularized tumor), Pattern 2 (indicative of hypoxia), and Pattern 3 (indicative of necrosis), respectively, while in Decision Map 2, Pattern 3 is displayed as dark blue, Pattern 1 as yellow, Pattern 2 as cyan, and corresponding mixtures as mixture colors.
Shorter acquisition times may affect the overall identifiable shape of the signal-versus-time curves, and thus, may affect their criteria for interpretation, e.g. if the acquisition time is so short as to measure no CA washout in well-vascularized areas. Further, tracer-kinetic modeling or semi-quantitative parameters may be affected as well by shortening of acquisition times, depending on model used. Association of patterns with clinically relevant tumor characteristics may also differ from their preclinical interpretation and requiring future research beyond the scope of this study.