Abstract
Background
Recently-developed indices of cellular age based on DNA methylation (DNAm) data, referred to as DNAm age, are being used to study factors that influence the rate of aging and the health correlates of these metrics of the epigenetic clock. This study evaluated associations between trauma exposure, posttraumatic stress disorder (PTSD) symptoms, and accelerated versus decelerated DNAm age among military veterans. We also examined if accelerated DNAm age predicted mortality over the course of a 6.5-year medical record review period.
Methods
339 genotype-confirmed white, non-Hispanic, middle-aged, trauma-exposed veterans underwent psychiatric assessment and genome-wide DNAm analysis. DNAm age was calculated using a previously validated algorithm. Medical records were available for a subset of 241 veterans and were reviewed approximately 6.5 years after DNA collection and PTSD assessment.
Results
PTSD hyperarousal symptoms were associated with accelerated DNAm age (β = .20, p = .009) but trauma exposure and total PTSD severity were not. Accelerated DNAm age was also associated with 13% increased risk for all-cause mortality (HR: 1.13, 95% CI: 1.01 – 1.26) during the medical record review period.
Conclusions
Findings of this study replicate the association between PTSD and accelerated DNAm age and suggest this effect may be specific to the hyperarousal symptom cluster. Results point to the potential utility of DNAm age algorithms for identifying individuals who are aging at an accelerated rate and for determining the factors that influence this process.
Keywords: DNA methylation, accelerated aging, PTSD, mortality, hyperarousal
Recent discoveries in the genetics of aging have led to a paradigm shift in approaches to quantifying cellular age. Specifically, newly developed DNA methylation (DNAm) age algorithms based on methylation loci from across the genome yield strong associations with chronological age (rs = .96; 1,2) that are unparalleled in comparison to other metrics of cellular age, such as telomeres (r with age ~ .30; 3). Two such algorithms, developed independently by Hannum et al. (1) and Horvath (2), for use with data from the Illumina 450k microarray have garnered the most attention to date. The algorithms yield DNAm age estimates that are highly correlated with each other (4) and are based on methylation loci in genes involved in DNA replication and repair, lipid metabolism, and other age-related processes (1, 2). These algorithms are now being used to identify factors that accelerate or decelerate cellular aging and the health correlates of these changes. This study evaluated associations between traumatic stress (trauma exposure and posttraumatic stress disorder [PTSD]) and accelerated DNAm age and examined the utility of DNAm age estimates for predicting mortality over a 6.5 year period in a sample of military veterans.
Emerging evidence suggests that traumatic stress may accelerate cellular aging (4–8). Recent research by our group (4) conducted in an independent sample of young military veterans (M age = 32 years) found that lifetime PTSD severity was associated with accelerated DNAm age using the Hannum et al. algorithm. Two other published studies have also examined associations between PTSD and DNAm age. The first reported, unexpectedly, that PTSD was associated with lower Horvath DNAm age (9), but did not model the relationship between DNAm age and chronological age. The second study found no association between PTSD and Horvath DNAm age estimates relative to chronological age, but did find an association between daily life stressors and accelerated DNAm age (10). Other studies are needed to evaluate the potential value of these algorithms as tools for studying the neurobiology of PTSD and related conditions.
In other fields, several studies have shown accelerated DNAm age to be associated with various age-related health and neurocognitive problems and this helps to demonstrate the applied utility and validity of this index of cellular aging. For example, Marioni et al. (11) found that advanced Horvath DNAm age (relative to chronological age) evidenced cross-sectional associations with reduced lung function and poorer performance on cognitive and motor tasks in epidemiological samples (11). Similarly, accelerated Horvath DNAm age has been associated with a multi-indicator measure of frailty (12). Our previous study showed that advanced Hannum DNAm age was associated with decreased microstructural integrity of the genu of the corpus callosum, a region important for communication across the prefrontal cortices (4). Furthermore, through this neuronal deficit, accelerated DNAm age was also associated with poorer performance on tests of executive function and working memory (4). Finally, consistent with the aforementioned associations with worse health and neurocognitive indices, evidence from multiple epidemiological birth cohorts of individuals ranging in age from 70 – 90 years, suggest that accelerated Horvath and Hannum DNAm age estimates are associated with mortality (13). Specifically, Marioni et al. (13) reported that every 5 year increase in DNAm age beyond chronological age was associated with 11% and 21% (respectively) increased odds for all-cause mortality during the study follow-up per the Horvath and Hannum et al. algorithms, respectively. Finally, a meta-analysis of time-to-death across 13 epidemiological cohorts with median ages in the late 50s to late 70s concluded that age acceleration, as defined by both the Horvath and Hannum algorithms, was associated with earlier death, even after controlling for a variety of relevant medical diagnoses (14).
Based on the foregoing, the primary aim of this study was to determine if the association between PTSD symptom severity and accelerated Hannum et al. DNAm age that we reported previously (4) would replicate in an independent dataset of veterans who were, on average, approximately 20 years older than those in our prior study. We also sought to extend this work by examining differential associations between DNAm age and the four DSM-IV PTSD symptom clusters: intrusive re-experiencing of the trauma; avoidance of trauma-related cues; emotional numbing; and hyperarousal symptoms (15). We hypothesized that accelerated DNAm age would be most strongly associated with the PTSD hyperarousal symptoms due to the physiological nature of these symptoms which include sleep disturbance, hypervigilance, and tonic hyperarousal (5, 6). We also examined whether accelerated Hannum et al. DNAm age predicted mortality over the course of an approximately 6.5 year medical record review period. Prior studies of the association between DNAm age and mortality have been conducted exclusively in epidemiological samples and it is unknown if this biomarker might be associated with mortality in a clinical sample with substantial health comorbidities.
Methods
Participants
Participants were drawn from two research studies with identical assessment procedures allowing data to be combined. The pooled sample included 852 participants who were veterans and a subset of their cohabitating partners (16). Participants were excluded from these analyses for the following reasons: 42 failed to fully complete the protocol; 25 were enrolled in both studies and data from only one study were included; 24 did not have successful blood draws; and 32 did not endorse trauma exposure on the Clinician Administered PTSD Scale (CAPS; 17). From this group of 729 participants, 491 had genotype data and were of white, non-Hispanic ancestry, as determined through principal components (PC) analysis of the DNA data (see below). DNAm data were available for 466 participants, 339 of whom were veterans. We limited analyses to this group of 339 trauma-exposed white, non-Hispanic veterans for a number of reasons. Specifically, we focused on trauma-exposed veterans given that an assessment of PTSD is only relevant secondary to trauma exposure and because we aimed to replicate our prior work conducted in a trauma-exposed veteran sample (4). We only obtained DNAm data from participants who were of white, non-Hispanic ancestry, given that this was the largest ancestrally homogenous subgroup in the sample and ancestry can influence DNAm data (18). Through there are valid scientific reasons for limiting analyses to this sub-sample, one consequence of this is that the sample did not include sufficient representation of the demographic groups known to have the greatest vulnerability to PTSD (racial and ethnic minorities and women;19).
The sample was comprised of 295 (87.0%) men and 44 (13.0%) women. The mean age at the time of the blood draw and PTSD assessment was 52.58 years (SD: 10.65, range: 23 – 72). The mean number of traumatic experiences in the sample was 21.71 (SD = 15.56). The two most common forms of trauma exposure were sudden death of a friend/loved one (59.6%), and combat exposure (48.7%). The majority (69.0%) met DSM-IV criteria for a lifetime diagnosis of PTSD, and 47.8% met criteria for current PTSD at the time of the blood draw, as determined by the CAPS (17). Participant demographics are listed in Table 1.
Table 1.
Participant Characteristics (N = 339)
| Variable | M (SD) | n (%) |
|---|---|---|
| Demographics | ||
| Sex (male) | 295 (87.0) | |
| Age | 52.58 (10.65) | |
| Psychiatric | ||
| Lifetime PTSD Dx | 234 (69.0) | |
| Lifetime PTSD Severity | 66.09 (30.62) | |
| # of Traumatic Experiences | 21.71 (15.56) | |
| Biological | ||
| CD8-T (%) | .04 (.04) | |
| CD4-T (%) | .15 (.05) | |
| NK (%) | .06 (.05) | |
| B cells (%) | .04 (.05) | |
| Monocytes (%) | .09 (.03) | |
| Body Mass Indexa | 31.48 (6.28) | |
| Mortality Relatedb | ||
| Deceased | 17 (7.1) | |
| # years to death or censor | 6.48 (1.47) | |
Note. White blood cell counts represent proportions of total white blood cell count estimates, derived from the DNA methylation data. M = mean; SD = standard deviation; PTSD = posttraumatic stress disorder; dx = diagnosis; NK = natural killer cells.
Sample size for BMI = 192. BMI = (weight [lbs])/height [in])2 * 703)
Sample size for analyses related to mortality = 241
Measures
Trauma exposure was assessed with the Traumatic Life Events Questionnaire (TLEQ; 20). This self-report measure assesses lifetime exposure to 22 different potentially traumatic experiences (DSM-IV PTSD Criterion A1) as well as peri-traumatic emotions of fear, helplessness or horror (DSM-IV PTSD Criterion A2); the number of times each event was experienced was rated on a 7-point scale (“never” to “more than 5 times”). As all subjects were trauma exposed per CAPS interview, analysis of associations between trauma and DNAm age focused on cumulative lifetime trauma exposure as indicated by the total number of traumatic experiences both within and across trauma types (and requiring PTSD criterion A2). Lifetime PTSD was indexed by the CAPS for DSM-IV (17), the gold-standard structured diagnostic interview that assesses frequency and intensity of the 17 DSM-IV PTSD criteria, each rated on a 0–4 scale. Frequency and intensity scales were summed within and across items to indicate total lifetime PTSD severity. Diagnostic determinations were made following the DSM-IV algorithm and the frequency ≥ 1 and intensity ≥ 2 rule to determine symptom presence (21). The CAPS was administered by MA- and PhD-level psychology trainees and professionals and was videotaped for reliability purposes. Approximately 30% of these tapes were viewed by an independent rater which resulted in an intraclass correlation coefficient for lifetime PTSD severity of r = .97. Mean lifetime PTSD severity was 66.09 (SD: 30.62). The Structured Clinical Interview for DSM-IV (SCID; 22) was also administered and for the purposes of this study, symptom summary scores on the major depressive episode and alcohol use disorder modules were examined as potential confounds of the PTSD effects.
Procedure
Details on the recruitment of subjects are available in Logue et al. (16) and included recruitment from a Veterans Affairs (VA)-generated registry, recruitment database, clinician referrals, and flyers. Participants completed a variety of self-report and interview measures and had their blood drawn for genetic analyses. Of these, a subset of the veterans (n = 241) had data available in the local VA electronic medical record. We examined if these veterans were still alive via medical record follow-up. The interval between the initial study assessment and medical record follow-up was 6.5 years on average. We focused on all-cause mortality as information was frequently not available concerning cause of death and to maintain consistency with prior work (13). Baseline data were collected from 2006 to 2011. All participants provided written informed consent and these procedures were reviewed and approved by the VA Boston Healthcare System Institutional Review Board.
Genotyping and Methylation
Peripheral blood samples were drawn and DNA was extracted from buffy coat. Genotypes, used here for ancestry analysis, were obtained using the Illumina HumanOmni 2.5–8 microarray and scanned with an Illumina iScan System (Illumina, San Diego, CA; see Supplementary Materials). DNAm data were obtained using the Illumina (Illumina, San Diego, CA) Human Methylation 450K microarray.
The integrity and quantity of the DNA samples were determined by TaqMan® RNase P Detection assay (Applied Biosystems Assay, Life Technologies, Carlsbad, CA) with fluorescence detection on a 7900 Fast Real Time PCR System (Applied Biosystems, Life Technologies, Carlsbad, CA) per the manufacturer’s protocol. For methylation, samples were bisulfite-modified using Zymo EZ-96 DNA Methylation Kits (D5004). The bisulfite-mediated conversion efficiency was determined by PCR with DAPK1 primers (Zymo) and gel electrophoresis of PCR-products. The bisulfite-modified DNA samples were whole-genome amplified, fragmented, precipitated, resuspended, and hybridized to Illumina Human Methylation 450 beadchips. They then underwent single-base extension and staining. The beadchips were scanned using the Illumina iScan System. Internal quality control and performance was determined using GenomeStudio v2011.1 software using Methylation module v1.9.0. Individual-level background-corrected methylation-probe data for 489 white non-Hispanic samples were output from GenomeStudio. Of these samples, 20 duplicates and three previously identified sample swaps were excluded, leaving 466 samples. Of these, 339 were obtained from veterans and included in these analyses. Cleaning was performed within the CpGassoc package Bioconductor package in R (23). All samples passed intensity thresholds (intensity > 50% of the experiment-wide and > 20,000 arbitrary units). Singleton probe values with detection-p greater than 0.001 were set to missing. Probes and subjects with > 10% missing values were excluded. Probe normalization was performed using beta mixture quantile dilation (BMIQ) method (24) as implemented in the watermelon package (25, 26). An empirical Bayes batch-correction method (ComBat; 27) as implemented in the Bioconductor sva package (28) was used to remove chip and position effects. Missing data were imputed using the Bioconductor impute package (http://www.bioconductor.org/packages/release/bioc/html/impute.html) using a k nearest neighbor method (KNN; 29), which “fills in” missing values based on methylation patterns for similarly behaving genes (30). As observed differential white-cell counts were not available for our samples, cell counts were estimated directly from the methylation data using the R minfi package (31).
Hannum DNAm age estimates were calculated from beta values (proportion of methylated DNA) after filtering quality control procedures. Of the 89 probes specified for use in the Hannum et al. (1) “all data” algorithm, two loci (cg25428494 and ch.13.39564907R) did not make it through cleaning and were not included in the analysis. Of the 87 remaining probes, 12 probes included had missing data for one or more subjects and imputed data was used to fill in these values for the DNAm age calculation. The maximum amount of missing data for one of these loci was 2.95%, as shown in Table S1. Hannum methylation age estimates were then computed via linear multiplication of the beta values from the 87 available probes and the supplied coefficient values from the Hannum et al. model using the R statistical analysis package.
Ancestry was determined using a Bayesian clustering analysis of 100,000 randomly chosen single nucleotide polymorphisms (SNPs) using the program STRUCTURE (32, 33) in comparison to self-reported race and ethnicity. PCs, which model the potentially confounding effects of population substructure (e.g., in this case, ancestral variation within the white, non-Hispanic sample), were computed using the program EIGENSTRAT (34). White blood cell counts (cd4 and cd8 t-cells, natural killer cells, b cells, and monocytes—indices of inflammation) were estimated from the DNAm data (31).
Data Analyses
We first evaluated the association between chronological age and Hannum et al. DNAm age. We did not evaluate Horvath DNAm age as we did not previously find an association between it and PTSD (4) and because of evidence of stronger effects of Hannum et al. DNAm age on mortality (13). We computed DNAm age residuals from a regression in which DNAm age estimates were regressed on chronological age and the unstandardized residuals from the equation were saved as a new variable, following prior work (4, 11). Over-predicted DNAm age estimates are captured by positive DNAm age residuals and reflect advanced estimated age compared to chronological age (i.e., accelerated age) while negative DNAm age residuals reflect under-predicted age compared to chronological age (i.e., decelerated cellular age).
We then conducted a series of multiple regressions focused on the associations between lifetime trauma exposure and lifetime PTSD severity in the prediction of DNAm age residuals. We controlled for ancestry (the top two PCs), sex, and white blood cell count proportional estimates, as these are potential confounds in DNAm data (18, 31). Total trauma exposure and total lifetime PTSD severity were the primary predictors in the first set of regressions. Total trauma exposure and the four lifetime DSM-IV PTSD symptom cluster severity scores (i.e., reexperiencing, avoidance, emotional numbing, and hyperarousal; 35) were examined as predictors of DNAm age residuals in the second round of regressions; trauma exposure was retained in this analysis to allow for equitable comparison across the two sets of regressions. Analyses focused on associations with lifetime, as opposed to current, PTSD severity because this replicated our prior work and because we hypothesized that the cumulative burden of PTSD symptoms across the lifespan would most strongly relate to cellular aging rather than symptom severity at a single random time point. Post-hoc t-tests evaluated differences in DNAm age residuals as a function of sex. Follow-up regression analyses evaluated financial stress (which has previously shown association with cellular age, 36), and major depression and alcohol use disorders (the latter of which was also previously associated with cellular age, 37) as possible confounds of PTSD effects.
We next conducted a Cox regression to examine the effects of PTSD severity and DNAm age residuals on all-cause mortality among veterans. We controlled for age (at baseline), sex, ancestry (top 2 PCs), and white blood cell count estimates. For these analyses, white blood cell count estimates and the PCs were transformed by a constant (multiplied by 100) to reduce convergence issues associated with variables with highly different scales. Veterans were censored at the date of death or on the date we accessed their medical record and determined they were living. The number of days between the PTSD assessment and the date of death or censor was included in the equation. Significant effects were further evaluated using post-hoc independent t-tests to characterize baseline DNAm age residual differences as a function of death during the study follow-up period. Finally, an additional analysis was conducted to begin to address the possibility that health conditions associated with elevated mortality risk (e.g., obesity) might confound the association between cellular aging and mortality. Specifically, we included body mass index (BMI) as a covariate in a follow-up Cox regression in a subset of 191 veterans with available BMI data. This trait was evaluated because it was the most common health indicator available in the medical record with data available within 6 months of the study medical record follow-up. As well, this variable was important to examine because obesity has previously been associated with accelerated DNAm age (38).
Results
Associations between DNAm Age and Chronological Age
The correlation between the DNAm age estimates and chronological age was r = .90, p < .001. After regressing chronological age out from DNAm age, the correlation between chronological age and the DNAm age residuals was r = −.039, p = .48. The average age residual (i.e., difference between predicted DNAm age and chronological age) was 0.58 years (SD: 4.73, range: −13.35 to 14.01).
Associations between Trauma, PTSD, and DNAm Age
The regression examining the main effects of lifetime PTSD severity and trauma exposure in association with DNAm age residuals revealed main effects of sex and CD4 t-cells, but no effects of trauma exposure nor total lifetime PTSD severity (Table 2). A post-hoc t-test revealed that on average, DNAm age residuals were 2.11 years (SD: 4.62) below chronological age among women and 0.98 years (SD: 4.62) greater than chronological age among men, t (337) = 4.14, p < .001. The subsequent regression evaluating the PTSD symptom clusters yielded an additional association between the hyperarousal symptoms and accelerated aging (standardized β = .20, p = .009; see Table 3 and Figure S1).
Table 2.
Trauma Exposure and Lifetime Total PTSD Severity as Predictors of Accelerated DNAm Age
| Variable | β | p | R2 | p |
|---|---|---|---|---|
| PC1 | .00 | .933 | ||
| PC2 | −.01 | .845 | ||
| CD8T | −.09 | .117 | ||
| CD4T | −.25 | .000 | ||
| NK | .02 | .722 | ||
| B cell | −.01 | .874 | ||
| Mono | −.04 | .508 | ||
| Sex | .21 | .000 | ||
| Total trauma count | .08 | .163 | ||
| Lifetime PTSD sev | .03 | .623 | ||
| Overall model | .12 | .000 |
Note. Results are from multiple regression equation. Significant associations are bolded. PC = principal component; CD8T = cd8 t-cells; CD4T = CD4 t-cells; NK = natural killer cells; mono = monocytes; sev = severity.
Table 3.
Trauma Exposure and Lifetime PTSD Symptom Cluster Severity as Predictors of Accelerated DNAm Age
| Variable | β | p | R2 | p |
|---|---|---|---|---|
| PC1 | .00 | .947 | ||
| PC2 | −.01 | .930 | ||
| CD8T | −.08 | .123 | ||
| CD4T | −.24 | .000 | ||
| NK | .02 | .702 | ||
| B cell | −.02 | .732 | ||
| Mono | −.05 | .404 | ||
| Sex | .21 | .000 | ||
| Total trauma count | .09 | .142 | ||
| Reexp sev | −.07 | .353 | ||
| Avoidance sev | −.03 | .649 | ||
| Emo. Numb sev | −.08 | .262 | ||
| Hyperarousal sev | .20 | .009 | ||
| Overall model | .14 | .000 |
Note. Results are from multiple regression equation. Significant associations are bolded. DNAm age residuals. PC = principal component; CD8T = cd8 t-cells; CD4T = CD4 t-cells; NK = natural killer cells; mono = monocytes; reexp = reexperiencing; sev = severity; emo numb = emotional numbing.
We evaluated a number of potential confounds of this association, including income, lifetime alcohol abuse and dependence symptoms, and lifetime major depression symptoms. None of these variables were significantly associated with DNAm age residuals and in each regression, PTSD hyperarousal symptoms remained significantly and positively associated with DNAm age residuals after inclusion of the potential confound (details available from first author).
Accelerated DNAm Age and Mortality
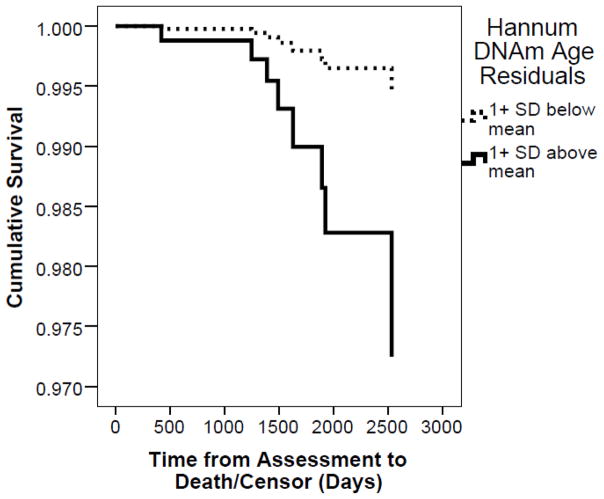
Among the cohort with available medical record data (n = 241), there were 17 deaths (7%). Death occurred an average of 3.42 years following the assessment (SD: 1.65 years, range: 0.28 to 6.9 years). In the Cox regression, the only significant predictor of mortality was DNAm age residuals (HR = 1.13, 95% CI: 1.01 to 1.26, p = .029; Table 4). This association is shown in Figure 1 with differential survival curves plotted for individuals who were at least 1 SD above and below the mean on DNAm age residuals at baseline. Veterans who died showed, on average, a 3.69 year (SD: 5.20) age acceleration at baseline relative to an average of 0.48 year age acceleration (SD: 4.58) among those still living at the time of the medical record review, t (239) = 2.77, p = .006. In the follow-up analysis of 191 veterans with available BMI data, we found that BMI was not associated with mortality, while the DNAm age residuals continued to evidence a significant association in this smaller cohort (HR: 1.24, 95% CI: 1.06 to 1.45, p = .006).
Table 4.
Results of Cox Regression Model Predicting Mortality During the 6.5 Year Follow-up (n = 241)
| Variable | HR | p | 95.0% CI
|
|
|---|---|---|---|---|
| Lower | Upper | |||
| Age | 1.02 | .437 | .964 | 1.089 |
| Sex | 0.53 | .652 | .034 | 8.29 |
| PC1 | 0.94 | .407 | .826 | 1.08 |
| PC2 | 0.99 | .803 | .884 | 1.10 |
| CD8T | 1.03 | .734 | .872 | 1.215 |
| CD4T | 0.96 | .570 | .851 | 1.093 |
| NK | 1.02 | .627 | .931 | 1.127 |
| B cell | 1.05 | .114 | .988 | 1.119 |
| Mono | 1.01 | .962 | .821 | 1.230 |
| Lifetime PTSD Sev | 0.99 | .260 | .975 | 1.007 |
| DNAm age residuals | 1.13 | .029 | 1.012 | 1.255 |
Note. Significant associations are bolded. PC = principal component; CD8T = cd8 t-cells; CD4T = CD4 t-cells; NK = natural killer cells; mono = monocytes; sev = severity; DNAm = DNA methylation; HR = hazard ratio; CI = confidence interval. Due to convergence issues associated with variables on very different scales, the PCs and white blood cell count estimates were changed by a constant (multiplied by 100), which does not affect statistical significance.
Figure 1.
Survival curves demonstrating differential survival as a function of accelerated DNAm age residuals. For the purposes of plotting the effects observed in the Cox regression, we show separate lines for veterans who were 1 SD above or below the mean on DNAm age residuals at baseline. DNAm = DNA methylation; SD = standard deviation.
Discussion
The development of a reliable and accurate epigenetic index of cellular age marks a turning point in the ability to identify factors that alter the rate of cellular aging and its many consequences. This could have far reaching implications for defining biological mechanisms of age-related disease and developing interventions to mitigate these processes. The results of this study are the first to replicate our previously reported finding of association between PTSD and accelerated DNAm age and further suggest that the association may be driven primarily by PTSD symptoms defined by heightened physiological arousal. Specifically, results suggested that the hyperarousal cluster of symptoms (which include hypervigilance, anger, startle, and sleep disturbance) were uniquely associated with advanced DNAm age relative to chronological age among this sample of white, non-Hispanic and primarily male veterans. These findings are consistent with the hypothesis that sleep disturbance and tonic physiological arousal (i.e., components of hyperarousal) play a central role in promoting cellular aging (5, 40). Though the specific biological mechanisms underlying hyperarousal and PTSD-related accelerated aging have yet to be identified, processes that may link symptoms to cellular aging include autonomic reactivity (40), hypothalamic-pituitary-adrenal (HPA) axis reactivity (10, 41), inflammation (41), and oxidative stress (5). Perturbations in these biological systems and processes are unlikely to be specific to either PTSD or cellular aging but may still represent important areas of study to better understand the biological consequences of PTSD and to develop interventions to slow the pace of cellular aging and health decline. Likewise, we suspect that accelerated DNAm age may be a sensitive, though perhaps not specific, biomarker of health decline. Critical next steps to elucidate mechanisms and correlates of epigenetic aging include: (a) in vivo and in vitro studies to examine responsivity of the DNAm age loci to various biological challenges (e.g., glucocorticoids, catecholamines); and (b) clinical studies to examine associations between DNAm age acceleration, physiological reactivity, and age-related health conditions frequently comorbid with PTSD, such as cardiometabolic (42, 43) and autoimmune disorders (44). As well, it will be critical to determine if biological mechanisms implicated in PTSD-related accelerated aging and pre-mature health decline vary by race, ethnicity, or sex. Taken together, results of our two studies provide preliminary evidence that PTSD-related accelerated DNAm age is found among both young (with an average age in the early 30s as per Wolf et al.; 4) and middle-aged (average age in mid-50s) veterans. This suggests the generalizability of PTSD-related epigenetic aging across early and middle adulthood, though longitudinal study is still needed in more heterogeneous samples.
Results also suggested that accelerated DNAm age was associated with increased risk for death during the 6.5 year study follow-up period. Though this has been shown previously in epidemiological samples (13, 14), to our knowledge, this is the first time an association between accelerated DNAm age and mortality has been observed in a clinical sample. This is important as clinical samples would be expected to have greater medical comorbidity that would independently increase the risk for mortality, potentially obscuring associations with DNAm age. Moreover, prior studies on DNAm age and mortality were conducted among elderly subjects (13, 14) wherein the effects of age acceleration might be expected to be more impactful with respect to mortality risk.
Finally, we also found a significant sex effect in our sample suggesting that female veterans had decreased DNAm age residuals relative to male veterans. Lower epigenetic age among women compared to men has been reported before (1, 10, 13), though the interpretation of the difference is unclear. One possibility is that women age slower than men, consistent with the common finding that women tend to outlive men (39). Alternative possibilities are that the DNAm age algorithm shows differential performance by sex (e.g., is biased in one group) or that the individual loci included in the algorithm contribute differently to the total score in men versus women. Further evaluation is critical for determining if sex-specific DNAm age indices are needed and for understanding the implications of DNAm age estimates that deviate from chronological age. Our sample was not optimal for evaluating this issue given the small number of women included in the study (consistent with the percentage of women veterans) and as a result, effects pertaining to sex should be interpreted with caution.
Limitations
Results should be considered in light of several important study limitations. To begin with, this was a cross-sectional study and as such, we were unable to determine causal associations. Future work using a longitudinal design is needed to address this. As noted above, the percentage of female veterans was small, and the sample was limited to a single racial and ethnic group, which limits the generalizability of results and underscores the need for replication. There are also a number of potentially confounding variables (e.g., tobacco use, health diagnoses) that we were unable to examine either because there was not sufficient representation (i.e., small sample size) of the comorbid health diagnosis for analytic purposes or because the temporally-linked data were not available in the record (e.g., for veterans who receive only part of their care within the system). Evaluation of mortality did not account for cause of death (as this information is typically not available in the medical record for veterans who died outside of the VA system) or control for other health factors (e.g., medical diagnoses) that could have accounted for effects of accelerated DNAm age on mortality. Further, the analysis predicting mortality was conducted in a relatively small sample whose medical records we had permission to access and with a small number of subject deaths, and should be interpreted in light of these limitations. That said, it was remarkable that 7% of this middle-aged sample died during the study follow-up period as this is much greater than would be expected based on large scale studies of mortality among middle-aged adults living in the United States. Specifically, recent estimates (45) suggest that the mortality rate among middle-aged, white, non-Hispanics is approximately 415 per 100,000 (i.e., 0.415%). This underscores the importance of identifying those at risk for early death in the vulnerable veteran population.
Conclusions
Understanding the health consequences of PTSD and the biological mechanisms that link PTSD to premorbid health decline is important. Though it has long been established that PTSD is associated with substantial medical comorbidities (46), and there have been numerous important models put forward to conceptualize the ways in which PTSD might alter biology (e.g., allostatic load; 6), it has not been until very recently that we have had the ability to index cellular aging directly. Here we show that among white, non-Hispanic and primarily male veterans, PTSD hyperarousal symptoms in veterans were associated with accelerated DNAm age and that accelerated DNAm age was associated with, on average, a 13% increased risk for mortality over the course of approximately 6.5 years. This, along with the results of prior work (4, 11–14), points to the potential applied utility of DNAm age calculators and suggests that it might be clinically useful to track DNAm age estimates over time. This could inform who is at greatest risk for development of premature disease and death and help determine if behavioral and pharmacological interventions improve the pace of cellular aging and align it with chronological age. If future studies continue to support the validity and utility of DNAm age calculators, then assessment of cellular age could be incorporated into standard wellness and preventative medical screenings. Findings could help pave the way for a new line of research focused on the use of epigenetic data to help individuals live healthier lives.
Supplementary Material
Acknowledgments
Research reported in this publication was supported in part by the National Institute on Aging of the National Institutes of Health award R03AG051877 to EJW and by National Institute on Mental Health award RO1 MH079806 to MWM. This work was also supported in part by Merit Review Award Number I01 CX-001276-01 to EJW from the United States (U.S.) Department of Veterans Affairs (VA) Clinical Sciences R&D (CSRD) Service, and by a Presidential Early Career Award for Scientists and Engineers (PECASE 2013A) to EJW as administered by U.S. Department of VA Office of Research and Development. This work was also supported by a U.S. Department of VA CSRD Merit Review award 5I01CX000431-02 and by U.S. Department of VA Biomedical Laboratory Research & Development Program award 1I01BX002150-01, both to MWM. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, or the United States Government.
Abbreviations
- BMI
Body mass index
- BMIQ
Beta mixture quantile dilation
- CAPS
Clinician-Administered PTSD Scale
- CI
Confidence Interval
- DNA
Deoxyribonucleic acid
- DNAm
DNA methylation
- DSM-IV
Diagnostic and Statistical Manual of Mental Disorders, Fourth edition
- Emo. Numb
Emotional numbing
- HPA
Hypothalamic-pituitary-adrenal
- HR
Hazard ratio
- KNN
k nearest neighbor
- M
mean
- MA-
Masters-level
- Mono
Monocytes
- NK
Natural killer
- PC
Principal component
- PhD
Doctor of Philosophy-level
- PTSD
Posttraumatic stress disorder
- Reexp
Re-experiencing
- SCID
Structured Clinical Interview for DSM-IV
- SD
Standard deviation
- Sev
Severity
- SNP
single nucleotide polymorphism
- TLEQ
Traumatic Life Events Questionnaire
- VA
Veterans Affairs
Footnotes
Financial Disclosures
All authors report no financial conflicts of interest.
References
- 1.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, Deconde R, Chen M, Rajapakse I, Friend S, Ideker T, Zhang K. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell. 2013;49:359–367. doi: 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Horvath S. DNA methylation age of human tissues and cell types. Genome Biol. 2013;14:R115. doi: 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Müezzinler A, Zaineddin AK, Brenner H. A systematic review of leukocyte telomere length and age in adults. Ageing Res Rev. 2013;12:509–519. doi: 10.1016/j.arr.2013.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Wolf EJ, Logue MW, Hayes JP, Sadeh N, Schichman SA, Stone A, Salat DH, Milberg W, McGlinchey R, Miller MW. Accelerated DNA methylation age: associations with PTSD and neural integrity. Psychoneuroendocrinology. 2016;63:155–162. doi: 10.1016/j.psyneuen.2015.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller MW, Sadeh N. Traumatic stress, oxidative stress and post-traumatic stress disorder: neurodegeneration and the accelerated-aging hypothesis. Mol Psychiatry. 2014;19:1156–1162. doi: 10.1038/mp.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman MJ, McEwen BS. Posttraumatic stress disorder, allostatic load, and medical illness. In: Schnurr PP, Green BL, editors. Trauma and health: Physical health consequences of exposure to extreme stress. Washington, DC: American Psychological Association; 2004. pp. 157–188. [Google Scholar]
- 7.Kiecolt-Glaser JK, Wilson SJ. Psychiatric disorders, morbidity, and mortality: tracing mechanistic pathways to accelerated aging. Psychosom Med. 2016;78:772–775. doi: 10.1097/PSY.0000000000000373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Darrow SM, Verhoeven JE, Révész D, Lindqvist D, Penninx BW, Delucchi KL, Wolkowitz OM, Mathews CA. The association between psychiatric disorders and telomere length: a meta-analysis involving 14,827 persons. Psychosom Med. 2016;78:776–787. doi: 10.1097/PSY.0000000000000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boks MP, van Mierlo HC, Rutten BP, Radstake TR, De Witte L, Geuze E, Horvath S, Schalkwyk LC, Vinkers CH, Broen JC, Vermetten E. Longitudinal changes of telomere length and epigenetic age related to traumatic stress and post-traumatic stress disorder. Psychoneuroendocrinology. 2015;51:506–512. doi: 10.1016/j.psyneuen.2014.07.011. [DOI] [PubMed] [Google Scholar]
- 10.Zannas AS, Arloth J, Carrillo-Roa T, Iurato S, Röh S, Ressler KJ, Nemeroff CB, Smith AK, Bradley B, Heim C, Menke A, Lange JF, Brückl T, Ising M, Wray NR, Erhardt A, Binder EB, Mehta D. Lifetime stress accelerates epigenetic aging in an urban, African American cohort: relevance of glucocorticoid signaling. Genome Biol. 2015;16:266. doi: 10.1186/s13059-015-0828-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marioni RE, Shah S, McRae AF, Ritchie SJ, Muniz-Terrera G, Harris SE, Gibson J, Redmond P, Cox SR, Pattie A, Corley J, Taylor A, Murphy L, Starr JM, Horvath S, Visscher PM, Wray NR, Deary IJ. The epigenetic clock is correlated with physical and cognitive fitness in the Lothian Birth Cohort 1936. Int J Epidemiol. 2015;44:1388–1396. doi: 10.1093/ije/dyu277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breitling LP, Saum KU, Perna L, Schöttker B, Holleczek B, Brenner H. Frailty is associated with the epigenetic clock but not with telomere length in a German cohort. Clin Epigenetics. 2016;8:21. doi: 10.1186/s13148-016-0186-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Marioni RE, Shah S, McRae AF, Chen BH, Colicino E, Harris SE, Gibson J, Henders AK, Redmond P, Cox SR, Pattie A, Corley J, Murphy L, Martin NG, Montgomery GW, Feinberg AP, Fallin MD, Multhaup ML, Jaffe AE, Joehanes R, Schwartz J, Just AC, Lunetta KL, Murabito JM, Starr JM, Horvath S, Baccarelli AA, Levy D, Visscher PM, Wray NR, Deary IJ. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 2015;16:25. doi: 10.1186/s13059-015-0584-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai PC, Roetker NS, Just AC, Demerath EW, Guan W, Bressler J, Fornage M, Studenski S, Vandiver AR, Moore AZ, Tanaka T, Kiel DP, Liang L, Vokonas P, Schwartz J, Lunetta KL, Murabito JM, Bandinelli S, Hernandez DG, Melzer D, Nalls M, Pilling LC, Price TR, Singleton AB, Gieger C, Holle R, Kretschmer A, Kronenberg F, Kunze S, Linseisen J, Meisinger C, Rathmann W, Waldenberger M, Visscher PM, Shah S, Wray NR, McRae AF, Franco OH, Hofman A, Uitterlinden AG, Absher D, Assimes T, Levine ME, Lu AT, Tsao PS, Hou L, Manson JE, Carty CL, LaCroix AZ, Reiner AP, Spector TD, Feinberg AP, Levy D, Baccarelli A, van Meurs J, Bell JT, Peters A, Deary IJ, Pankow JS, Ferrucci L, Horvath S. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging (Albany NY) 2016;8:1844–1865. doi: 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.American Psychiatric Association. Diagnostic and statistical manual of mental disorders: DSM-IV. 4. Washington, DC: American Psychiatric Association; 1994. [Google Scholar]
- 16.Logue MW, Baldwin C, Guffanti G, Melista E, Wolf EJ, Reardon AF, Uddin M, Wildman D, Galea S, Koenen KC, Miller MW. A genome-wide association study of post-traumatic stress disorder identifies the retinoid-related orphan receptor alpha (RORA) gene as a significant risk locus. Mol Psychiatry. 2013;18:937–942. doi: 10.1038/mp.2012.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Blake DD, Weathers FW, Nagy LM, Kaloupek DG, Gusman FD, Charney DS, Keane TM. The development of a Clinician-Administered PTSD Scale. J Trauma Stress. 1995;8:75–90. doi: 10.1007/BF02105408. [DOI] [PubMed] [Google Scholar]
- 18.Barfield RT, Almli LM, Kilaru V, Smith AK, Mercer KB, Duncan R, Klengel T, Mehta D, Binder EB, Epstein MP, Ressler KJ, Conneely KN. Accounting for population stratification in DNA methylation studies. Genet Epidemiol. 2014;38:231–241. doi: 10.1002/gepi.21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roberts AL, Gilman SE, Breslau J, Breslau N, Koenen KC. Race/ethnic differences in exposure to traumatic events, development of post-traumatic stress disorder, and treatment-seeking for post-traumatic stress disorder in the United States. Psychol Med. 2011;41:71–83. doi: 10.1017/S0033291710000401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kubany ES, Haynes SN, Leisen MB, Owens JA, Kaplan AS, Watson SB, Burns K. Development and preliminary validation of a brief broad-spectrum measure of trauma exposure: the Traumatic Life Events Questionnaire. Psychol Assess. 2000;12:210–224. doi: 10.1037//1040-3590.12.2.210. [DOI] [PubMed] [Google Scholar]
- 21.Weathers FW, Ruscio AM, Keane TM. Psychometric properties of nine scoring rules for the Clinician-Administered Posttraumatic Stress Disorder Scale. Psychol Assess. 1999;11:124–133. [Google Scholar]
- 22.First MB, Gibbon M. The Structured Clinical Interview for DSM-IV Axis I Disorders (SCID-I) and the Structured Clinical Interview for DSM-IV Axis II Disorders (SCID-II) 2004. [Google Scholar]
- 23.R Development Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- 24.Teschendorff AE, Marabita F, Lechner M, Bartlett T, Tegner J, Gomez-Cabrero D, Beck S. A beta-mixture quantile normalization method for correcting probe design bias in Illumina Infinium 450k DNA methylation data. Bioinformatics. 2013;29:189–196. doi: 10.1093/bioinformatics/bts680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Touleimat N, Tost J. Complete pipeline for Infinium((R)) Human Methylation 450K BeadChip data processing using subset quantile normalization for accurate DNA methylation estimation. Epigenomics. 2012;4:325–341. doi: 10.2217/epi.12.21. [DOI] [PubMed] [Google Scholar]
- 26.Pidsley R, Wong CC, Volta M, Lunnon K, Mill J, Schalkwyk LC. A data-driven approach to preprocessing Illumina 450K methylation array data. BMC Genomics. 2013;14:293. doi: 10.1186/1471-2164-14-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 28.Leek JT, Johnson WE, Parker HS, Jaffe AE, Storey JD. R package version 3. 2013. Sva: Surrogate Variable Analysis. [Google Scholar]
- 29.Hastie T, Tibshirani R, Sherlock G, Eisen M, Brown P, Botstein D. Imputing missing data for gene expression arrays. 1999:1–7. [Google Scholar]
- 30.Troyanskaya O, Cantor M, Sherlock G, Brown P, Hastie T, Tibshirani R, Botstein D, Altman RB. Missing value estimation methods for DNA microarrays. Bioinformatics. 2001;17:520–525. doi: 10.1093/bioinformatics/17.6.520. [DOI] [PubMed] [Google Scholar]
- 31.Jaffe AE, Irizarry RA. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 2014;15:R31. doi: 10.1186/gb-2014-15-2-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–959. doi: 10.1093/genetics/155.2.945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–909. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 35.King DW, Leskin GA, King LA, Weathers FW. Confirmatory factor analysis of the clinician-administered PTSD scale: evidence for the dimensionality of posttraumatic stress disorder. Psychol Assess. 1998;10:90. [Google Scholar]
- 36.Simons RL, Lei MK, Beach SR, Philibert RA, Cutrona CE, Gibbons FX, Barr A. Economic hardship and biological weathering: the epigenetics of aging in a U.S. sample of black women. Soc Sci Med. 2016;150:192–200. doi: 10.1016/j.socscimed.2015.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beach SR, Dogan MV, Lei MK, Cutrona CE, Gerrard M, Gibbons FX, Simons RL, Brody GH, Philibert RA. Methylomic aging as a window onto the influence of lifestyle: tobacco and alcohol use alter the rate of biological aging. J Am Geriatr Soc. 2015;63:2519–2525. doi: 10.1111/jgs.13830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Horvath S, Erhart W, Brosch M, Ammerpohl O, von Schönfels W, Ahrens M, Heits N, Bell JT, Tsai PC, Spector TD, Deloukas P, Siebert R, Sipos B, Becker T, Röcken C, Schafmayer C, Hampe J. Obesity accelerates epigenetic aging of human liver. Proc Natl Acad Sci U S A. 2014;111:15538–15543. doi: 10.1073/pnas.1412759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beltrán-Sánchez H, Finch CE, Crimmins EM. Twentieth century surge of excess adult male mortality. Proc Natl Acad Sci U S A. 2015;112:8993–8998. doi: 10.1073/pnas.1421942112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Williamson JB, Porges EC, Lamb DG, Porges SW. Maladaptive autonomic regulation in PTSD accelerates physiological aging. Front Psychol. 2015;5:1571. doi: 10.3389/fpsyg.2014.01571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lohr JB, Palmer BW, Eidt CA, Aailaboyina S, Mausbach BT, Wolkowitz OM, Thorp SR, Jeste DV. Is post-traumatic stress disorder associated with premature senescence? A review of the literature. Am J Geriatr Psychiatry. 2015;23:709–725. doi: 10.1016/j.jagp.2015.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vaccarino V, Goldberg J, Rooks C, Shah AJ, Veledar E, Faber TL, Votaw JR, Forsberg CW, Bremner JD. Post-traumatic stress disorder and incidence of coronary heart disease: a twin-study. J Am Coll Cardiol. 2013;62:970–978. doi: 10.1016/j.jacc.2013.04.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kubzansky LD, Koenen KC. Is posttraumatic stress disorder related to development of heart disease? An update. Cleve Clin J Med. 2009;76(Suppl 2):S60–S65. doi: 10.3949/ccjm.76.s2.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.O’Donovan A, Cohen BE, Seal KH, Bertenthal D, Margaretten M, Nishimi K, Neylan TC. Elevated risk for autoimmune disorders in Iraq and Afghanistan veterans with posttraumatic stress disorder. Biol Psychiatry. 2015;77:365–374. doi: 10.1016/j.biopsych.2014.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Case A, Deaton A. Rising morbidity and mortality in midlife among white non-Hispanic Americans in the 21st century. PNAS. 2015;112:15078–15083. doi: 10.1073/pnas.1518393112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schnurr PP, Spiro A, 3rd, Paris AH. Physician-diagnosed medical disorders in relation to PTSD symptoms in older male military veterans. Health Psychol. 2000;19:91–97. doi: 10.1037//0278-6133.19.1.91. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.