Abstract
To better understand the molecular and cellular differences in brain organization between human and non-human primates, we performed transcriptome sequencing of sixteen regions of adult human, chimpanzee, and macaque brains. Integration with human single-cell transcriptomic data revealed global, regional, and cell-type specific species expression differences in genes representing distinct functional categories. We validated and further characterized the human specificity of genes enriched in distinct cell types through histological and functional analyses, including rare subpallial-derived interneurons expressing dopamine biosynthesis genes enriched in the human striatum and absent in the non-human African ape neocortex. Our integrated analysis of the generated data revealed diverse molecular and cellular features of the phylogenetic reorganization of the human brain across multiple levels of organization, with relevance for brain function and disease.
Introduction
Although the human brain is approximately three times larger than those of our closest living relatives, the non-human African great apes (chimpanzee, bonobo, and gorilla), increased size and neural cell counts fail to explain its unique functionalities (1–5). The brain has also undergone microstructural, connectional, and molecular changes in the human lineage (1–5), changes likely mediated by divergent spatiotemporal gene expression (6–17).
Here, we profiled the mRNA and small noncoding RNA transcriptomes of sixteen adult brain regions involved in higher-order cognition and behavior of human (H, Homo sapiens), chimpanzee (C, Pan troglodytes) - our closest extant relative - and rhesus macaque (M, Macaca mulatta) - a commonly studied non-human primate. We integrated these profiles with single-cell transcriptomic data from the human brain (18, 19), histological data from adult and developmental brains of these and other primates (bonobo, gorilla, orangutan, pig-tailed macaque, baboon, and capuchin), and multimodal data from human primary and induced pluripotent stem cell (iPSC)-derived neural cultures. In doing so, we have investigated the evolutionary, cellular, and developmental framework that makes the human brain unique.
Overview of regional transcriptome profiling
We generated transcriptional profiles of 247 tissue samples representing hippocampus, amygdala, striatum, mediodorsal nucleus of thalamus, cerebellar cortex, and eleven areas of the neocortex from six humans, five chimpanzees, and five macaques (figs. S1–S5; table S1). To minimize biases in comparative transcriptome analyses, we used the XSAnno pipeline to create a common annotation set of 26,514 orthologous mRNAs, including 16,531 protein-coding genes and 3,253 long intergenic non-coding (linc) RNAs (fig. S2). We re-annotated all chimpanzee and macaque micro RNAs (miRNAs) based on annotated human precursor sequences (fig. S3). Assessment of global correlation between regions and species by unsupervised hierarchical clustering (fig. S5) revealed clustering of the miRNA dataset primarily by species. In contrast, cerebellar mRNA samples from all species formed a distinct cluster separated from other brain regions (fig. S5), indicating that the various cerebella are more similar to each other than to other brain regions within the same species. Within each species, hierarchical clustering of mRNA or miRNA datasets were calculated based on pairwise correlation matrices of brain regions and confirmed by multiscale bootstrap resampling and intra-species genetic distance measurements (figs. S6–7). This revealed a similar pattern of inter-regional hierarchical clustering, reflecting known topographical proximity and functional overlap (11, 14).
Species differences in regional gene expression
Differentially expressed genes were identified (False Discovery Rate [FDR] < 0.01) in each region by comparing generalized linear models with species as the main factor and batch as a cofactor. We found 25.9% of mRNAs (6,866 of 26,514) and 40.6% of miRNAs (603 of 1,485 mature miRNAs included in the analysis) were differentially expressed between at least two species in one or more regions. 11.9% of mRNAs (3,154) and 13.6% of miRNAs (202), representing distinct functional categories, exhibited human-specific upregulation (H>C=M) or downregulation (H<C=M; Figs. 1A–B; tables S2–4), with the highest number of differentially expressed genes observed in striatum followed by thalamus, primary visual cortex, and dorsolateral prefrontal cortex (fig. S8; table S3). These observations were not attributable to variations in the ratio of major cell types among species (fig. S9).
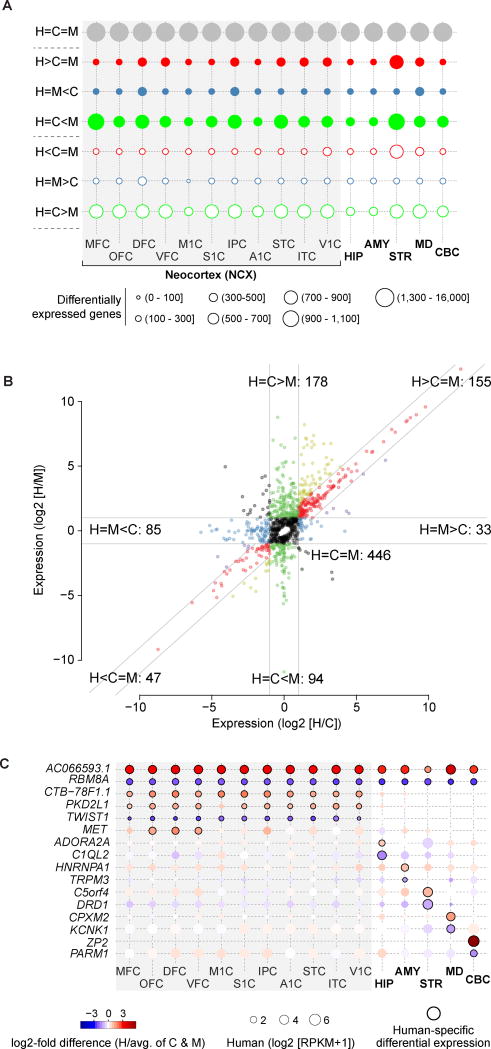
Fig. 1. Inter-species differential gene expression across sixteen brain regions.
(A) Bubble matrix showing the number of mRNA genes with conserved expression (grey circles), species-specific upregulation (filled circles), or downregulation (open circles). Post-hoc comparisons described in table S2. H – human; C – chimpanzee; M – macaque. (B) Inter-species patterns of normalized miRNA expression across all regions. Guidelines indicate ± 2-fold difference. (C) Examples of protein-coding and non-coding genes exhibiting global and regional human-specific upregulation (red and circles with black borders) or downregulation (blue and circles with black borders). Additional information and validations are provided in figs. S10–S12. OFC – orbital prefrontal cortex; DFC – dorsolateral prefrontal cortex; VFC – ventrolateral prefrontal cortex; MFC – medial prefrontal cortex; M1C – primary motor cortex; S1C – primary somatosensory cortex; IPC – inferior posterior parietal cortex; A1C – primary auditory cortex; STC – superior temporal cortex; ITC – inferior temporal cortex; V1C – primary visual cortex; HIP – hippocampus; AMY – amygdala; STR – striatum; MD – mediodorsal nucleus of the thalamus; CBC – cerebellar cortex.
Among the 3,154 mRNA genes with human-specific differential expression, only 22 were upregulated and 9 down-regulated across all analyzed regions (fig. S10). Only 3 genes were differentially expressed across analyzed neocortical areas: TWIST1 (down-regulated), a transcriptional regulator of neural genes that is mutated in Saethre–Chotzen syndrome, a disorder associated with intellectual disability (20) (Fig. 1C; fig. S11A–B), and two functionally uncharacterized lincRNAs (RP11-364P22.1 and CTB-78F1.1; upregulated). The remaining 3,120 mRNA genes displayed human-specific differential expression in one or a subset of brain regions or neocortical areas (Fig. 1C; fig. S10; table S3). Among miRNAs, 10.4% (155) and 3.2% (47) were upregulated or downregulated, respectively, in the human brain with many displaying region-specific patterns (Fig. 1B; fig. S12). Independently validated examples include: PKD2L1 (upregulated in neocortical areas except primary motor cortex), a gene encoding an ion channel (21); MET (upregulated in prefrontal cortex), a gene implicated in autism spectrum disorder (22); ZP2 (upregulated in cerebellum), a gene encoding a protein mediating sperm-egg recognition (23); and several miRNAs (Fig. 1C; figs. S11–S12).
Species differences in gene co-expression patterns
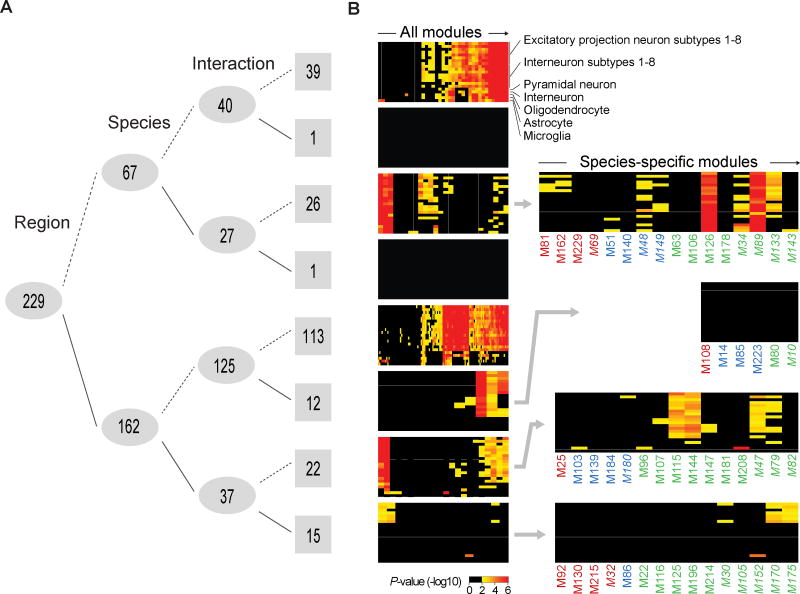
To extract additional biologically relevant information, we applied weighted gene co-expression correlation network analysis (WGCNA) to generate modules of genes with similar variation across regions and/or species. We identified 229 mRNA modules, many of which exhibited regional and/or species-specific expression patterns (Figs. 2A–B; table S5). For example, genes in module 92 (M92) and M32 are respectively upregulated and downregulated in human neocortex, and M130 genes are upregulated in human striatum, hippocampus, and amygdala (fig. S13). M130 includes tyrosine hydroxylase (TH) and DOPA decarboxylase (DDC), both involved in dopamine biosynthesis (fig. S13F). Human-specific modules were enriched for genes associated with categories and pathways such as “Thrombospondin N-terminal-like domains” and “alternative splicing” (table S5).
Fig. 2. Conserved and species-specific gene co-expression modules.
(A) Number of WGCNA modules (numbers on grey; see table S5) clustered by differential expression across brain regions, species, and inter-species differences across regions (interaction) (ANOVA of eigengene Bonferroni adjusted P < 0.01, solid line; ≥ 0.01, dashed line). (B) Left panel: Enrichment of gene expression for modules (columns) in several cell types (rows) based on human single-cell transcriptome data (18, 19), sorted by unsupervised hierarchical clustering to show similarities among modules. Right panel: Species-specific modules showing human (red), chimpanzee (blue), or macaque (green) upregulation (normal font) or downregulation (italics) relative to the other two species exhibit distinct patterns of cell-type associated gene expression.
We also clustered all miRNAs based on their individual correlations to the average expression profile of each mRNA module (fig. S14A; table S6). Because the expression of each miRNA might correlate with multiple mRNA modules, module pairings were refined using a transcriptome-wide HITS-CLIP map of miRNA binding sites in the human brain (24) (fig. S14B; table S7). We identified 37 stable miRNA modules, with several pairs of miRNA/mRNA modules exhibiting opposing regional and/or species-specific enrichment for potential miRNA-mRNA target predictions (fig. S14C–E).
Cell-type specificity of differentially expressed genes
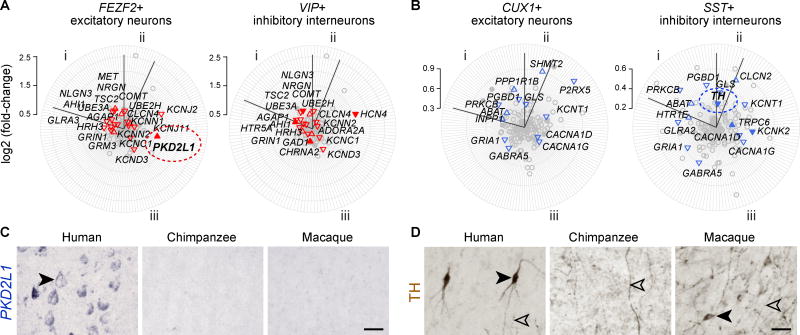
To investigate differential gene expression patterns at the cellular level, we integrated our datasets with single-cell RNA-seq data generated from the human neocortex (18, 19) and validated findings via immunohistochemistry or in situ hybridization. We found that many of the genes displaying species- and/or region-specific patterns also exhibited cell-type specific expression. For example, PKD2L1 is enriched in excitatory projection neurons (Figs. 3A and 3C), TH is expressed in a subset of somatostatin (SST)-expressing inhibitory interneurons in human and macaque neocortical deep layers and white matter (Figs. 3B and 3D), and ZP2 is upregulated in the granule cells of the human cerebellum (figs. S11C–D). Additionally, we found cell-type specific enrichment among WGCNA modules, including human-specific M81 and M162, which were composed of genes enriched in a subset of neocortical excitatory projection neurons (Fig. 2B, right panels; table S5).
Fig. 3. Cellular specificity of neocortical human and chimpanzee-specific differential expression.
(A–B) Radar plots depicting neocortical neuron cell-type enrichments of (A) human- or (B) chimpanzee-specific differences of genes associated with (i) neuropsychiatric disorders, (ii) neurotransmitter biosynthesis, degradation, and transport proteins, and (iii) encoding ion channels (table S10). Only genes expressed in the respective cell type are plotted. The distance of each gene from the center represents differential expression between human and the average of chimpanzee and macaque (red) or between chimpanzee and the other two species (blue). The direction of triangles denotes up- or downregulation; filled triangles represent cell-type specific expression (Pearson correlations > 0.5). (C) In situ hybridization shows that PKD2L1 is expressed in pyramid-shaped cell bodies of excitatory projection neurons of human, but not chimpanzee or macaque, neocortex. (D) TH-immunopositive interneurons (filled arrowheads) are present in neocortex of human and macaque, but not chimpanzee, where only TH+ midbrain dopaminergic axons (open arrowheads) are present. Scale bar represents 30 µm.
Species differences in neurotransmitter receptor gene expression
The species- and region-specific expression patterns of several genes associated with neurotransmission prompted us to investigate whether there were broad inter-species differences in the co-expression networks and genomic sequences of genes encoding receptors underlying excitatory, inhibitory, or modulatory signaling (figs. S15A–D). Gene co-expression networks of the cholinergic and serotonergic systems differed among the three species (figs. S15E–F). While the dopaminergic system did not have enough genes for reliable network construction, we found that DRD1, DRD2, and DRD3, genes encoding dopamine receptors, exhibited human-specific downregulation in striatum (fig. S10). By contrast, excitatory glutamatergic and inhibitory GABAergic systems’ genes exhibited conserved networks among species and their coding sequences were more conserved than the coding sequences of genes with similar expression levels (figs. S15–17; table S8).
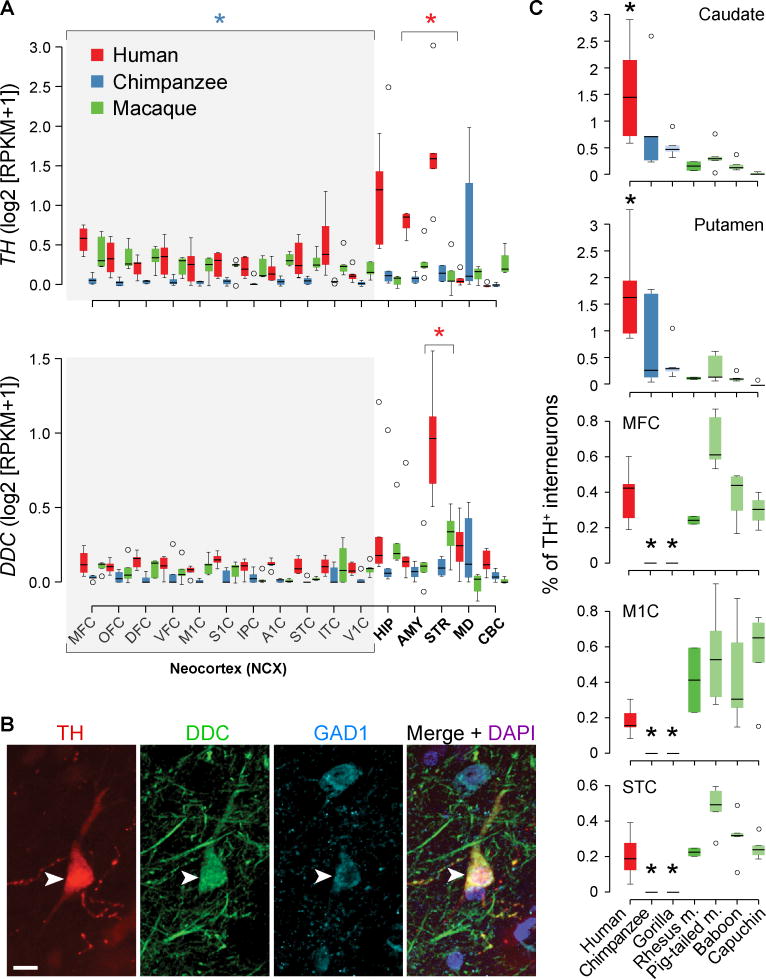
Species differences in dopamine biosynthesis gene expression
We next investigated dopamine biosynthesis and signaling genes. TH and DDC displayed human-specific (H>C=M) upregulation in the striatum (Fig. 4A). TH also displayed chimpanzee-specific downregulation (C<H=M) in the neocortex (Fig. 4A). An extended analysis of RNA-seq data (25) independently validated the downregulation of TH mRNA in chimpanzee neocortex compared to human, as well as the downregulation of TH expression in the neocortex of bonobo and gorilla, but not of orangutan (fig. S18A).
Fig. 4. Human-specific expression of genes encoding dopamine biosynthesis enzymes.
(A) TH and DDC, respectively, showing higher expression in the human striatum (STR). TH is also downregulated in the chimpanzee neocortex. Boxes represent quartiles and whiskers 1.5 times interquartile range. Red and blue asterisks represent human-specific differential expression in striatum and chimpanzee-specific differential expression combining all neocortical areas, respectively (FDR < 0.01). (B) Immunofluorescence shows co-localization of TH, DDC, and GAD1 in adult human neocortical interneurons (arrowheads). Scale bar represents 10 µm. (C) STR (caudate and putamen) shows an enrichment of TH+ interneurons in human. MFC, M1C, and STC show a complete depletion of TH+ interneurons in chimpanzee and gorilla. Asterisk represents Tukey's honest significance test P < 0.05 comparing human or chimpanzee/gorilla with all other species.
Analyses of cis-regulatory elements active near the TH gene in the adult human, chimpanzee, and macaque brain (26) revealed no differences that would explain observed TH expression patterns. We hypothesized that the species-specific TH expression patterns might be explained by changes in the number and distribution of TH-expressing interneurons, which have been previously identified in telencephalic regions and shown to vary in distribution across species (27–29), including depletion in the prefrontal cortex of non-human great apes (28). Therefore, we quantified TH-immunopositive (TH+) interneurons (figs. S19A–C) on an independent set of 45 adult brains from 9 primate species (table S9). Consistent with our transcriptome data, humans have a higher number (Tukey's honest significance test all P < 0.05) of TH+ interneurons in both the dorsal caudate nucleus and putamen (striatum) when compared to all other analyzed non-human primates (Fig. 4C; fig. S20A). Furthermore, we found neocortical TH+ interneurons in all analyzed areas of human, all monkey species, and orangutan (Fig. 4C, figs. S20B; S20D), but only TH+ fibers in all analyzed neocortical areas of chimpanzee, bonobo, and gorilla (Fig. 4C; fig. S20B). We found no differences in the number of TH+ interneurons in human, chimpanzee, gorilla, and macaque olfactory bulbs (fig. S19D).
Molecular profiling of human TH+ interneurons
To further explore the phenotype of adult human neocortical TH+ interneurons, we performed immunohistochemistry and in situ hybridization. TH+ interneurons expressed GAD1, the GABA synthesis enzyme (Fig. 4B), but were lacking canonical markers of neocortical interneuron subtypes such as SST, PVALB, NPY, NOS1, CALB2, and VIP (fig. S21), as well as ETV1, which is required for differentiation of dopaminergic neurons in multiple species (30), or its homolog, ETV5 (figs. S21H–I). Most TH+/GAD1+ interneurons co-expressed DDC (62.54 ± 1.01%; Fig. 4B), the enzyme that converts L-DOPA to dopamine, but not DBH (fig. S18; 31), the enzyme that converts dopamine to noradrenaline, indicating that a subset of TH+ interneurons are able to produce dopamine, but not noradrenaline.
Developmental origin of human TH+ interneurons
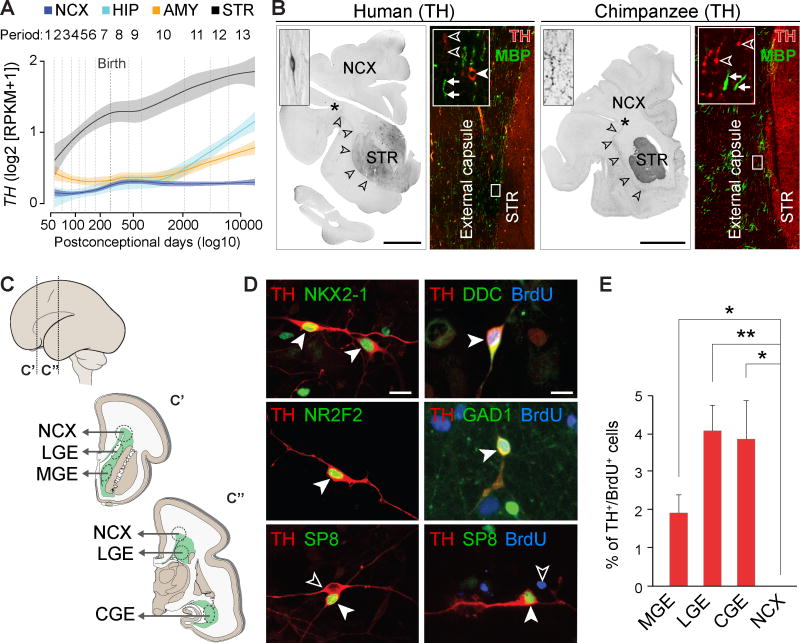
To gain insight into the development of TH+ interneurons, we analyzed the regional expression of TH across human brain development using the BrainSpan RNA-seq dataset (www.brainspan.org). The highest TH expression is observed in striatum and increases steadily from early fetal development (period 2, as defined in ref. 11) to young adulthood (period 13; Fig. 5A). Lower TH expression is observed in neocortex, hippocampus, and amygdala and increases perinatally (periods 7 [late fetal development] and 8 [early infancy]) and remains stable in neocortex. In addition, TH expression increases from early childhood (period 10) to young adulthood in the amygdala and hippocampus (Fig. 5A).
Fig. 5. Human telencephalic TH+ interneurons are of subpallial origin and start to express TH protein perinatally.
(A) TH expression in human neocortex (NCX), HIP, AMY, and STR throughout development. The shaded area corresponds to a confidence interval of 50%. (B) Immunohistochemistry reveals TH+ axons in external capsule (arrowheads), STR, and NCX of newborn (38 pcw) human and chimpanzee brains. Bipolar TH+ interneurons (filled arrowhead) are present in parallel with MBP+/TH− (arrows) and TH+/MBP− (open arrowheads) fibers in the external capsule. No TH+ cells were detected in chimpanzee external capsule. Scale bar represents 1 cm. (C) Schematic of dissection of ganglionic eminences (lateral [LGE], medial [MGE], and caudal [CGE]) and neocortical proliferative zones (NCX) from mid-fetal brain for primary cell culture. (D) TH+ cells from ganglionic eminences also express NKX2-1, NR2F2, or SP8, and were BrdU+, DDC+, and GAD1+. TH+ interneurons in the neocortical culture were SP8+, but BrdU− (bottom right panel). Scale bar represents 20 µm. (E) Percentage of TH+/BrdU+ cells in culture from MGE, LGE, CGE, and NCX. Error bars represent SEM. Pairwise t-tests were performed and corrected for multiple testing using Bonferroni correction. * P < 0.05; ** P < 0.01.
Using immunohistochemistry, we detected TH+ axons in striatum as early as late midfetal development (fig. S22), and occasional bipolar TH+ interneurons were first observed in the external capsule and neocortical white matter in the newborn human (Fig. 5B). The neonatal chimpanzee brain displayed the same pattern of unmyelinated TH+ fibers in the external capsule (Fig. 5B), but no TH+ interneurons were detected in the neocortex.
To identify the birthplace of TH+ interneurons, we prepared primary cell cultures from 17–18 postconceptional week (pcw)-old human brains (Fig. 5C) of lateral, medial, and caudal ganglionic eminences (LGE, MGE, and CGE) of the ventral forebrain (subpallium), known to generate interneurons (32–34), and neocortical proliferative zones, which may also generate interneurons in humans (13). We found TH+ interneurons co-expressing canonical markers of distinct progenitor lineages within ganglionic eminences (NKX2-1, NR2F2, or SP8) (Fig. 5D). BrdU birthdating confirmed that TH+ interneurons are generated by ganglionic eminence, but not neocortical, progenitors (Fig. 5D–E), indicating that TH+ interneurons are derived from diverse subpallial lineages and are developmentally heterogeneous. Similar to adult neocortical TH+ interneurons, subpallial-derived TH+ interneurons also co-expressed GAD1 and DDC (Fig. 5D). Neocortical TH+ interneurons were mainly SP8+ (77.68 ± 12.11%), with a smaller NR2F2+ (22.22 ± 8.78%) subpopulation, and were all BrdU−, indicating that they began to migrate into neocortex before 17 pcw, but express TH protein later in development (Fig. 5D–E).
In vitro characterization of human TH+ interneurons
To further characterize TH+ interneuron development and properties, we asked whether TH+ interneurons could be generated from human iPSCs using a differentiation protocol for cortical excitatory projection neurons and inhibitory interneurons (fig. S23; Methods). Immunofluorescence confirmed the presence of TH+ cells co-expressing GAD1 and SP8, but not SST, PVALB, NR2F2, or NKX2-1, confirming that iPSC-derived TH+ interneurons display a similar molecular profile to TH+ interneurons from the adult neocortex and neocortical primary culture (fig. S24). Complementary analysis of a single-cell RNA-seq dataset from human embryonic stem cell-derived cortical interneurons (35) revealed that many TH-expressing cells co-express GABAergic marker genes GAD1/2 at all time points, as well as SST, ETV1, and ETV5 transiently at early time points (fig. S25).
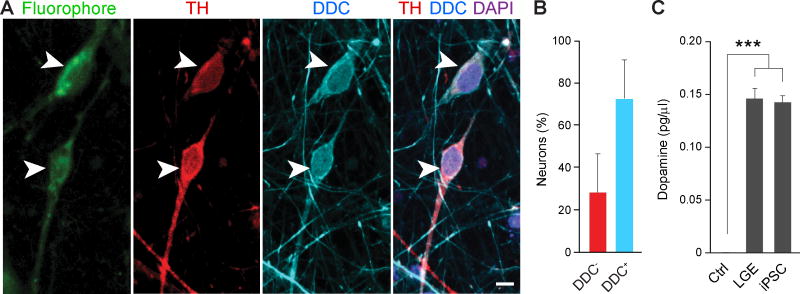
We characterized human iPSC-derived TH+ interneurons by assessing their ability to produce and transport dopamine using immunofluorescence, a monoamine uptake assay, and high-performance liquid chromatography. We found that 72.1 ± 10.0% of 80 DIV TH+ interneurons that had taken up a monoamine-imitating fluorophore were DDC+ (Fig. 6A–B), and consequently could produce and transport dopamine in vitro. Commensurate with these observations, we detected dopamine in conditioned culture media from iPSC-derived and LGE primary neural cultures, both of which contained TH+/DDC+ interneurons, but not in control culture media (Fig. 6C).
Fig. 6. Human telencephalic TH+ interneurons synthesize and transport dopamine in vitro.
Human iPSC-derived neurons were incubated with a fluorophore-labeled synthetic monoamine. (A) TH+ (red) and DDC+ (blue) immunolabeled interneurons (arrowheads) that transported monoamine-imitating fluorophore (green) in vitro. Scale bar represents 10 µm. (B) Percentage of neurons that took up the fluorophore and were positive for both the uptake assay and TH. This population is composed of DDC+ (blue) or DDC− (red) interneurons. (C) Concentration of dopamine detected by HPLC in the unused (control, Ctrl) cell culture medium, and the conditioned media from LGE and iPSC-derived cultures. Error bars represent SEM. Dunnett’s test. *** P < 0.001.
Discussion
Our analysis of transcriptomic data revealed global, regional, and cell-type specific species expression differences in protein-coding and non-coding genes. Genes with human-specific differential expression patterns include those encoding transcription factors, ion channels, and neurotransmitter biosynthesis enzymes and receptors. Changes in the regional and cellular expression patterns of these genes could affect function of neural circuits by altering transcription of other genes, intrinsic electrophysiological properties, or synaptic transmission.
Neuromodulatory systems show broad expression differences between species. One example includes a rare and molecularly heterogeneous subpopulation of interneurons expressing dopamine biosynthesis genes TH and DDC, which are enriched in the human striatum and neocortex as compared to non-human African apes. These cells originate in the subpallial ganglionic eminences and likely migrate into the striatum and neocortex during late prenatal and early postnatal development. We also observed an increase in TH expression during postnatal development and young adulthood, suggesting that TH expression and/or the migration of TH+ interneurons may be dynamically regulated and protracted.
The absence of TH+ interneurons from the cortex of non-human African apes (see also 28), and their decreased density in the striatum of non-human primates, may result from several mechanisms. First, these cells could have been lost due to genetic disruptions affecting interneuron migration, differentiation, or survival (32–34). These disruptions may have occurred in the common ancestor of African apes prior to being reversed in the human lineage (homoplasy) or, in a less-likely scenario, may have occurred independently in the Gorilla and Pan lineages. A second possibility is that these interneurons are present in the non-human African ape cortex but do not express TH, do so only transiently, or die prior to our ability to detect them. Commensurate with this possibility, the molecular profile of mouse cortical SST-positive interneurons is malleable (36) and sensory stimuli can cause a switch from the production of TH and dopamine to SST in rat hypothalamic interneurons (37). Finally, TH+ interneurons of non-human African apes may have lost their ability to deviate to cortex from the rostral migratory stream. Indeed some human TH+ interneurons migrating via the rostral migratory stream to the olfactory bulb divert to the prefrontal cortex (38) and our observation of SP8+/TH+ co-expression is consistent with a rostral migratory stream origin. However, other routes of migration are possible, as suggested by our observation of TH+ interneurons in the external capsule of newborn human brain.
Neuromodulatory transmitters, in particular dopamine, are involved in distinctly human aspects of cognition and behavior, such as working memory, reasoning, reflective exploratory behavior, and overall intelligence. By analyzing brain regions involved in these processes, we show that evolutionary modifications in gene expression and the distribution of neurons associated with neuromodulatory systems may underlie cognitive and behavioral differences between species. Cortical TH+ interneurons are depleted in patients affected by Parkinson’s disease (39) or dementia with Lewy bodies (40), and these alterations may contribute to cognitive impairments.
As these results demonstrate, the resource we present here may aid future studies on the evolution and neuroscience of primates.
Supplementary Material
Acknowledgments
We thank A. Bauernfeind, M. Horn, D. Singh, G. Terwilliger, I.B. Toxopeus, B. Wicinski, and S. Wilson for assistance with tissue acquisition and processing, and the Alamogordo Primate Facility and the Primate Brain Bank, Netherlands Institute for Neuroscience, for providing primate tissue. Data was generated as part of the PsychENCODE Consortium, supported by MH103339, MH106934, and MH110926. Additional support was provided by the NIH grant MH109904, the Kavli Foundation, the James S. McDonnell Foundation, NSF grant BCS-1316829, and ERC Starting Grant 260372, and MICINN BFU2011-28549. Supplement contains additional data.
Footnotes
This manuscript has been accepted for publication in Science. This version has not undergone final editing. Please refer to the complete version of record at https://urldefense.proofpoint.com/v2/url?u=http-3A__www.sciencemag.org_&d=DwICaQ&c=cjytLXgP8ixuoHflwc-poQ&r=beTYUUmtVBGN7sWB6Odzf6Zp4VK7uAsjknV9yJ1FpUI&m=dBUXAQnL3LiZxAQGbj3tErIISbb8dw2xBr7iX9aROlc&s=1V7mVoAllCR1DYbYiPubU-L9x885Iz2lEn4_lDbwyaI&e=. The manuscript may not be reproduced or used in any manner that does not fall within the fair use provisions of the Copyright Act without the prior, written permission of AAAS.
Data is available at NCBI BioProjects (accession number: PRJNA236446).
List of Supplementary Materials:
References (41–77)
References
- 1.Preuss TM. What is it like to be a human. In: Gazzaniga MS, editor. The Cognitive Neurosciences. MIT Press; Cambridge, MA: 2004. pp. 5–22. [Google Scholar]
- 2.Sherwood CC, Subiaul F, Zawidzki TW. A natural history of the human mind: tracing evolutionary changes in brain and cognition. J. Anat. 2008;212:426–454. doi: 10.1111/j.1469-7580.2008.00868.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Teffer K, Semendeferi K. Human prefrontal cortex: evolution, development, and pathology. Prog. Brain Res. 2012;195:191–218. doi: 10.1016/B978-0-444-53860-4.00009-X. [DOI] [PubMed] [Google Scholar]
- 4.Gabi M, et al. No relative expansion of the number of prefrontal neurons in primate and human evolution. Proc. Nat.l Aca.d Sci. U.S.A. 2016;113:9617–9622. doi: 10.1073/pnas.1610178113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sousa AMM, Meyer KA, Santpere G, Gulden FO, Sestan N. Evolution of the human nervous system function, structure, and development. Cell. 2017;170:226–247. doi: 10.1016/j.cell.2017.06.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- 7.Cáceres M, et al. Elevated gene expression levels distinguish human from non-human primate brains. Proc. Natl. Acad. Sci. U.S.A. 2003;100:13030–13035. doi: 10.1073/pnas.2135499100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Uddin M, et al. Sister grouping of chimpanzees and humans as revealed by genome-wide phylogenetic analysis of brain gene expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2004;101:2957–2962. doi: 10.1073/pnas.0308725100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khaitovich P, et al. Parallel patterns of evolution in the genomes and transcriptomes of humans and chimpanzees. Science. 2005;309:1850–1854. doi: 10.1126/science.1108296. [DOI] [PubMed] [Google Scholar]
- 10.Babbitt CC, et al. Both noncoding and protein-coding RNAs contribute to gene expression evolution in the primate brain. Genome Biol. Evol. 2010;2:67–79. doi: 10.1093/gbe/evq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kang HJ, et al. Spatio-temporal transcriptome of the human brain. Nature. 2011;478:483–489. doi: 10.1038/nature10523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Konopka G, et al. Human-specific transcriptional networks in the brain. Neuron. 2012;75:601–617. doi: 10.1016/j.neuron.2012.05.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Geschwind DH, Rakic P. Cortical evolution: judge the brain by its cover. Neuron. 2013;80:633–647. doi: 10.1016/j.neuron.2013.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pletikos M, et al. Temporal specification and bilaterality of human neocortical topographic gene expression. Neuron. 2014;81:321–332. doi: 10.1016/j.neuron.2013.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bae BI, Jayaraman D, Walsh CA. Genetic changes shaping the human brain. Dev. Cell. 2015;32:423–434. doi: 10.1016/j.devcel.2015.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Silver DL. Genomic divergence and brain evolution: how regulatory DNA influences development of the cerebral cortex. Bioessays. 2016;38:162–171. doi: 10.1002/bies.201500108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lein ES, Belgard TG, Hawrylycz M, Molnar Z. Transcriptomic perspectives on neocortical structure, development, evolution, and disease. Annu. Rev. Neurosci. 2017;40:629–652. doi: 10.1146/annurev-neuro-070815-013858. [DOI] [PubMed] [Google Scholar]
- 18.Darmanis S, et al. A survey of human brain transcriptome diversity at the single cell level. Proc. Natl. Acad. Sci. U.S.A. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lake BB, et al. Neuronal subtypes and diversity revealed by single-nucleus RNA sequencing of the human brain. Science. 2016;352:1586–1590. doi: 10.1126/science.aaf1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Howard TD, et al. Mutations in TWIST, a basic helix-loop-helix transcription factor, in Saethre-Chotzen syndrome. Nat. Genet. 1997;15:36–41. doi: 10.1038/ng0197-36. [DOI] [PubMed] [Google Scholar]
- 21.Huang AL, et al. The cells and logic for mammalian sour taste detection. Nature. 2006;442:934–938. doi: 10.1038/nature05084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell DB, et al. A genetic variant that disrupts MET transcription is associated with autism. Proc. Natl. Acad. Sci. U.S.A. 2006;103:16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoodbhoy T, Dean J. Insights into the molecular basis of sperm-egg recognition in mammals. Reproduction. 2004;127:417–422. doi: 10.1530/rep.1.00181. [DOI] [PubMed] [Google Scholar]
- 24.Boudreau RL, et al. Transcriptome-wide discovery of microRNA binding sites in human brain. Neuron. 2014;81:294–305. doi: 10.1016/j.neuron.2013.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brawand D, et al. The evolution of gene expression levels in mammalian organs. Nature. 2011;478:343–348. doi: 10.1038/nature10532. [DOI] [PubMed] [Google Scholar]
- 26.Vermunt MW, et al. Epigenomic annotation of gene regulatory alterations during evolution of the primate brain. Nat. Neurosci. 2016;19:494–503. doi: 10.1038/nn.4229. [DOI] [PubMed] [Google Scholar]
- 27.Berger B, Verney C, Gaspar P, Febvret A. Transient expression of tyrosine hydroxylase immunoreactivity in some neurons of the rat neocortex during postnatal development. Brain Res. 1985;355:141–144. doi: 10.1016/0165-3806(85)90013-6. [DOI] [PubMed] [Google Scholar]
- 28.Raghanti MA, et al. Species-specific distributions of tyrosine hydroxylase-immunoreactive neurons in the prefrontal cortex of anthropoid primates. Neuroscience. 2009;158:1551–1559. doi: 10.1016/j.neuroscience.2008.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Benavides-Piccione R, DeFelipe J. Distribution of neurons expressing tyrosine hydroxylase in the human cerebral cortex. J. Anat. 2007;211:212–222. doi: 10.1111/j.1469-7580.2007.00760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Flames N, Hobert O. Gene regulatory logic of dopamine neuron differentiation. Nature. 2009;458:885–889. doi: 10.1038/nature07929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaspar P, Berger B, Febvret A, Vigny A, Henry JP. Catecholamine innervation of the human cerebral-cortex as revealed by comparative immunohistochemistry of tyrosine-hydroxylase and dopamine-beta-hydroxylase. J. Comp. Neurol. 1989;279:249–271. doi: 10.1002/cne.902790208. [DOI] [PubMed] [Google Scholar]
- 32.Hansen DV, Lui JH, Parker PRL, Kriegstein AR. Neurogenic radial glia in the outer subventricular zone of human neocortex. Nature. 2010;464:554–561. doi: 10.1038/nature08845. [DOI] [PubMed] [Google Scholar]
- 33.Ma T, et al. Subcortical origins of human and monkey neocortical interneurons. Nat. Neurosci. 2013;16:1588–1597. doi: 10.1038/nn.3536. [DOI] [PubMed] [Google Scholar]
- 34.Kepecs A, Fishell G. Interneuron cell types are fit to function. Nature. 2014;505:318–326. doi: 10.1038/nature12983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Close JL, et al. Single-cell profiling of an in vitro model of human interneuron development reveals temporal dynamics of cell type production and maturation. Neuron. 2017;93:1035–1048. doi: 10.1016/j.neuron.2017.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dehorter N, et al. Tuning of fast-spiking interneuron properties by an activity-dependent transcriptional switch. Science. 2015;349:1216–1220. doi: 10.1126/science.aab3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dulcis D, Jamshidi P, Leutgeb S, Spitzer NC. Neurotransmitter switching in the adult brain regulates behavior. Science. 2013;340:449–453. doi: 10.1126/science.1234152. [DOI] [PubMed] [Google Scholar]
- 38.Sanai N, et al. Corridors of migrating neurons in the human brain and their decline during infancy. Nature. 2011;478:382–386. doi: 10.1038/nature10487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fukuda T, Takahashi J, Tanaka J. Tyrosine hydroxylase-immunoreactive neurons are decreased in number in the cerebral cortex of Parkinson's disease. Neuropathology. 1999;19:10–13. doi: 10.1046/j.1440-1789.1999.00196.x. [DOI] [PubMed] [Google Scholar]
- 40.Marui W, Iseki E, Kato M, Kosaka K. Degeneration of tyrosine hydroxylase-immunoreactive neurons in the cerebral cortex and hippocampus of patients with dementia with Lewy bodies. Neurosci. Lett. 2003;340:185–188. doi: 10.1016/s0304-3940(03)00101-0. [DOI] [PubMed] [Google Scholar]
- 41.USDA. 7 USC, Title 7-AGRICULTURE. CHAPTER 54 - TRANSPORTATION, SALE, AND HANDLING OF CERTAIN ANIMALS. Washington, D.C.: U.S. Goverment Printing Office; 2013. Animal Welfare Act and Animal Welfare Regulations. http://awic.nal.usda.gov/government-and-professional-resources/federal-laws/animal-welfare-act. [Google Scholar]
- 42.NIH/OLAW. U.S. Government Principles for the Utilization and Care of Vertebrate Animals Used in Testing, Research, and Training. 2002 http://grants.nih.gov/grants/olaw/references/phspol.htm#PublicHealthServicePolicyonHumaneCareandUseofLaboratory.
- 43.National Research Council. Guide for the Care and Use of Laboratory Animals. 8. The National Academies Press; 2011. [PubMed] [Google Scholar]
- 44.DeLucchi MR, Dennis BJ, Adey WR. A stereotaxic atlas of the chimpanzee brain (Pan satyrus) University of California Press; Berkeley and Los Angeles: 1965. [Google Scholar]
- 45.Saleem KS, Logothetis N. A combined MRI and histology atlas of the rhesus monkey brain in stereotaxic coordinates. Academic Press; San Diego: 2007. [Google Scholar]
- 46.Jiang L, et al. Synthetic spike-in standards for RNA-seq experiments. Genome Res. 2011;22:1543–1551. doi: 10.1101/gr.121095.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu Y, Li M, Sousa AMM, Sestan N. XSAnno: A framework for annotating orthologs in cross-species transcriptome comparisons. BMC Genomics. 2014;15:343. doi: 10.1186/1471-2164-15-343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrow J, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012;22:1760–1774. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hinrichs AS, et al. The UCSC Genome Browser Database: update 2006. Nucl. Acids Res. 2006;34:D590–D598. doi: 10.1093/nar/gkj144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Habegger L, et al. RSEQtools: a modular framework to analyze RNA-Seq data using compact, anonymized data summaries. Bioinformatics. 2011;27:281–283. doi: 10.1093/bioinformatics/btq643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nature Meth. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 53.Team RC. R: A language and environment for statistical computing. R Foundation for Statistical Computing; 2013. [Google Scholar]
- 54.Johnson WE, Li C, Rabinovic A. Adjusting batch effects in microarray expression data using empirical Bayes methods. Biostatistics. 2007;8:118–127. doi: 10.1093/biostatistics/kxj037. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Y, et al. Purification and characterization of progenitor and mature human astrocytes reveals transcriptional and functional differences with mouse. Neuron. 2016;89:37–53. doi: 10.1016/j.neuron.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suzuki R, Shimodaira H. Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
- 58.Langfelder P, Horvath S. WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics. 2008;9:559. doi: 10.1186/1471-2105-9-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zhang B, Horvath S. A general framework for weighted gene co-expression network analysis. Stat. Applic. Gen. Mol. Biol. 2005;4 doi: 10.2202/1544-6115.1128. Article17. [DOI] [PubMed] [Google Scholar]
- 60.Langfelder P, Luo R, Oldham MC, Horvath S. Is my network module preserved and reproducible? PLoS Comput. Biol. 2011;7:e1001057. doi: 10.1371/journal.pcbi.1001057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cunningham F, et al. Ensembl 2015. Nucl. Acids Res. 2015;43:D662–669. doi: 10.1093/nar/gku1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Smedley D, et al. The BioMart community portal: an innovative alternative to large, centralized data repositories. Nucl. Acids Res. 2015;43:W589–598. doi: 10.1093/nar/gkv350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Baev V, Daskalova E, Minkov I. Computational identification of novel microRNA homologs in the chimpanzee genome. Comput. Biol. Chem. 2009;33:62–70. doi: 10.1016/j.compbiolchem.2008.07.024. [DOI] [PubMed] [Google Scholar]
- 64.Yue J, Sheng Y, Orwig KE. Identification of novel homologous microRNA genes in the rhesus macaque genome. BMC Genomics. 2008;9:8. doi: 10.1186/1471-2164-9-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hu HY, et al. MicroRNA expression and regulation in human, chimpanzee, and macaque brains. PLoS Genet. 2011;7:e1002327. doi: 10.1371/journal.pgen.1002327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hackenberg M, Rodríguez-Ezpeleta N, Aransay AM. miRanalyzer: an update on the detection and analysis of microRNAs in high-throughput sequencing experiments. Nucl. Acids Res. 2011;39:W132–W138. doi: 10.1093/nar/gkr247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hofacker IL. Vienna RNA secondary structure server. Nucl. Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gentleman RC, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Li H, et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Okita K, et al. An Efficient Nonviral Method to Generate Integration-Free Human-Induced Pluripotent Stem Cells from Cord Blood and Peripheral Blood Cells. Stem Cells. 2013;31:458–466. doi: 10.1002/stem.1293. [DOI] [PubMed] [Google Scholar]
- 74.Chambers SM, et al. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nature Biotechnol. 2009;27:275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Onorati M, et al. Molecular and functional definition of the developing human striatum. Nat. Neurosci. 2014;17:1804–1815. doi: 10.1038/nn.3860. [DOI] [PubMed] [Google Scholar]
- 76.Stutz B, et al. Murine dopaminergic Müller cells restore motor function in a model of Parkinson's disease. J. Neurochem. 2014;128:829–840. doi: 10.1111/jnc.12475. [DOI] [PubMed] [Google Scholar]
- 77.Prado-Martinez J, et al. Great ape genetic diversity and population history. Nature. 2013;499:471–475. doi: 10.1038/nature12228. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.