Abstract
The mammalian basal forebrain (BF) plays key roles in controlling sleep and wakefulness, but the underlying neural circuit remains poorly understood. Here we delineate the BF circuit by recording and optogenetically perturbing the activity of four genetically defined cell types across sleep-wake cycles and by comprehensively mapping their synaptic connections. Recordings from channelrhodopsin-2 (ChR2)-tagged neurons showed that three BF cell types - cholinergic, glutamatergic, and parvalbumin-positive (PV+) GABAergic neurons - were more active during wakefulness and rapid eye movement (REM) sleep than non-REM (NREM) sleep (wake/REM-active), and activation of each cell type rapidly induced wakefulness. By contrast, activation of somatostatin-positive (SOM+) GABAergic neurons promoted NREM sleep, although only some of them were NREM-active. Synaptically, the wake-promoting neurons were organized hierarchically by glutamatergic → cholinergic → PV+ neuron excitatory connections, and they all received inhibition from SOM+ neurons. Together, these findings reveal the basic organization of BF circuit for sleep-wake control.
The sleep-wake cycle is a fundamental biological process observed throughout the animal kingdom1, and its disruption causes a variety of detrimental effects. Following the landmark studies by Von Economo2 and Nauta3, multiple brain regions involved in sleep-wake control have been identified, including the brainstem, hypothalamus, and the BF4–7. The neuronal activity in these regions changes between brain states8–14, and their roles in sleep-wake regulation have been demonstrated using lesion, electrical stimulation, and pharmacological manipulations2, 3, 7, 8, 15–23. In particular, lesion or inactivation of the BF was found to increase delta electroencephalogram (EEG) activity and decrease behavioral arousal in some studies16, 20, 21, but to reduce sleep in others8, 15, 18. These studies suggest that the BF is crucial for both sleep and wakefulness, but it is unclear which BF neurons promote each brain state, and how they interact with each other.
There are three major neuronal types in the BF: cholinergic, glutamatergic, and GABAergic. The cholinergic neurons are known to be active during both wakefulness and REM sleep but silent during NREM sleep24, and their activation enhances arousal, attention, and memory23, 25–29. The glutamatergic and GABAergic neurons are also likely to play key roles in sleep-wake control21. However, their functional properties are much less understood. Furthermore, anatomical studies have shown that both cholinergic and non-cholinergic neurons make extensive synaptic contacts within the BF30, but little is known about the rules of connectivity between cell types and how they contribute to sleep-wake control. In this study, we used optogenetic methods to characterize the functional properties of individual BF cell types in vivo and to map their synaptic connections in brain slices, thus providing the first comprehensive BF circuit diagram for sleep-wake control.
RESULTS
Genetically defined cell types
To target different cell types for recording and optogenetic activation, we used choline acetyltransferase (ChAT)-Cre, vesicular glutamate transporter 2 (VGLUT2)-Cre, PV-Cre and SOM-Cre mice for cell-type-specific expression of fluorescent proteins and ChR2. These mouse lines were chosen because ChAT is a reliable, widely used marker for cholinergic neurons, and among VGLUT1, 2, and 3 (specific markers for glutamatergic neurons) VGLUT2 is the predominant marker in the BF31 (in situ hybridization data are available in Allen Mouse Brain Atlas, Allen Institute for Brain Science: http://mouse.brain-map.org/experiment/show/70436317, http://mouse.brain-map.org/experiment/show/73818754, and http://mouse.brain-map.org/experiment/show/71587918 for VGLUT1, 2 and 3, respectively). The GABAergic neurons in the BF are known to be functionally diverse13, and PV and SOM have been used as markers for different subtypes of GABAergic neurons in both the BF11, 30 and other brain regions32.
To visualize these genetically defined cell types, we crossed each Cre driver mouse with a tdTomato reporter mouse (Ai14)33. Comparison of tdTomato expression with immunohistochemical staining or in situ hybridization of the four markers showed high specificity of labeling in each Cre line and low overlap between neurons expressing different markers (Fig. 1; Supplementary Table 1; in most cases the overlap was < 1%, with the exception of PV+ neurons: ~5% of tdTomato-labeled neurons in PV-Cre mice also expressed VGLUT2, and ~10% immunohistochemically identified PV+ neurons were labeled by tdTomato in SOM-Cre mice). Thus, ChAT+, VGLUT2+, PV+ and SOM+ neurons constitute largely distinct BF populations.
Figure 1.
Genetically defined BF cell types. (a) Fluorescence image of BF (box in coronal diagram) showing tdTomato-expressing cholinergic neurons in ChAT-Cre × Ai14 mouse. (b) Fluorescence images of tdTomato (red) and immunohistochemical (IHC) staining or in situ hybridization for ChAT, VGLUT2, PV, and SOM (green) in ChAT-Cre × Ai14 mouse. (c–e) Similar to b, but for VGLUT2-Cre (c), PV-Cre (d) and SOM-Cre (e) mice crossed with Ai14 mouse. Red, green, and yellow arrows indicate cells that are tdTomato+, immunostaining/in situ hybridization+, and both, respectively. Scale bar, 20 μm.
Sleep-wake activity of different cell types
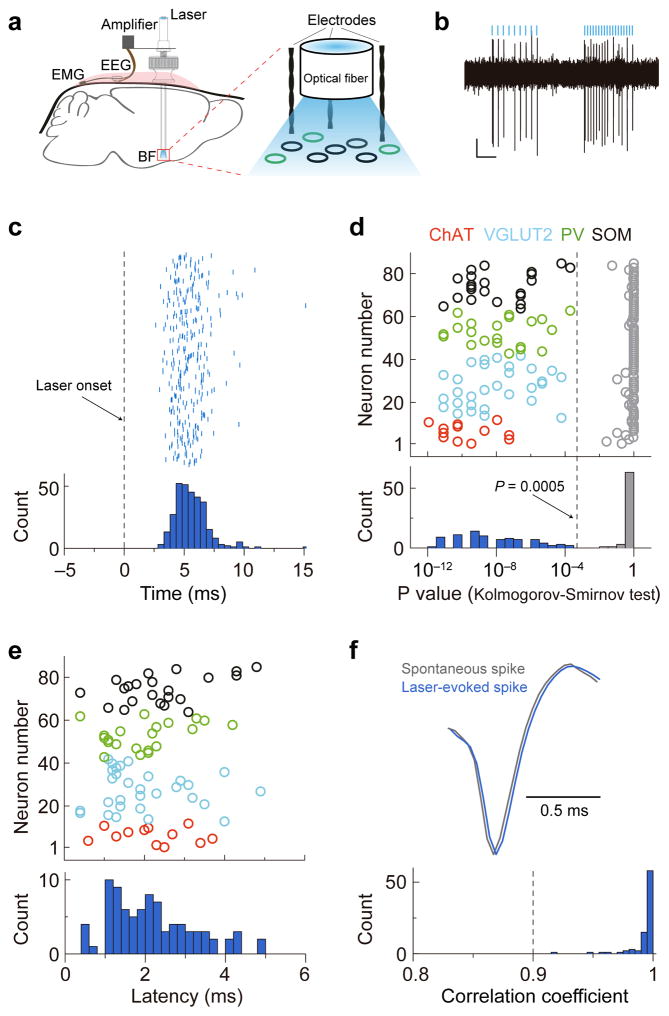
To measure the firing rates of each cell type across natural sleep-wake cycles, we tagged the neurons with ChR2 by crossing the respective Cre driver mouse with a ChR2 reporter mouse (Ai32)34 or by injecting Cre-inducible adeno-associated virus (AAV) expressing ChR2 into BF of the Cre mouse. Recordings were made in freely moving mice using optrodes, consisting of an optic fiber surrounded by several stereotrodes (Fig. 2a). High-frequency laser pulse trains (16 or 33 Hz, 5 ms/pulse, Fig. 2b) were applied intermittently, and single units exhibiting reliable laser-evoked spiking at short latencies were identified as ChAT+, VGLUT2+, PV+, or SOM+ neurons in the respective mouse line35 (see Methods, Fig. 2c–f).
Figure 2.
Identification of BF cell types using ChR2 tagging and optrode recording. (a) Schematic of optrode recording in freely moving mice. (b) Example recording of laser-evoked spiking in ChAT-ChR2 mice. Blue lines, laser pulses (5 ms, 16 and 33Hz). Scale bars, 50 μV, 200 ms. (c) Raster plot and peri-stimulus time histogram (PSTH, bin size, 1 ms) for laser-evoked spikes of an example ChR2-tagged neuron. Dashed line, laser onset time. (d) Distribution of P values of Kolmogorov-Smirnov (K-S) test for all 85 optogenetically identified BF neurons. Colored circle, K-S test of spike timing distribution before and after laser onset for each neuron (cell types are color coded). Gray circles, K-S test after random shuffling of laser onset time. Dashed line, P = 0.0005. (e) Distribution of latency of laser-evoked spiking for the 85 neurons. The latency was measured by change point analysis (see Methods). (f) Waveform comparison between laser-evoked and spontaneous spikes. Upper panel, averaged waveforms of laser-evoked spikes (blue) and spontaneous spikes (gray) of an example neuron. Lower panel, distribution of correlation coefficient between laser-evoked and spontaneous spike waveforms for all 85 optogenetically identified BF neurons.
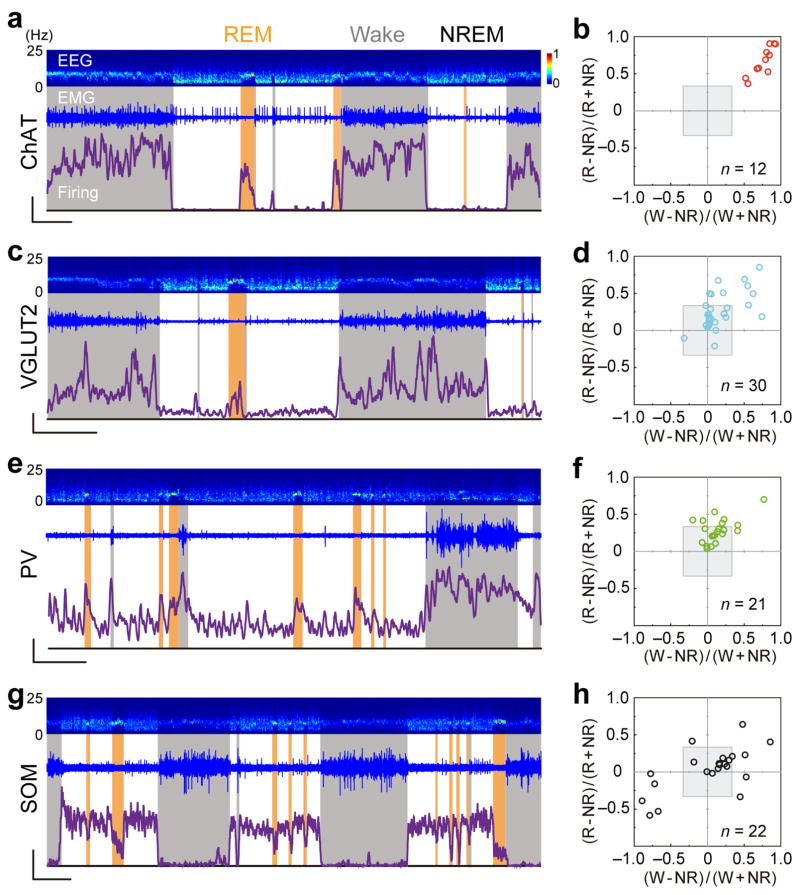
We recorded from 85 identified neuron, each for 36–136 (median 105) min, encompassing multiple cycles of wake, NREM, and REM states classified by EEG and electromyogram (EMG) recordings (see Methods, Fig. 3a). Cholinergic neurons typically fired a few spikes per second during wakefulness (3.9 ± 0.6 spikes/s, s.e.m.) and REM sleep (2.7 ± 0.4 spikes/s) but at much lower rates during NREM sleep (0.4 ± 0.1 spikes/s; Supplementary Movie 1, Supplementary Fig. 1a), consistent with previous studies using juxtacellular recording from head-fixed rats24. To quantify the relative firing rates of each neuron in different brain states, we plotted its REM-NREM modulation [(RREM− RNREM)/(RREM+ RNREM), where R is the mean firing rate within each state] vs. wake-NREM modulation [(Rwake− RNREM)/(Rwake+ RNREM)] (Fig. 3b). The gray shading indicates < 2 fold firing rate change (|modulation| < 0.33), which was considered ‘state indifferent’ by previous investigators8. All the identified cholinergic neurons (n = 12) fell into quadrant I (top-right) outside of the gray shading, indicating that they were strongly modulated, wake/REM-active neurons.
Figure 3.
Firing rates of identified BF cell types across natural sleep-wake cycles. (a) Firing rates of an example ChAT+ neuron over 108 min. Top panel, EEG power spectrogram (0–25 Hz). Middle panel, EMG trace. Bottom panel, firing rate of the ChAT+ neuron. Scale bars, 3 spikes/s, 500 s. Brain states are color coded (wake, gray; REM, orange; NREM, white). (b) Summary of firing rate modulation of 12 ChAT+ neurons (from 6 mice). Gray shading, <2 fold firing rate change between brain states. Note that in this 2D plot, firing rates are compared explicitly between wake and NREM and between REM and NREM, but not between wake and REM states. Neurons that are maximally active during REM sleep are located in the upper left quadrant and part of the upper right quadrant (closer to the vertical than horizontal axis). (c, d), Similar to a & b, for VGLUT2+ neurons (6 mice). (e, f), PV+ neurons (5 mice). (g, h), SOM+ neurons (5 mice). Scale bars (c, e, g), 10 spikes/s, 500 s.
We then measured the firing rates of non-cholinergic cell types, which were much less characterized in previous studies. Of the 30 glutamatergic neurons identified in VGLUT2-ChR2 mice, the vast majority also fell into quadrant I (Fig. 3c, d). The few neurons found in other quadrants (4/30) were all within the gray-shaded area, indicating weak firing rate modulation. Thus, glutamatergic BF neurons were also primarily wake/REM-active. Unlike the ChAT+ neurons, however, many VGLUT2+ neurons were only weakly modulated across brain states (20/30 in gray-shaded area). The firing rates of the VGLUT2+ neurons (wake, 19.5 ± 3.2 spikes/s; NREM, 15.9 ± 3.0; REM, 19.8 ± 3.2, s.e.m.; Supplementary Fig. 1b) were also much higher than the cholinergic neurons (P < 0.003 for all three states, t-test).
The PV+ GABAergic neurons exhibited even higher firing rates than glutamatergic neurons across all brain states (Supplementary Fig. 1c; wake, 30.7 ± 4.3 spikes/s; NREM, 25.2 ± 4.2; REM, 33.9 ± 4.6, s.e.m., P < 0.04, t-test). Most of them were weakly modulated (Fig. 3e, f), with the majority found in quadrant I (wake/REM-active) and a few in quadrant II (REM-active), consistent with a previous finding in anesthetized rats that PV+ neurons were more active during desynchronized EEG11.
Recording from the SOM+ GABAergic neurons, we observed a number of strongly modulated NREM-active units (Fig. 3g, h, Supplementary Fig. 1d, e). Apart from neurons within the gray shading (11/22), 5/11 strongly modulated neurons were highly active during NREM sleep (10 ± 6.4 spikes/s, s.e.m.) with low firing rates during wake (1.6 ± 1.3 spikes/s) and REM (3.8 ± 1.9 spikes/s) periods (Supplementary Fig. 1d, red lines). Such a firing pattern was not observed in any of the cholinergic, glutamatergic, or PV+ GABAergic BF neurons.
Effects of BF neuron activation on sleep-wake states
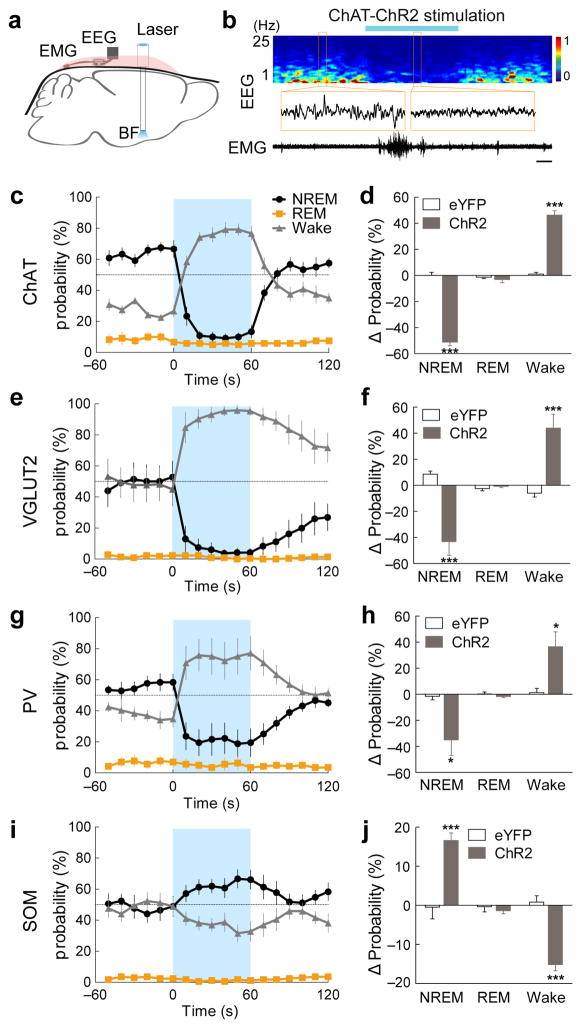
We next tested the causal role of each cell type in sleep-wake control using ChR2-mediated activation. To ensure localized ChR2 expression, we injected Cre-inducible AAV expressing ChR2-eYFP into the BF of each Cre mouse (see Methods, Supplementary Fig. 2a). Three weeks after injection, laser stimulation was applied through optic fibers implanted bilaterally into the BF (4–8 mW, 10 ms/pulse, 10 Hz, 60 s/trial, Fig. 4a).
Figure 4.
Effects of BF neuron activation on sleep-wake states. (a) Schematic of optogenetic stimulation experiment. (b) An example trial of ChAT+ neuron activation. Shown are EEG power spectrum, EEG traces during selected periods (indicated by boxes) and EMG trace during the whole trial. Blue bar, period of laser stimulation (10 ms pulses, 10 Hz, 60 s). Scale bar, 10 s. (c) Probability of wake, NREM or REM states before, during, and after laser stimulation of ChAT+ neurons (n = 5 mice). Error bar, ± s.e.m. Blue shading, period of laser stimulation. (d) Laser-induced change in the probability of each state (difference between the 60s periods before and during laser stimulation) in ChAT-ChR2 (filled bar) and ChAT-eYFP (open bar, n = 3) mice. (e, f) Similar to (c, d), for VGLUT2-ChR2 (n = 6) and VGLUT2-eYFP (n = 5) mice. (g, h) PV-ChR2 (n = 6) and PV-eYFP (n = 4) mice. (i, j) SOM-ChR2 (n = 7) and SOM-eYFP (n = 4) mice. The number of trials per mouse was 24–36. *P ≤ 0.05, ***P ≤ 0.001 (difference between ChR2 and eYFP mice, two-way ANOVA followed by Bonferroni post-hoc test).
We found that activation of cholinergic BF neurons induced a rapid desynchronization of the EEG and an increase of EMG power (Fig. 4b). Compared to the baseline period, laser stimulation caused a significant increase in wakefulness (Fig. 4c, P = 1.6×10−4, paired t-test) and decrease in NREM sleep (P = 4.8×10−5) both by increasing the NREM → wake transition and by maintaining the wake state (Supplementary Fig. 3a). In control mice expressing eYFP without ChR2, laser stimulation had no effect (Fig. 4d, P = 0.95, 0.51 and 0.35 for wake, NREM, and REM, respectively), and the laser-induced increase in wakefulness was significantly greater in ChR2 than eYFP mice (P = 6.4×10−5, t-test). Such a wake-promoting effect is consistent with previous findings based on cell-type-specific lesion20 or pharmacological manipulation19.
For glutamatergic neurons, optogenetic activation also induced an immediate transition from NREM sleep to wakefulness (Fig. 4e, f, Supplementary Movie 2; Supplementary Fig. 3b, P = 0.009, paired t-test). During laser stimulation, the probability of wakefulness approached 100%, attesting to the high efficiency of glutamatergic neurons in inducing wakefulness. Activation of PV+ GABAergic neurons also caused a significant increase of wakefulness (P = 0.02, paired t-test) and decrease of NREM sleep (P = 0.03, Fig. 4g, h, Supplementary Fig. 3c), although the efficacy of these neurons appeared lower.
Activation of the SOM+ neurons, in contrast, caused a significant increase of NREM sleep (P = 0.00014, paired t-test) and decrease of wakefulness (P = 6×10−5, Fig. 4i, j, Supplementary Fig. 3d). Thus, among the four BF cell types tested, SOM+ neurons were unique in their NREM-promoting effect, mirroring the finding that NREM-active neurons were only found in the SOM+ population (Fig. 3h).
Local connectivity among BF cell types
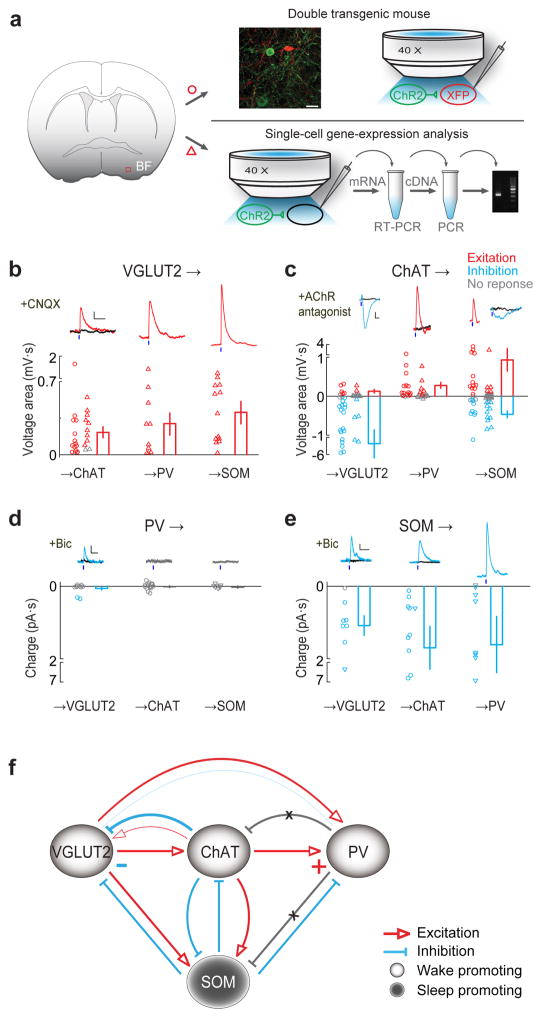
Ultrastructural studies have revealed numerous local synaptic contacts among BF neurons30, which are likely to play important roles in sleep-wake regulation. We next mapped the local connectivity between the four BF cell types. ChR2 was expressed in the presynaptic cell type, and the postsynaptic cell type was either labeled with tdTomato or eGFP or identified using single cell RT-PCR (Fig. 5a, see Methods). For example, to test whether glutamatergic BF neurons innervate cholinergic neurons, we crossed the VGLUT2-Cre mouse with the ChAT-eGFP mouse, and injected Cre-inducible AAV expressing ChR2-mCherry into the BF (see Supplementary Table 2 for other pre- and postsynaptic pairs). We then made whole-cell recordings from the eGFP-labeled postsynaptic cells in acute BF slices while activating the ChR2-expressing presynaptic neurons with blue light (5 ms).
Figure 5.
Local connectivity of BF cell types. (a) Schematic of slice experiment using two strategies. The first strategy (upper right) is to use double transgenic mice. Shown are fluorescence image of a small BF area (red box in coronal diagram) showing ChR2-eYFP-expressing ChAT+ neurons (green) and tdTomato-expressing PV+ neurons (red) in an example experiment. Scale bar, 30 μm. Blue light was used to activate ChR2-expressing (presynaptic) neurons and whole-cell recordings were made from fluorescently labeled postsynaptic neurons. The second strategy (lower right) requires single-cell gene-expression analysis. Recordings were made from unlabeled neurons and the cell type is identified using RT-PCR. (b–e) Synaptic interactions between multiple pairs of pre- and postsynaptic cell types. (b) VGLUT2+ to ChAT+, PV+ and SOM+ neuron connections. Top, example light-evoked excitatory responses (red) recorded under current clamp, blocked by AMPA receptor antagonist CNQX (10 μM, black traces). Short blue bar, light pulse (5 ms). Scale bars, 1 mV, 200 ms. Bottom, population summary of input strength (measured by voltage area, integral of EPSP), each circle (using double transgenic mice) or triangle (based on single-cell gene-expression analysis, performed in the presence of mAChR, nAChR antagonists) represents one cell. Red, significant excitatory response (P < 0.05, t-test); Gray, no significant response. Error bar, ± s.e.m. (c) Similar to b, for ChAT+ to VGLUT2+, PV+ and SOM+ connections (recorded under current clamp). Blue, significant inhibitory response (P < 0.05); black, after application of AChR antagonists. ChAT→VGLUT2 excitatory response was blocked by nAChR antagonists MLA (methyllycaconitine, α7-containing nAChR antagonist, 5 nM) and DhβE (dihydro-β-erythroidine, non-α7 nAChR antagonist, 500 nM), inhibitory response blocked by mAChR antagonist scopolamine (20 μM). ChAT→PV response was blocked by MLA + DhβE. ChAT→SOM excitatory responses were blocked by MLA, DhβE and scopolamine, and inhibitory responses were blocked by scopolamine. Scale bars, 1 mV, 200 ms. All experiments indicated by triangle were performed in the presence of glutamate and GABA receptor antagonists. (d) PV+ to VGLUT2+, ChAT+ and SOM+ connections (voltage clamp). Among all recorded neurons (VGLUT2+, n = 11; ChAT+, n = 17; SOM+, n = 8), inhibitory responses were detected only in two VGLUT2+ neurons, which were blocked by GABAA receptor antagonist bicuculline (‘bic’, 20 μM, black trace). Scale bars, 10 pA, 50 ms. (e) SOM+ to VGLUT2+, ChAT+ and PV+ connections (voltage clamp). All inhibitory responses (blue) were blocked by bicuculline (black). Scale bars, 10 pA, 50 ms. (f) Diagram of BF local circuit. Light circles, wake-promoting neurons. Dark circle, sleep-promoting SOM+ neurons (containing both wake/REM-active and NREM-active neurons). Excitatory and inhibitory connections are indicated by red and blue lines, respectively. Gray line with cross indicates tested connection with no detectable response. Connection strength is represented qualitatively by line thickness.
As summarized in Fig. 5b, we detected extensive synaptic interactions among the BF cell types. Glutamatergic neurons excited cholinergic and both PV+ and SOM+ GABAergic neurons (Fig. 5b). Interestingly, cholinergic neurons excited PV+ neurons primarily through nicotinic ACh receptors (nAChRs), but provided strong inhibition to glutamatergic neurons through muscarinic AChRs (with very weak excitation, Fig. 5c, Supplementary Fig. 4a). Their innervations of SOM+ neurons were heterogeneous, consisting of excitatory and/or inhibitory inputs with a wide range of excitation/inhibition ratios (Supplementary Fig. 4b), likely related to the functional diversity of SOM+ neurons observed in vivo (Fig. 3h). PV+ neurons only weakly inhibited glutamatergic neurons, and no input was detected in cholinergic or SOM+ neurons (Fig. 5d), even though the light pulse activated PV+ neurons reliably, and spontaneous inhibitory currents were frequently observed in cholinergic neurons (Supplementary Fig. 4c). By contrast, SOM+ neurons provided strong GABAA-mediated inhibition to glutamatergic, cholinergic, and PV+ neurons (Fig. 5e).
Based on the functional characterization of each BF cell type (Figs. 3 and 4) and the connectivity mapping experiments (Fig. 5), we constructed a simple circuit diagram (Fig. 5f, the relative strengths of different connections are represented qualitatively by line thickness). The three wake-promoting cell types (light circles) are organized in a hierarchical chain of excitatory connections (glutamatergic → cholinergic → PV+ neurons), with the feedback connections either absent (PV+ → cholinergic) or primarily inhibitory (cholinergic → glutamatergic, PV+ → glutamatergic). Importantly, the sleep-promoting SOM+ neuron population (dark circle) provides inhibition to all three wake-promoting cell types.
DISCUSSION
The BF is known to be important for both sleep and wakefulness8, 15, 16, 18, 20, 21, and it contains spatially intermingled wake- and sleep-active neurons8–10, 12–14, 36. By recording and manipulating the activity of ChR2-expressing neurons in various Cre mice, we have demonstrated the functional distinction between genetically defined cell types, especially for non-cholinergic neurons. This is a key step in dissecting the BF circuit for sleep-wake control.
The circuit mapping experiments suggest that the wake/REM-active property of cholinergic and PV+ neurons partly originates from the local glutamatergic input (Fig. 5b). Compared to the cholinergic neurons with well characterized functions in wakefulness and arousal16, 23, 25, 27–29, we found that BF glutamatergic neurons showed an even stronger wake-promoting effect (Fig. 4e, Supplementary Movie 2). In addition to the larger number of light-activated VGLUT2+ neurons as compared to ChAT+ and PV+ neurons (Supplementary Fig. 2b), this strong effect is also consistent with the BF circuit diagram (Fig. 5f): optogenetic activation of the glutamatergic neurons can excite both cholinergic and PV+ GABAergic neurons, each of which promoted wakefulness (Fig. 4c–h). In fact, the relative efficacy of glutamatergic, cholinergic, and PV+ GABAergic neurons in inducing wakefulness is consistent with their positions in the hierarchical chain of excitation (Fig. 5f).
Notably, while optogenetic activation of BF cholinergic neurons caused a significant increase in wakefulness, we found no significant change in REM sleep (P = 0.28, paired t-test; Fig. 4d). This is similar to the finding in one recent study37, but different from another study using optogenetic activation38. In addition to differences in the stimulation protocol, there may be differences in the brain-state classification criteria. In our experiment, although laser stimulation in some trials induced EEG desynchronization without EMG change (Supplementary Fig. 5b), the EEG power spectrum in these trials was different from that in natural REM sleep (Supplementary Fig. 5c); thus the brain state was left unclassified in our analysis (Supplementary Fig. 5d).
We found that activation of PV+ neurons in the BF evoked no detectable response in cholinergic or SOM+ neurons and only weak responses in glutamatergic neurons (Fig. 5d), which may be partly related to the relatively small number of PV+ neurons in the BF. On the other hand, their activation in vivo caused a significant increase in wakefulness, which may be mediated by long-range projections to outside of the BF. For example, in the cortex, axons of BF PV+ neurons were shown to innervate GABAergic interneurons39 and may thus disinhibit pyramidal neurons. In a recent study, optogenetic stimulation of BF PV+ neurons was also found to enhance gamma oscillations in the cortex40. Although in the current study we did not observe any prominent, specific increase in gamma band oscillation in the EEG, the difference may be due to the different experimental protocols used in the two studies (e.g., since the focus of our study is not on gamma oscillation, we did not use 40 Hz stimulation to test whether it is more effective than other frequencies in entraining cortical oscillations). In addition to the PV+ neurons, other BF cell types also send extensive long-range projections41. For example, cholinergic transmission in the cortex by BF ChAT+ neurons can cause rapid EEG desynchronization and enhanced behavioral performance27–29, 42.
Compared to the wake-promoting network, the circuits promoting sleep are much less understood5. Sleep-active neurons have been observed in the BF and several subregions of the hypothalamus43. In particular, the ventrolateral preoptic area (VLPO) has been shown to be a key region promoting NREM sleep, as cFos staining and electrophysiological recording revealed a high density of sleep-active neurons in VLPO44, 45, and lesion of VLPO significantly reduced NREM sleep46. Here we found that optogenetic activation of BF SOM+ neurons rapidly increased the probability of NREM sleep (Fig. 4i, j), and some of these neurons were strongly NREM active (Fig. 3g, h). Whereas the GABAergic neurons in VLPO inhibit the monoaminergic neurons in the ascending arousal pathway47, 48, SOM+ neurons in the BF inhibit all three types of wake-promoting neurons (Fig. 5e). Thus, our results reveal a new pathway that promotes NREM sleep via broad inhibition of multiple wake-promoting cell types within the BF local circuit. Given the functional diversity of SOM+ neurons, it would be important for future studies to further divide them into different subgroups using additional molecular markers. In addition, as more effective optogenetic tools for neuronal silencing are developed, it would be important to test the effect of inactivating each cell type on brain states.
Complete understanding of sleep-wake control mechanisms requires elucidation of the functional roles of different cell types in relevant brain regions as well as the synaptic connections between them. Our study illustrates the power of combining optogenetic manipulation with in vivo and in vitro electrophysiology in mapping the activity, causality, and synaptic connectivity of specific cell types in sleep-wake control.
Methods
Virus preparation
AAV2-EF1α-FLEX-ChR2-eYFP and AAV2-EF1α-FLEX-eYFP were produced by the University of North Carolina (UNC) Vector Core. The titer was estimated to be ~1012 gc/mL. AAV-DJ-EF1α-FLEX-ChR2-mCherry and AAV-DJ-EF1α-FLEX-eNpHR3.0-eYFP was produced by Neuroscience Gene Vector and Virus Core of Stanford University. The titer was estimated to be ~1013 gc/mL.
Animals
All experimental procedures were approved by the Animal Care and Use Committee at the University of California, Berkeley. In vivo optogenetic manipulation and recording experiments were performed on adult mice (include both genders, singly housed, naive before experiments, > P40, body weight 25–35g). Slice recording were performed in young mice (> 3 weeks).
The following mouse lines were used in the current study: Mice from Jackson Laboratory (JAX# in parenthesis): ChAT-ChR2(H134R)-eYFP (014546); ChAT-IRES-Cre (006410); VGLUT2-IRES-Cre (016963); PV-IRES-Cre (008069); SOM-IRES-Cre (013044); ChAT-eGFP (007902); Ai14 tdTomato reporter (007914); Ai32 ChR2-eYFP reporter (012569); Mice from MMRRC: VGLUT2-eGFP (MMRRC#011835-UCD).
Surgery
To implant electroencephalogram (EEG) and electromyogram (EMG) recording electrodes, mice were anesthetized with 1.5–2% isoflurane. Two stainless steel screws were inserted into the skull 1.5 mm from midline and 1.5 mm anterior to the bregma, and two others were inserted 3 mm from midline and 3.5 mm posterior to the bregma. Two EMG electrodes were inserted into the neck musculature. Insulated leads from the EEG and EMG electrodes were soldered to a 2 × 3 pin header, which was secured to the skull using dental cement.
For all the experiments, we targeted the caudal portion of the BF (including the horizontal limb of the diagonal band of Broca, magnocellular preoptic nucleus, and substantia innominata) rather than the rostral nuclei (medial septum and the vertical limb of the diagonal band of Broca). For optogenetic activation experiments (Fig. 4), a craniotomy (~0.5 mm in diameter) was made 0.1 mm anterior to bregma and 1.2–1.5 mm from midline (in the same surgery as for EEG and EMG implant), and 1 μl AAV (0.5 μl per hemisphere) was injected bilaterally into the BF (5.2–5.4 mm from cortical surface). We then implanted optic fibers bilaterally into the BF. Dental cement was applied to cover the exposed skull completely and to secure the implants to the screws. After surgery, mice were allowed to recover for at least 2 weeks before experiments. For optrode recording experiments, a custom-made optrode (see below) was implanted unilaterally into the BF using similar procedure as described above.
Polysomnographic recordings and analysis
Animals were housed on a 12-h dark/12-h light cycle (light on between 7:00 and 19:00). All optogenetic manipulation experiments were performed between 11:00 and 15:00. Most of the optrode recording experiments were carried out between 9:00 and 19:00, but a few were performed till 23:00. EEG and EMG electrodes were connected to flexible recording cables via a mini-connector. The signals were recorded and amplified using AM-Systems amplifiers, filtered (0.1–1000 Hz or 10–1000 Hz for EEG and EMG recordings, respectively) and digitized at 600 Hz using LabView. Spectral analysis was carried out using fast Fourier transform (FFT) and NREM, REM and wake states were semi-automatically classified using a sleep analysis software (SleepSign for Animal, Kissei Comtec America) for each 10 s epoch (wake: desynchronized EEG and high EMG activity; NREM sleep: synchronized EEG with high power at 0.5–4 Hz and low EMG activity; REM sleep: desynchronized EEG with high power at theta frequencies (6–9 Hz) and low EMG activity).
Optogenetic manipulation
Since crossing the Ai32 reporter mouse with a given Cre mouse causes ChR2 expression in multiple brain regions containing the corresponding cell type, local laser stimulation within the BF may activate passing axons from non-BF neurons. To ensure localized ChR2 expression, we injected a Cre-inducible AAV expressing ChR2-eYFP into the BF of each Cre mouse for all the optogenetic activation experiments in Fig. 4.
Two fiber optic cables (200 μm diameter; ThorLabs) were each attached through an FC/PC adaptors to a 473 nm blue laser diode (Shanghai Laser & Optics Century Co. Ltd.), and light pulses were controlled by a Master 8 pulse stimulator (A.M.P.I.) that provided synchronous inputs to both lasers. The two fiber optic cables were connected to bilaterally implanted optic fibers 2 hrs before each experiment. During experiments, laser pulses (10 ms/pulse, 10 Hz, 4–8 mW, 60 s) were applied every 5 min. Each experimental session lasted for 2–3 hrs.
Optrode recording
To tag genetically defined BF neurons with ChR2, we used the following strategies: (1) transgenic mice expressing ChR2 in a specific neuronal type (ChAT-ChR2-eYFP), (2) crossing a Cre driver line with a ChR2 reporter line (PV-Cre × Ai32, VGLUT2-Cre × Ai32, SOM-Cre × Ai32), or (3) injecting Cre-inducible AAV expressing ChR2 into a Cre mouse (VGLUT2-Cre, SOM-Cre). To identify ChR2-expressing neurons in the BF of freely moving mice, custom-made optrodes (consisting of a 200 μm optical fiber and 6 pairs of stereotrodes) were used. The stereotrodes were made by twisting together two 25-μm diameter FeNiCr wires (Stablohm 675, California Fine Wire), and the wires were cut with sharp scissors and electroplated with platinum (H2PtCl6, Sigma-Aldrich) to an impedance of ~300 kΩ with a custom-built MSP430 MCU-controlled multi-channel plating device. The optrode assembly was mounted on a light-weight driver to allow vertical movement the optrode assembly to search for light-responsive neurons. Cortical EEG and EMG (see above) were also recorded for brain state classification. All signals were acquired using TDT system-3 workstation (RA16 pre-amplifier + RX5 base processor) controlled by OpenEx software (TDT). Extracellular signals were filter at 0.3–8 kHz and sampled at 25 kHz for offline spike detection and sorting. EEG and EMG were filtered at 0–300 Hz and sampled at 1.5 kHz. Recording location and virus expression were verified after recording though standard histology procedures.
To identify ChR2+ neurons, high-frequency laser pulse trains (0.5–8 mW, 473 nm) were delivered intermittently. In the majority of recording sessions, each trial consisted of three components that were 500 ms apart: (1) 5 ms pulse train at 16 Hz for 500 ms; (2) 5 ms pulse train at 33 Hz for 500 ms; (3) 200 ms step pulse. The stimulation was applied every 45 s throughout the recording to monitor unit stability. To analyze the firing rates of each neuron during different brain states, spikes recorded 0 – 4 s from onset of laser stimulation were excluded. For each cell type, the laser power was optimized to identify light-responsive neurons without changing the brain state. However, in VGLUT2-ChR2 mice, laser power sufficient for activating the recorded units often promoted wakefulness. In such cases, laser pulse trains were applied every 10 s during a period separate from the recording session.
To identify neurons with ChR2-mediated responses, we performed the following three analyses:
Distribution of spike timing
To determine whether each laser pulse caused a significant change in spike timing, we computed the PSTH triggered on each laser pulse (0.5 ms bin, from 15 ms before to 15 ms after each pulse, Fig. 2c), and used Kolmogorov–Smirnov (K-S) test to compare the spike timing distributions before and after the pulse. Laser-responsive neurons were identified at P < 0.0005 (Fig. 2d). To validate the statistical method used here, we performed the same procedure after random shuffling the timing of laser pulses and found that the P value was typically > 0.5 (Fig. 2d, gray). Thus the units satisfying our statistical criterion showed highly significant laser-evoked responses.
Latency
To assess whether these units were driven directly through ChR2 or indirectly through synaptic inputs, we analyzed the spike latency relative to each laser pulse. We first identified the time of maximal firing rate (tmax) of the up-sampled PSTH (0.1 ms resolution, linearly interpolated from 0.5 ms bins). We then tested the difference between the firing rates in each 5-point sliding window (from laser onset to tmax) and that before laser onset (t-test). The latency was measured by the change point (tchg), defined as the point following the last point with P > 0.01 (in all cases, P was found to be < 0.01 for all the windows between tchg and tmax). We found that the latencies of the identified neurons were typically ~2 ms (2.1±1.1 ms, s.d., Fig. 2e), indicating that they were directly driven.
Spike-waveform correlation
In principle, stringent single unit sorting procedure should ensure that the spontaneous and laser-evoked spikes originate from the same neuron. Nevertheless, we tested the waveform similarity between laser-evoke and spontaneous spikes for all identified neurons. We first identified the peak of PSTH after laser pulse, and fitted the peak with a Gaussian function to determine the mean (tmean) and standard deviation (σ). We then searched for spikes between tmean − σ and tmean + σ after each laser pulse as laser evoked spikes. The correlation coefficient was computed between the average waveforms of these laser-evoked spikes and the spontaneous spikes. The correlation coefficients of most of the identified neurons in this study were >0.97 (0.99±0.01, s.d., Fig. 2f).
The optrode recording experiments were performed over multiple circadian windows, because to obtain stable recordings electrodes were moved slowly to search for units, and stabilization of the recording could take up to several hours. To test whether the sleep-wake related firing rate modulation differs significantly between different circadian times, we divided the neurons of each type into 3 groups according to the time of recording and performed two-way ANOVA analysis. We found that the circadian time did not significantly affect brain state modulation in any of the four cell types (ChAT: PBS = 1.0 × 10−6, PCT = 0.60, Pinteraction = 0.34; VGLUT2+: PBS = 0.0027, PCT = 0.73, Pinteraction = 0.69; PV+: PBS = 2.4 × 10−5, PCT = 0.43, Pinteraction = 0.50; SOM+: PBS = 3.0 × 10−4, PCT = 0.19, Pinteraction = 0.14; BS: brain state; CT: circadian time).
Slice recording
To map the local connectivity between the four genetically defined cell types in the BF, ChR2 was selectively expressed in presynaptic neurons and tdTomato or eGFP was expressed in postsynaptic neurons using crossed transgenic mice and virus (Supplementary Table 2).
Slice recordings were made at P23–P30. When virus injection was needed, AAV-DJ-EF1α-FLEX-ChR2-mCherry (400–600 nl) was injected into the BF, and recording was made 1 week after injection. Slice preparation was according to procedures described previously49. Mouse was deeply anaesthetized with 5% isoflurane. After decapitation, the brain was dissected rapidly and placed in ice-cold oxygenated HEPES buffered ACSF (in mM: NaCl 92, KCl 2.5, NaH2PO4 1.2, NaHCO3 30, HEPES 20, glucose 25, sodium ascorbate 5, thiourea 2, sodium pyruvate 3, MgSO4·7H2O 10, CaCl2·2H2O 0.5 and NAC 12, at pH 7.4, adjusted with 10 M NaOH), and coronal sections of the BF were made with a vibratome (Leica). Slices (350 μm thick) were recovered in oxygenated NMDG-HEPES solution (in mM: NMDG 93, KCl 2.5, NaH2PO4 1.2, NaHCO3 30, HEPES 20, glucose 25, sodium ascorbate 5, thiourea 2, sodium pyruvate 3, MgSO4·7H2O 10, CaCl2·2H2O 0.5 and NAC 12, at pH 7.4, adjusted with HCl) at 32 °C for 10 min and then maintained in an incubation chamber with oxygenated standard ACSF (in mM: NaCl 125, KCl 3, CaCl2 2, MgSO4 2, NaH2PO4 1.25, sodium ascorbate 1.3, sodium pyruvate 0.6, NaHCO3 26, glucose 10 and NAC 10, at pH 7.4, adjusted by 10 M NaOH) at 25 °C for 1–4 hr before recording. All chemicals were from Sigma.
Whole-cell recordings were made at 30 °C in oxygenated solution (in mM: NaCl 125, KCl 4, CaCl2 2, MgSO4 1, NaH2PO4 1.25, sodium ascorbate 1.3, sodium pyruvate 0.6, NaHCO3 26 and glucose 10, at pH 7.4). EPSPs and IPSPs were recorded using a potassium-based internal solution (in mM: K-gluconate 135, KCl 5, HEPES 10, EGTA 0.3, MgATP 4, Na2GTP 0.3, and Na2-phosphocreatine 10, at pH 7.3, adjusted with KOH, 290–300 mOsm). IPSCs were recorded using a cesium-based internal solution (in mM: CsMeSO4 125, CsCl 2, HEPES 10, EGTA 0.5, MgATP 4, Na2GTP 0.3, Na2-phosphocreatine 10, TEACl 5, QX-314 3.5, at pH 7.3, adjusted with CsOH, 290–300 mOsm) and isolated by clamping the membrane potential of the recorded neuron at the reversal potential of the excitatory synaptic currents. The resistance of patch pipette was 3–5 MΩ. The cells were excluded if the series resistance exceeded 40 MΩ or varied by more than 20% during the recording period. To activate ChR2, we used a mercury arc lamp (Olympus) coupled to the epifluorescence light path and bandpass filtered at 450–490 nm (Semrock), gated by an electromagnetic shutter (Uniblitz). Blue light pulse (5 ms) was delivered through a 40× 0.8 NA water immersion lens (Olympus) at a power of 1–2 mW. Data were recorded with a Multiclamp 700B amplifier (Axon instruments) filtered at 0 – 2 kHz and digitized with a Digidata 1440A (Axon instruments) at 4 kHz. Recordings were analyzed using Clampfit (Axon instruments).
Single cell RT-PCR
At the end of each recording, cytoplasm was aspirated into the patch pipette, expelled into a PCR tube as described previously50. The single cell RT-PCR protocol was designed to detect the presence of mRNAs coding for ChAT, VGLUT2, PV and SOM. First, reverse transcription and the first round of PCR amplification were performed with gene-specific multiplex primer (Supplementary Table 2) using the SuperScript® III One-Step RT-PCR kit (12574-018, Invitrogen) according to the manufacturer’s protocol. Second, nested PCR was carried out using AmpliTaq Gold® 360 PCR kit (4398886, Invitrogen) with nested primers for each gene (Extended Data Table 2). All multiplex primers were designed to target two different exons to differentiate mRNAs from genomic DNA51–53. The final PCR products were sequenced and verified. Amplification products were visualized via electrophoresis using 2% agarose gel. During single cell RT-PCR procedures, care was taken to minimize RNA degradation and contamination.
Immunohistochemistry and in situ hybridization
For immunohistochemistry, mice were deeply anaesthetized with isoflurane and immediately perfused using 0.1M PBS followed by 4% paraformaldehyde in PBS. The brains were removed, and post-fixed in the same fixative overnight at 4 °C. For cryoprotection, brains were stored in 30% sucrose in PBS overnight. Brains were embedded and mounted with Tissue-Tek OCT compound (Sakura finetek) and 20 μm sections were cut using a cryostat (Leica). Brain slices were washed using PBS, permeabilized using PBST (0.3% Triton X-100 in PBS) for 30 min and then incubated with blocking solution (2% normal goat serum or normal donkey serum in the PBST) for 1 hr followed by primary antibody incubation overnight at 4 °C using following antibodies: anti-ChAT antibody (AB144P, Millipore; 1:1000), anti-PV antibody (24428, Immunostar; 1:1,000), anti-SOM antibody (MAB354, Millipore; 1:200). The next day, slices were washed three times with PBS and then incubated with appropriate secondary antibody (A 11008, Alexa Fluor 488 Goat Anti-Rabbit IgG or A-11055, Alexa Fluor 488 Donkey Anti-goat IgG, Invitrogen; 1:500) for 2 hrs and coverslipped.
In situ hybridization for VGLUT2, GAD1 or GAD2 was performed as previously described54,55. Cryosections (50 μm thick) were collected into a 24-well plate, fixed in 4% PFA, rinsed with PBS, and incubated with Proteinase K buffer. After fixing again with 4% PFA and rinsing with PBS, the sections were incubated with acetylation buffer, washed with PBS, and then incubated with hybridization buffer (without the probe) for 1 hr. Probes were added and sections were incubated at 50–65°C for 16 – 20 hrs. After hybridization, sections were washed, first with 2× SSC-50% formamide, then with 2× SSC, and then treated with RNase buffer. After blocking with 5% NGS, sections were incubated with AP-conjugated anti-DIG antibody (1093274, Roche Applied Science, 1:1000), and anti-DsRed (632496, Clontech, 1:500) or anti-GFP antibodies (GFP-1020, Aves Labs, 1:500) overnight at 4°C. After washing, anti-DsRed or GFP antibodies were detected by Alexa Fluor® 488 conjugated secondary antibodies (Invitrogen), and the DIG-labeled probe was detected by Fast Red TR/Naphthol AS-MX (F4523, Sigma-Aldrich). All probes were synthesized using published primers (Website: © 2015 Allen Institute for Brain Science. Allen Brain Atlas. Available from: http://www.brain-map.org), following the procedure described previously54.
Fluorescence images were taken using a confocal microscope (LSM 710 AxioObserver Inverted 34-Channel Confocal, Zeiss) and Nanozoomer (Hamamatsu).
Statistics
Samples size
We did not perform a calculation on sample size. Our sample size is comparable to many studies using similar techniques and animal models.
Evaluation of brain states modulation upon optogenetic manipulations
All statistical tests were two-sided. Significant modulation by optogenetic manipulation for each cell type was assessed with a paired t-test, t-test or two-way repeated measures ANOVA with Bonferroni post-hoc comparisons. For t-tests, data distributions were assumed to be normal, but this was not formally tested. Data collection and analysis were not performed blindly. Randomization was not performed, as majority of the experiments primarily involved within-animal comparisons.
Data exclusion criteria
Data from animals used in behavior experiments were excluded based on histological criteria that included injection sites, virus expression and optical fiber placement. Only animals with injection sites in the region of interest were included as shown in Fig. S2. A supplementary methods checklist is available.
Supplementary Material
1. Expression of neuronal markers in genetically defined BF cell types.
ChAT-, VGLUT2-, PV-, and SOM-Cre mice were crossed with Ai14 tdTomato reporter mouse. Shown in each box are numbers of BF neurons expressing tdTomato+ (red), a neuronal marker (ChAT, VGLUT2, PV, or SOM; shown in green) detected by either immunostaining or in situ hybridization (indicated by §), or both (orange). The two data sets based on in situ hybridization (indicated by *) were from the Allen Mouse Brain Atlas (*1http://connectivity.brain-map.org/transgenic/experiment/81657984; *2http://connectivity.brain-map.org/transgenic/experiment/167643437). Neurons were counted in the caudal portion of the BF (including the horizontal limb of the diagonal band of Broca, magnocellular preoptic nucleus, and substantia innominata), where all the electrophysiology and optogenetic experiments were performed. Note that >99.5% of tdTomato+ neurons in ChAT-, VGLUT2, PV-, or SOM-Cre mice were positive for their respective markers (gray-shaded cells). For each genetically defined cell type, only a small percentage of neurons expressed other markers.
2. Mouse lines and virus used to map local connectivity among BF cell types.
Primers (5′>3′) for single-cell RT-PCR:
ChAT (sense/anti-sense):
multiplex, atggccattgacaaccatcttctg/ccttgaactgcagaggtctctcat; nested, same as multiplex.
VGLUT2 (sense/anti-sense):
multiplex, gccgctacatcatagccatc/gctctctccaatgctctcctc; nested, acatggtcaacaacagcactatc/ataagacaccagaagccagaaca.
PV (sense/anti-sense):
multiplex, ggatgtcgatgacagacgtg/cagccaccagagtggagaat; nested, cagcgctgaggacatcaag/ctgaggagaagcccttcaga.
SOM (sense/anti-sense):
multiplex, agatgctgtcctgccgtct/gggccaggagttaaggaaga; nested, cccagactccgtcagtttct/gaagttcttgcagccagctt.
Each box in the middle column describes the crossing of mouse lines and virus injected into the BF, for testing the synaptic interaction between each pair of presynaptic (left column) and postsynaptic (right column) cell types.
Acknowledgments
We thank J. Cox, F. Weber, and N. Hirai for the help with data analysis, K. Kao for technical assistance, Dr. T. Hnasko for sharing VGLUT2-eGFP mice. Stanford Neuroscience Gene Vector and Virus Core for AAV-DJ supply.
Footnotes
AUTHOR CONTRIBUTIONS
M.X., S.C., and Y.D. conceived and designed the experiments. M.X. performed all optrode recording experiments, some of the in situ hybridization experiments, and some of the slice recording experiments. S.C. performed histological characterization of BF cell types and all the optogenetic activation experiments. S.Z. performed some of the slice experiments. P.Z performed some of the slice recording experiments. C.M., W.-C.C. performed some of the in situ hybridization experiments. N.S and S.N. helped to establish sleep recording and data analysis. B.W. and L.L. provided reagents and helped to establish in situ hybridization. M.X., S.C., and Y.D. wrote the manuscript, and all authors participated in revision of the manuscript.
COMPETING FINANCIAL INTERESTS
The authors declare no competing financial interests.
References
- 1.Siegel JM. Sleep viewed as a state of adaptive inactivity. Nat Rev Neurosci. 2009;10:747–753. doi: 10.1038/nrn2697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Von Economo C. Sleep as a problem of localization. J Nerv Ment Dis. 1930;71:249–259. [Google Scholar]
- 3.Nauta WJ. Hypothalamic regulation of sleep in rats; an experimental study. J Neurophysiol. 1946;9:285–316. doi: 10.1152/jn.1946.9.4.285. [DOI] [PubMed] [Google Scholar]
- 4.Moruzzi G, Magoun HW. Brain stem reticular formation and activation of the EEG. Electroencephalogr Clin Neurophysiol. 1949;1:455–473. [PubMed] [Google Scholar]
- 5.Saper CB, Fuller PM, Pedersen NP, Lu J, Scammell TE. Sleep State Switching. Neuron. 2010;68:1023–1042. doi: 10.1016/j.neuron.2010.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jones BE. Neurobiology of waking and sleeping. Handb Clin Neurol. 2011;98:131–149. doi: 10.1016/B978-0-444-52006-7.00009-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brown RE, Basheer R, McKenna JT, Strecker RE, McCarley RW. Control of sleep and wakefulness. Physiol Rev. 2012;92:1087–1187. doi: 10.1152/physrev.00032.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Szymusiak R, McGinty D. Sleep suppression following kainic acid-induced lesions of the basal forebrain. Exp Neurol. 1986;94:598–614. doi: 10.1016/0014-4886(86)90240-2. [DOI] [PubMed] [Google Scholar]
- 9.Szymusiak R, McGinty D. Sleep-waking discharge of basal forebrain projection neurons in cats. Brain Res Bull. 1989;22:423–430. doi: 10.1016/0361-9230(89)90069-5. [DOI] [PubMed] [Google Scholar]
- 10.Alam MN, McGinty D, Szymusiak R. Thermosensitive neurons of the diagonal band in rats: relation to wakefulness and non-rapid eye movement sleep. Brain Res. 1997;752:81–89. doi: 10.1016/s0006-8993(96)01452-7. [DOI] [PubMed] [Google Scholar]
- 11.Duque A, Balatoni B, Detari L, Zaborszky L. EEG correlation of the discharge properties of identified neurons in the basal forebrain. J Neurophysiol. 2000;84:1627–1635. doi: 10.1152/jn.2000.84.3.1627. [DOI] [PubMed] [Google Scholar]
- 12.Lee MG, Manns ID, Alonso A, Jones BE. Sleep-wake related discharge properties of basal forebrain neurons recorded with micropipettes in head-fixed rats. J Neurophysiol. 2004;92:1182–1198. doi: 10.1152/jn.01003.2003. [DOI] [PubMed] [Google Scholar]
- 13.Hassani OK, Lee MG, Henny P, Jones BE. Discharge profiles of identified GABAergic in comparison to cholinergic and putative glutamatergic basal forebrain neurons across the sleep-wake cycle. J Neurosci. 2009;29:11828–11840. doi: 10.1523/JNEUROSCI.1259-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Takahashi K, Lin JS, Sakai K. Characterization and mapping of sleep-waking specific neurons in the basal forebrain and preoptic hypothalamus in mice. Neuroscience. 2009;161:269–292. doi: 10.1016/j.neuroscience.2009.02.075. [DOI] [PubMed] [Google Scholar]
- 15.McGinty DJ, Sterman MB. Sleep suppression after basal forebrain lesions in the cat. Science. 1968;160:1253–1255. doi: 10.1126/science.160.3833.1253. [DOI] [PubMed] [Google Scholar]
- 16.Buzsaki G, et al. Nucleus basalis and thalamic control of neocortical activity in the freely moving rat. J Neurosci. 1988;8:4007–4026. doi: 10.1523/JNEUROSCI.08-11-04007.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sallanon M, et al. Long-lasting insomnia induced by preoptic neuron lesions and its transient reversal by muscimol injection into the posterior hypothalamus in the cat. Neuroscience. 1989;32:669–683. doi: 10.1016/0306-4522(89)90289-3. [DOI] [PubMed] [Google Scholar]
- 18.Lin JS, Sakai K, Vanni-Mercier G, Jouvet M. A critical role of the posterior hypothalamus in the mechanisms of wakefulness determined by microinjection of muscimol in freely moving cats. Brain Res. 1989;479:225–240. doi: 10.1016/0006-8993(89)91623-5. [DOI] [PubMed] [Google Scholar]
- 19.Nishino S, et al. Muscle atonia is triggered by cholinergic stimulation of the basal forebrain: implication for the pathophysiology of canine narcolepsy. J Neurosci. 1995;15:4806–4814. doi: 10.1523/JNEUROSCI.15-07-04806.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kaur S, Junek A, Black MA, Semba K. Effects of ibotenate and 192IgG-saporin lesions of the nucleus basalis magnocellularis/substantia innominata on spontaneous sleep and wake states and on recovery sleep after sleep deprivation in rats. J Neurosci. 2008;28:491–504. doi: 10.1523/JNEUROSCI.1585-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fuller PM, Sherman D, Pedersen NP, Saper CB, Lu J. Reassessment of the structural basis of the ascending arousal system. J Comp Neurol. 2011;519:933–956. doi: 10.1002/cne.22559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Deurveilher S, Semba K. Basal forebrain regulation of cortical activity and sleep-wake states: Roles of cholinergic and non-cholinergic neurons. Sleep Biol Rhythms. 2011;9:65–70. [Google Scholar]
- 23.Jones BE. The Organization of Central Cholinergic Systems and Their Functional Importance in Sleep-Waking States. Prog Brain Res. 1993;98:61–71. doi: 10.1016/s0079-6123(08)62381-x. [DOI] [PubMed] [Google Scholar]
- 24.Lee MG, Hassani OK, Alonso A, Jones BE. Cholinergic basal forebrain neurons burst with theta during waking and paradoxical sleep. J Neurosci. 2005;25:4365–4369. doi: 10.1523/JNEUROSCI.0178-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Everitt BJ, Robbins TW. Central cholinergic systems and cognition. Annu Rev Psychol. 1997;48:649–684. doi: 10.1146/annurev.psych.48.1.649. [DOI] [PubMed] [Google Scholar]
- 26.Sarter M, Hasselmo ME, Bruno JP, Givens B. Unraveling the attentional functions of cortical cholinergic inputs: interactions between signal-driven and cognitive modulation of signal detection. Brain research Brain Res Rev. 2005;48:98–111. doi: 10.1016/j.brainresrev.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 27.Pinto L, et al. Fast modulation of visual perception by basal forebrain cholinergic neurons. Nat Neurosci. 2013;16:1857–1863. doi: 10.1038/nn.3552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu Y, et al. A cortical circuit for gain control by behavioral state. Cell. 2014;156:1139–1152. doi: 10.1016/j.cell.2014.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eggermann E, Kremer Y, Crochet S, Petersen CC. Cholinergic signals in mouse barrel cortex during active whisker sensing. Cell Rep. 2014;9:1654–1660. doi: 10.1016/j.celrep.2014.11.005. [DOI] [PubMed] [Google Scholar]
- 30.Zaborszky L, Duque A. Local synaptic connections of basal forebrain neurons. Behav Brain Res. 2000;115:143–158. doi: 10.1016/s0166-4328(00)00255-2. [DOI] [PubMed] [Google Scholar]
- 31.Lein ES, et al. Genome-wide atlas of gene expression in the adult mouse brain. Nature. 2007;445:168–176. doi: 10.1038/nature05453. [DOI] [PubMed] [Google Scholar]
- 32.Xu X, Roby KD, Callaway EM. Immunochemical characterization of inhibitory mouse cortical neurons: three chemically distinct classes of inhibitory cells. J Comp Neurol. 2010;518:389–404. doi: 10.1002/cne.22229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Madisen L, et al. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Madisen L, et al. A toolbox of Cre-dependent optogenetic transgenic mice for light-induced activation and silencing. Nat Neurosci. 2012;15:793–802. doi: 10.1038/nn.3078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anikeeva P, et al. Optetrode: a multichannel readout for optogenetic control in freely moving mice. Nat Neurosci. 2012;15:163–170. doi: 10.1038/nn.2992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Modirrousta M, Mainville L, Jones BE. Gabaergic neurons with alpha2-adrenergic receptors in basal forebrain and preoptic area express c-Fos during sleep. Neuroscience. 2004;129:803–810. doi: 10.1016/j.neuroscience.2004.07.028. [DOI] [PubMed] [Google Scholar]
- 37.Irmak SO, de Lecea L. Basal forebrain cholinergic modulation of sleep transitions. Sleep. 2014;37:1941–1951. doi: 10.5665/sleep.4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Han Y, et al. Selective activation of cholinergic basal forebrain neurons induces immediate sleep-wake transitions. Curr Biol. 2014;24:693–698. doi: 10.1016/j.cub.2014.02.011. [DOI] [PubMed] [Google Scholar]
- 39.Freund TF, Meskenaite V. gamma-Aminobutyric acid-containing basal forebrain neurons innervate inhibitory interneurons in the neocortex. P Natl Acad Sci USA. 1992;89:738–742. doi: 10.1073/pnas.89.2.738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim T, et al. Cortically projecting basal forebrain parvalbumin neurons regulate cortical gamma band oscillations. P Natl Acad Sci USA. 2015;112:3535–3540. doi: 10.1073/pnas.1413625112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Steriade M, McCarley RW. Brain control of wakefulness and sleep. Plenum Press; New York: 2005. [Google Scholar]
- 42.Goard M, Dan Y. Basal forebrain activation enhances cortical coding of natural scenes. Nat Neurosci. 2009;12:1444–1449. doi: 10.1038/nn.2402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Szymusiak R, Gvilia I, McGinty D. Hypothalamic control of sleep. Sleep Med. 2007;8:291–301. doi: 10.1016/j.sleep.2007.03.013. [DOI] [PubMed] [Google Scholar]
- 44.Sherin JE, Shiromani PJ, McCarley RW, Saper CB. Activation of ventrolateral preoptic neurons during sleep. Science. 1996;271:216–219. doi: 10.1126/science.271.5246.216. [DOI] [PubMed] [Google Scholar]
- 45.Szymusiak R, Alam N, Steininger TL, McGinty D. Sleep-waking discharge patterns of ventrolateral preoptic/anterior hypothalamic neurons in rats. Brain Res. 1998;803:178–188. doi: 10.1016/s0006-8993(98)00631-3. [DOI] [PubMed] [Google Scholar]
- 46.Lu J, Greco MA, Shiromani P, Saper CB. Effect of lesions of the ventrolateral preoptic nucleus on NREM and REM sleep. J Neurosci. 2000;20:3830–3842. doi: 10.1523/JNEUROSCI.20-10-03830.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sherin JE, Elmquist JK, Torrealba F, Saper CB. Innervation of histaminergic tuberomammillary neurons by GABAergic and galaninergic neurons in the ventrolateral preoptic nucleus of the rat. J Neurosci. 1998;18:4705–4721. doi: 10.1523/JNEUROSCI.18-12-04705.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Steininger TL, Gong H, McGinty D, Szymusiak R. Subregional organization of preoptic area/anterior hypothalamic projections to arousal-related monoaminergic cell groups. J Comp Neurol. 2001;429:638–653. [PubMed] [Google Scholar]
- 49.Zhang S, et al. Selective attention. Long-range and local circuits for top-down modulation of visual cortex processing. Science. 2014;345:660–665. doi: 10.1126/science.1254126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lambolez B, Audinat E, Bochet P, Crepel F, Rossier J. AMPA receptor subunits expressed by single Purkinje cells. Neuron. 1992;9:247–258. doi: 10.1016/0896-6273(92)90164-9. [DOI] [PubMed] [Google Scholar]
- 51.Pfeffer CK, Xue M, He M, Huang ZJ, Scanziani M. Inhibition of inhibition in visual cortex: the logic of connections between molecularly distinct interneurons. Nat Neurosci. 2013;16:1068–1076. doi: 10.1038/nn.3446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Danik M, Puma C, Quirion R, Williams S. Widely expressed transcripts for chemokine receptor CXCR1 in identified glutamatergic, gamma-aminobutyric acidergic, and cholinergic neurons and astrocytes of the rat brain: a single-cell reverse transcription-multiplex polymerase chain reaction study. J Neurosci Res. 2003;74:286–295. doi: 10.1002/jnr.10744. [DOI] [PubMed] [Google Scholar]
- 53.Nordenankar K, et al. Increased hippocampal excitability and impaired spatial memory function in mice lacking VGLUT2 selectively in neurons defined by tyrosine hydroxylase promoter activity. Brain Struct Funct. 2014;220:2171–2190. doi: 10.1007/s00429-014-0778-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weissbourd B, et al. Presynaptic partners of dorsal raphe serotonergic and GABAergic neurons. Neuron. 2014;83:645–662. doi: 10.1016/j.neuron.2014.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Watakabe A, Komatsu Y, Ohsawa S, Yamamori T. Fluorescent in situ hybridization technique for cell type identification and characterization in the central nervous system. Methods. 2010;52:367–374. doi: 10.1016/j.ymeth.2010.07.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
1. Expression of neuronal markers in genetically defined BF cell types.
ChAT-, VGLUT2-, PV-, and SOM-Cre mice were crossed with Ai14 tdTomato reporter mouse. Shown in each box are numbers of BF neurons expressing tdTomato+ (red), a neuronal marker (ChAT, VGLUT2, PV, or SOM; shown in green) detected by either immunostaining or in situ hybridization (indicated by §), or both (orange). The two data sets based on in situ hybridization (indicated by *) were from the Allen Mouse Brain Atlas (*1http://connectivity.brain-map.org/transgenic/experiment/81657984; *2http://connectivity.brain-map.org/transgenic/experiment/167643437). Neurons were counted in the caudal portion of the BF (including the horizontal limb of the diagonal band of Broca, magnocellular preoptic nucleus, and substantia innominata), where all the electrophysiology and optogenetic experiments were performed. Note that >99.5% of tdTomato+ neurons in ChAT-, VGLUT2, PV-, or SOM-Cre mice were positive for their respective markers (gray-shaded cells). For each genetically defined cell type, only a small percentage of neurons expressed other markers.
2. Mouse lines and virus used to map local connectivity among BF cell types.
Primers (5′>3′) for single-cell RT-PCR:
ChAT (sense/anti-sense):
multiplex, atggccattgacaaccatcttctg/ccttgaactgcagaggtctctcat; nested, same as multiplex.
VGLUT2 (sense/anti-sense):
multiplex, gccgctacatcatagccatc/gctctctccaatgctctcctc; nested, acatggtcaacaacagcactatc/ataagacaccagaagccagaaca.
PV (sense/anti-sense):
multiplex, ggatgtcgatgacagacgtg/cagccaccagagtggagaat; nested, cagcgctgaggacatcaag/ctgaggagaagcccttcaga.
SOM (sense/anti-sense):
multiplex, agatgctgtcctgccgtct/gggccaggagttaaggaaga; nested, cccagactccgtcagtttct/gaagttcttgcagccagctt.
Each box in the middle column describes the crossing of mouse lines and virus injected into the BF, for testing the synaptic interaction between each pair of presynaptic (left column) and postsynaptic (right column) cell types.