Abstract
AIM
To investigate micro (mi)R-34a-antagonizing circular (circ)RNA that underlies hepatocellular steatosis.
METHODS
The effect of circRNA on miR-34a was recognized by the miRNA response element (MRE), and validated by the dual-luciferase reporter assay. Its association with hepatocellular steatosis was investigated in HepG2-based hepatocellular steatosis induced by free fatty acids (FFAs; 2:1 oleate:palmitate) stimulation. After normalization of the steatosis-related circRNA by expression vector, analysis of miR-34a activity, peroxisome proliferator-activated receptor (PPAR)α level, and expression of downstream genes were carried out so as to reveal its impact on the miR-34a/PPARα regulatory system. Both triglyceride (TG) assessment and cytopathological manifestations uncovered the role of circRNA in miR-34a-dependent hepatosteatogenesis.
RESULTS
Bioinformatic and functional analysis verified circRNA_0046366 to antagonize the activity of miR-34a via MRE-based complementation. In contrast to its lowered level during FFA-induced hepatocellular steatosis, circRNA_0046366 up-regulation abolished the miR-34a-dependent inhibition of PPARα that played a critical role in metabolic signaling pathways. PPARα restoration exerted transcriptional improvement to multiple genes responsible for lipid metabolism. TG-specific lipolytic genes [carnitine palmitoyltransferase 1A (CPT1A) and solute-carrier family 27A (SLC27A)] among these showed significant increase in their expression levels. The circRNA_0046366-related rebalancing of lipid homeostasis led to dramatic reduction of TG content, and resulted in the ameliorated phenotype of hepatocellular steatosis.
CONCLUSION
Dysregulation of circRNA_0046366/miR-34a/PPARα signaling may be a novel epigenetic mechanism underlying hepatocellular steatosis. circRNA_0046366 serves as a potential target for the treatment of hepatic steatosis.
Keywords: Hepatocytes, Steatosis, circRNA_0046366, miR-34a, Peroxisome proliferator-activated receptor α
Core tip: circRNA_0046366, which demonstrated expression loss in HepG2-based hepatocellular steatosis, exerts antagonistic effect on miR-34a activity. miR-34a inactivation abrogates its inhibitory role against peroxisome proliferator-activated receptor (PPAR)α, and then rescues the PPARα level. PPARα restoration further improves the expression of downstream genes [i.e. carnitine palmitoyltransferase 1A (CPT1A) and solute-carrier family 27A (SLC27A)], at both transcriptional and translational levels, which are associated to triglyceride metabolism. In conclusion, the rebalancing of lipid homeostasis down-regulates triglyceride content, and attenuates the hepatocellular steatosis.
INTRODUCTION
Hepatic steatosis is associated with hepatocyte-specific accumulation of lipid droplets (≥ 5% of volume or weight), and has increased dramatically worldwide with the growing incidence of obesity[1,2]. Hepatic steatosis is one of the most common chronic liver diseases in western countries and the Asia-Pacific area[2-6]. It is a critical step in the development of nonalcoholic fatty liver disease, which ranges from simple steatosis to nonalcoholic steatohepatitis, liver fibrosis/cirrhosis, and finally hepatocellular carcinoma[7,8]. Patients with hepatic steatosis are also susceptible to some aspects of metabolic syndrome (e.g., type 2 diabetes and hyperlipidemia)[9,10] and related diseases (i.e. cardiovascular events, cerebrovascular diseases, and extrahepatic cancer)[11,12]. Although it is being recognized as a public health burden, clinical intervention for hepatic steatosis is deficient because of the limited understanding of its underlying mechanisms.
Micro (mi)RNAs have been identified as critical regulators of various physiological processes and diseases[13,14]. miR-34a is significantly up-regulated in different models of rodent hepatic steatosis[15,16]. Consistently, miR-34a expression correlates well with the clinical occurrence of hepatic steatosis in the Chinese, Japanese and Philippine populations[17-19]. The increased level of circulating miR-34a discriminates patients with hepatic steatosis from healthy controls with area under the curve (AUC) of 0.781[19]. Targeted repression of peroxisome proliferator-activated receptor (PPAR)α, which maintains homeostasis of lipid metabolism in hepatocytes, highlights the steatosis-related effect of miR-34a[20]. Antagonism of miR-34a, therefore, has potential in the therapy of hepatic steatosis.
Circular (circ)RNAs are a novel class of non-coding RNAs containing miRNA response elements (MREs)[21] that can be used to investigate miRNA-specific antagonists. circRNA_000203 in angiotensin-II-induced cardiac fibroblasts inactivates miR-26b-5p by the complementation between MREs and miRNAs[22]. circRNA_010567 in diabetic mice abrogates the effect of miR-141 in a same manner[23]. The miRNA-dependent inhibition of target mRNAs (transforming growth factor-β1, collagen 1a2 and connective tissue growth factor) is then abolished, which results in myocardial fibrosis[22,23]. By serving as a natural “miR-223 sponge”, heart-related circRNA (HRCR) sequesters and antagonizes its inhibitory role against activity-regulated cytoskeleton-associated protein (ARC)[24]. Thus, pathological hypertrophic responses and heart failure are prevented by the HRCR/miR-223/ARC axis[24]. However, both miR-34a-targeting circRNA and its actions are rarely reported in hepatic steatosis.
Employing databases of non-coding RNA (circBase and miRBase) and algorithms of circRNA-miRNA interaction, circRNA_0046366 is now recognized to be a specific antagonist of miR-34a. To reveal the circRNA-miRNA interaction underlying hepatocellular steatosis, expression patterns of circRNA_0046366 and miR-34a were investigated in a model of HepG2-based steatosis model in the presence of high-fat stimulation. The antagonistic effect of circRNA_0046366 on miR-34a was evaluated by bioinformatic analysis and dual-luciferase reporter assay. After the up-regulation of hepatocellular circRNA_0046366, functional experiments exhibited its impact on miR-34a, key miRNA-target (PPARα), and downstream genes responsible for lipid metabolism. Phenotypic identification finally revealed the pathophysiological role of circRNA_0046366/miR-34a/PPARα signaling in hepatocellular steatogenesis.
MATERIALS AND METHODS
Establishment of hepatocellular steatosis by high-fat stimulation
Exponentially growing HepG2 cells (Cell Bank of Type Culture Collection, Shanghai, China) were seeded in a 6-well plate at 2 × 105/well, and randomized into groups of normal and steatosis (9 wells/group). In contrast to those cultured in Dulbecco’s modified Eagle’s medium, penicillin-streptomycin, and 10% fetal bovine serum (control group), HepG2 cells in the model group were subjected to additional exposure to 0.5 mmol/L FFA (oleate:palmitate = 2:1; Sigma-Aldrich, St. Louis, MO, United States) for 24 h[25].
Assessment of circRNA_0046366 expression
Total RNA of each sample was extracted by the phenol/chloroform method, and then treated by the ExScript RT Reagent Kit (TaKaRa, Kusatsu, Japan) for reverse transcription (RT). Quantitative real-time polymerase chain reaction (PCR) was performed, with primers specific to cDNA of circRNA_0046366 (Table 1)[24], using SYBR Premix ExTaq (TaKaRa) on the Applied Biosystems 7500 Real-Time PCR Detection Systems (Bio-Rad Laboratories, Hercules, CA, United States). The expression level of hepatocellular circRNA_0046366 was evaluated against U6 by the 2-ΔΔCt method.
Table 1.
Primers for real-time PCR
| Gene | Primer sequence, 5'-3' | Product, nt | |
| hsa_circ_000366 | F: CGTCCATTCGTTTGTGAGCC | R: CTTCACAGCCTCATCGGAGC | 126 |
| APOA1 | F: GCCAGGCTCGGCATTTCTG | R: GCCGCTGTCTTTGAGCACATCCA | 104 |
| APOC3 | F: CGGGTACTCCTTGTTGTTGC | R: TTGTCCTTAACGGTGCTCCA | 230 |
| CPT1A | F: CCAGACGAAGAACGTGGTCA | R: ATCTTGCCGTGCTCAGTGAA | 132 |
| FASN | F: ATGAGCACCAACGACACGAT | R: CTATAGGCCGCAGCCTTCTC | 140 |
| HMGCS | F: TGGTTCCCTTGCATCTGTTC | R: TTATCAAGAGCAGACCCCGG | 150 |
| LPL | F: CATTCCCGGAGTAGCAGAGT | R: GGACACTGGGTAATGCTCCT | 205 |
| PPARα | F: CCCCTCCTCGGTGACTTATC | R: ATTCGTCCAAAACGAATCGCGT | 297 |
| SLC27A | F: GGCCCAACGACATCGTCTAT | R: TAGCGGCACAGTTCACCAAT | 190 |
| VLDLR | F: GTAGGCAAAGAGCCAAGTC | R: GTACACCCAATCAACAGCA | 264 |
| U6 | F: ATTGGAACGATACAGAGAAGATT | R: GGAACGCTTCACGAATTTG | 70 |
| GAPDH | F: CTGAACGGGAAGCTCACTGG | R: AAAGTGGTCGTTGAGGGCAA | 252 |
Bioinformatic analysis
The effect of circRNA_0046366 on miRNAs was analyzed by the CircInteractome method according to MRE-based circRNA/miRNA complementation[26], and then the set of circRNA_0046366-targeting miRNAs was constructed. On the other side, the experiment-proven, hepatic steatosis-inducing miRNAs were pooled to establish another miRNA set[27]. Key target miRNAs that play an essential part in circRNA-related hepatocellular steatosis were successively filtered by the intersection of circRNA_0046366-targeting miRNA set and hepatic steatosis-inducing set.
To shed light on their actions associated with hepatocellular steatosis, a miRNA-specific targetome was constructed using genes obtained from miRBase[28]. According to the annotations of Kyoto Encyclopedia of Genes and Genomes (KEGG) database[29], enrichment scoring, Fisher’s exact test and false discovery rate (FDR) analysis were further employed to recognize the miRNA-regulating signal pathways[30]. Finally, gene interaction was illustrated within the top-enrichment pathways using databases of KEGG and PubMed[31].
Luciferase reporter assays
hsa-circRNA_0046366 (circBase; Rajewsky Laboratory, Berlin, Germany) with putative target sites for miR-34a was synthesized and cloned into the pMIR-REPORT vector (Thermo Fisher Scientific, Waltham, MA, United States) for the purpose of constructing recombinant reporter vector (pMIR-REPORT-circRNA_0046366-wildtype)[32]. Reporter vector with mutant circRNA_0046366 (pMIR-REPORT-circRNA_0046366-mutant) was also generated by deletion of an MRE sequence. As defined by the relative activity of firefly luciferase against Renilla luciferase, cotransfection of reporter vector (pMIR-REPORT-circRNA_0046366-wildtype or pMIR-REPORT-circRNA_0046366-mutant) and oligonucleotides (miR-34a mimics or negative control) revealed circRNA-miRNA interaction using a dual-luciferase assay kit (Promega, Madison, WI, United States).
circRNA administration
cDNA of circRNA_0046366 was synthesized according to the sequence obtained from circBase database (www.circbase.org), and then cloned into H3790 pcDNA3.1 plasmid using TA Cloning™ Kit (Thermo Fisher Scientific) as per the manufacturer’s instructions[33]. DNA sequencing verified the construction of recombinant vector (pcDNA3.1(+)-GFP-circRNA_0046366) expressing circRNA_0046366.
HepG2 cells in the exponential phase were randomly divided into 6 groups, including normal, steatosis, control, circRNA, circRNA+mimics, and circRNA+mimics normal control (NC) group (2 × 105 cells/well, 9 wells/group). In contrast to those without circRNA_0046366 regulation (normal and steatosis groups), the circRNA, circRNA+mimics, and circRNA+mimics NC groups were exposed to 12-h transfection of pcDNA3.1(+)-GFP-circRNA_0046366 (4 μg/well), pcDNA3.1(+)-GFP-circRNA_0046366 (4 μg/well) + miR-34a mimics (Genechem, Shanghai, China; 50 pmol/well), and pcDNA3.1(+)-GFP-circRNA_0046366 (4 μg/well) + miR-34a mimics, negative control (50 pmol/well; Genechem, Shanghai, China), respectively, using Lipofectamine 2000[34]. Transfection of blank plasmid (pcDNA3.1(+)-GFP at 4 μg/well) was applied to the control group. All groups, with the exception of the normal group, received FFA treatment for a further 24 h as mentioned above. After the transfection of circRNA-containing plasmid, the expression level of circRNA_0046366 was dynamically assessed in the circRNA group at the time points of 24, 48, and 72 h.
Real-time RT-PCR
cDNA of miR-34a was generated by the Mir-X miRNA First Strand Synthesis Kit (TaKaRa) using total RNA samples from each group. After normalization against an internal control (U6), real-time PCR demonstrated expression of hepatocellular miR-34a by the SYBR Fast qPCR Mix (TaKaRa)[20]. RT and real-time PCR for PPARα, apolipoprotein A1 (APOA1), apolipoprotein C3 (APOC3), fatty acid synthase (FASN), lipoprotein lipase (LPL), 3-hydroxy-3-methylglutaryl-CoA synthase 1 (HMGCS), very low-density lipoprotein receptor (VLDLR), carnitine palmitoyltransferase 1A (CPT1A), solute-carrier family 27A (SLC27A), and GAPDH were carried out using the ExScript RT Reagent Kit (TaKaRa), and SYBR Premix ExTaq (TaKaRa), respectively, using standard procedures. Gene-specific primers of these reactions were designed by Premier 5.0 software (PREMIER Biosoft, Palo Alto, CA, United States) (Table 1). According to the results obtained from the Applied Biosystems 7500 Real-Time PCR Detection Systems (Bio-Rad Laboratories), expression of these genes was assessed based on the 2-ΔΔCt method.
Western blotting
Total protein of each sample was prepared using RIPA lysis buffer, and quantified using the Pierce BCA Protein Assay Kit (Thermo Fisher Scientific). After separation by SDS-PAGE, protein samples were electrophoretically transferred to polyvinylidene difluoride membranes. These nonfat dry milk-blocked membranes were incubated with anti-PPARα (1:500; Santa Cruz Biotechnology, Dallas, TX, United States), anti-CPT1A (1:1000; Santa Cruz Biotechnology), anti-SLC27A (1:1000; Santa Cruz Biotechnology), and anti-GAPDH (1:1000; Santa Cruz Biotechnology) overnight at 4 °C, and reacted with horseradish-peroxidase-conjugated secondary antibody (1:1500; Jackson ImmunoResearch Laboratories, West Grove, PA, United States) for 1 h at room temperature. Chemiluminescent signals were visualized by ECL detection system, and scanned densitometrically by Image Lab Software 5.1 software (Bio-Rad Laboratories).
Phenotypic evaluation for hepatocellular steatosis
According to the above-mentioned subgrouping, HepG2 cells in normal, steatosis, control, circRNA, circRNA+ mimics, and circRNA+mimics NC groups were plated on cover glasses, and fixed in 4% paraformaldehyde. After washing in distilled water, each group was treated with 0.5% Oil Red O for 30 min, 60% isopropanol for 10-20 s, and hematoxylin counterstaining for 10-20 s so as to uncover the lipid droplets with neutral fat (triglyceride (TG)). Those HepG2 cells with cytoplasmic enrichment of positive-staining droplets were defined to be steatotic. Quantitatively, the TG level in each group was enzymatically analyzed using the TG Assay Kit (Applygen Technologies, Shanghai, China)[35]. Lysed HepG2 cells were subjected to centrifugation at 12000 rpm for 5 min, followed by coculture of supernatant and working solution for 10 min at 37 °C. OD550 indicated the hepatocellular TG concentration, which was normalized against the protein content of HepG2 cells evaluated by BCA method.
Statistical analysis
The present results are expressed as mean ± SD. Statistical analysis was performed by Student’s t-test or one-way analysis of variance with GraphPad Prism Software (GraphPad, La Jolla, CA, United States)[36]. Fisher’s exact test was used to filter the significant pathways using R software 3.3.1 (R Development Core Team, Vienna, Austria). Differences with P < 0.05 (two-tailed) were considered statistically significant.
RESULTS
circRNA_0046366 deficiency characterized high fat-induced hepatocellular steatosis
In contrast to the steatosis-free phenotype of the normal group, enrichment of lipid droplets dominated the steatosis group. Oil Red O staining specific to lipid droplets verified accumulation of neutral fat (TG) under FFA stimulation (Figure 1A). Consistent with these pathological observations, an enzymatic assay showed that the TG level of the steatosis group was higher than that of the normal group (Figure 1B). Compared to the normal group, the steatosis group exhibited significant loss of circRNA_0046366 expression (Figure 1C). FASN, which shares the same precursor mRNA (pre-mRNA) with circRNA_0046366, showed much higher mRNA level (7.64 ± 0.54) in the steatosis group in comparison to that of the normal group (0.98 ± 0.06, P < 0.0001). An inverse correlation between circRNA_0046366 expression and hepatocellular TG level (r = -0.73, P = 0.03; Figure 1D) suggested an essential role in the occurrence of hepatocellular steatosis.
Figure 1.

circRNA_0046366 loss demonstrates the epigenetic characteristic of FFA-induced hepatocellular steatosis. A: Oil Red O staining identified HepG2 cells with (steatosis group) or without (normal group) FFA-induced steatosis (200 ×); B: The steatosis group exhibited significant up-regulation of TG content; C: circRNA_0046366 quantification revealed inhibition of its expression after hepatocellular steatosis; D: circRNA_0046366 expression was inversely correlated with hepatocellular TG level in the steatosis group. Results are expressed as mean ± SD. bP < 0.01. FFA: Free fatty acid; TG: Triglyceride.
circRNA_0046366 functioned as antagonist of miR-34a
Five miRNAs (miR-34a, miR-513a-5p, miR-646, miR-892a and miR-1265) were predicted to be the targets of circRNA_0046366, using the algorithms of base complementation between MRE of circRNA and seed sequence of miRNA (Figure 2A). The intersection of circRNA_0046366-targeted miRNAs and hepatic-steatosis-related miRNAs recognized those that mediated the regulatory effect of circRNA_0046366 on hepatic steatogenesis. miR-34a was the only target of circRNA_0046366 and had a critical role in hepatocellular lipid dysmetabolism (Figure 2A).
Figure 2.
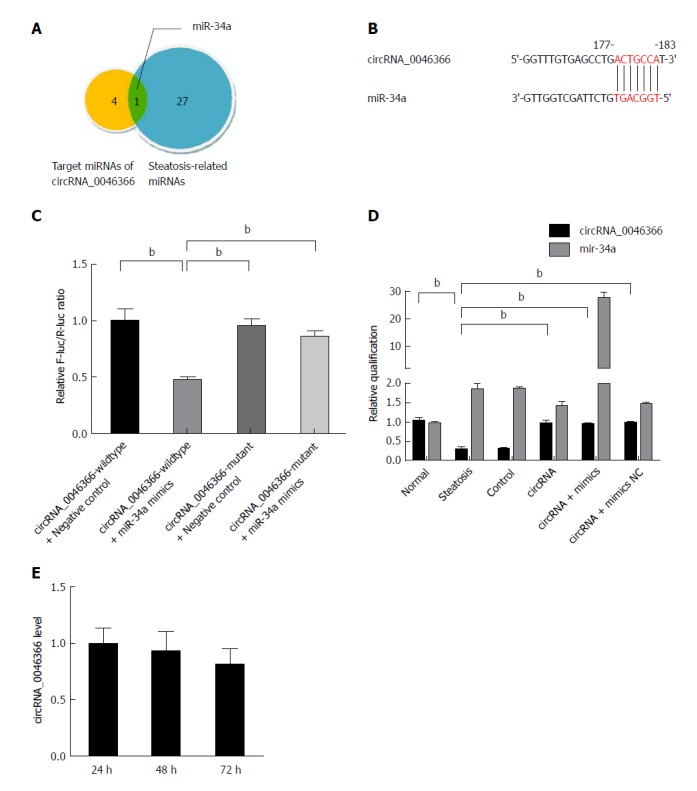
circRNA_0046366 functions as the antagonist of miR-34a. A: Set intersection filters circRNA_0046366-targeting miRNAs associated with hepatocellular steatosis; B: circRNA-miRNA interaction recognized by the base complementation between MRE of circRNA_0046366 and seed sequence of miR-34a; C: Dual-luciferase reporter assay verified the antagonistic effect of circRNA_0046366 on miR-34a; D: Treatment of circRNA-carrying vector normalized expression level of circRNA_0046366 with counteracting impact on miR-34a; E: Results are expressed as mean ± SD. bP < 0.01.
Dual-luciferase reporter assay was performed to verify the MRE-based circRNA-miRNA interaction (Figure 2B). After cotransfection of pMIR-REPORT vector containing wild-type circRNA_0046366 and miR-34a mimics, the activity ratio of firefly/Renilla luciferases was down-regulated in the pMIR-REPORT-circRNA_0046366-wildtype-treated group (Figure 2C). On the contrary, there was no reduction in relative luciferase activity in the pMIR-REPORT-circRNA_0046366-mutant-treated group with the MRE-lacking vector (Figure 2C). This suggests that circRNA_0046366 is an antagonist of miR-34a via targeted, complementary binding.
circRNA_0046366 up-regulation rescued PPARα expression via miR-34a inactivation
HepG2 cells were exposed to transfection of circRNA_0046366-carrying expression vector, which led to normalization of circRNA level, on the condition of FFA culture (Figure 2D). miR-34a was inactivated by circRNA_0046366-dependent antagonism. Because of the approximately stable level of intracellular circRNA_0046366 (Figure 2E), pharmacological effect could be yielded from the miR-34a inactivation.
Bioinformatic analysis showed that miR-34a had a major impact on metabolic signaling pathways (hsa01100), pathways in cancer (hsa05200), Rap1 signaling pathway (hsa04015), PI3K/Akt signaling pathway (hsa04151), and miRNAs in cancer (hsa05206) (Figure 3A). Some of these metabolic pathways were shown to mediate the steatosis-inducing action of miR-34a. Gene interaction analysis subsequently revealed that PPARα, the high-affinity target of miR-34a, controlled multiple lipometabolic genes by transcriptional induction (Figure 3B). Thus, circRNA_0046366 is proposed to modulate hepatocellular steatosis through the miR-34a/PPARα regulatory system.
Figure 3.
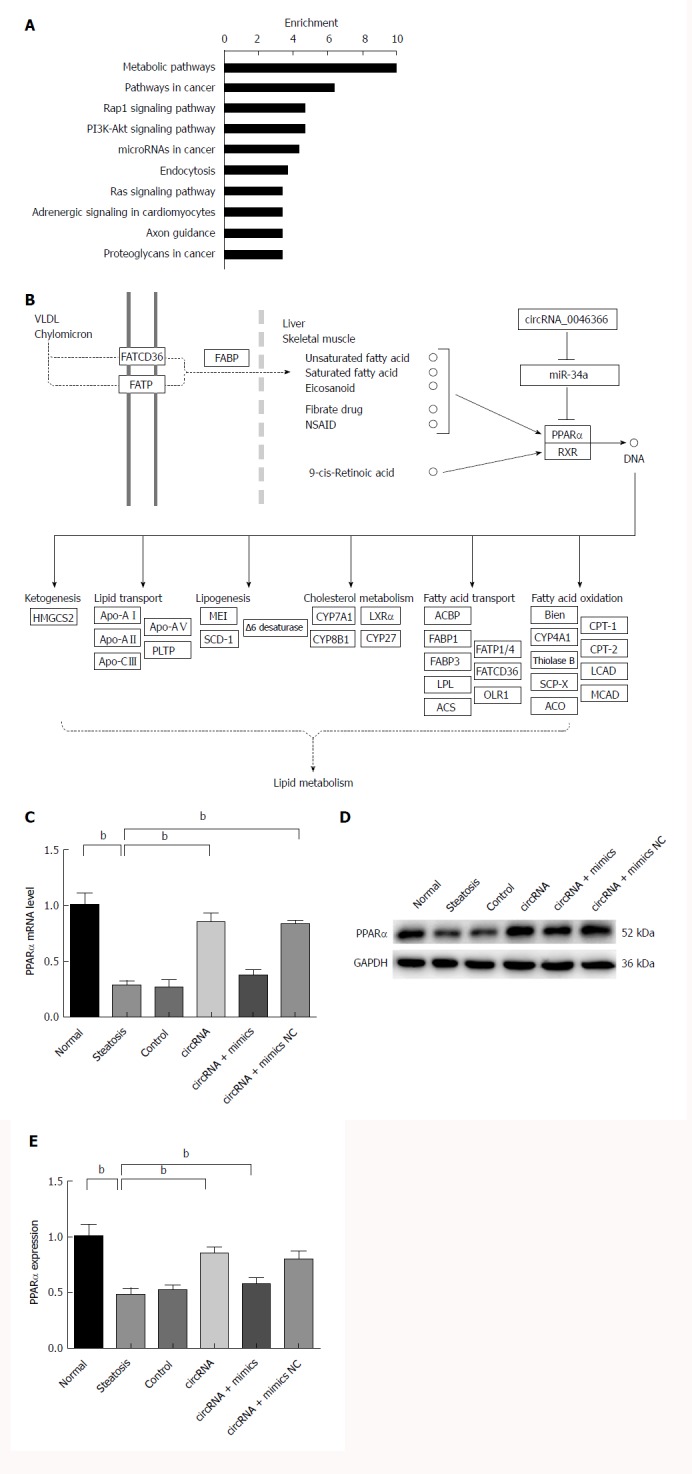
circRNA_0046366 restoration rescued PPARα expression by miR-34a inactivation. A: Metabolic pathway (hsa01100) was the top-enriched pathway that was affected by circRNA_0046366-induced miR-34a antagonism; B: PPARα mediated the regulatory role of miR-34a in lipid metabolism signaling by controlling expression of multiple lipometabolic genes (adapted from the KEGG database). miR-34a inactivation leads to the up-regulated expression of PPARα at transcriptional (C) and translational (D) levels. Results are expressed as mean ± SD. bP < 0.01. KEGG: Kyoto Encyclopedia of Genes and Genomes; PPAR: Peroxisome proliferator-activated receptor.
miR-34a-induced PPARα inhibition was one of the critical manifestations of hepatocellular steatosis (Figure 3C and D). A significant increase in PPARα, at both transcriptional and translational levels, characterized the circRNA and circRNA+mimics NC groups with circRNA_0046366 normalization (Figure 3C and D). However, saturated binding of circRNA_0046366 and miR-34a prevented PPARα restoration in the circRNA+mimics group (Figure 3C and D). These findings showed that circRNA_0046366 rescued PPARα expression, mainly by abolishing the inhibitory effect of miR-34a.
PPARα restoration promoted transcriptional activation of lipometabolic genes
Resulting from the circRNA_0046366 loss, PPARα repression took place in steatotic cells upon miR-34a activation. The PPARα deficiency abrogated its transcriptional regulation of multiple genes associated with lipid metabolism, especially CPT1A and SLC27A (Figure 4A-I). Their abnormal expression contributed to the lipometabolic imbalance, which was largely responsible for the occurrence of hepatocellular steatosis.
Figure 4.
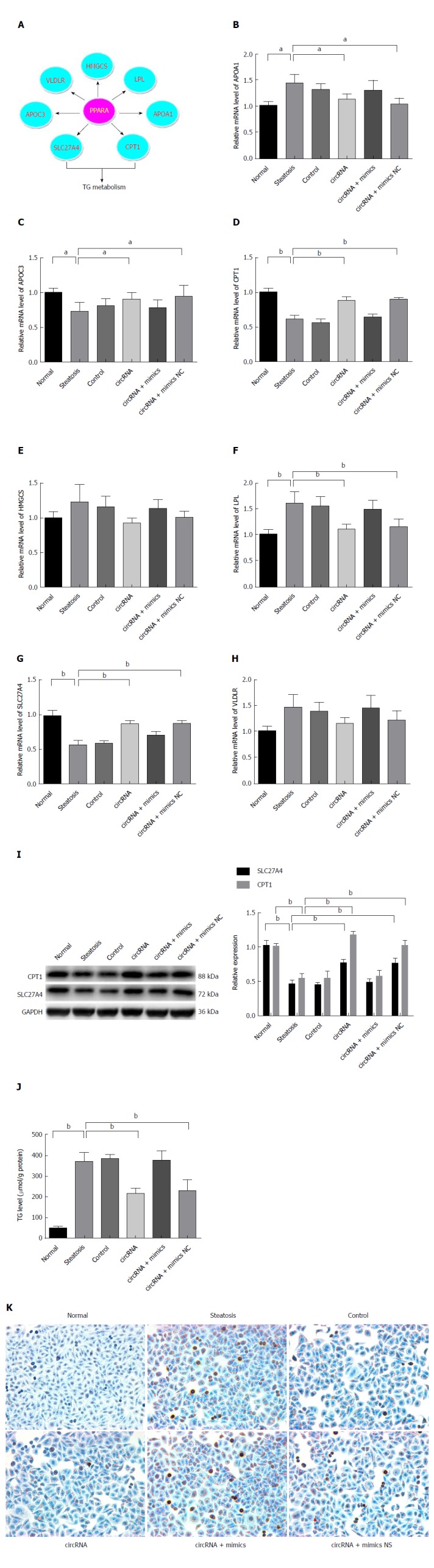
PPARα normalization improved hepatocellular steatosis by transcriptional promotion of lipometabolic genes. A: PPARα restoration by circRNA-0046366 promoted expression of CPT1A and SLC27A at the transcriptional level; B: Transcriptional activation of CPT1A and SLC27A resulted in their significant up-regulation. Improved expression of lipometabolic genes reduced the TG level (C) and attenuated hepatocellular steatosis (D) (200 ×). Results are expressed as mean ± SD. aP < 0.05, bP < 0.01. PPAR: Peroxisome proliferator-activated receptor; TG: Triglyceride.
In contrast to the abnormal transcription of downstream genes after FFA exposure (steatosis, control, and circRNA+mimics groups), circRNA_0046366-based PPARα restoration brought about improvement in their expression in both circRNA and circRNA+mimics NC groups (Figure 4B-H). TG metabolic genes (CPT1A and SLC27A) among these exhibited significant up-regulation of mRNA levels (Figure 4G and H). Consequently, the transcription-promoting effect of PPARα gave rise to obvious elevation of hepatocellular CPT1A and SLC27A, with similar levels to those in the normal group (Figure 4I). Thereby, a resolution of the steatosis-related lipid dysmetabolism could be achieved.
Improvement of lipid metabolism ameliorated hepatocellular steatosis by TG reduction
Biochemical analysis highlighted an overview of the lipometabolic improvements that were in agreement with the gene expression. Compared to the normal group, reduction of hepatocellular TG level dominated the groups with regaining of lipid homeostasis (steatosis group vs circRNA group: 368.73 ± 46.16 μmol/g vs 215.13 ± 26.42 μmol/g protein, P < 0.01; steatosis group vs circRNA+mimics NC group: 368.73 ± 46.16 μmol/g vs 227.40 ± 54.06 μmol/g protein, P < 0.05) (Figure 4J).
Pathophysiological alleviation was achieved from disruption of TG deposition. The groups with diminished TG content (circRNA and circRNA+mimics NC), instead of those without alteration (steatosis, control, and circRNA+mimics), lacked cytoplasmic lipid droplets under treatment with oleate and palmitate (Figure 4K). Therefore, attenuation of hepatocellular steatosis was verified after circRNA_0046366-based miR-34a antagonism and restoration of lipid metabolism via the miR-34a-related PPAR signaling.
DISCUSSION
circRNA has recently been discovered to serve as a novel, yet crucial, component of epigenetic regulation of miRNA-mRNA interaction[14]. Its antagonistic effect against miRNA is one of its most important mechanisms related to physiological functions (e.g., insulin production and cartilage degradation)[37,38] and various diseases (e.g., Alzheimer’s disease, colorectal cancer, ischemia-reperfusion injury, and hypertrophic heart failure)[24,34,39-41]. In the present study, prominent loss of circRNA_0046366 expression characterized the steatotic HepG2 cells under stimulation with saturated (palmitate) and polyunsaturated (oleate) fatty acids. In contrast, the mRNA level of FASN, one of the major determinants in de novo lipogenesis[42], experienced obvious up-regulation on condition of fatty acids exposure. Being the products of alternative splicing, circRNA_0046366 and FASN share a common pre-mRNA. Thus, increase in FASN transcription hampers the circRNA_0046366 production by the competitive effect in alternative splicing. Some surprising results even confirmed that circRNA_0046366 level significantly correlated with total TG content in an inverse manner. circRNA_0046366 dysregulation is, therefore, suggested to play an essential, perhaps miRNA-antagonizing, role in FFA-induced hepatocellular steatosis.
To investigate the actions of circRNA_0046366 underlying hepatocellular steatosis, target prediction and key-miRNA mining were performed on the basis of base-complementation algorithms and miRNA-set intersection, respectively[28,43]. miR-34a is a well-established miRNA with lipogenic properties[20], and it was the only candidate target of circRNA_0046366 that was associated with steatosis-related lipid dysmetabolism. Dual-luciferase reporter assay provided further evidence for the circRNA_0046366/miR-34a interaction. In contrast to cotransfection of miR-34a with reporter vector containing mutant circRNA_0046366, miR-34a demonstrated a complementary effect on wild-type circRNA_0046366 by a decrease in firefly/Renilla luciferase activity. The results of bioinformatic and functional analysis highlight that circRNA_0046366 is a miR-34a-specific antagonist. Because of its multiple-targeted characteristics, miR-34 facilitates a subtle antagonism of steatosis-related miRNA/mRNA interaction by relative limited circRNA_0046366 on a basis of competitive binding[44-46].
Targetome and targetome-based pathway analyses of miR-34a were integrated to uncover the downstream signaling and related functions of circRNA_0046366 in a miRNA-dependent way[47,48]. As a result of the top-ranking enrichment, metabolic pathway hsa01100 was identified as a key signaling pathway that was influenced by circRNA_0046366. circRNA_0046366 focuses its impact on metabolic process, especially lipid metabolism. PPARα, a transcription factor of the NR1C nuclear receptor subfamily, has recently been identified as a direct target of miR-34a[20]. This kind of liver-specific, ligand-activated isoform of the PPAR family induces the expression of multiple genes with lipometabolic characteristics[29]. Lack of PPARα in obese patients confers a high risk of insulin resistance, n-3 long-chain polyunsaturated fatty acid depletion, and liver steatogenesis[49]. Increasing evidence suggests a miR-34a/PPARα regulatory system, which acts as the crucial mediator of the actions of circRNA_0046366 in hepatocellular lipid metabolism (Figure 5).
Figure 5.
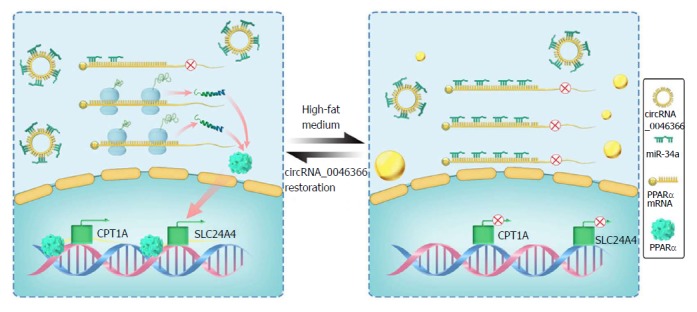
Schematic representation exhibits the circRNA_0046366/miR-34a/PPARα axis underlying occurrence and resolution of hepatocellular steatosis. In contrast to the hepatic steatogenesis that occurs upon its deficiency, circRNA_0046366 restoration abrogates the inhibitory effect of miR-34a on PPARα by antagonizing the miRNA-mRNA interaction. PPARα normalization transcriptionally activates the lipometabolic genes, which further ameliorates hepatic steatosis by regaining lipid homeostasis. mi: Micro; PPAR: Peroxisome proliferator-activated receptor.
With the relative stability in expressive dynamics, normalized circRNA_0046366 brought about antagonistic inactivation of miR-34a in the circRNA and circRNA+mimics NC groups in our study. PPARα up-regulation within these groups occurred as a result of diminished miR-34a-induced gene inhibition. This counteracting role of circRNA_0046366 against the miR-34a/PPARα regulatory system is supposed to expressively activate the lipometabolic genes and, in turn, rescue hepatic lipid homeostasis. Indeed, circRNA_0046366 treatment dramatically increased both CPT1A and SLC27A, the key genes downstream to PPARα[50], at transcriptional and translational levels.
CPT1a is located on the liver mitochondrial outer membrane and forms a hexamer fatty acid transfer complex[51], and functions as the rate-limiting enzyme of fatty acid β-oxidation[52]. CPT1A deficiency gives rise to impairment of long-chain fatty acid oxidation and ketogenesis, which are involved in the occurrence of hepatic steatosis[53,54]. In contrast to the insufficient expression of CPT1A during steatogenesis, PPARα agonists reduce lipid accumulation in close association with up-regulated CPT1A concentration[55]. SLC27A, also known as fatty acid transport protein (FATP)4, reflects another liver-dominant, evolutionarily conserved PPARα target that takes central place in the cellular uptake and metabolism of long-chain and very long-chain fatty acids[56,57]. Lowered SLC27A level facilitates fat deposition via the reduction of CPT1A-mediated fatty acid oxidation[58]. A similar phenomenon has been verified in FATP4-deficient animals with aggravated liver fatty degeneration under high-fat diet[59]. Therefore, restoration of CPT1A and SLC27A seen in the present study could counterbalance lipid metabolism by increasing activity of lipolytic catalyzation.
As defined by the colorimetric detection assay, regaining of lipid homeostasis with reduced intracellular TG content was validated in groups with expression-rescued lipolytic enzymes (circRNA and circRNA+mimics NC) compared with those without enough catalytic lipid oxidation (steatosis, control and circRNA+mimics). Disruption of glycerol-esterified neutral fat accumulation improved the pathophysiological outcome in conditions of high-fat culture. In contrast to those in the TG-enriched groups, HepG2 cells in the circRNA and circRNA+mimics NC groups showed a dramatic decrease in cytoplasmic lipid droplets in parallel with TG down-regulation. Finally, they were protected from lipotoxicity-stimulated phenotypic transition toward hepatocellular steatosis.
ARTICLE HIGHLIGHTS
Research background
Hepatic steatosis reflects one of the most common chronic liver diseases with hepatocyte-specific lipid dysmetabolism and triglyceride (TG) accumulation. Its close association to steatohepatitis, metabolic syndrome, and extrahepatic diseases (i.e., cardiovascular events, cerebrovascular diseases, cancers) indicates an importance for clinical interference. Micro (mi)R-34a is now confirmed to underlie the hepatic steatosis. However, the ambiguity in miR-34a-specific antagonist keeps hepatic steatosis from effective therapy. Circular (circ)RNA has recently been determined to interact with miRNA, mainly on the basis of complementation between miRNA response element (MRE) of circRNA and ‘seed sequence’ of miRNA. This circRNA/miRNA interaction abolishes the inhibitory effect of miRNA on its targets. circRNA, therefore, is highlighted to function in a miRNA-antagonizing manner.
Research motivation
Because of its importance in hepatic steatosis, miR-34a represents a critical target of clinical intervention. miR-34a-targeting antagonist, therefore, is assessed in our experiments so as to cure hepatosteatotic degeneration on the basis of miR-34a inactivation.
Research objectives
Serving as the selective sponge of miRNA, circRNA is exposed to bioinformatical and functional analysis for a purpose of uncovering the antagonist specific to miR-34a.
Research methods
To shed light on the antagonistic effect of circRNA against miR-34a, investigation of miR-34a-targeting circRNA was carried out by MRE recognization and dual-luciferase reporter assay. The filtered circRNA was then subjected to functional study in HepG2-based experimental steatosis induced high-fat stimulation. In detail, rescue experiment, real-time quantitative PCR, and western blot demonstrated the impact of circRNA on miR-34a, peroxisome proliferator-activated receptor (PPAR)α, and transcriptional downstream genes. Both triglyceride (TG) quantification and cytopathologic assessment revealed the steatosis-related outcome of circRNA administration.
Research results
circRNA_0046366 loss reflects the epigenetic characteristics of high fat-induced hepatocellular steatosis. Both bioinformatical and functional proofs indicate a circRNA_0046366-dependent miR-34a inactivation by the complementary antagonism. Dramatically, circRNA_0046366 up-regulation abolishes the inhibitory effect of miR-34a on PPARα. PPARα restoration further promotes the transcriptional activity of downstream genes, which improves the steatosis-related TG metabolism. In conclusion, the circRNA_0046366 administration leads to a significant attenuation of TG accumulation, and finally alleviates the hepatosteatotic phenotype with decreased cytoplasmic lipid droplets.
Research conclusions
The present study identifies great importance of circRNA_0046366/miR-34a/PPARα signaling in hepatocellular steatosis. circRNA_0046366 may act as a potential agent in the clinical interference of hepatic steatosis.
Research perspectives
The role of circRNA_0046366 in hepatocellular steatosis qualifies it for further evaluation in experimental hepatic steatosis with different etiologies (i.e., high-fat high-cholesterol diet, high-fat high-fructose diet, methionine and choline-deficient diet). These results could provide substantial evidence for circRNA_0046366-related prevention and therapy of hepatic steatosis.
Footnotes
Manuscript source: Unsolicited manuscript
Specialty type: Gastroenterology and hepatology
Country of origin: China
Peer-review report classification
Grade A (Excellent): 0
Grade B (Very good): 0
Grade C (Good): C, C
Grade D (Fair): 0
Grade E (Poor): 0
Supported by National Key Research and Development Plan ‘Precision Medicine Research’, No. 2017YFSF090203; National Natural Science Foundation of China, No. 81070346, No. 81270492, No. 81470859, No. 81270491 and No. 81470840; State Key Development Program for Basic Research of China, No. 2012CB517501; 100 Talents Program, No. XBR2011007h; and Program of the Committee of Science and Technology, No. 09140903500.
Institutional review board statement: This paper was approved by the Xinhua Hospital Ethics Committee Affiliated to Shanghai Jiaotong University School of Medicine.
Conflict-of-interest statement: No conflict of interest is declared for each author of the manuscript.
Data sharing statement: Technical appendix, statistical code, and dataset available from the authors at fanjiangao@xinhuamed.com.cn or panqin@xinhuamed.com.cn. Participants gave informed consent for data sharing.
Peer-review started: October 9, 2017
First decision: October 25, 2017
Article in press: November 27, 2017
P- Reviewer: Inoue K, Laguna JC S- Editor: Gong ZM L- Editor: Filipodia E- Editor: Huang Y
Contributor Information
Xing-Ya Guo, Department of Gastroenterology, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200092, China.
Fang Sun, Department of Gastroenterology, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200092, China.
Jian-Neng Chen, Department of Hepatology, Zhengxing Hospital, Zhangzhou 363000, Fujian Province, China.
Yu-Qin Wang, Department of Gastroenterology, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200092, China.
Qin Pan, Department of Gastroenterology, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200092, China. panqin@xinhuamed.com.cn.
Jian-Gao Fan, Department of Gastroenterology, Xinhua Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200092, China; Shanghai Key Laboratory of Children’s Digestion and Nutrition, Shanghai 200092, China.
References
- 1.Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 2.Wang FS, Fan JG, Zhang Z, Gao B, Wang HY. The global burden of liver disease: the major impact of China. Hepatology. 2014;60:2099–2108. doi: 10.1002/hep.27406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Williams CD, Stengel J, Asike MI, Torres DM, Shaw J, Contreras M, Landt CL, Harrison SA. Prevalence of nonalcoholic fatty liver disease and nonalcoholic steatohepatitis among a largely middle-aged population utilizing ultrasound and liver biopsy: a prospective study. Gastroenterology. 2011;140:124–131. doi: 10.1053/j.gastro.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 4.Caballería L, Pera G, Auladell MA, Torán P, Muñoz L, Miranda D, Alumà A, Casas JD, Sánchez C, Gil D, et al. Prevalence and factors associated with the presence of nonalcoholic fatty liver disease in an adult population in Spain. Eur J Gastroenterol Hepatol. 2010;22:24–32. doi: 10.1097/MEG.0b013e32832fcdf0. [DOI] [PubMed] [Google Scholar]
- 5.Fung J, Lee CK, Chan M, Seto WK, Lai CL, Yuen MF; Hong Kong Liver Health Census Study Group. High prevalence of non-alcoholic fatty liver disease in the Chinese - results from the Hong Kong liver health census. Liver Int. 2015;35:542–549. doi: 10.1111/liv.12619. [DOI] [PubMed] [Google Scholar]
- 6.Goh GB, Kwan C, Lim SY, Venkatanarasimha NK, Abu-Bakar R, Krishnamoorthy TL, Shim HH, Tay KH, Chow WC. Perceptions of non-alcoholic fatty liver disease - an Asian community-based study. Gastroenterol Rep (Oxf) 2016;4:131–135. doi: 10.1093/gastro/gov047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Calzadilla Bertot L, Adams LA. The Natural Course of Non-Alcoholic Fatty Liver Disease. Int J Mol Sci. 2016;17:pii: E774. doi: 10.3390/ijms17050774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ascha MS, Hanouneh IA, Lopez R, Tamimi TA, Feldstein AF, Zein NN. The incidence and risk factors of hepatocellular carcinoma in patients with nonalcoholic steatohepatitis. Hepatology. 2010;51:1972–1978. doi: 10.1002/hep.23527. [DOI] [PubMed] [Google Scholar]
- 9.Marchesini G, Marzocchi R. Metabolic syndrome and NASH. Clin Liver Dis. 2007;11:105–117, ix. doi: 10.1016/j.cld.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 10.Richard J, Lingvay I. Hepatic steatosis and Type 2 diabetes: current and future treatment considerations. Expert Rev Cardiovasc Ther. 2011;9:321–328. doi: 10.1586/erc.11.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gaggini M, Morelli M, Buzzigoli E, DeFronzo RA, Bugianesi E, Gastaldelli A. Non-alcoholic fatty liver disease (NAFLD) and its connection with insulin resistance, dyslipidemia, atherosclerosis and coronary heart disease. Nutrients. 2013;5:1544–1560. doi: 10.3390/nu5051544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mikolasevic I, Orlic L, Stimac D, Hrstic I, Jakopcic I, Milic S. Non-alcoholic fatty liver disease and colorectal cancer. Postgrad Med J. 2017;93:153–158. doi: 10.1136/postgradmedj-2016-134383. [DOI] [PubMed] [Google Scholar]
- 13.Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15:429. doi: 10.2174/138920101505140828161335. [DOI] [PubMed] [Google Scholar]
- 14.Mitra CK, Korla K. Functional, structural, and sequence studies of microRNA. Methods Mol Biol. 2014;1107:189–206. doi: 10.1007/978-1-62703-748-8_11. [DOI] [PubMed] [Google Scholar]
- 15.Feng YY, Xu XQ, Ji CB, Shi CM, Guo XR, Fu JF. Aberrant hepatic microRNA expression in nonalcoholic fatty liver disease. Cell Physiol Biochem. 2014;34:1983–1997. doi: 10.1159/000366394. [DOI] [PubMed] [Google Scholar]
- 16.Pogribny IP, Starlard-Davenport A, Tryndyak VP, Han T, Ross SA, Rusyn I, Beland FA. Difference in expression of hepatic microRNAs miR-29c, miR-34a, miR-155, and miR-200b is associated with strain-specific susceptibility to dietary nonalcoholic steatohepatitis in mice. Lab Invest. 2010;90:1437–1446. doi: 10.1038/labinvest.2010.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu XL, Pan Q, Zhang RN, Shen F, Yan SY, Sun C, Xu ZJ, Chen YW, Fan JG. Disease-specific miR-34a as diagnostic marker of non-alcoholic steatohepatitis in a Chinese population. World J Gastroenterol. 2016;22:9844–9852. doi: 10.3748/wjg.v22.i44.9844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamada H, Suzuki K, Ichino N, Ando Y, Sawada A, Osakabe K, Sugimoto K, Ohashi K, Teradaira R, Inoue T, et al. Associations between circulating microRNAs (miR-21, miR-34a, miR-122 and miR-451) and non-alcoholic fatty liver. Clin Chim Acta. 2013;424:99–103. doi: 10.1016/j.cca.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 19.Salvoza NC, Klinzing DC, Gopez-Cervantes J, Baclig MO. Association of Circulating Serum miR-34a and miR-122 with Dyslipidemia among Patients with Non-Alcoholic Fatty Liver Disease. PLoS One. 2016;11:e0153497. doi: 10.1371/journal.pone.0153497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding J, Li M, Wan X, Jin X, Chen S, Yu C, Li Y. Effect of miR-34a in regulating steatosis by targeting PPARα expression in nonalcoholic fatty liver disease. Sci Rep. 2015;5:13729. doi: 10.1038/srep13729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388. doi: 10.1038/nature11993. [DOI] [PubMed] [Google Scholar]
- 22.Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, Zhang Z, Lin QX, Zheng XL, -Yang M, et al. CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts. Sci Rep. 2017;7:40342. doi: 10.1038/srep40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res Commun. 2017;487:769–775. doi: 10.1016/j.bbrc.2017.04.044. [DOI] [PubMed] [Google Scholar]
- 24.Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al. A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223. Eur Heart J. 2016;37:2602–2611. doi: 10.1093/eurheartj/ehv713. [DOI] [PubMed] [Google Scholar]
- 25.Gómez-Lechón MJ, Donato MT, Martínez-Romero A, Jiménez N, Castell JV, O’Connor JE. A human hepatocellular in vitro model to investigate steatosis. Chem Biol Interact. 2007;165:106–116. doi: 10.1016/j.cbi.2006.11.004. [DOI] [PubMed] [Google Scholar]
- 26.Dudekula DB, Panda AC, Grammatikakis I, De S, Abdelmohsen K, Gorospe M. CircInteractome: A web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13:34–42. doi: 10.1080/15476286.2015.1128065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sobolewski C, Calo N, Portius D, Foti M. MicroRNAs in fatty liver disease. Semin Liver Dis. 2015;35:12–25. doi: 10.1055/s-0034-1397345. [DOI] [PubMed] [Google Scholar]
- 28.Griffiths-Jones S, Grocock RJ, van Dongen S, Bateman A, Enright AJ. miRBase: microRNA sequences, targets and gene nomenclature. Nucleic Acids Res. 2006;34:D140–D144. doi: 10.1093/nar/gkj112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yi M, Horton JD, Cohen JC, Hobbs HH, Stephens RM. WholePathwayScope: a comprehensive pathway-based analysis tool for high-throughput data. BMC Bioinformatics. 2006;7:30. doi: 10.1186/1471-2105-7-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marriott AS, Vasieva O, Fang Y, Copeland NA, McLennan AG, Jones NJ. NUDT2 Disruption Elevates Diadenosine Tetraphosphate (Ap4A) and Down-Regulates Immune Response and Cancer Promotion Genes. PLoS One. 2016;11:e0154674. doi: 10.1371/journal.pone.0154674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang HM, Do HJ, Kim DK, Park JK, Chang WK, Chung HM, Choi SY, Kim JH. Transcriptional regulation of human Oct4 by steroidogenic factor-1. J Cell Biochem. 2007;101:1198–1209. doi: 10.1002/jcb.21244. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22:256–264. doi: 10.1038/nsmb.2959. [DOI] [PubMed] [Google Scholar]
- 34.Lin SP, Ye S, Long Y, Fan Y, Mao HF, Chen MT, Ma QJ. Circular RNA expression alterations are involved in OGD/R-induced neuron injury. Biochem Biophys Res Commun. 2016;471:52–56. doi: 10.1016/j.bbrc.2016.01.183. [DOI] [PubMed] [Google Scholar]
- 35.Zhang XQ, Pan Y, Yu CH, Xu CF, Xu L, Li YM, Chen WX. PDIA3 Knockdown Exacerbates Free Fatty Acid-Induced Hepatocyte Steatosis and Apoptosis. PLoS One. 2015;10:e0133882. doi: 10.1371/journal.pone.0133882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Upreti D, Pathak A, Kung SK. Development of a standardized flow cytometric method to conduct longitudinal analyses of intracellular CD3ζ expression in patients with head and neck cancer. Oncol Lett. 2016;11:2199–2206. doi: 10.3892/ol.2016.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Q, Zhang X, Hu X, Dai L, Fu X, Zhang J, Ao Y. Circular RNA Related to the Chondrocyte ECM Regulates MMP13 Expression by Functioning as a MiR-136 ‘Sponge’ in Human Cartilage Degradation. Sci Rep. 2016;6:22572. doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Xu H, Guo S, Li W, Yu P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci Rep. 2015;5:12453. doi: 10.1038/srep12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Alexandrov PN, Jaber V, Lukiw WJ. Deficiency in the Ubiquitin Conjugating Enzyme UBE2A in Alzheimer’s Disease (AD) is Linked to Deficits in a Natural Circular miRNA-7 Sponge (circRNA; ciRS-7) Genes (Basel) 2016;7:pii: E116. doi: 10.3390/genes7120116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Xie H, Ren X, Xin S, Lan X, Lu G, Lin Y, Yang S, Zeng Z, Liao W, Ding YQ, et al. Emerging roles of circRNA_001569 targeting miR-145 in the proliferation and invasion of colorectal cancer. Oncotarget. 2016;7:26680–26691. doi: 10.18632/oncotarget.8589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng Q, Bao C, Guo W, Li S, Chen J, Chen B, Luo Y, Lyu D, Li Y, Shi G, et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7:11215. doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Dorn C, Riener MO, Kirovski G, Saugspier M, Steib K, Weiss TS, Gäbele E, Kristiansen G, Hartmann A, Hellerbrand C. Expression of fatty acid synthase in nonalcoholic fatty liver disease. Int J Clin Exp Pathol. 2010;3:505–514. [PMC free article] [PubMed] [Google Scholar]
- 43.Glažar P, Papavasileiou P, Rajewsky N. circBase: a database for circular RNAs. RNA. 2014;20:1666–1670. doi: 10.1261/rna.043687.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li WQ, Chen C, Xu MD, Guo J, Li YM, Xia QM, Liu HM, He J, Yu HY, Zhu L. The rno-miR-34 family is upregulated and targets ACSL1 in dimethylnitrosamine-induced hepatic fibrosis in rats. FEBS J. 2011;278:1522–1532. doi: 10.1111/j.1742-4658.2011.08075.x. [DOI] [PubMed] [Google Scholar]
- 45.Koufaris C, Wright J, Currie RA, Gooderham NJ. Hepatic microRNA profiles offer predictive and mechanistic insights after exposure to genotoxic and epigenetic hepatocarcinogens. Toxicol Sci. 2012;128:532–543. doi: 10.1093/toxsci/kfs170. [DOI] [PubMed] [Google Scholar]
- 46.Pok S, Wen V, Shackel N, Alsop A, Pyakurel P, Fahrer A, Farrell GC, Teoh NC. Cyclin E facilitates dysplastic hepatocytes to bypass G1/S checkpoint in hepatocarcinogenesis. J Gastroenterol Hepatol. 2013;28:1545–1554. doi: 10.1111/jgh.12216. [DOI] [PubMed] [Google Scholar]
- 47.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42:D68–D73. doi: 10.1093/nar/gkt1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huang DW, Sherman BT, Tan Q, Kir J, Liu D, Bryant D, Guo Y, Stephens R, Baseler MW, Lane HC, et al. DAVID Bioinformatics Resources: expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007;35:W169–W175. doi: 10.1093/nar/gkm415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettinelli P, Del Pozo T, Araya J, Rodrigo R, Araya AV, Smok G, Csendes A, Gutierrez L, Rojas J, Korn O, et al. Enhancement in liver SREBP-1c/PPAR-alpha ratio and steatosis in obese patients: correlations with insulin resistance and n-3 long-chain polyunsaturated fatty acid depletion. Biochim Biophys Acta. 2009;1792:1080–1086. doi: 10.1016/j.bbadis.2009.08.015. [DOI] [PubMed] [Google Scholar]
- 50.Janssen AW, Betzel B, Stoopen G, Berends FJ, Janssen IM, Peijnenburg AA, Kersten S. The impact of PPARα activation on whole genome gene expression in human precision cut liver slices. BMC Genomics. 2015;16:760. doi: 10.1186/s12864-015-1969-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee K, Kerner J, Hoppel CL. Mitochondrial carnitine palmitoyltransferase 1a (CPT1a) is part of an outer membrane fatty acid transfer complex. J Biol Chem. 2011;286:25655–25662. doi: 10.1074/jbc.M111.228692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Orellana-Gavaldà JM, Herrero L, Malandrino MI, Pañeda A, Sol Rodríguez-Peña M, Petry H, Asins G, Van Deventer S, Hegardt FG, Serra D. Molecular therapy for obesity and diabetes based on a long-term increase in hepatic fatty-acid oxidation. Hepatology. 2011;53:821–832. doi: 10.1002/hep.24140. [DOI] [PubMed] [Google Scholar]
- 53.Tan L, Narayan SB, Chen J, Meyers GD, Bennett MJ. PTC124 improves readthrough and increases enzymatic activity of the CPT1A R160X nonsense mutation. J Inherit Metab Dis. 2011;34:443–447. doi: 10.1007/s10545-010-9265-5. [DOI] [PubMed] [Google Scholar]
- 54.Kirpich I, Ghare S, Zhang J, Gobejishvili L, Kharebava G, Barve SJ, Barker D, Moghe A, McClain CJ, Barve S. Binge alcohol-induced microvesicular liver steatosis and injury are associated with down-regulation of hepatic Hdac 1, 7, 9, 10, 11 and up-regulation of Hdac 3. Alcohol Clin Exp Res. 2012;36:1578–1586. doi: 10.1111/j.1530-0277.2012.01751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Litherland NB, Bionaz M, Wallace RL, Loor JJ, Drackley JK. Effects of the peroxisome proliferator-activated receptor-alpha agonists clofibrate and fish oil on hepatic fatty acid metabolism in weaned dairy calves. J Dairy Sci. 2010;93:2404–2418. doi: 10.3168/jds.2009-2716. [DOI] [PubMed] [Google Scholar]
- 56.Herrmann T, Buchkremer F, Gosch I, Hall AM, Bernlohr DA, Stremmel W. Mouse fatty acid transport protein 4 (FATP4): characterization of the gene and functional assessment as a very long chain acyl-CoA synthetase. Gene. 2001;270:31–40. doi: 10.1016/s0378-1119(01)00489-9. [DOI] [PubMed] [Google Scholar]
- 57.Gallardo D, Amills M, Quintanilla R, Pena RN. Mapping and tissue mRNA expression analysis of the pig solute carrier 27A (SLC27A) multigene family. Gene. 2013;515:220–223. doi: 10.1016/j.gene.2012.11.029. [DOI] [PubMed] [Google Scholar]
- 58.Qiu F, Xie L, Ma JE, Luo W, Zhang L, Chao Z, Chen S, Nie Q, Lin Z, Zhang X. Lower Expression of SLC27A1 Enhances Intramuscular Fat Deposition in Chicken via Down-Regulated Fatty Acid Oxidation Mediated by CPT1A. Front Physiol. 2017;8:449. doi: 10.3389/fphys.2017.00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lenz LS, Marx J, Chamulitrat W, Kaiser I, Gröne HJ, Liebisch G, Schmitz G, Elsing C, Straub BK, Füllekrug J, et al. Adipocyte-specific inactivation of Acyl-CoA synthetase fatty acid transport protein 4 (Fatp4) in mice causes adipose hypertrophy and alterations in metabolism of complex lipids under high fat diet. J Biol Chem. 2011;286:35578–35587. doi: 10.1074/jbc.M111.226530. [DOI] [PMC free article] [PubMed] [Google Scholar]


