Abstract
Esophageal carcinoma is a common malignancy worldwide, with a low 5-year survival rate. As the majority of cases are diagnosed at an advanced stage, there is an urgent need for an effective biomarker for early diagnosis of esophageal cancer patients. Surface-enhanced laser desorption ionization time-of-flight mass spectrometry (SELDI-TOF-MS) was applied to detect the serum protein expression in esophageal cancer patients using ProteinChip software, and the results were analyzed and screened using Biomarker Patterns and SPSS16.0 software. The ELISA method was conducted to determine the concentration of anaphylatoxin C3a, which is one of the complement proteins, in the serum of esophageal cancer patients and non-esophageal cancer participants. A total of 144 effective differential expression protein peaks in the window of 1–10 kDa were obtained (P<0.05). M/Z 8,926.478 (P<10−6) protein peak was employed as the diagnostic biomarker for esophageal carcinoma. This established diagnostic biomarker has a sensitivity of 95% (19/20) and an accuracy of 100% (19/19) for positive prediction. The results suggested that anaphylatoxin C3a may be a promising biomarker in the diagnosis of esophageal carcinoma, and may play a key role in promoting esophageal carcinogenesis.
Keywords: esophageal carcinoma, surface-enhanced laser desorption ionization time-of-flight mass spectrometry, anaphylatoxin C3a, diagnostic biomarker, tumor heterogeneity, immune escape
Introduction
Esophageal carcinoma is a common malignancy worldwide, with a 5-year survival rate of ~30%, even with chemotherapy, surgery and radiation therapy, due to tumor heterogeneity (1–9). It is well-known that early diagnosis and timely treatment may improve the survival rate in early-stage cancer patients, with a 5-year survival rate as high as 90% (10,11). Cancer Research UK (http://www.cancerresearchuk.org/about-cancer/cancer-symptoms/why-is-early-diagnosis-important) clearly reported the association of survival rate with stage at diagnosis for several cancers, such as breast, ovarian, lung and bowel cancer. However, the majority of esophageal cancer patients are often diagnosed at an advanced stage, as there are no obvious symptoms in the early stages of the disease. Therefore, it is crucial to identify an effective biomarker for early diagnosis. Recently, the matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MOLDI-TOF-MS) and surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-TOF-MS) techniques have been widely employed in the search for cancer biomarkers in brain cancer (12), oral squamous cell carcinoma (13), pancreatic cancer (14), lung cancer (15), esophageal squamous cell carcinoma (16) and breast cancer (17,18), among others. There have been several attempts to identify esophageal carcinoma biomarkers using these techniques (16,19–22). However, those studies failed to identify any single biomarker for the diagnosis of esophageal cancer, but rather indicated several proteins. In addition, the identified proteins differed among different research groups. This makes it difficult to establish a unified standard for rapid and accurate diagnosis of this type of cancer. In the present study, a single protein, anaphylatoxin C3a, which is one of the complement proteins, was investigated as a diagnostic biomarker to distinguish between esophageal cancer patients and healthy individuals.
Materials and methods
Research subjects
A total of 40 serum samples were included in this study, 20 of which were collected from esophageal carcinoma patients from Yancheng First People's Hospital (group A), whereas the others were collected from non-esophageal carcinoma participants recruited from the First Hospital of Nanjing Medical University (group B) between November 2014 and May 2015. The participants were aged 25–76 years, with a mean age of 55.7 years. In group A, there were 8 cases of patients who were undergoing therapy. All the blood collections were performed an overnight fast. Blood was collected in 5-ml blood collection tubes without anticoagulant and the serum samples were stored at −80°C for further use.
Ethics statement
All patients signed an informed consent form and the study protocol was approved by the Institutional Review Board of the First People's Hospital of Yancheng (Yancheng, China). The study was reviewed and approved by Yancheng Medical Ethics Committee.
SELDI-TOF-MS assay
SELDI-TOF-MS (Ciphergen Biosystems, Fremont, CA, USA) was applied to collect the raw mass spectrometry data. Biomarker Wizard software (Ciphergen Biosystems, Fremont, CA, USA) was used to export the raw data into a digital format, according to the standard Excel format. A t-test was conducted with the Excel data using SPSS 17.0 software (SPSS Inc., Chicago, IL, USA).
Enzyme-linked immunoabsorbent assay (ELISA)
The concentrations of C3a in the serum samples were quantified by ELISA according to the manufacturer's protocol (cat. no. ab133037; Abcam, Cambridge, MA, USA). Briefly, 50 µl standard samples and 50 µl serum samples were diluted 5 times with phosphate-buffered saline, placed into a 96-well plate and cultured at 37°C for 30 min. The plate was washed 5 times, after which time 50 µl reagent A was added into each well, followed by 50 µl reagent B. The samples were mixed well and cultured for a further 15 min at 37°C in the dark; stop solution was then added into each well. An ELISA plate reader (Synergy HXT; BioTek Instruments Inc., Winooski, VT, USA) was used at a wavelength of 450 nm; the inter-assay and intra-assay coefficients of variation of the ELISA kits for C3a were <10%.
Statistical analysis
The SPSS 19.0 software package (SPSS Inc., Chicago, IL, USA) was used for data analysis, and the data are expressed as means ± standard deviation. The comparison between the groups was performed using one-way analysis of variance. P<0.05 indicates that the difference was statistically significant.
Results
Serum proteomic profiles
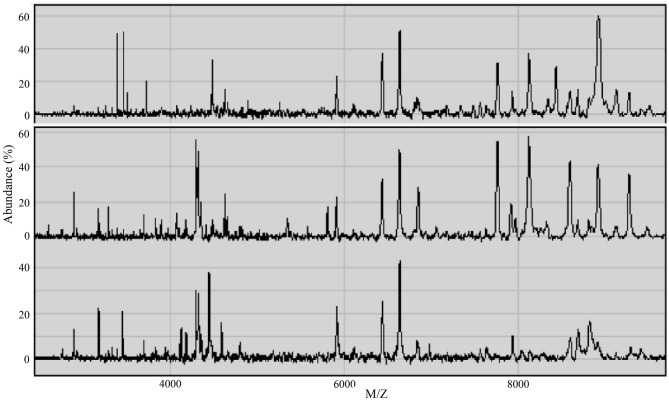
SELDI-TOF-MS was applied to examine the serum samples within a window of 1–10 kDa, and 253 protein peaks were detected in the sera of esophageal cancer patients. There were 144 statistically significant difference peaks (P<0.05): 56 peaks had higher protein expression and 88 had lower protein expression in esophageal cancer patients. The mass spectra (MS) of the serum proteins of esophageal cancer patients and non-esophageal cancer participants are shown in Fig. 1.
Figure 1.
Protein mass spectrum peaks in the sera of esophageal cancer patients prior to treatment (upper) and after treatment (middle), and of non-esophageal carcinoma participants (bottom).
Establishment of diagnostic biomarker
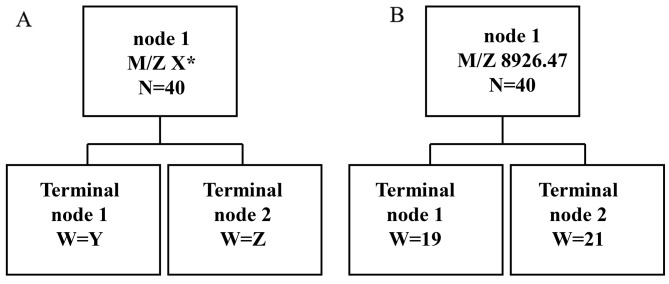
In order to establish the diagnostic biomarker, 6 MS peaks, i.e., M/Z 2,748.87 (P=2.38×10−7), M/Z 4,119.31 (P=3.18×10−7), M/Z 4,425.94 (P=2.38×10−7), M/Z 4,798.62 (P=3.67×10−7), M/Z 9,136.76 (P=6.45×10−7) and M/Z 8,926.47 (P=7.33×10−8), exhibiting statistically significant differences, were further analyzed. The classical tree model was employed to examine the predictor variables (Fig. 2A). The protein M/Z 8,926.47 had the best prediction result (Fig. 2B). The sensitivity and specificity of the biomarker were 95% (19/20) and 97.5% (39/40), respectively. The positive predictive accuracy was 100% (19/19) and the negative predictive accuracy was 95.2% (20/21). Thus, the protein at M/Z 8,926.47 may be an optimal biomarker for distinguishing the esophageal cancer and non-esophageal cancer sera with high accuracy and high specificity.
Figure 2.
(A) The classical tree model; (B) the predicted results of M/Z 8,926.47.
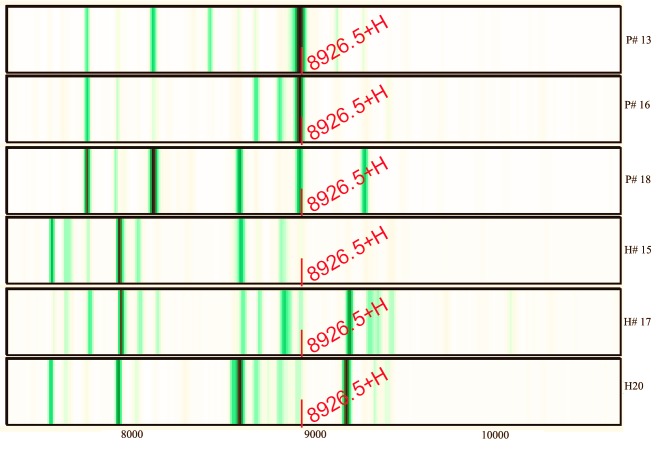
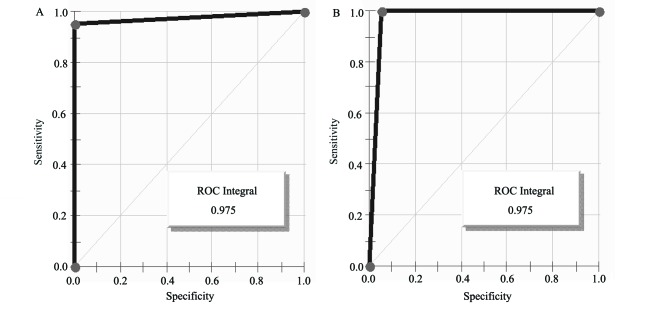
Further study indicated that the abundance of M/Z 8,926.47 was reduced from 60.69 prior to therapy to 43.39 after therapy (the mean abundance of this peak was 6.81 for non-esophageal carcinoma participants). Three of the serum protein MS of esophageal cancer patients and non-esophageal carcinoma participants are shown in Fig. 3. The intensities of M/Z 8,926.47 in the non-esophageal carcinoma participants were distinctly lower compared with those in esophageal carcinoma patients. The areas under the receiver operating characteristic curves of the diagnostic biomarker were 97.5% (Fig. 4).
Figure 3.
Serum protein mass spectra. Upper three, esophageal carcinoma patients (P); bottom three, non-esophageal carcinoma participants (healthy; H).
Figure 4.
Receiver operating characteristics (ROC) curves of diagnostic biomarker. (A) ROC curve of diagnostic biomarker of patients; (B) ROC curve of diagnostic biomarker of non-esophageal carcinoma participants.
Determination of the concentration of C3a via ELISA
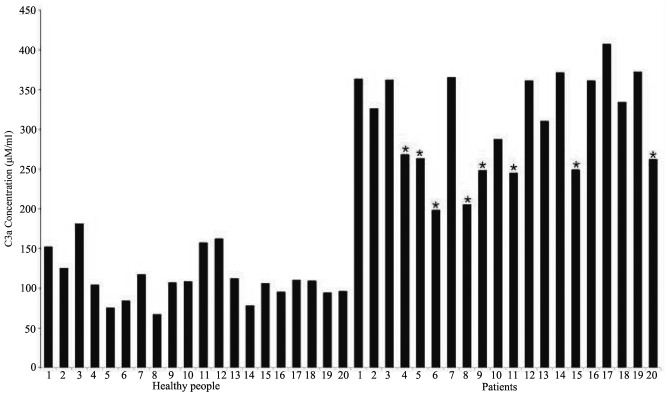
The human C3a ELISA kit was used to determine the concentration of anaphylatoxin C3a using the obtained serum samples. The ELISA experimental results (Fig. 5) revealed that the concentration of anaphylatoxin C3a in the sera of esophageal cancer patients (mean, 308±60 ng/ml) were significantly higher compared with those in healthy participants (mean, 112±30 ng/ml). Among esophageal carcinoma patients, the mean concentration of C3a in the sera of those without therapy and those undergoing therapy was 352±31 and 242±25 ng/ml, respectively.
Figure 5.
Concentrations of C3a in different serum samples determined by ELISA. The first 20 bars represent the C3a concentrations in healthy participants, whereas the remaining bars represent those in esophageal cancer patients (P<0.01). *sera from esophageal carcinoma patients undergoing chemotherapy and surgery.
Discussion
Regarding the SELDI-TOF MS results, significant differences were observed in the serum samples between the esophageal carcinoma patients and healthy participants for 6 protein peaks (P<10−6). Among those, the abundance of the peak M/Z 8,926.47 (P=7.33×10−8) markedly increased from 6.81 (non-esophageal carcinoma) to 60.69 (esophageal carcinoma), suggesting that the expression of M/Z 8,926.47 was significantly increased in the plasma of esophageal carcinoma patients. This particular peak distinguished between the sera of esophageal carcinoma patients and non-esophageal carcinoma participants with high accuracy (100%) and high efficiency (97.5%), indicating that M/Z 8,926.47 may be a promising biomarker for early diagnosis. Based on our experience, this peak is likely to be complementary to the C3a protein.
The ELISA results demonstrated that the concentrations were statistically significantly different (P<0.01) and the mean concentration of anaphylatoxin C3a in esophageal carcinoma and non-esophageal carcinoma samples was 308±60 and 112±30 ng/ml, respectively, which was in agreement with the MS peak abundance of 53.98 and 6.81. Furthermore, among esophageal cancer samples, the C3a concentration was 352±31 and 242±25 ng/ml for samples before and after treatment, respectively, which was also in agreement with the MS peak abundances of 60.69 and 43.39. This finding indicates that, after treatment, the concentration of C3a in the plasma was markedly decreased. According to the ELISA results, it may be concluded that, when the serum C3a concentration is >120 ng/ml, the risk of esophageal cancer is increased.
C3a is a serum protein first discovered in 1896, which plays a key role in either antitumor immune response (23–27), or in promoting tumor growth and progression (28,29). However, the role of C3a in esophageal cancer has not been determined to date. According to the MS and ELISA results, the anaphylatoxin C3a concentrations in the sera of treated patients are significantly lower compared with those without treatment. It may be hypothesized that C3a plays a key role in promoting esophageal tumorigenesis. As previously reported, anaphylatoxin C3a may contribute to cancer cell immune escape via promoting local immunosuppression (30,31). However, more studies should be conducted to elucidate the mechanisms through which C3a promotes esophageal tumorigenesis.
Acknowledgements
The authors would like to acknowledge the Department of Human Resources and Social Security of Jiangsu Province (swyy-030) and Foundation of Jiangsu Educational Committee (16KJB180031) for financially supporting the present study. Esophageal carcinoma serum samples were provided by Dr Zhou Xiaoning (Oncology Department, Yancheng First People's Hospital). Healthy participants' serum samples were provided by Dr Shao Qianwen (Oncology Department, the First Hospital of Nanjing Medical University).
References
- 1.Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: Implications for targeted therapeutics. Br J Cancer. 2013;108:479–485. doi: 10.1038/bjc.2012.581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graff BA, Kvinnsland Y, Skretting A, Rofstad EK. Intratumour heterogeneity in the uptake of macromolecular therapeutic agents in human melanoma xenografts. Br J Cancer. 2003;88:291–297. doi: 10.1038/sj.bjc.6600680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim JH, Ko ES, Lim Y, Lee KS, Han BK, Ko EY, Hahn SY, Nam SJ. Breast cancer heterogeneity: MR imaging texture analysis and survival outcomes. Radiology. 2017;282:665–675. doi: 10.1148/radiol.2016160261. [DOI] [PubMed] [Google Scholar]
- 4.Beca F, Polyak K. Intratumor heterogeneity in breast cancer. Adv Exp Med Biol. 2016;882:169–189. doi: 10.1007/978-3-319-22909-6_7. [DOI] [PubMed] [Google Scholar]
- 5.Aleskandarany MA, Green AR, Ashankyty I, Elmouna A, Diez-Rodriguez M, Nolan CC, Ellis IO, Rakha EA. Impact of intratumoural heterogeneity on the assessment of Ki67 expression in breast cancer. Breast Cancer Res Treat. 2016;158:287–295. doi: 10.1007/s10549-016-3893-x. [DOI] [PubMed] [Google Scholar]
- 6.Suda K, Murakami I, Sakai K, Tomizawa K, Mizuuchi H, Sato K, Nishio K, Mitsudomi T. Heterogeneity in resistance mechanisms causes shorter duration of epidermal growth factor receptor kinase inhibitor treatment in lung cancer. Lung Cancer. 2016;91:36–40. doi: 10.1016/j.lungcan.2015.11.016. [DOI] [PubMed] [Google Scholar]
- 7.Bashir U, Siddique MM, Mclean E, Goh V, Cook GJ. Imaging heterogeneity in lung cancer: Techniques, applications, and challenges. AJR Am J Roentgenol. 2016;207:534–543. doi: 10.2214/AJR.15.15864. [DOI] [PubMed] [Google Scholar]
- 8.Obulkasim A, Ylstra B, van Essen HF, Benner C, Stenning S, Langley R, Allum W, Cunningham D, Inam I, Hewitt LC, et al. Reduced genomic tumor heterogeneity after neoadjuvant chemotherapy is related to favorable outcome in patients with esophageal adenocarcinoma. Oncotarget. 2016;7:44084–44095. doi: 10.18632/oncotarget.9857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao W, Wu W, Yan M, Tian F, Liu HS, Wang JW, Zhang QW, Li YJ, Li M. Multiregion sequencing reveals intratumor heterogeneity in esophageal squamous cell carcinoma. Zhonghua Zhong Liu Za Zhi. 2016;38:660–666. doi: 10.3760/cma.j.issn.0253-3766.2016.09.005. (In Chinese) [DOI] [PubMed] [Google Scholar]
- 10.Chen LQ, Hu CY, Ghadirian P, Duranceau A. Early detection of esophageal squamous cell carcinoma and its effects on therapy: An overview. Dis Esophagus. 1999;12:161–167. doi: 10.1046/j.1442-2050.1999.00039.x. [DOI] [PubMed] [Google Scholar]
- 11.Schweigert M, Dubecz A, Stein HJ. Oesophageal cancer-an overview. Nat Rev Gastroenterol Hepatol. 2013;10:230–244. doi: 10.1038/nrgastro.2012.236. [DOI] [PubMed] [Google Scholar]
- 12.Spalding K, Board R, Dawson T, Jenkinson MD, Baker MJ. A review of novel analytical diagnostics for liquid biopsies: Spectroscopic and spectrometric serum profiling of primary and secondary brain tumors. Brain Behav. 2016;6:e00502. doi: 10.1002/brb3.502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xie X, Jiang Y, Yuan Y, Wang P, Li X, Chen F, Sun C, Zhao H, Zeng X, Jiang L, et al. MALDI imaging reveals NCOA7 as a potential biomarker in oral squamous cell carcinoma arising from oral submucous fibrosis. Oncotarget. 2016;7:59987–60004. doi: 10.18632/oncotarget.11046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Potjer TP, Mertens BJ, Nicolardi S, van der Burgt YE, Bonsing BA, Mesker WE, Tollenaar RA, Vasen HF. Application of a serum protein signature for pancreatic cancer to separate cases from controls in a pancreatic surveillance cohort. Transl Oncol. 2016;9:242–247. doi: 10.1016/j.tranon.2016.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ren J, Zhang D, Liu Y, Zhang R, Fang H, Guo S, Zhou D, Zhang M, Xu Y, Qiu L, Li Z. Simultaneous quantification of serum nonesterified and esterified fatty acids as potential biomarkers to differentiate benign lung diseases from lung cancer. Sci Rep. 2016;6:34201. doi: 10.1038/srep34201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jia K, Li W, Wang F, Qu H, Qiao Y, Zhou L, Sun Y, Ma Q, Zhao X. Novel circulating peptide biomarkers for esophageal squamous cell carcinoma revealed by a magnetic bead-based MALDI-TOFMS assay. Oncotarget. 2016;7:23569–23580. doi: 10.18632/oncotarget.8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Chen X, Luan H, Gao D, Lin S, Cai Z, Liu J, Liu H, Jiang Y. Matrix-assisted laser desorption/ionization mass spectrometry imaging of cell cultures for the lipidomic analysis of potential lipid markers in human breast cancer invasion. Rapid Commun Mass Spectrom. 2016;30:533–542. doi: 10.1002/rcm.7466. [DOI] [PubMed] [Google Scholar]
- 18.Kawaguchi-Sakita N, Kaneshiro-Nakagawa K, Kawashima M, Sugimoto M, Tokiwa M, Suzuki E, Kajihara S, Fujita Y, Iwamoto S, Tanaka K, Toi M. Serum immunoglobulin G Fc region N-glycosylation profiling by matrix-assisted laser desorption/ionization mass spectrometry can distinguish breast cancer patients from cancer-free controls. Biochem Biophys Res Commun. 2016;469:1140–1145. doi: 10.1016/j.bbrc.2015.12.114. [DOI] [PubMed] [Google Scholar]
- 19.Zhao J, Fan YX, Yang Y, Liu DL, Wu K, Wen FB, Zhang CY, Zhu DY, Zhao S. Identification of potential plasma biomarkers for esophageal squamous cell carcinoma by a proteomic method. Int J Clin Exp Pathol. 2015;8:1535–1544. [PMC free article] [PubMed] [Google Scholar]
- 20.Schwacke J, Millar TP, Hammond CE, Saha A, Hoffman BJ, Romagnuolo J, Hill EG, Smolka AJ. Discrimination of normal and esophageal cancer plasma proteomes by MALDI-TOF mass spectrometry. Dig Dis Sci. 2015;60:1645–1654. doi: 10.1007/s10620-014-3513-8. [DOI] [PubMed] [Google Scholar]
- 21.Fan NJ, Gao CF, Wang XL. Tubulin beta chain, filamin A alpha isoform 1, and cytochrome b-c1 complex subunit 1 as serological diagnostic biomarkers of esophageal squamous cell carcinoma: A proteomics study. OMICS. 2013;17:215–223. doi: 10.1089/omi.2012.0133. [DOI] [PubMed] [Google Scholar]
- 22.Zhai XH, Yu JK, Lin C, Wang LD, Zheng S. Combining proteomics, serum biomarkers and bioinformatics to discriminate between esophageal squamous cell carcinoma and pre-cancerous lesion. J Zhejiang Univ Sci B. 2012;13:964–971. doi: 10.1631/jzus.B1200066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weiner LM, Surana R, Wang S. Monoclonal antibodies: Versatile platforms for cancer immunotherapy. Nat Rev Immunol. 2010;10:317–327. doi: 10.1038/nri2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rutkowski MJ, Sughrue ME, Kane AJ, Mills SA, Parsa AT. Cancer and the complement cascade. Mol Cancer Res. 2010;8:1453–1465. doi: 10.1158/1541-7786.MCR-10-0225. [DOI] [PubMed] [Google Scholar]
- 25.Sliwkowski MX, Mellman I. Antibody therapeutics in cancer. Science. 2013;341:1192–1198. doi: 10.1126/science.1241145. [DOI] [PubMed] [Google Scholar]
- 26.Taylor RP, Lindorfer MA. The role of complement in mAb-based therapies of cancer. Methods. 2014;65:18–27. doi: 10.1016/j.ymeth.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 27.Baig NA, Taylor RP, Lindorfer MA, Church AK, LaPlant BR, Pettinger AM, Shanafelt TD, Nowakowski GS, Zent CS. Induced resistance to ofatumumab-mediated cell clearance mechanisms, including complement-dependent cytotoxicity, in chronic lymphocytic leukemia. J Immunol. 2014;192:1620–1629. doi: 10.4049/jimmunol.1302954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Markiewski MM, DeAngelis RA, Benencia F, Ricklin-Lichtsteiner SK, Koutoulaki A, Gerard C, Coukos G, Lambris JD. Modulation of the antitumor immune response by complement. Nat Immunol. 2008;9:1225–1235. doi: 10.1038/ni.1655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan MA, Assiri AM, Broering DC. Complement and macrophage crosstalk during process of angiogenesis in tumor progression. J Biomed Sci. 2015;22:58. doi: 10.1186/s12929-015-0151-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sayegh ET, Bloch O, Parsa AT. Complement anaphylatoxins as immune regulators in cancer. Cancer Med. 2014;3:747–758. doi: 10.1002/cam4.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Denko NC, Fontana LA, Hudson KM, Sutphin PD, Raychaudhuri S, Altman R, Giaccia AJ. Investigating hypoxic tumor physiology through gene expression patterns. Oncogene. 2003;22:5907–5914. doi: 10.1038/sj.onc.1206703. [DOI] [PubMed] [Google Scholar]