Abstract
Bacterial genomes are rife with orphan biosynthetic gene clusters (BGCs) associated with secondary metabolism of unrealized natural product molecules. Often up to a tenth of the genome is predicted to code for the biosynthesis of diverse metabolites with mostly unknown structures and functions. This phenomenal diversity of BGCs coupled with their high rates of horizontal transfer raise questions about whether they are really active and beneficial, whether they are neutral and confer no advantage, or whether they are carried in genomes because they are parasitic or addictive. We previously reported that Salinispora bacteria broadly use the desferrioxamine family of siderophores for iron acquisition. Herein we describe a new and unrelated group of peptidic siderophores called salinichelins from a restricted number of Salinispora strains in which the desferrioxamine biosynthesis genes have been lost. We have reconstructed the evolutionary history of these two different siderophore families and show that the acquisition and retention of the new salinichelin siderophores co-occurs with the loss of the more ancient desferrioxamine pathway. This identical event occurred at least three times independently during the evolution of the genus. We surmise that certain BGCs may be extraneous because of their functional redundancy and demonstrate that the relative evolutionary pace of natural pathway replacement shows high selective pressure against retention of functionally superfluous gene clusters.
Introduction
Bacteria produce a vast array of natural product chemicals that are largely known for their essential use as life-saving medicinal drugs (Cragg and Newman, 2013). The ecological roles of these specialized metabolites are often as diverse as their chemical structures, and include defense, communication and nutrient acquisition (Davies, 2013). Indeed certain genera dedicate up to 10% of their genomes to secondary metabolism (Nett et al., 2009), and the genes that are responsible for production of these metabolites usually reside in biosynthetic gene clusters (BGCs). A variety of computer programs and web tools (Medema et al., 2011; Ziemert et al., 2012; Skinnider et al., 2015; Weber et al., 2015) now allow rapid and straightforward detection of BGCs, with recent surveys of bacterial genomes predicting the existence of large numbers of different gene cluster families (Cimermancic et al., 2014; Schorn et al., 2016). However, despite the wealth of knowledge concerning the existence of these BGCs, most have yet to be linked to the small molecules they encode. Bioinformatic analysis, molecular biology and organic chemistry approaches have been established in recent years to connect unknown BGCs to molecules with the goal of discovering new drug leads and furthering our understanding of the natural world (Nett, 2014; Medema and Fischbach, 2015; Ziemert et al., 2016).
With the identification of so many BGC families came the realization that there was enormous diversity of BGCs, even within closely related species (Ziemert et al., 2014). In addition, high rates of horizontal gene transfer and recombination (Medema et al., 2014) have been observed, and these findings raise questions concerning how many of these clusters are really functional in the genome in which they are found, and how many actually encode a chemical product. The ratio of functional to non-functional BGCs and how fast selective processes work on loss or maintenance of horizontally acquired BGCs is currently unknown (Jensen, 2016).
Population genetic theory suggests that free-living prokaryotes have sufficiently large long-term effective population sizes so that natural selection can overcome random genetic drift, even for small selective effects (Eyre-Walker and Keightley, 2007). This theory suggests that even slightly deleterious variations would be quickly purged from a population, yet enormous amounts of variation, recombination and gene transfer persists. Maintaining non-expressed BGCs seems difficult for a free-living prokaryote. Although the cost of replication of a small section of a genome is not so high in terms of energy (Lynch and Marinov, 2015), it would still be subject to selection in organisms with large long-term effective population sizes. We would therefore expect genomes containing non-functional BGCs to lose out to genomic variants where the non-functioning BGC had been deleted. However, if BGCs were ‘selfish’, as has been suggested (Fischbach et al., 2008) or addictive in some way, as for example in the case of toxin–antitoxin systems (Rocker and Meinhart, 2016) then their retention may not be because of increased fitness of the host genome, rather BGCs would behave more like parasites. In order to explore these concepts, it is imperative to understand the functional roles of BGCs and to reconstruct their evolutionary history. Here, we uncover a case of two functionally related pathways that appear to be mutually exclusive in the same genome. We demonstrate that in three separate instances the acquisition of one pathway by horizontal transfer results in the displacement of another.
Siderophores are important iron-chelating molecules (Saha et al., 2013). Widespread within the bacterial kingdom, a variety of siderophore chemical families with diverse biosynthetic origins have evolved to capture, solubilize and deliver essential Fe(III) ions into the cytoplasm (Sandy and Butler, 2009). Siderophores often contain specific functional groups, mainly catecholates and hydroxamates, to chelate iron and other metals with high affinity. Their BGCs usually contain special transporter genes for the active export and uptake of the siderophore molecules in addition to genes encoding the biosynthetic machinery. In the model marine actinomycete bacterium Salinispora (Jensen et al., 2015), genome mining studies revealed that the well-known siderophore desferrioxamine is common among the three distinct but closely related species. We report herein that some Salinispora strains, however, replace the desferrioxamine BGC (des) with a functionally equivalent yet chemically unrelated siderophore, encoded by non-homologous genes.
Following the evolutionary history of both pathways within the genus revealed that acquisition of one siderophore correlates to the loss of the other. Three times independently in the history of the genus, a whole biosynthetic pathway has been deleted when a new siderophore pathway was acquired horizontally. This observation shows not only that strong selection pressure on genome size exists, but also that functionally redundant segments of DNA are rapidly excised. This in turn suggests that a majority of BGCs are likely to be functional even if they remain resistant to being expressed under laboratory conditions. It also demonstrates a case of mutual exclusivity of biosynthetic pathways, in this case because the presence of both is functionally redundant.
Materials and methods
Genome mining analysis
Salinispora genomes were analyzed with antiSMASH 2.0 (Blin et al., 2013), the JGI IMG (integrated microbial genomes database expert review (https://img.jgi.doe.gov/er)) and NaPDoS (Ziemert et al., 2012). Bioinformatic predictions of adenylation domain specificities were carried out with nonribosomal peptide synthetase (NRPS) predictor 2.0 (Röttig et al., 2011).
Culture and extraction of strains and conversion into Ga/Fe complexes
For analytical purposes, Salinispora strains were cultured at 30 °C with continuous shaking at 220 r.p.m. in iron-limited media (Roberts et al., 2012), supplemented with 36 μM FeSO4, when required. The extraction solvent was n-BuOH. The extract, dissolved in MeOH, was often treated with 100 mM FeCl3 or GaBr3.
For the isolation of siderophores, Salinispora pacifica CNY-331 was grown in 1 liter of iron-limited media supplemented with 0.2 mM nalidixic acid for 7–12 days. The supernatant was treated with 100 mg l–1 GaBr3. After addition of activated XAD-7 resin (15 g l–1), the resin was filtered off, washed and extracted with acetone. After evaporation of acetone, lyophilization yielded a dark brown residue, which was dissolved in water, filtered and subjected to HPLC chromatography.
For HPLC coupled with mass spectrometry (HPLC-MS) screening of salinichelin production, Salinispora strains were grown on iron-limited agar (Roberts et al., 2012). When cells started to sporulate, a plug of agar and cells was extracted with n-BuOH by sonication, filtered and subjected to HPLC-MS analysis.
HPLC analysis of salinichelins
Analytical HPLC was carried out on an Agilent (Agilent, Santa Clara, CA, USA) 1200 instrument. Culture extracts were analyzed by analytical HPLC on a Luna C18 column, (5 μm, 100 A, 150 × 4.6 mm, Phenomenex, Torrence, CA, USA) with a flow rate of 0.7 ml min–1 and an isocratic flow of 98% H2O/2% MeCN for 10 min and a gradient to 100% MeCN from 10 to 30 min.
HPLC analysis, purification and isolation of Ga-salinichelins
The analysis and purification of the crude extract was carried out on a Dionex Ultimate 3000 (Thermo Fisher Scientific, Waltham, MA, USA). For analysis, the instrument was equipped with a Nucleodur C18 HTec column (5 μm, 5.6 × 250 mm, Macherey Nagel, Dueren, Germany) and run in isocratic mode with 97% A (H2O with 0.05% trifluoroacetic acid) and 3% B (MeCN with 0.05% trifluoroacetic acid) at a flow of 1 ml min–1 for 26 min with ultraviolet detection at 230 nm.
For purification, a semi-preparative Nucleodur C18 HTec column (5 μm, 10 × 250 mm, Macherey Nagel) was used. The same method was applied with a flow of 6.25 ml min–1. Fractions were collected with a fraction collector. Overall six fractions were collected corresponding to salinichelins A-F with retention times of 4–21 min. Based on amount and purity, salinichelins A to C were subjected to nuclear magnetic resonance spectroscopy (NMR) analysis.
Mass spectrometry analysis
HPLC-MS experiments were carried out on an Agilent (Santa Clara, CA, USA) Q-TOF 6530 coupled with an Agilent 1260 HPLC in positive ion mode. The mass range was set to 300–1700 amu with auto MS2 acquisition (MS scan rate 2/s, MSMS scan rate 3/s) and active exclusion after three spectra and static exclusion from 300 to 400 m/z. Gas temperature was 300 °C with a gas flow of 11 l min–1 and nebulizer at 35 psig. Collision energies were set to 20 keV. Data were analyzed with MassHunter software (Agilent).
For fourier transform mass spectrometry measurements, samples were injected by a nanomate-electrospray ionization robot (Advion, Ithaca, New York, USA) for consecutive electrospray into the MS inlet of a LTQ 6.4 T Fourier transform ion cyclon resonance (FT-ICR) mass spectrometer (Thermo Finnigan, Thermo Fisher Scientific). MS and MS2 data were acquired in the positive ion mode. Fourier transform mass spectrometry data were acquired in 400–2000 m/z scans. Selected peptide mass signals were manually isolated and fragmented by CID. MSn data were collected either in ion trap or FT detection mode. All data were analyzed using QualBrowser, which is part of the Xcalibur LTQ-FT software package (Thermo Fisher, Waltham, MA, USA).
NMR analysis
1H NMR and 2D NMR spectra were recorded on a Bruker (Billerica, MA, USA) Avance II 600 and referenced to residual proton signals of DMSO-d6 (Deutero GmbH, Kastellaun, Germany).
Stereochemical analysis (Marfey analysis)
Marfey analysis was carried out applying the modified method by Kodani et al. (2015). Of the Ga-siderophores, 0.10–0.18 mg were dissolved in 50 μl water (0.14–0.25 μmol) and after addition of 200 μl concentrated HI (57 wt% in H2O) heated to 106 °C for 24 h. The solvent was evaporated under N2 stream, and the residue dissolved in 100 μl water. After addition of 10 μl (3.73 μmol) of a 1% Marfey solution in acetone (1-fluoro-2,4-dinitrophenyl-5-l-alaninamide, 10 mg ml–1) and 40 μl of a 1 m NaHCO3 solution, the mixture was heated for 1 h at 40 °C. After addition of 40 μl of 1 m HCl, the solvent was evaporated under N2 stream. The residue was dissolved in 200 μl of a 1:1 MeCN/H2O mixture, filtered and subjected to HPLC analysis.
For preparation of the standards of d- and l-arginine, d- and l-ornithine, and d- and l-lysine, and 7 μl of the 1% Marfey solution (2.6 μmol, 1.05 eq.) were added to 50 μl (2.5 μmol) of a 50 mM amino-acid solution in water. Subsequent treatment was as described above.
The HPLC analysis was carried out on a Dionex Ultimate 3000 equipped with a Nucleodur C18 HTec column (5 μm, 10 × 250 mm, Macherey Nagel). The solvent composition was 95% A (H2O with 0.05% trifluoroacetic acid) and 5% B (MeCN with 0.05% trifluoroacetic acid) for 5 min and then 67.5% A and 32.5% B for 35 min at a flow of 0.25 ml min–1 and ultraviolet detection at 340 nm. Analysis was carried out with the pure Marfey derivatives of the salinichelins A-F as well as co-injections with the six standards each.
Removal of gallium from Ga-siderophore complexes
The removal of Ga3+ from Ga-salinichelin complex was done according to the procedure of Lautru et al. (Stephan et al., 1993; Lautru et al., 2005). The Ga-salinichelin complex (1 mg, about 1.40 mol) was dissolved in 600 μl H2O and 150 μl of 8-hydroxyquinoline solution (0.015 g in 1.5 ml MeOH:H2O 2:1, 100 μmol, 71 eq.) was added. The solution was heated at 70 °C for 1 h. The color changed from yellow to greenish because of the Ga-8-hydroxyquinoline complex, which was subsequently removed by extraction with dichloromethane (5 × 1 ml). The free siderophore was obtained after lyophilization.
Determination of iron-binding constant pM(FeIII)
The binding constant pM(FeIII) of salinichelin A (2) and desferrioxamine B mesylate were determined via spectrophotometric competition tritration of the Fe-siderophore (FeSid) complex against EDTA at pH 7.4 in 0.01 m HEPES (2-[4-(2-hydroxyethyl)piperazine-1-yl]ethanesulfonic acid) buffer with 0.1 m KCl previously described (Abergel et al., 2008; Böttcher and Clardy, 2014). Three independent titrations were performed. The extinction coefficients needed to calculate the concentrations were determined separately for both FeSid complexes with concentrations at 0.01, 0.05, 0.1 and 0.15 mM at identical pH and buffer concentration. To quantify the amount of FeSid and FeEDTA complexes, the absorption at 430 nm was used. The obtained ΔpM values were used to calculate the pM(FeIII) from the known pM(FeIII) of EDTA (23.4). (Abergel et al., 2008).
Salinispora genome sequences
All Salinispora genomes have been sequenced by the Joint Genome Institute as previously described (Ziemert et al., 2014) and are publically available at the Joint Genome Institute’s Integrated Microbial Genomes (IMG) database, http://img.jgi.doe.gov/cgi-bin/w/main.cgi.
Species and phylogenetic trees
Species and gene trees were inferred from codon alignments of 14 single copy genes (Table 1) and slcE genes, respectively. Selections of genes were based on those identified as single copy from a reference set of 191 complete actinobacterial genomes, filtered by nearest Robinsons Foulds difference. Alignments were made with MAFFT (Katoh and Standley, 2013) with iterations set to 1000 on protein sequences and then back translated using the pal2nal (Suyama et al., 2006) perl script to obtain the final DNA alignment. Ultrametric trees were built using BEAST v2 (Bouckaert et al., 2014) with MCMC chains set to 10 million and 30 million for gene and species tree, respectively. A strict clock, and Yule speciation prior with default parameters were used for each run. Evolutionary models for each gene were chosen based on those selected by jmodeltest2 (Table 1) and site substitution and invariant rates were estimated in BEAST.
Table 1. Genes and models used for phylogenetic analysis.
| Gene | Model | Partition | -lnL | p | BIC |
|---|---|---|---|---|---|
| TIGR00012 | HKY+I | 10 010 | 447, 7861 | 241 | 2210, 3046 |
| TIGR00060 | HKY+G | 10 010 | 1087, 7826 | 241 | 3611, 5456 |
| TIGR00064 | GTR+I | 12 345 | 4830, 304 | 245 | 11 392, 1013 |
| TIGR00166 | HKY+I | 10 010 | 672, 6171 | 241 | 2710, 0077 |
| TIGR00436 | GTR+G | 12 345 | 4045, 8933 | 245 | 9757, 282 |
| TIGR00468 | GTR+I | 12 345 | 4002, 9428 | 245 | 9713, 7144 |
| TIGR00496 | GTR+G | 12 345 | 2048, 4794 | 245 | 5645, 106 |
| TIGR00631 | HKY+G | 10 010 | 9093, 5085 | 241 | 20 030, 593 |
| TIGR00952 | HKY+G | 10 010 | 689, 3193 | 241 | 2725, 1656 |
| TIGR01066 | GTR+I | 12 345 | 1055, 8994 | 245 | 3603, 6147 |
| TIGR01164 | HKY+I | 10 010 | 897, 8683 | 241 | 3253, 1532 |
| TIGR01308 | HKY+I | 10 010 | 423, 0226 | 241 | 2097, 5478 |
| TIGR01455 | GTR+G | 12 345 | 5535, 952 | 245 | 12 838, 3735 |
| TIGR02012 | GTR+I+G | 12 345 | 10 450, 698 | 246 | 22 893, 7369 |
| NRPS16 | HKY+I+G | 10 010 | 32 670, 0197 | 56 | 65 880, 3644 |
List of single copy genes used (TigrFam IDs) and associated model selection results from jmodeltest2. Models shown, HKY: Hasegawa–Kishino–Yano, GTR: general time reversible, G: gamma distribution, I: invariant sites. Parameters shown, BIC: Bayesian information criterion, -lnL: negative log likelihood, partition: substitution code of the model, p: number of model parameters (K).
Horizontal gene transfer analysis
Transfers were identified by species and gene tree reconciliation using the Ranger-DTL-D program (Bansal et al., 2012) on ultrametric trees obtained from BEAST in addition to manual inspection (Supplementary Figures S19–S21).
Ancestral state analysis
The ancestral node of the des and slc pathways were inferred in the species tree using the trace character history function implemented in Mesquite v2.75 (Maddison and Maddison, 2015, 2010). A categorical character matrix was created, and likelihood calculations were performed using the Mk1 model. Likelihood scores >50% were used to infer the points of OBU acquisition (ancestral nodes) in the species tree. OBU ML phylogenies were used to corroborate points of acquisition based on congruence with the species tree.
Results
Genome mining of siderophore pathways in 118 closely related Salinispora genomes
Salinispora is a marine actinomycete genus specifically known for its rich biosynthetic capacity to produce bioactive secondary metabolites, including the potent proteasome inhibitor salinosporamide A (Gulder and Moore, 2010). Initial genome analysis of S. tropica CNB-440 revealed that about 10% of its genome is dedicated to secondary metabolism, including four BGCs predicted to code for siderophore-like compounds (Udwary et al., 2007; Penn et al., 2009). However, only members of the desferrioxamine family, particularly desferrioxamine E (Figure 1), were shown to be produced under iron-depleting conditions in the laboratory (Roberts et al., 2012). The desferrioxamines are widespread tris-hydroxamate siderophores assembled from alternating units of succinic acid and monohydroxylated diamines by four enzymes (Sandy and Butler, 2009).
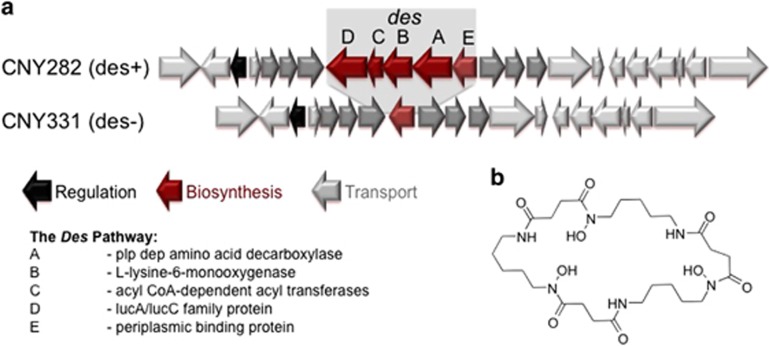
Figure 1.
Deletion of a siderophore cluster. (a) Comparative genomic analysis showed that the majority of analyzed Salinispora genomes contain the full des cluster for the production of desferrioxamine under iron-limiting conditions (example S. pacifica CNY282). However, almost the whole biosynthesis cluster is deleted in 19 of the analyzed strains, whereas the surrounding genes (light gray) are highly conserved. Only the periplasmic binding protein and metal transporters (dark gray) remain. (b) Structure of desferrioxamine E.
During a recent sequencing project of 118 Salinispora genomes, comparative analyses revealed that the majority of the sequenced genomes contained the des cluster at the same relative location in the genome irrespective of the species type. However, in 20 S. pacifica and 6 S. arenicola strains, the des genes were incomplete (Figure 1). Interestingly, it was always the four structural genes responsible for biosynthesis that were absent in all strains, whereas the periplasmatic binding proteins and ATP binding cassette transporter required for iron uptake were still present. One possible explanation of this observation is that desferrioxamines exogenously produced by other organisms are pirated by the des-minus Salinispora strains. In other words, selection favored the evolution of cheater genomes that do not use the energy to synthesize the iron chelators (West et al., 2006). These so-called cheaters (Griffin et al., 2004), however, would be dependent on cooperating with related producer organisms within bacterial communities (Cordero et al., 2012). We sought to test this hypothesis experimentally.
Discovery of salinichelins, new siderophores in Salinispora spp.
To test whether the des- strains are so-called ‘cheaters’ and suffer from growth deficiencies relative to des+ strains, we grew 24 des- strains in iron-deficient media. To our surprise, we did not observe any major growth differences, suggesting the possible production of other siderophore-type compounds. When we queried the genomes for additional siderophore BGCs, we identified two orphan hydroxamate siderophore pathways, an aerobactin-like pathway and the orphan NRPS16 pathway (Figure 2). A closer look at the siderophore pathway distribution revealed that the des and NRPS16 pathways were mutually exclusive. This orphan BGC is located on genomic island 16, which is distantly removed from the genomic location of the des gene cluster (Ziemert et al., 2014).
Figure 2.
Organization of the NRPS16/salinichelin BGC. Comparative genome mining revealed an orphan BGC only present in strains without the desferrioxamine gene cluster. The salinichelin (slc) gene cluster consists of 13 genes (slcA-slcM). Highlighted is the NRPS domain architecture of slcE. A, adenylation domain; C, condensation domain; E, epimerization domain; T, thiolation domain.
Detailed bioinformatic examination of the NRPS16 gene cluster (Figure 2, Supplementary Table S1) revealed a four-domain NRPS with bioinformatic predictions for three ornithine-derived amino-acid residues, which are commonly found in hydroxamate-type siderophores. The fourth amino-acid residue could not be predicted. Instead of a thioesterase domain, a terminal condensation domain was encoded, which has been shown to be responsible for peptide cyclization (Gao et al., 2012). In addition, genes for the formation of hydroxamate groups, including an ornithine N-hydroxylase, an N-acetyltransferase, and a formyltransferase were identified (Bosello et al., 2012). Furthermore, six iron-related ATP binding cassette transporters were encoded in the gene cluster.
To determine whether NRPS16 indeed functions as a siderophore-encoding BGC in the desferrioxamine missing (des-) Salinispora strains, three NRPS16-containing strains were cultivated in iron-deficient media. Compared with cultures that were supplemented with iron, we identified a series of polar compounds by reversed phase HPLC that is produced by all three strains (Figure 3). For simplification and handling purposes, we chose S. pacifica CNY-331 as a representative strain for detailed chemical analysis. High resolution HPLC-MS revealed parent masses differing by 14 Da each corresponding to methylene units (Supplementary Figure S1). When iron (III) salts were added to the extracts, the solution turned red, with an ultraviolet maximum of 435 nm consistent with iron binding. In addition, the binding of iron could be observed by MS (Supplementary Figure S2). Examination of the MS2 spectra of the desferri compounds displayed high similarities to a recently discovered siderophore called peucechelin from Streptomyces peucetius (Park et al., 2013; Kodani et al., 2015), which fits the bioinformatically predicted structures based on the BGC.
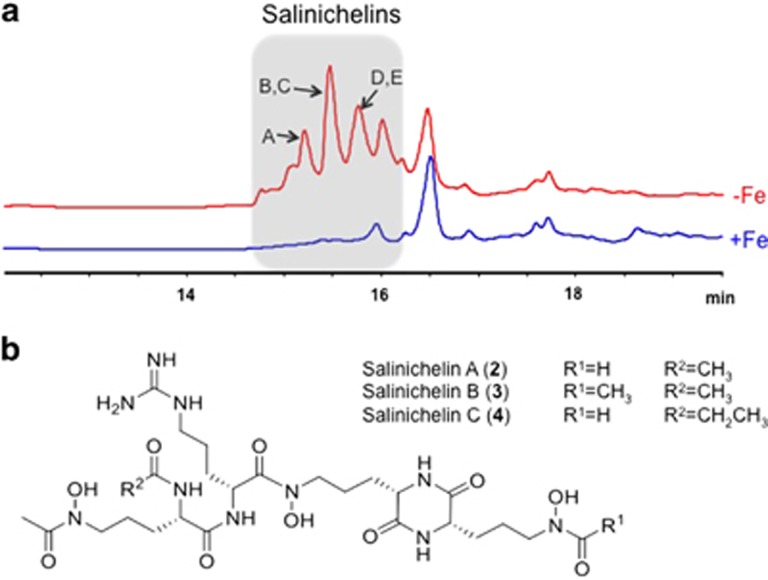
Figure 3.
Production and structures of the salinichelins. (a) HPLC chromatograms of extracts of S. pacifica CNY-331 grown under Fe-limiting conditions leads to the production of a series of salinichelin siderophores (red trace) compared with the Fe-supplemented culture (blue trace). HPLC was monitored at 210 nm. (b) The structures of salinichelins A-C (2–4) is shown.
To isolate and structurally characterize the NRPS16 siderophores, we cultured S. pacifica CNY-331 in large scale. During the extraction and purification process, the peptides appeared to be highly unstable when not bound to a metal. Thus, we added GaBr3 to the culture broth before extraction. The highly polar gallium complexes were detected by mass spectrometry via their distinct isotopic patterns (Supplementary Figure S3). The gallium-bound siderophores were isolated from the crude extract by HPLC (Supplementary Figure S4). Hydrophilic interaction chromatography-HPLC-MS experiments showed that the three heavier analogs exist in isomeric forms, suggesting that seven analogs are expressed in total (Supplementary Figure S5).
Comprehensive NMR analysis (1H, DQF-COSY, TOCSY, HSQC, HMBC and NOESY) of the most abundant gallium complex with m/z 725.26, which was characterized as an inseparable mixture with the less abundant isomeric complex, revealed a previously undescribed linear tetrapeptide consisting of N1-acetyl-N5-acetyl-N5-hydroxyornithine, arginine, N5-hydroxyornithine and N5-formyl-N5-hydroxyornithine residues, which we named salinichelin A (2) (Figure 3, Supplementary Figures S6–S11, Supplementary Table S2). The connectivity of the building blocks was deduced by HMBC and NOESY experiments (Supplementary Figure S12). The stereochemistry of the amino-acid building blocks was determined using Marfey’s method to be L for all ornithine derivatives and D for arginine (Supplementary Figures S13–S15), which was consistent with the NRPS module organization containing an epimerase domain in module 2 (Figure 2). The structure elucidation of the heavier analogs salinichelin B (3) and C (4) was achieved by combination of MS, Marfey and NMR analysis (Supplementary Figures S16–S18). Owing to low amounts the structure elucidation of the heavier analogs, salinichelin D-G could not be fully achieved. However, MS and Marfey analyses show that the modifications take place only at the ornithine residues, presumably with propionylation and/or butyrylation instead of acetylation for the first ornithine residue and acetylation instead of formylation for the last (Supplementary Figure S15). Overall modifications were only observed in the acylation pattern but not via incorporation of lysine instead of ornithine. As expected, the stereochemistry of all isomers was determined to be D for arginine and L for ornithine residues (Supplementary Figure S19). The presence of several salinichelin analogs with high abundance suggests a loose substrate specificity of the salinichelin biosynthetic enzymes, especially of the acetyltransferase SlcM.
We next compared the iron-binding capacities of the two siderophores desferrioxamine B mesylate and salinichelin A. We determined their pM(FeIII) values using a photometric approach (Supplementary Figure S20). Both siderophores exhibited very similar pM(FeIII) values at pH 7.4 with 24.5 for salinichelin A and 24.3 for desferrioxamine B mesylate.
Evolution and functional pathway replacement of two siderophore families
In order to reconstruct the evolutionary history of both siderophore pathways within the genus Salinispora, we performed a detailed phylogenetic analysis including constructing a species tree, gene trees of key genes within both pathways, and ancestral reconstructions (Figure 4, Supplementary Figures S21–S23). The ancestral state reconstruction of the des pathway showed that the des pathway was likely present in the common ancestor of the genus (Supplementary Figure S18). The slc pathway on the other hand was acquired horizontally once in S. pacifica and twice in S. arenicola and subsequently inherited vertically within these specific branches (Figure 4a). These same branches lost the desferrioxamine biosynthesis genes, the most parsimonious explanation being that the two siderophore molecules have similar iron-binding capabilities and the presence of the two BGCs would therefore be functionally redundant. Fascinatingly, the des-minus strains retained the genes associated with desferrioxamine transport.
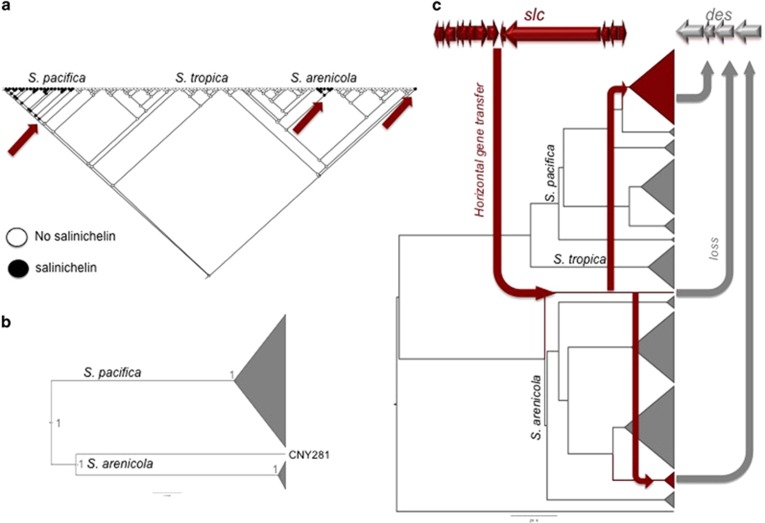
Figure 4.
Siderophore evolution in Salinispora. (a) The character trace history analysis of slc (formerly NRPS16) was performed in Mesquite v2.75 (45). Each strain in the phylogenetic tree is depictured as circle, black marked strains contain the gene cluster, white strains do not. Arrows indicated where the gene cluster was likely acquired via horizontal gene transfer (HGT) independently in S. pacifica and twice in S. arenicola. (b) The phylogenetic gene tree of slc shows two main clades: one for S. pacifica and one for S. arenicola, indicating that the pathway was exchanged once between the two species and once within S. arenicola. (c) Evolutionary scenario of HGT of slc and the concurrent deletion of the more ancient des gene cluster as deviated from the comparison of the gene tree and species tree with Ranger-DTL (Supplementary Figures S17–S20; Bansal et al., 2012) and the character trace history.
The slc genes in S. arenicola and S. pacifica are closely related and are very likely horizontally transferred between the species. The gene tree of the slc pathway (Figure 4b) proves the high relatedness of the slc BGCs from S. pacifica and S. arenicola, suggesting horizontal gene transfer from one species to another. Based on comparison of gene tree and species tree, as well as Ranger-DTL (Bansal et al., 2012) and BEAST (Bouckaert et al., 2014) analyses (Supplementary Figures S21–S24), the most likely scenario is that the slc BGC was transferred from S. arenicola to S. pacifica (Figure 4, Supplementary Figure S22). Interestingly, no strain was found containing both siderophore BGCs. Considering the high relatedness of the Salinispora strains and the fact that there were three separate slc acquisition events, the des genes must have been lost relatively quickly on an evolutionary timeline.
An alternative evolutionary scenario would be that the des pathway was lost first, before the slc genes were acquired. Cheaters have been demonstrated to have a fitness advantage in mixed populations (Cordero et al., 2012). In the case that not producing any of these siderophores would be of advantage for survival, we would expect to detect strains that encode neither the des nor the slc pathway. However, as all strains contained at least one of the two gene clusters, we believe that the ability to produce at least one siderophore is advantageous to the fitness of the strain. Therefore, we think that it is more likely that the slc gene cluster was acquired, before the des genes were lost.
In summary, every genome in our collection has the ability to produce one of the two siderophores, but each genome has precisely one gene cluster that can confer this function. Therefore, we suggest that the two BGCs are incompatible in the same genome. This incompatibility appears to stem from the similarity in function, but not in gene content of the BGC nor the BGC location in the genomes. We also infer that the organisms do not benefit from the dosage effect of having two clusters that produce such similar products.
Discussion
Siderophores are widespread, structurally diverse deriving from different biosynthetic origins. They encompass structurally diverse molecules with different biosynthetic origins and are one of the few examples of bacterial secondary metabolites where the ecological function is known (Seyedsayamdost et al., 2012). These combined traits make them ideal model systems to follow and understand the evolutionary history of secondary metabolite genes. Our example shows how the acquisition of one siderophore pathway correlates with the deletion of another. The latter encodes a structurally different but functionally equivalent molecule made by an unrelated biosynthetic mechanism located elsewhere in the genome. This pathway exchange has happened at least three times within the history of the genus.
It is relatively common for closely related species to produce multiple and different siderophores (Barona-Gómez et al., 2006). It is also known that piracy of siderophore molecules is quite common. So-called cheaters contain all the necessary binding proteins and transporters to be able to detect and import iron chelators without actually producing the molecules themselves (Griffin et al., 2004). According to the ‘public goods’ hypothesis, this actually increases the chances for surviving of a population by increasing diversity without the high cost for individual strains (Cordero et al., 2012). However, in these examples, cheater strains have been shown to suffer a significant decrease in growth rate when grown in pure culture under iron-limiting conditions. In our case, a different siderophore cluster has been acquired around the same time that the desferrioxamine biosynthesis genes were lost. As a consequence, we have identified a system that may function as a de novo siderophore biosynthesizer for its ability to produce salinichelin and possibly as a cheater for its ability to use exogenous desferrioxamine. Although, it needs to be clarified in the future if the des- strains are actually able to function as cheaters.
Secondary metabolites are known to be prone to horizontal gene transfer (Ziemert et al., 2014). However, to the best of our knowledge, this is the first example that shows how fast and exact evolutionary mechanisms work on secondary metabolite pathways. Relatively simple mutations of important amino acids within one of the required genes or partial deletion within the cluster would have been sufficient to interrupt desferrioxamine production. However, in both cases the exact same four genes responsible and needed for biosynthesis are completely lost. This indicates a strong selection pressure on deletion of non-functional or functionally redundant genes associated with secondary metabolism. Keeping non-functional genes or even clusters within the genomes would probably increase genome size and replication costs. This, in turns, strongly implies that BGCs within bacterial genomes are largely functional and their retention is because of purifying selection on genomes to preserve these functions.
It is not clear yet if salinichelin provides a selective advantage over desferrioxamine in certain ecological niches or maintaining one siderophore over the other is coincidental. The differences in iron-binding capacities measured at pH 7 are probably too small to explain improved siderophore activity. However, it has been shown that iron-chelating activities are highly dependent on pH and other environmental factors (Ahmed and Holmström, 2014). Thus, it could be possible that salinichelin works better in specific environmental conditions and that cluster replacement is not neutral, but instead is selectively advantageous.
Also unclear is the ecological role of the other predicted but orphan siderophores encoded within the Salinispora genomes. Despite multiple efforts, we have not detected any of the predicted products under iron-limiting conditions (Roberts et al., 2012). This could indicate that they are just produced under very specific growth conditions. Those siderophores could have different characteristics such as different iron-binding capacities at different pHs, or different solubility, hence differing in their specific ecological function. Another explanation would be that these BGCs encode other ionophores that respond to limitations of different metals such as copper or zinc (Spohn et al., 2015). In general, it is also important to distinguish between bioactivities of secondary metabolites and ecological functions. It is likely that bacteria can produce multiple metabolites acting synergistically or contingently as has been shown for certain antibiotics and siderophores in Streptomyces species (Challis and Hopwood, 2003). However, in these cases the siderophores are not redundant but need to be maintained for other reasons. Desferrioxamines from Streptomyces, for example, have been shown to mediate ecological relationships with other bacteria and plants (Yamanaka et al., 2005; Barona-Gómez et al., 2006; Traxler et al., 2012).
In conclusion, here we present the first example of functional replacement of a secondary metabolite BGC. The acquisition of a pathway encoding a compound with the same ecological function is associated with the deletion of a more ancient BGC and displays how relatively fast obsolete gene clusters are deleted. In the past, we were able to show that BGCs with homologous core genes can be exchanged within genomic islands in a process that we called pathway swapping (Ziemert et al., 2014). However, here we show that pathway replacement can occur independent of sequence homology and location but instead based on the functions of the compounds they encode.
Acknowledgments
We thank Dr Brendan M Duggan for help in NMR measurements and Kimberly Chang for HPLC assistance. Dr Pieter C Dorrestein is acknowledged for providing access to FT-ICR measurements. This work was supported by NIH grants R01-GM085770 to BSM and PRJ and a postdoctoral fellowship CR464-1 by the Deutsche Forschungsgemeinschaft (DFG) to MC. NZ thanks the German Center for Infection Biology (DZIF) for funding. Further funding was provided by the Deutsche Forschungsgemeinschaft (DFG) to HB and SS (SFB/TRR 51/2 C 02).
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Abergel RJ, Zawadzka AM, Raymond KN. (2008). Petrobactin-mediated iron transport in pathogenic bacteria: coordination chemistry of an unusual 3,4-catecholate/citrate siderophore. J Am Chem Soc 130: 2124–2125. [DOI] [PubMed] [Google Scholar]
- Ahmed E, Holmström SJM. (2014). Siderophores in environmental research: roles and applications. Microb Biotechnol 7: 196–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bansal MS, Alm EJ, Kellis M. (2012). Efficient algorithms for the reconciliation problem with gene duplication, horizontal transfer and loss. Bioinformatics 28: i283–i291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barona-Gómez F, Lautru S, Francou F-X, Leblond P, Pernodet J-L, Challis GL. (2006). Multiple biosynthetic and uptake systems mediate siderophore-dependent iron acquisition in Streptomyces coelicolor A3(2) and Streptomyces ambofaciens ATCC 23877. Microbiology 152: 3355–3366. [DOI] [PubMed] [Google Scholar]
- Blin K, Medema MH, Kazempour D, Fischbach MA, Breitling R, Takano E et al. (2013). antiSMASH 2.0—a versatile platform for genome mining of secondary metabolite producers. Nucleic Acids Res 41: W204–W212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosello M, Mielcarek A, Giessen TW, Marahiel MA. (2012). An enzymatic pathway for the biosynthesis of the formylhydroxyornithine required for rhodochelin iron coordination. Biochemistry 51: 3059–3066. [DOI] [PubMed] [Google Scholar]
- Böttcher T, Clardy J. (2014). A chimeric siderophore halts swarming Vibrio. Angew Chem Int Ed Engl 53: 3510–3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D et al. (2014). BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput Biol 10: e1003537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Challis GL, Hopwood DA. (2003). Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species. Proc Natl Acad Sci USA 100: 14555–14561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cimermancic P, Medema MH, Claesen J, Kurita K, Wieland Brown LC, Mavrommatis K et al. (2014). Insights into secondary metabolism from a global analysis of prokaryotic biosynthetic gene clusters. Cell 158: 412–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordero OX, Ventouras L-A, DeLong EF, Polz MF. (2012). Public good dynamics drive evolution of iron acquisition strategies in natural bacterioplankton populations. Proc Natl Acad Sci USA 109: 20059–20064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cragg GM, Newman DJ. (2013). Natural products: a continuing source of novel drug leads. Biochim Biophys Acta - Gen Subj 1830: 3670–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. (2013). Specialized microbial metabolites: functions and origins. J Antibiot (Tokyo) 66: 361–364. [DOI] [PubMed] [Google Scholar]
- Eyre-Walker A, Keightley PD. (2007). The distribution of fitness effects of new mutations. Nat Rev Genet 8: 610–618. [DOI] [PubMed] [Google Scholar]
- Fischbach MA, Walsh CT, Clardy J. (2008). The evolution of gene collectives: how natural selection drives chemical innovation. Proc Natl Acad Sci USA 105: 4601–4608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao X, Haynes SW, Ames BD, Wang P, Vien LP, Walsh CT et al. (2012). Cyclization of fungal nonribosomal peptides by a terminal condensation-like domain. Nat Chem Biol 8: 823–830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin AS, West SA, Buckling A. (2004). Cooperation and competition in pathogenic bacteria. Nature 430: 1024–1027. [DOI] [PubMed] [Google Scholar]
- Gulder TAM, Moore BS. (2010). Salinosporamide natural products: potent 20 S proteasome inhibitors as promising cancer chemotherapeutics. Angew Chemie - Int Ed 49: 9346–9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PR. (2016). Natural products and the gene cluster revolution. Trends Microbiol 24: 968–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PR, Moore BS, Fenical W. (2015). The marine Actinomycete Genus Salinispora: a model organism for secondary metabolite discovery. Nat Prod Rep 32: 738–751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30: 772–780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kodani S, Komaki H, Suzuki M, Kobayakawa F, Hemmi H. (2015). Structure determination of a siderophore peucechelin from Streptomyces peucetius. BioMetals 28: 791–801. [DOI] [PubMed] [Google Scholar]
- Lautru S, Deeth RJ, Bailey LM, Challis GL. (2005). Discovery of a new peptide natural product by Streptomyces coelicolor genome mining. Nat Chem Biol 1: 265–269. [DOI] [PubMed] [Google Scholar]
- Lynch M, Marinov GK. (2015). The bioenergetic costs of a gene. Proc Natl Acad Sci USA 112: 15690–15695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddison W, Maddison D. (2010), Mesquite 2. Manual 1–258.
- Maddison WP, Maddison DR. (2015), Mesquite: a modular system for evolutionary analysis. Version 2.75. 2011. URLhttp//mesquiteprojectorg.
- Medema MH, Blin K, Cimermancic P, de Jager V, Zakrzewski P, Fischbach Ma et al. (2011). antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res 39: W339–W346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Cimermancic P, Sali A, Takano E, Fischbach MA. (2014). A systematic computational analysis of biosynthetic gene cluster evolution: lessons for engineering biosynthesis. PLoS Comput Biol 10: e1004016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medema MH, Fischbach MA. (2015). Computational approaches to natural product discovery. Nat Chem Biol 11: 639–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nett M. (2014). Genome mining: concept and strategies for natural product discovery. Prog Chem Org Nat Prod 99: 199–245. [DOI] [PubMed] [Google Scholar]
- Nett M, Ikeda H, Moore BS. (2009). Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat Prod Rep 26: 1362–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park HM, Kim BG, Chang D, Malla S, Joo HS, Kim EJ et al. (2013). Genome-based cryptic gene discovery and functional identification of NRPS siderophore peptide in Streptomyces peucetius. Appl Microbiol Biotechnol 97: 1213–1222. [DOI] [PubMed] [Google Scholar]
- Penn K, Jenkins C, Nett M, Udwary DW, Gontang EA, McGlinchey RP et al. (2009). Genomic islands link secondary metabolism to functional adaptation in marine Actinobacteria. ISME J 3: 1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts AA, Schultz AW, Kersten RD, Dorrestein PC, Moore BS. (2012). Iron acquisition in the marine actinomycete genus Salinispora is controlled by the desferrioxamine family of siderophores. FEMS Microbiol Lett 335: 95–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocker A, Meinhart A. (2016). Type II toxin: antitoxin systems. More than small selfish entities? Curr Genet 62: 287–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Röttig M, Medema MH, Blin K, Weber T, Rausch C, Kohlbacher O. (2011). NRPSpredictor2—a web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res 39: W362–W367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha R, Saha N, Donofrio RS, Bestervelt LL. (2013). Microbial siderophores: a mini review. J Basic Microbiol 53: 303–317. [DOI] [PubMed] [Google Scholar]
- Sandy M, Butler A. (2009). Microbial iron acquisition: marine and terrestrial siderophores. Chem Rev 109: 4580–4595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schorn MA, Alanjary MM, Aguinaldo K, Korobeynikov A, Podell S, Patin N et al. (2016). Sequencing rare marine actinomycete genomes reveals high density of unique natural product biosynthetic gene clusters. Microbiology 162: 2075–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seyedsayamdost MR, Cleto S, Carr G, Vlamakis H, João Vieira M, Kolter R et al. (2012). Mixing and matching siderophore clusters: structure and biosynthesis of serratiochelins from serratia sp. V4. J Am Chem Soc 134: 13550–13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skinnider MA, Dejong CA, Rees PN, Johnston CW, Li H, Webster ALHLH et al. (2015). Genomes to natural products prediction informatics for secondary metabolomes (PRISM). Nucleic Acids Res 43: 9645–9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spohn M, Wohlleben W, Stegmann E. (2015). Elucidation of the zinc dependent regulation in Amycolatopsis japonicum enabled the identification of the ethylenediamine-disuccinate ([S,S]-EDDS) genes. Environ Microbiol 18: 1249–1263. [DOI] [PubMed] [Google Scholar]
- Stephan H, Freund S, Meyer J-M, Winkelmann G, Jung G. (1993). Structure elucidation of the gallium–ornibactin complex by 2D-NMR spectroscopy. Liebigs Ann der Chemie 1993: 43–48. [Google Scholar]
- Suyama M, Torrents D, Bork P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res 34: W609–W612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Traxler MF, Seyedsayamdost MR, Clardy J, Kolter R. (2012). Interspecies modulation of bacterial development through iron competition and siderophore piracy. Mol Microbiol 86: 628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udwary DW, Zeigler L, Asolkar RN, Singan V, Lapidus A, Fenical W et al. (2007). Genome sequencing reveals complex secondary metabolome in the marine actinomycete Salinispora tropica. Proc Natl Acad Sci USA 104: 10376–10381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R et al. (2015). antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43: W237–W243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West SA, Griffin AS, Gardner A, Diggle SP. (2006). Social evolution theory for microorganisms. Nat Rev Micro 4: 597–607. [DOI] [PubMed] [Google Scholar]
- Yamanaka K, Oikawa H, Ogawa HO, Hosono K, Shinmachi F, Takano H et al. (2005). Desferrioxamine E produced by Streptomyces griseus stimulates growth and development of Streptomyces tanashiensis. Microbiology 151: 2899–2905. [DOI] [PubMed] [Google Scholar]
- Ziemert N, Alanjary M, Weber T. (2016). The evolution of genome mining in microbes - a review. Nat Prod Rep 33: 988–1005. [DOI] [PubMed] [Google Scholar]
- Ziemert N, Lechner A, Wietz M, Millán-Aguiñaga N, Chavarria KL, Jensen PR. (2014). Diversity and evolution of secondary metabolism in the marine actinomycete genus Salinispora. Proc Natl Acad Sci USA 111: E1130–E1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziemert N, Podell S, Penn K, Badger JH, Allen E, Jensen PR. (2012). The natural product domain seeker NaPDoS: a phylogeny based bioinformatic tool to classify secondary metabolite gene diversity De Crécy-Lagard V (ed). PLoS One 7: e34064. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.