Abstract
Understanding how microbial diversity influences ecosystem properties is of paramount importance. Cellular traits—which determine responses to the abiotic and biotic environment—may help us rigorously link them. However, our capacity to measure traits in natural communities has thus far been limited. Here we compared the predictive power of trait richness (trait space coverage), evenness (regularity in trait distribution) and divergence (prevalence of extreme phenotypes) derived from individual-based measurements with two species-level metrics (taxonomic richness and evenness) when modelling the productivity of natural phytoplankton communities. Using phytoplankton data obtained from 28 lakes sampled at different spatial and temporal scales, we found that the diversity in individual-level morphophysiological traits strongly improved our ability to predict community resource-use and biomass yield. Trait evenness—the regularity in distribution of individual cells/colonies within the trait space—was the strongest predictor, exhibiting a robust negative relationship across scales. Our study suggests that quantifying individual microbial phenotypes in trait space may help us understand how to link physiology to ecosystem-scale processes. Elucidating the mechanisms scaling individual-level trait variation to microbial community dynamics could there improve our ability to forecast changes in ecosystem properties across environmental gradients.
Introduction
Functional traits can help illuminate the relationship between biodiversity and ecosystem processes (e.g. Norberg et al., 2001; Norberg, 2004; Hillebrand and Matthiessen, 2009; Reiss et al., 2009). Most research in this area has thus far largely focused on taxonomic or phylogenetic richness as measures of biodiversity (see reviews by Cardinale et al., 2011; Krause et al., 2014). However, the importance of relative species densities for ecological processes has been neglected (Mulder et al., 2004; Hillebrand et al., 2008; Zhang et al., 2012), as well as the functional redundancy or diversity of organisms (including intraspecific variation) that may partially decouple the number of species from the functions supported by a community (Chase and Leibold, 2003).
Traits, which are often analysed in terms of species mean values, may be better defined as the features of individual organisms’ phenotypes that determine fitness and life history (McGill et al., 2006; Violle et al., 2007). They influence ecological interactions and dynamics (e.g. Bolnick et al., 2011; de Bello et al., 2011; Violle et al., 2012) and have important consequences for population demography and ecosystem processes (Norberg, 2004; McGill et al., 2006; Cadotte et al., 2011). Empirical evidence for the importance of individual trait variation for ecosystem properties is still lacking, despite the fact that intraspecific trait variance contributes significantly to plant community functional responses to environmental change (Siefert and Ritchie, 2016; Volf et al., 2016). The importance of intraspecific variation for explaining community properties has mostly been explored theoretically in ecology (Bolnick et al., 2011; Albert et al., 2012; Violle et al., 2012; De Laender et al., 2014; Barabás and D’Andrea, 2016; Hart et al., 2016). We expect that, for example, changes in trait diversity (TD) should reflect natural selection mechanisms, which affect processes at all scales of community and ecosystem organisation (Matthews et al., 2011). Additionally, competition for resources, predation, environmental change and rates of mutation and plasticity will all affect individual phenotypes and consequently TD within and between taxa, which will in turn influence population demography, community dynamics and ecosystem properties (Norberg et al., 2001). The link between individual-level TD and large-scale processes is underexplored in real communities, particularly in microorganisms (Gsell et al., 2013; Ackermann, 2015; Schreiber et al., 2016).
Individual-level traits in microbial assemblages have become easier to measure owing to recent technological developments (Shade et al., 2009; Pomati et al., 2011; Fontana et al., 2014; Krause et al., 2014). Indices that quantify community TD using individual-level data (thereby integrating inter- and intraspecific trait variation) have also been recently tested and developed (Fontana et al., 2016). Hereafter, we refer to them as ‘individual-level TD indices’ because, despite quantifying community properties, they are calculated using traits measured on individual organisms. These indices jointly describe the three independent components of TD (richness, evenness and divergence, respectively): the trait onion peeling (TOP; Fontana et al., 2016) index is the sum of all successive convex hulls’ areas touching the individuals of a community in multidimensional trait space, the trait even distribution (TED; Fontana et al., 2016) index is the regularity in the distribution of individuals when compared with a perfectly even reference distribution, and the functional dispersion index (FDis; Laliberté and Legendre, 2010) is the mean distance of individuals to the centroid of trait distribution. The TOP index has been successfully applied to high-throughput individual-level data to study the role of phenotypic variation over time in the adaptation of microbial populations to environmental perturbations (Krismer et al., 2017). It reflects changes in the trait space coverage, which may happen as a consequence of environmental filtering both within and at the edges of trait distribution. TED is related to the reciprocal distances between neighbour individuals in the trait space and is likely to reflect biotic interactions (Fontana et al., 2016). FDis distinguishes communities where individuals are closer to the centroid of the multidimensional trait distribution from communities where extreme phenotypes are found (Fontana et al., 2016).
In this study, we focus on natural phytoplankton communities, whose diversity and dynamics are tightly linked to large-scale biogeochemical processes. These microorganisms form the basis of aquatic food webs and are responsible for almost 50% of total global primary production (Field et al., 1998). The TD metrics described above have allowed us to explore the relative strength of individual-level trait variation and taxonomic diversity metrics (species richness and evenness), as predictors of important ecosystem properties—phytoplankton community biomass and resource-use efficiency.
We studied these ecosystem properties in 28 lakes from two European regions (Switzerland and Danube Delta, Romania), sampled at different temporal and spatial scales (Supplementary Table S1). We related these properties to the TD and taxonomic diversity metrics, while accounting for variation in water chemistry and physics. This heterogeneity across sites allowed us to sample environmental responses in terms of individual-level TD change or turnover of species, and their effects on community properties. We used microscopy for classification of algal species and scanning flow cytometry (SFC) to estimate total biovolume (hereafter biomass) and morphological and physiological traits measured on individual cells/colonies (Table 1, more details in Supplementary Table S2). We tested all combinations of species richness, evenness, TOP, TED and FDis using linear mixed-effects model averaging to quantify and compare their predictive ability on biomass and resource-use efficiency, while controlling for environmental variables.
Table 1. Ecological relevance of the measured phytoplankton traits.
| Measure | Ecological importance |
|---|---|
| Length of the particle | Reproduction, resource acquisition, predator avoidance |
| Frontal shape of the particle | Resource acquisition |
| Fluorescence of chlorophyll a | Resource acquisition |
| Fluorescence of phycoerythrina | Resource acquisition |
| Fluorescence of accessory and decaying pigments | Resource acquisition |
| Evenness in the distribution of pigments within cell/colony | Resource acquisition |
| Cell rugosity/internal structure/gas vesicles/thylacoids | Resource acquisition, predator avoidance |
List of the seven traits used for calculating trait diversity indices (Litchman and Klausmeier, 2008; Pomati et al., 2013).
For Lake Zurich data (only one laser), this measure is missing.
We expected that the inclusion of individual-level TD would improve the variance explained by statistical models predicting phytoplankton productivity. In our SFC data, we cannot distinguish between inter- and intraspecific trait variation because we are unable to classify cells into species groups. Therefore, our assessment of the importance of intraspecific trait variation is only indirect: if interspecific trait variation is more important in influencing ecosystem properties, models including TD indices (i.e. trait richness) should not improve on those that incorporate species-level metrics (i.e. species richness), which are expected to be good proxies of TD in this case. However, if intraspecific trait variation has an important role, standard biodiversity metrics will be poorer predictors of ecosystem properties than TD indices.
Materials and methods
Phytoplankton communities at three study sites
To test relationships between biodiversity indices and ecosystem properties, we used monitoring data from three different lake systems, covering a wide range of temporal and spatial scales. Lake Greifensee, Lake Zurich and Danube Delta (with 26 lakes), in this order, represent a gradient of increasing sampling time span (3 months, 7 months, 2 years), decreasing sampling frequency (weekly, monthly, seasonally) and increasing spatial scale (6 depths from 1 to 8.5 m, 11 depths from 0 to 40 m, 26 lakes in a geographical region). All data sets include microscopy data (phytoplankton classification, enumeration and biomass calculation) obtained using the Utermöhl method (Utermöhl, 1931), as well as phytoplankton morphological and physiological traits acquired by SFC, in addition to physical and chemical water parameters. Supplementary Table S1 summarises the characteristics of the three lake systems.
In lake Greifensee (Switzerland), monitoring data were collected from a single location at the North end of the lake by an automated system that integrates physical, chemical (Idronaut, Brugherio, Italy; http://www.idronaut.it) and biological analyses (Pomati et al., 2011). From the 31st of July to the 24th of October 2014, SFC measurements were collected every 4 h from samples collected at six different depths (1, 2.5, 4, 5.5, 7 and 8.5 m). Additional chemical analyses and microscopy measurements (to obtain species diversity metrics) were performed on water samples collected manually at the same location, depths and time (average time interval between successive samples=58.3 h, standard deviation=67.7 h). Total phosphorus (TP) and total nitrogen (TN) were measured using DIN Standards (German Institute for Standardisation). The sample size is 207 (36 time points x 6 depths, with some missing data points due to technical problems).
In lake Zurich (Switzerland), monitoring data were collected from a single location in the centre of the lake by the Zurich drinking water company (WVZ), which also performed microscopy and chemical analyses as described in Pomati et al. (2012). Water was sampled at 11 different depths (0, 1, 2.5, 5, 7.5, 10, 12.5, 15, 20, 30 and 40 m) from the 6th of May to the 2nd of December 2009, with a time interval of 1 month. The same water samples were analysed with SFC. The sample size is 82 (8 months x 11 depths, with some missing data points due to technical problems).
Monitoring data were collected from 26 shallow lakes of the Danube Delta (Romania). These lakes form a complex system in a region of hundreds of km2 (Supplementary Table S1). From a single location in each lake, an integrated sample over the water column was collected in spring, summer and autumn of two following years (May, July and September 2013; May, July and October 2014). The same water samples were used for microscopy and chemical analyses, as well as SFC measurements. TP and TN were measured using standard colorimetric methods (Tartari and Mosello, 1997). The sample size is 136 (6 months x 26 lakes, with some missing data points due to technical problems).
SFC measurements
The scanning flow cytometer Cytobuoy (Woerden, The Netherlands; http://www.cytobuoy.com) was used for counting and characterising phytoplankton single cells and colonies (e.g. Dubelaar et al., 2004; Pomati et al., 2013; Fontana et al., 2014). The present instrument contains two laser beams (coherent solid-state sapphire, wavelengths 488 and 635 nm), but the samples from lake Zurich were measured with a previous version of the instrument containing only one laser (wavelength 488 nm). Light, both from forward scattering (FWS) and sideward scattering, provides information on particle morphology. The fluorescence emitted by photosynthetic pigments was measured by three different detectors referred to as red (668–734 nm range; FL.Red from the 488 nm laser and FL.2.Red from the 635 nm laser), orange (601–668 nm range; FL.Orange) and yellow (536–601 nm range; FL.Yellow). Cytobuoy measurements provide time-resolved pulse signals, from which many descriptors of morphology, internal structure and fluorescence profile of each particle were extracted. More details on the instrument can be found elsewhere (Dubelaar et al., 2004; Pomati et al., 2011, 2013; Fontana et al., 2014).
Individual-level TD indices
For all phytoplankton samples, we calculated the TD indices describing the three components of TD defined in Mason et al. (2005): richness, evenness and divergence. We quantified these three TD components using distinct indices: TOP, TED and FDis, respectively (Laliberté and Legendre, 2010; Fontana et al., 2016). These TD indices fulfil theoretical requirements that make them suitable for application to individual-based measurements, when every organism constitutes a unique combination of traits and therefore influences the TD (Fontana et al., 2014). Thus, in the present study we did not identify neither taxonomic nor functional groups in the SFC data, and we calculated TD metrics without classifying particles (single cells represent the fundamental unit in this study). These multivariate indices were calculated using seven Cytobuoy-derived traits, selected on the basis of their ecological and physiological relevance for phytoplankton (Table 1), and to avoid trait multicollinearity: length (maximum value between length by sideward scattering and length by FWS), average FWS, average FL.Red, average FL2.Red, average FL.Yellow, fill factor FL.Red, ratio between average sideward scattering and average FWS. Owing to this a priori selection of traits, there was no need for dimensionality reduction. In all data sets the absolute value of pairwise Pearson’s correlation coefficients between the selected traits were below 0.7, which we chose as the maximal acceptable limit to reduce collinearity issues (Dormann et al., 2013). Before calculating TD indices, the selected traits were standardised (mean=0; standard deviation=1) so that each trait has equal weight (Petchey and Gaston, 2006). This was necessary because the selected traits have different units and vary in value by orders of magnitude. Details about the interpretation of the selected traits and their ecological relevance are reported in Supplementary Table S2. These seven traits provide information on phytoplankton three-dimensional structure, fluorescence properties, cell/colony size and distribution of pigments and other structures within cells (Pomati et al., 2013). Although they do not cover all relevant dimensions of trait space in phytoplankton (e.g. life history, nutrient uptake kinetics and mixotrophy), these traits relate to photosynthesis, resource acquisition (surface-to-volume ratio), reproduction and predator avoidance (Litchman and Klausmeier, 2008; Pomati et al., 2013; Table 1).
Species-level biodiversity metrics
We calculated species richness (number of species) and Pielou’s evenness using microscopic counts of the phytoplankton community in each sample. Effort was made to standardise the counting method and taxonomic identification (at the species level) across all data sets. The same sample preparation method was used (Utermöhl, 1931) and taxonomists exchanged knowledge and information. Pielou’s evenness was calculated based on the biovolume of each species present, derived by multiplying the abundance data from microscopy by the best available estimate of species-specific biovolume.
Biomass calculation
Biomass was defined as the sum of the biovolumes of all phytoplankton cells or colonies in the samples, divided by the sample volume analysed. The biovolume of each single particle was estimated assuming an ellipsoid shape with the formula: (Biovolume2=0.0017 × FWS.Total−0.013) (Foladori et al., 2008; Pomati and Nizzetto, 2013). It is important to note that total FWS was derived by SFC, but it is not one of the traits included in the calculation of the TD indices (see above).
Model selection and averaging
We formulated linear mixed-effects models to explain variation in ecosystem properties. We included all possible combinations of five explanatory variables as fixed effects: species richness, Pielou’s evenness and the three metrics of individual-level TD (TOP, TED and FDis). TD estimates are not independent from taxonomy-based biodiversity measures. However, we found these two groups of predictors to be only weakly correlated (a maximum Pearson’s r of 0.50 across all data sets, between TED and Pielou’s evenness in lake Zurich). In addition, we accounted for temporal and spatial autocorrelation in abiotic (physics and chemistry) and biotic environmental variables (e.g. grazers and parasites). First, we included the date of sampling as a fixed effect, with a quadratic term. In the case of lake Greifensee (high temporal sampling frequency), the exact time of sampling and the date were combined in a single variable. Second, random intercepts were also included in all models to account for spatial ecosystem heterogeneity: depth of sampling for lakes Greifensee and Zurich, and lake identity for the shallow Danube Delta lakes. This approach resulted in a list of 32 models for each data set. The variance inflation factors of all variables of interest were lower than 4 in all models, indicating that multicollinearity was not a problem.
We calculated marginal R2 values (Nakagawa and Schielzeth, 2013) to estimate the proportion of variance explained by the fixed effects alone in each model. Models with delta correction to the Akaike information criterion <7 were defined as the strongest set of models (Burnham and Anderson, 2002; Richards, 2005). These were subsequently used for model averaging to minimise the dependence of the regression coefficients on single models and to assess overall predictive power. Model selection and averaging were performed using the MuMIn R-package (R Core Team, 2013).
Additional analyses
We also performed all the above analyses using resource use efficiency (RUE, sensu Ptacnik et al., 2008) as ecosystem property instead of biomass, which allowed us to study the predictive power of biodiversity metrics on the ability of a community to take up limiting resources. We defined RUE as the amount of standing phytoplankton biomass per unit of limiting resource present (Ptacnik et al., 2008). We used phytoplankton total biovolume (calculated with SFC data) as a proxy for biomass and TP as the main limiting resource in temperate lakes (Ptacnik et al., 2008), which in turn is a proxy for potential system productivity. Therefore, RUE was calculated as (biomass/TP).
Additionally, we ran a full set of analyses including in all models the number of particles measured by SFC as a fixed effect, to account for variation in biomass or RUE caused by SFC sample size.
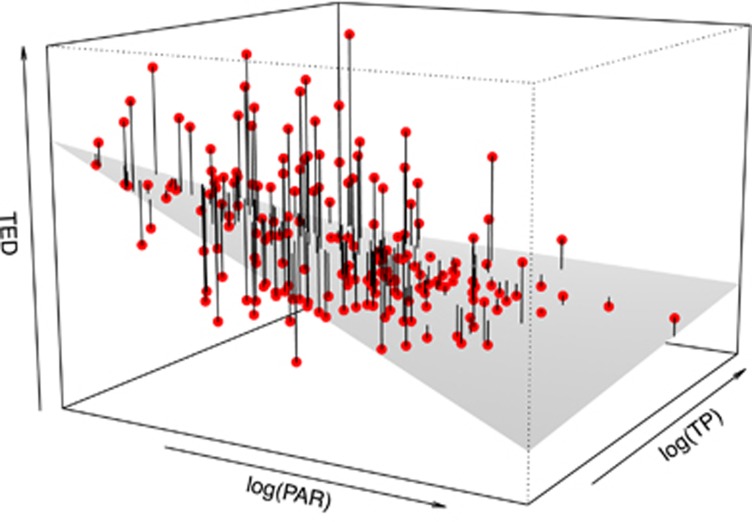
To examine the influence of phytoplankton limiting resources on TED, we used high-resolution data from lake Greifensee including photosynthetic active radiation (PAR, from high-frequency profiles) and TP (representing potential availability), the key limiting factors for algal growth in freshwater environments (Ptacnik et al., 2008). We performed a multiple linear regression analysis (N=195) with PAR, TP (both log-transformed) and their interaction as explanatory variables, and TED as response variable.
Results
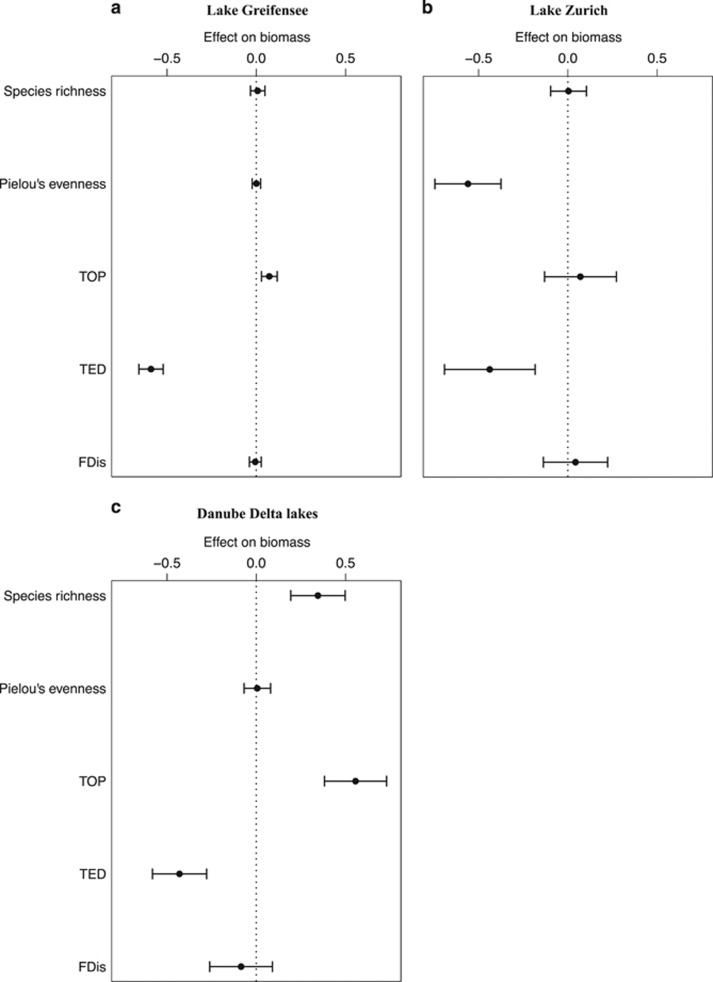
The best models retained for model averaging explained more than 60% of the variance in phytoplankton biomass, with a large proportion contributed by the biodiversity metrics (Figure 1, Table 2 and Supplementary Tables S3–S5). Temporal structure in the data contributed variable amounts depending on the lake system (Table 2). Note that marginal R2 values represent the variance explained by the fixed effects alone, and therefore do not include the variance attributable to spatial autocorrelation (random factor). Models containing individual-level TD metrics improved predictions of microbial biomass and RUE. Our results show a consistent negative correlation between TED and community biomass across data sets. The other components of TD (richness and divergence), after correcting for the number of cells/colonies, had a nonsignificant effect on ecosystem properties (Supplementary Figures S2 and S3, and Supplementary Tables S10–S17), except TOP in lake Greifensee where it had, however, a weaker effect than TED (Supplementary Figures S2a and S3a).
Figure 1.
Effect sizes of biodiversity metrics on biomass. TED is the strongest predictor of biomass among the five biodiversity metrics. Values are standardised model-averaged regression coefficients with 95% confidence intervals, derived from models accounting for temporal and spatial heterogeneity.
Table 2. Summary of the results with biomass as response variable.
| Greifensee (N=207) | Lake Zurich (N=82) | Danube delta lakes (N=136) | |
|---|---|---|---|
| Number of models used for model averaging (out of 32) | 8 | 8 | 4 |
| Sum of weights | 98.8% | 99.5% | >99.9% |
| Species richness estimate (95% CI) | 0.008 (−0.033, 0.048) | 0.004 (−0.097, 0.104) | 0.345 (0.193, 0.497) |
| Pielou's evenness estimate (95% CI) | 0.001 (−0.023, 0.024) | −0.559 (−0.744, −0.374) | 0.006 (−0.068, 0.080) |
| TOP estimate (95% CI) | 0.073 (0.029, 0.117) | 0.070 (−0.131, 0.271) | 0.556 (0.382, 0.730) |
| TED estimate (95% CI) | −0.590 (−0.658, −0.521) | −0.437 (−0.691, −0.184) | −0.430 (−0.581, −0.278) |
| FDis estimate (95% CI) | −0.006 (−0.039, 0.027) | 0.043 (−0.137, 0.223) | −0.085 (−0.260, 0.091) |
| Mean R2 (null modela) | 0.65 (0.02) | 0.63 (0.24) | 0.65 (0.17) |
Abbreviations: CI, confidence interval; FDis, functional dispersion index; TED, trait even distribution; TOP, trait onion peeling.
The estimates of the five explanatory variables (with 95% CIs) represent standardised model-averaged regression coefficients. Values in bold are significant at the P<0.05 level.
Accounting for temporal and spatial heterogeneity.
Species richness was not a significant predictor of biomass. A positive relationship in the Danube Delta lakes disappeared when the models accounted for the number of individuals measured by SFC (Supplementary Figure S2c and Supplementary Tables S10 and S13), suggesting a sampling effect.
The different lake systems showed some idiosyncratic patterns, but the main results were consistent. In lake Greifensee, TED was the strongest predictor of biomass, while TOP was the second (Table 2 and Figure 1a). Species richness, Pielou’s evenness and FDis were nonsignificant predictors of biomass (Figure 1a). In Lake Zurich, Pielou’s evenness was the strongest predictor of biomass, while TED was the second (Table 2 and Figure 1b). Species richness, TOP and FDis were nonsignificant predictors of biomass (Figure 1b). In the Danube Delta lakes, TOP was the strongest predictor of biomass, whereas TED and species richness were the second and third, respectively (Table 2 and Figure 1c). Pielou’s evenness and FDis were nonsignificant predictors of biomass (Figure 1c).
Identical analyses using RUE instead of biomass as the ecosystem property of interest yielded almost identical patterns as those presented in Figure 1 (Supplementary Figure S1 and Supplementary Table S6). Including in all models the number of individuals measured by SFC as a fixed effect, to correct for potential biases associated with the fact that some biodiversity metrics, especially TOP (Fontana et al., 2016), are increasing functions of sampled abundance, also did not change the results (Supplementary Figures S2 and S3 and Supplementary Tables S10–S17). These additional steps had the effect of reducing the predictive strength of species richness and TOP, when analysing both biomass and RUE in the Danube Delta lakes (Supplementary Figures S2c and S3c). TED was, however, the strongest predictor of ecosystem properties (Supplementary Figures S2 and S3) under all analytical conditions.
A multiple linear regression to test the influence of the main limiting resources on TED revealed a negative, significant effect of PAR and TP (P=0.03 and P<0.001, respectively; R2=0.25). TED was highest under limitation by both light and nutrients, with a marginal effect (P=0.07) of their interaction (Figure 2).
Figure 2.
Relationship between phytoplankton limiting resources and TED. Individual-level trait evenness (TED) increases with decrease in PAR and TP in lake Greifensee (N=195). The grey surface represents the fitted linear model relating TED to log(PAR), log(TP) and their interaction.
Discussion
This study demonstrates that individual-level trait metrics may help link biodiversity in natural microbial communities to essential ecosystem properties. In particular, we found that TED (regularity in the distribution of individual phenotypes in trait space) was the most important predictor of community productivity. Previous studies have explored the correlation between TD measures and ecosystem properties in macro- and microorganisms, but trait evenness has rarely emerged as being important (Tilman et al., 1997; Mouillot et al., 2011; Bílá et al., 2014). Gagic et al. (2015) found contrasting effects of trait evenness, whereas Santos et al. (2014) showed a negative correlation between trait evenness and phytoplankton productivity, consistent with our results. These differences are probably due to the fact that the tested relationships are dependent on environmental heterogeneity (Norberg et al., 2001; Hodapp et al., 2016). It has to be noted, however, that the concept of trait evenness used in previous studies is radically different from the one used here, where regularities in trait distances among individuals represent the unit of measure. Previous studies calculated TD using species-level data, disregarding the individual-level trait differences that we find to be the most important predictors.
Individual-level trait evenness, as characterised by TED, is also conceptually different from species evenness, as, by measuring the distances of individual organisms in multidimensional trait space, it does not include any information about relative abundances (Supplementary Figure S4; Fontana et al., 2016). To understand the negative correlation between TED and community biomass in our data, it is necessary to refer to environmental effects on both, although they cannot be fully disentangled given the correlative nature of our analyses. Our interpretation of the negative relationship between TED and total biomass is that resource scarcity indirectly affected both in opposite ways—decreasing total community biomass and increasing TED (Figure 2). The pattern that we observed may be explained by plasticity or selection for particular phenotypic characters under conditions of resource limitation. This may induce individual microbes to be more evenly spaced in a multidimensional landscape defined by their functional traits (thereby minimising the overlap of ‘individual niches’). Our analyses confirmed that low levels of both light and nutrients maximised TED values in microbial communities (Figure 2).
We believe that resource limitation constrained the total productivity of the phytoplankton communities in our study, inducing individual phenotypes to spread more evenly along trait axes (to better exploit available resources), leading to the negative correlation between TED and community biomass. This interpretation does not contradict the expected outcomes of competition under resource limitation, which leads to the dominance (at the population level) of the species with the highest affinity for resources (Tilman, 1982). Variation in TED signals individual organisms converging around certain trait combinations (low TED) or spreading more regularly among trait space (Supplementary Figure S4); this happens as a consequence of co-occurring physiological and ecological processes. While both acclimation and competitive dominance could explain the above patterns, the fact that individual-level TD dominates the predictive power of our statistical models relative to species-based metrics (considered here as proxies of trait change due to taxa turnover) suggests that variation in TED may be explainable mainly by physiological responses at the single-cell level or genetic variation within species.
Light limitation can induce changes in the distribution of individual phytoplankton fluorescence traits because both the absolute and relative intracellular abundance of different pigments can be regulated, thereby changing absorbance across the light spectrum. This has been previously shown in the cyanobacterium Tolypothrix tenuis, in which algal cells regulated the relative amounts of different pigments to fill gaps in the prevailing light spectrum that were not already exploited by competitors (Stomp et al., 2004). And in plant communities, intraspecific trait variability is influenced by light acquisition traits of neighbour individuals (Le Bagousse-Pinguet et al., 2015). These studies showed that a complementary effect caused by the differential use of light is not always a consequence of increased taxa richness (as in Striebel et al., 2009a, 2009b), but is in principle possible at the intraspecific level. Light represents a spectrum of wavelengths, being a potentially infinite set of resources that can be exploited by photosynthetic organisms able to regulate pigment composition (Stomp et al., 2007a, 2007b, 2008).
The relationship between TP and TED is more difficult to interpret as many phytoplankton traits might be involved in optimising phosphorus uptake rate and use (Litchman and Klausmeier, 2008; Table 1). In this context, cell size represents a key trait, which reflects different nutrient acquisition strategies by determining surface-to-volume ratio (Litchman and Klausmeier, 2008). The fact that patterns observed for RUE are very similar to those of biomass (Supplementary Figure S1 and Supplementary Tables S6–S9) suggests, however, that the TP–TED relationship might be less important than the PAR–TED relationship. If the relationship between ecosystem properties and TED was driven by limiting TP, then it would have disappeared when biomass was corrected for TP (i.e. considering RUE), which was not the case.
In our data, species richness, which represents the most commonly used metric in studies correlating biodiversity with ecosystem properties (Cardinale et al., 2011), did not show a strong relationship with biomass. Interestingly, species richness showed no clear correlation with TD metrics across our data sets (maximum absolute value of Pearson’s r was 0.40). Our results seem to contradict previous studies (e.g. Hector et al., 1999; Cardinale et al., 2011) and the common assumption of a positive correlation between species richness and primary production. However, nonsignificant as well as negative correlations have been already reported previously (Cardinale et al., 2011). One possible reason for the lack of importance of species richness in our study is that we focused on natural communities shaped by natural selection gradients, whereas studies correlating biodiversity with ecosystem properties typically deal with communities assembled randomly from a species pool (Hooper et al., 2005; Krause et al., 2014). Natural communities are the result of assembly processes over abiotic and biotic gradients and their species composition reflects the adaptation to a specific environment. In this context, phenotypic composition might have a more important role compared with the number of species present. However, Ptacnik et al. (2008) found a significant positive relationship between genus richness and RUE in natural algal communities. These authors investigated a very large spatial (almost continental) and temporal scale (over several years), whereas our largest scale is regional (Danube Delta) with a generally shorter temporal frequency. This suggests that the scale of analysis might also be important when considering the relative importance of biodiversity metrics in explaining ecosystem processes (Farnsworth, 1998; Cadotte et al., 2011; Chalmandrier et al., 2017).
The importance of species evenness for ecosystem properties has been intensively tested in the past few years using experimental, observational and modelling approaches, with partially contradictory results (positive effect of evenness through niche partitioning vs dominance of high productive species). Most studies have focussed on grassland and forest communities (e.g. Mulder et al., 2004; Zhang et al., 2012; Orwin et al., 2014; Dooley et al., 2015), but some microbial communities have also been investigated (Filstrup et al., 2014; Powell et al., 2015). In this study, species evenness emerged as important only in lake Zurich (Figure 1), where it was the strongest predictor of biomass. This result likely reflects the specific characteristics of this lake’s phytoplankton community: in the past few decades, the ecosystem has been dominated by a single cyanobacterial species (Planktothrix rubescens) that accounts for ~40% of the total annual phytoplankton biomass (Posch et al., 2012). Variation in species evenness is principally driven by growth of this cyanobacterium, which causes fluctuations in community biomass (blooms). This is consistent with patterns observed in many other lakes (Filstrup et al., 2014).
Ecosystem processes depend upon guilds of interacting organisms and their aggregated responses to their immediate environment. Theoretically, individual-level trait variation is important to maintain ecosystem processes only in a spatially or temporally heterogeneous environment (under constant environmental conditions the fittest phenotype would eventually prevail) (Norberg et al., 2001; Norberg, 2004; Hodapp et al., 2016). The importance of individual-level TD in our analyses suggests that (1) there is significant environmental heterogeneity in the studied aquatic ecosystems, which may lead to the emergence and maintenance of TD (Ackermann, 2015; Stark et al., 2017), and (2) this heterogeneity (characterised by our TD indices) is important to understand and predict ecosystem processes. As TD can characterise phenotypic variation among individual organisms, aggregating physiological (plastic) responses of phenotypes, evolution of populations and ecological turnover of species, it has been long hypothesised to perform better than species-level metrics in explaining ecosystem properties (Norberg et al., 2001; Petchey and Gaston, 2006; Fontana et al., 2014, 2016). Despite variation between systems, including TD metrics in statistical or mechanistic models explaining phytoplankton productivity may allow us to improve our predictive power over a range of environmental gradients. The relationship between trait evenness and both environment and ecosystem properties deserves further investigation, both theoretical and empirical. Experimental work can help disentangle the mechanisms (physiology/evolution) that determine variation in the regularity of algal phenotypes in trait space, its dependency on environmental conditions and its importance for ecological interactions.
Data accessibility
The data sets analysed during the current study are available in the Zenodo repository at https://doi.org/10.5281/zenodo.848273.
Acknowledgments
This research is supported by the Swiss National Science Foundation Projects 31003A_144053, CRSII2_147654 and IZERZ0_142165 (Enlargement Contribution Project ‘CyanoArchive’, in the framework of the Romanian-Swiss Research Programme). We thank D Steiner, E Keller, C Tellenbach, A Schildknecht, I Nichersu, G Cosor and D Fontana. We also thank the CyanoArchive team: A Dumitrache, I Moza, I Enache and C Sandu for all their help. This manuscript was improved by the helpful comments of A Narwani, C Vorburger, M Ackermann and the three anonymous reviewers.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Ackermann M. (2015). A functional perspective on phenotypic heterogeneity in microorganisms. Nat Rev Microbiol 13: 497–508. [DOI] [PubMed] [Google Scholar]
- Albert CH, de Bello F, Boulangeat I, Pellet G, Lavorel S, Thuiller W. (2012). On the importance of intraspecific variability for the quantification of functional diversity. Oikos 121: 116–126. [Google Scholar]
- Barabás G, D’Andrea R. (2016). The effect of intraspecific variation and heritability on community pattern and robustness. Ecol Lett 19: 977–986. [DOI] [PubMed] [Google Scholar]
- Bílá K, Moretti M, de Bello F, Dias ATC, Pezzatti GB, Van Oosten AR et al. (2014). Disentangling community functional components in a litter–macrodetritivore model system reveals the predominance of the mass ratio hypothesis. Ecol Evol 4: 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolnick DI, Amarasekare P, Araújo MS, Bürger R, Levine JM, Novak M et al. (2011). Why intraspecific trait variation matters in community ecology. Trends Ecol Evol 26: 183–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnham KP, Anderson DR. (2002) Model Selection and Multimodel Inference. Springer: New York, NY, USA. [Google Scholar]
- Cadotte MW, Carscadden K, Mirotchnick N. (2011). Beyond species: functional diversity and the maintenance of ecological processes and services. J Appl Ecol 48: 1079–1087. [Google Scholar]
- Cardinale BJ, Matulich KL, Hooper DU, Byrnes JE, Duffy E, Gamfeldt L et al. (2011). The functional role of producer diversity in ecosystems. Am J Bot 98: 572–592. [DOI] [PubMed] [Google Scholar]
- Chalmandrier L, Münkemüller T, Colace M-P, Renaud J, Aubert S, Carlson BZ et al. (2017). Spatial scale and intraspecific trait variability mediate assembly rules in alpine grasslands. J Ecol 105: 277–287. [Google Scholar]
- Chase JM, Leibold MA. (2003) Ecological Niches. University of Chicago Press: Chicago, IL, USA. [Google Scholar]
- de Bello F, Lavorel S, Albert CH, Thuiller W, Grigulis K, Dolezal J et al. (2011). Quantifying the relevance of intraspecific trait variability for functional diversity. Methods Ecol Evol 2: 163–174. [Google Scholar]
- De Laender F, Melián CJ, Bindler R, Van den Brink PJ, Daam M, Roussel H et al. (2014). The contribution of intra- and interspecific tolerance variability to biodiversity changes along toxicity gradient. Ecol Lett 17: 72–81. [DOI] [PubMed] [Google Scholar]
- Dooley Á, Isbell F, Kirwan L, Connolly J, Finn JA, Brophy C. (2015). Testing the effects of diversity on ecosystem multifunctionality using a multivariate model. Ecol Lett 18: 1242–1251. [Google Scholar]
- Dormann CF, Elith J, Bacher S, Buchmann C, Carl G, Carré G et al. (2013). Collinearity: a review of methods to deal with it and a simulation study evaluating their performance. Ecography 36: 27–46. [Google Scholar]
- Dubelaar GBJ, Geerders PJF, Jonker RR. (2004). High frequency monitoring reveals phytoplankton dynamics. J Environ Monit 6: 946–952. [DOI] [PubMed] [Google Scholar]
- Farnsworth EJ. (1998). Issues of spatial, taxonomic and temporal scale in delineating links between mangrove diversity and ecosystem function. Glob Ecol Biogeogr Lett 7: 15–25. [Google Scholar]
- Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. (1998). Primary production of the biosphere: integrating terrestrial and oceanic components. Science 281: 237–240. [DOI] [PubMed] [Google Scholar]
- Filstrup CT, Hillebrand H, Heathcote AJ, Harpole WS, Downing JA. (2014). Cyanobacteria dominance influences resource use efficiency and community turnover in phytoplankton and zooplankton communities. Ecol Lett 17: 464–474. [DOI] [PubMed] [Google Scholar]
- Foladori P, Quaranta A, Ziglio G. (2008). Use of silica microspheres having refractive index similar to bacteria for conversion of flow cytometric forward light scatter into biovolume. Water Res 42: 3757–3766. [DOI] [PubMed] [Google Scholar]
- Fontana S, Jokela J, Pomati F. (2014). Opportunities and challenges in deriving phytoplankton diversity measures from individual trait-based data obtained by scanning flow-cytometry. Front Microbiol 5: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana S, Petchey OL, Pomati F. (2016). Individual-level trait diversity concepts and indices to comprehensively describe community change in multidimensional trait space. Funct Ecol 30: 808–818. [Google Scholar]
- Gagic V, Bartomeus I, Jonsson T, Taylor A, Winqvist C, Fischer C et al. (2015). Functional identity and diversity of animals predict ecosystem functioning better than species-based indices. Proc R Soc Ser B 282: 20142620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gsell AS, de Senerpont Domis LN, Verhoeven KJF, van Donk E, Ibelings BW. (2013). Chytrid epidemics may increase genetic diversity of a diatom spring-bloom. ISME J 7: 2057–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart SP, Schreiber SJ, Levine JM. (2016). How variation between individuals affects species coexistence. Ecol Lett 19: 825–838. [DOI] [PubMed] [Google Scholar]
- Hector A, Schmid C, Beierkuhnlein C, Caldeira MC, Diemer M, Dimitrakopoulos PG et al. (1999). Plant diversity and productivity experiments in European grasslands. Science 286: 1123–1127. [DOI] [PubMed] [Google Scholar]
- Hillebrand H, Bennett DM, Cadotte MW. (2008). Consequences of dominance: a review of eveness effects on local and regional ecosystem processes. Ecology 89: 1510–1520. [DOI] [PubMed] [Google Scholar]
- Hillebrand H, Matthiessen B. (2009). Biodiversity in a complex world: consolidation and progress in functional biodiversity research. Ecol Lett 12: 1405–1419. [DOI] [PubMed] [Google Scholar]
- Hodapp D, Hillebrand H, Blasius B, Ryabov AB. (2016). Environmental and trait variability constrain community structure and the biodiversity–productivity relationship. Ecology 97: 1463–1474. [DOI] [PubMed] [Google Scholar]
- Hooper DU, Chapin FS, Ewel JJ, Hector A, Inchausti P, Lavorel S et al. (2005). Effects of biodiversity on ecosystem functioning: a consensus of current knowledge. Ecol Monogr 75: 3–35. [Google Scholar]
- Krause S, Le Roux X, Niklaus PA, Van Bodegom PM, Lennon JT, Bertilsson S et al. (2014). Trait-based approaches for understanding microbial biodiversity and ecosystem functioning. Front Microbiol 5: 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krismer J, Tamminen M, Fontana S, Zenobi R, Narwani A. (2017). Single-cell mass spectrometry reveals the importance of genetic diversity and plasticity for phenotypic variation in nitrogen-limited Chlamydomonas. ISME J 11: 988–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laliberté E, Legendre P. (2010). A distance-based framework for measuring functional diversity from multiple traits. Ecology 91: 299–305. [DOI] [PubMed] [Google Scholar]
- Le Bagousse-Pinguet Y, Börger L, Quero JL, García-Gómez M, Soriano S, Maestre FT et al. (2015). Traits of neighbouring plants and space limitation determine intraspecific trait variability in semi-arid shrublands. J Ecol 103: 1647–1657. [Google Scholar]
- Litchman E, Klausmeier CA. (2008). Trait-based community ecology of phytoplankton. Annu Rev Ecol Evol Syst 39: 615–639. [Google Scholar]
- Mason NWH, Mouillot D, Lee WG, Wilson JB. (2005). Functional richness, functional evenness and functional divergence: the primary components of functional diversity. Oikos 1: 112–118. [Google Scholar]
- Matthews B, Narwani A, Hausch S, Nonaka E, Peter H, Yamamichi M et al. (2011). Toward an integration of evolutionary biology and ecosystem science. Ecol Lett 14: 690–701. [DOI] [PubMed] [Google Scholar]
- McGill BJ, Enquist BJ, Weiher E, Westoby M. (2006). Rebuilding community ecology from functional traits. Trends Ecol Evol 21: 178–185. [DOI] [PubMed] [Google Scholar]
- Mouillot D, Villéger S, Scherer-Lorenzen M, Mason NWH. (2011). Functional structure of biological communities predicts ecosystem multifunctionality. PLoS One 6: e17476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder CPH, Bazeley-White E, Dimitrakopoulos PG, Hector A, Scherer-Lorenzen M, Schmid B. (2004). Species evenness and productivity in experimental plant communities. Oikos 107: 50–63. [Google Scholar]
- Nakagawa S, Schielzeth H. (2013). A general and simple method for obtaining R2 from generalized linear mixed-effects models. Methods Ecol Evol 4: 133–142. [Google Scholar]
- Norberg J. (2004). Biodiversity and ecosystem functioning: a complex adaptive systems approach. Limnol Oceanogr 49: 1269–1277. [Google Scholar]
- Norberg J, Swaney DP, Dushoff J, Lin J, Casagrandi R, Levin SA. (2001). Phenotypic diversity and ecosystem functioning in changing environments: a theoretical framework. Proc Natl Acad Sci USA 98: 11376–11381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orwin KH, Ostle N, Wilby A, Bardgett RD. (2014). Effects of species evenness and dominant species identity on multiple ecosystem functions in model grassland communities. Oecologia 174: 979–992. [DOI] [PubMed] [Google Scholar]
- Petchey OL, Gaston KJ. (2006). Functional diversity: back to basics and looking forward. Ecol Lett 9: 741–758. [DOI] [PubMed] [Google Scholar]
- Pomati F, Jokela J, Simona M, Veronesi M, Ibelings BW. (2011). An automated platform for phytoplankton ecology and aquatic ecosystem monitoring. Environ Sci Technol 45: 9658–9665. [DOI] [PubMed] [Google Scholar]
- Pomati F, Kraft NJB, Posch T, Eugster B, Jokela J, Ibelings BW. (2013). Individual cell based traits obtained by scanning flow-cytometry show selection by biotic and abiotic environmental factors during a phytoplankton spring bloom. PLoS ONE 8: e71677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomati F, Matthews B, Jokela J, Schildknecht A, Ibelings BW. (2012). Effects of re-oligotrophication and climate warming on plankton richness and community stability in a deep mesotrophic lake. Oikos 121: 1317–1327. [Google Scholar]
- Pomati F, Nizzetto L. (2013). Assessing triclosan-induced ecological and trans-generational effects in natural phytoplankton communities: a trait-based field method. Ecotoxicology 22: 779–794. [DOI] [PubMed] [Google Scholar]
- Posch T, Köster O, Salcher MM, Pernthaler J. (2012). Harmful filamentous cyanobacteria favoured by reduced water turnover with lake warming. Nat Clim Chang 2: 1–5. [Google Scholar]
- Powell JR, Welsh A, Hallin S. (2015). Microbial functional diversity enhances predictive models linking environmental parameters to ecosystem properties. Ecology 96: 1985–1993. [DOI] [PubMed] [Google Scholar]
- Ptacnik R, Solimini AG, Andersen T, Tamminen T, Brettum P, Lepistö L et al. (2008). Diversity predicts stability and resource use efficiency in natural phytoplankton communities. Proc Natl Acad Sci USA 105: 5134–5138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Reiss J, Bridle JR, Montoya JM, Woodward G. (2009). Emerging horizons in biodiversity and ecosystem functioning research. Trends Ecol Evol 24: 505–514. [DOI] [PubMed] [Google Scholar]
- Richards SA. (2005). Testing ecological theory using the information-theoretic approach: examples and cautionary results. Ecology 86: 2805–2814. [Google Scholar]
- Santos AMC, Carneiro FM, Cianciaruso MV. (2014). Predicting productivity in tropical reservoirs: the roles of phytoplankton taxonomic and functional diversity. Ecol Indic 48: 428–435. [Google Scholar]
- Schreiber F, Littmann S, Lavik G, Escrig S, Meibom A, Kuypers MMM et al. (2016). Phenotypic heterogeneity driven by nutrient limitation promotes growth in fluctuating environments. Nat Microbiol 1: 16055. [DOI] [PubMed] [Google Scholar]
- Shade A, Carey CC, Kara E, Bertilsson S, McMahon KD, Smith MC. (2009). Can the black box be cracked? The augmentation of microbial ecology by high-resolution, automated sensing technologies. ISME J 3: 881–888. [DOI] [PubMed] [Google Scholar]
- Siefert A, Ritchie ME. (2016). Intraspecific trait variation drives functional responses of old-field plant communities to nutrient enrichment. Oecologia 181: 245–255. [DOI] [PubMed] [Google Scholar]
- Stark J, Lehman R, Crawford L, Enquist BJ, Blonder B. (2017). Does environmental heterogeneity drive functional trait variation? A test in montane and alpine meadows. Oikos doi:10.1111/oik.04311.
- Stomp M, Huisman J, De Jongh F, Veraart AJ, Gerla D, Rijkeboer M et al. (2004). Adaptive divergence in pigment composition promotes phytoplankton biodiversity. Nature 432: 104–107. [DOI] [PubMed] [Google Scholar]
- Stomp M, Huisman J, Stal LJ, Matthijs HCP. (2007. a). Colorful niches of phototrophic microorganisms shaped by vibrations of the water molecule. ISME J 1: 271–282. [DOI] [PubMed] [Google Scholar]
- Stomp M, van Dijk MA, van Overzee HMJ, Wortel MT, Sigon CAM, Egas M et al. (2008). The timescale of phenotypic plasticity and its impact on competition in fluctuating environments. Am Nat 172: 169–185. [DOI] [PubMed] [Google Scholar]
- Stomp M, Huisman J, Vörös L, Pick FR, Laamanen M, Haverkamp T et al. (2007. b). Colourful coexistence of red and green picocyanobacteria in lakes and seas. Ecol Lett 10: 290–298. [DOI] [PubMed] [Google Scholar]
- Striebel M, Behl S, Diehl S, Stibor H. (2009. a). Spectral niche complementarity and carbon dynamics in pelagic ecosystems. Am Nat 174: 141–147. [DOI] [PubMed] [Google Scholar]
- Striebel M, Behl S, Stibor H. (2009. b). The coupling of biodiversity and productivity in phytoplankton communities: consequences for biomass stoichiometry. Ecology 90: 2025–2031. [DOI] [PubMed] [Google Scholar]
- Tartari GA, Mosello R. (1997). Metodologie analitiche e controlli di qualità nel laboratorio chimico dell’Istituto Italiano di Idrobiologia. Doc Ist Ital Idrobiol 60: 160. [Google Scholar]
- Tilman D. (1982) Resource Competition And Community Structure. Princeton University Press: Princeton, NJ, USA. [Google Scholar]
- Tilman D, Knops J, Wedin D, Reich P. (1997). The influence of functional diversity and composition on ecosystem processes. Science 277: 1300–1302. [Google Scholar]
- Utermöhl vH. (1931). Neue Wege in der quantitativen Erfassung des Planktons. (Mit besonderer Berücksichtigung des Ultraplanktons). Verh Int Ver Theoret Angew Limnol 5: 567–595. [Google Scholar]
- Violle C, Enquist BJ, McGill BJ, Jiang L, Albert CH, Hulshof C et al. (2012). The return of the variance: intraspecific variability in community ecology. Trends Ecol Evol 27: 244–252. [DOI] [PubMed] [Google Scholar]
- Violle C, Navas M-L, Vile D, Kazakou E, Fortunel C, Hummel I et al. (2007). Let the concept of trait be functional!. Oikos 116: 882–892. [Google Scholar]
- Volf M, Redmond C, Albert ÁJ, Le Bagousse‐Pinguet Y, Biella P, Götzenberger L et al. (2016). Effects of long- and short-term management on the functional structure of meadows through species turnover and intraspecific trait variability. Oecologia 180: 941–950. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Chen HYH, Reich PB. (2012). Forest productivity increases with evenness, species richness and trait variation: a global meta-analysis. J Ecol 100: 742–749. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data sets analysed during the current study are available in the Zenodo repository at https://doi.org/10.5281/zenodo.848273.