Abstract
Soil microbes are essential for soil fertility. However, most studies focus on bacterial and/or fungal communities, while the top-down drivers of this microbiome composition, protists, remain poorly understood. Here, we investigated how soil amendments affect protist communities and inferred potential interactions with bacteria and fungi. Specific fertilization treatments impacted both the structure and function of protist communities. Organic fertilizer amendment strongly reduced the relative abundance of plant pathogenic protists and increased bacterivorous and omnivorous protists. The addition of individual biocontrol bacteria and fungi further altered the soil protist community composition, and eventually function. Network analysis integrating protist, bacterial and fungal community data, placed protists as a central hub in the soil microbiome, linking diverse bacterial and fungal populations. Given their dynamic response to soil management practices and key position in linking soil microbial networks, protists may provide the leverage between soil management and the enhancement of bacterial and fungal microbiota at the service of improved soil health.
Protists are an often overlooked component of the soil microbiome. They are abundant and extremely diverse in soil, where they carry out a range of functions (Foissner, 1997; Geisen, 2016a). Protists are among the main consumers of soil bacteria and fungi, but also algae and nematodes (Geisen, 2016a, b; Seppey et al., 2017). Protist activity directly increases plant performance by enhancing the microbial loop (Bonkowski, 2004) and stimulating plant growth-promoting rhizobacteria (Rosenberg et al., 2009; Jousset, 2012). Further, soil protist communities encompass a range of plant- and animal-pathogenic species (Geisen et al., 2015b). Given their functional diversity, protists exert control over various soil organisms and are likely of critical importance for soil fertility. However, we lack an understanding of how protist communities are structured, how targeted soil management can alter protist communities, how such changes might affect protist functioning, and how they are in turn linked to their potential bacterial and fungal prey. Here, we experimentally examined under controlled greenhouse conditions if applications of organic versus conventional fertilizers could modify the taxonomic and functional composition of protists, as well as their putative interactions within the soil microbiome. Furthermore, we added either a bacterial (Bacillus amyloliquefaciens) or fungal (Trichoderma guizhouense) plant-protective agent (Wang et al., 2013; Zhang et al., 2016) to organic fertilizer (subsequently termed OF+B and OF+T, respectively) to study if these biocontrol agents also impact protist communities (Xiong et al., 2017 and Supplementary Information for additional details).
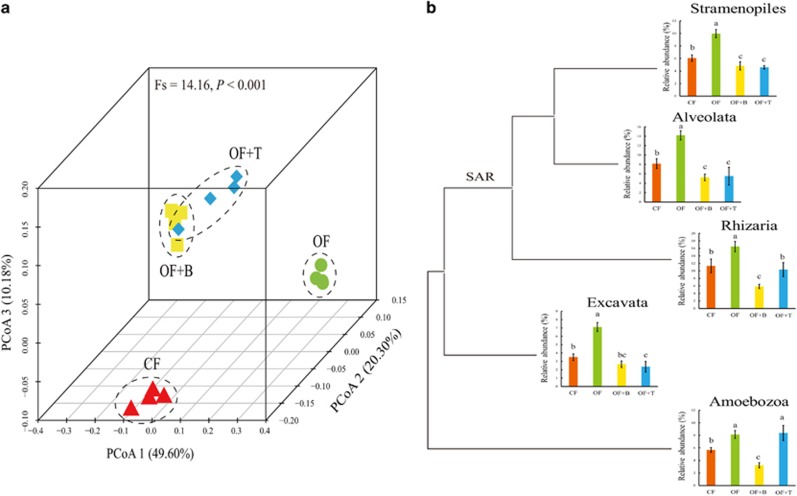
Protist communities differed significantly between all treatments one-year post application. Organic fertilizer-treated soil contained a fundamentally different protist community structure compared with the chemical fertilizer (CF) treatment (UniFrac-weighted and -unweighted PCoA: Figure 1a; Supplementary Figure 2; RDA: Supplementary Figure 6), which is in line with former studies (Heger et al., 2012; Murase et al., 2015). Organic fertilizer (OF) amendment enhanced the relative abundance of the most abundant protist taxonomic groups, that is, Stramenopiles, Alveolata, Rhizaria, Excavata and Amoebozoa (Figure 1b), most of which are predators of other microbes. This can be explained by the fact that organic fertilizer provides a wider resource spectrum than chemical fertilizer, which thereby promotes a higher biomass and diversity of bacteria and fungi (Xiong et al., 2017). This impact on primary consumers may foster diverse and active protist consumers, as previously observed in studies of paddy rice (Murase et al., 2015). Organic fertilizer enriched with beneficial microbes (Bacillus and Trichoderma) caused a further shift in the protist community composition by reducing Stramenopiles, Alveolata and Excavata, and most strongly Rhizaria and Amoebozoa in the OF+B treatment (Figure 1b). As a result, protist richness (observed OTUs), diversity (phylogenetic and Shannon) and evenness (Shannon) were lower in the OF+B and OF+T treatments as compared with the OF treatment (Supplementary Figure 1). These results suggest that the added microbes differentially affected specific protist taxonomic groups (Jousset et al., 2006; Pedersen et al., 2011), possibly by producing inhibitory compounds. Bacillus species are known to produce a broad range of secondary metabolites such as cyclic lipopeptide antibiotics (Alvarez et al., 2012) and various volatile compounds that can deter or even kill potential protist predators (Mazzola et al., 2009; Schulz-Bohm et al., 2017). Trichoderma may also have affected protists via the production of antimicrobial compounds (Cai et al., 2013) or by reducing fungal prey, but information on protist-fungal interactions is too scarce for reliably interpreting these data (Geisen, 2016a). Introduced microbes may also exert indirect effects; the lower diversity of protists and lower relative abundances of Stramenopiles, Alveolata and Excavata in the two microbe-enriched organic fertilizer treatments may be linked to the bacterial genus Lysobacter, which increased ~6-fold in OF+B and OF+T treatments compared with the CF treatment (Xiong et al., 2017). Members of this genus are known to produce a broad range of bioactive secondary metabolites that can inhibit soil organisms (Expósito et al., 2015).
Figure 1.
Protist community changes induced by fertilizations. (a) UniFrac-weighted principle coordinate analysis (PCoA) of soil protist community in the four fertilizer treatments and (b) relative abundance of five main protist taxonomic groups (Stramenopiles, Alveolata, Rhizaria, Excavata and Amoebozoa) with coarse phylogenetic affinities in the four fertilizer treatments. CF, chemical fertilizer; OF, organic matter fertilizer; OF+B, Bacillus enriched organic fertilizer; OF+T, Trichoderma enriched organic fertilizer. Different letters above the bars indicate a significant difference at the 0.05 probability level according to the Tukey’s test.
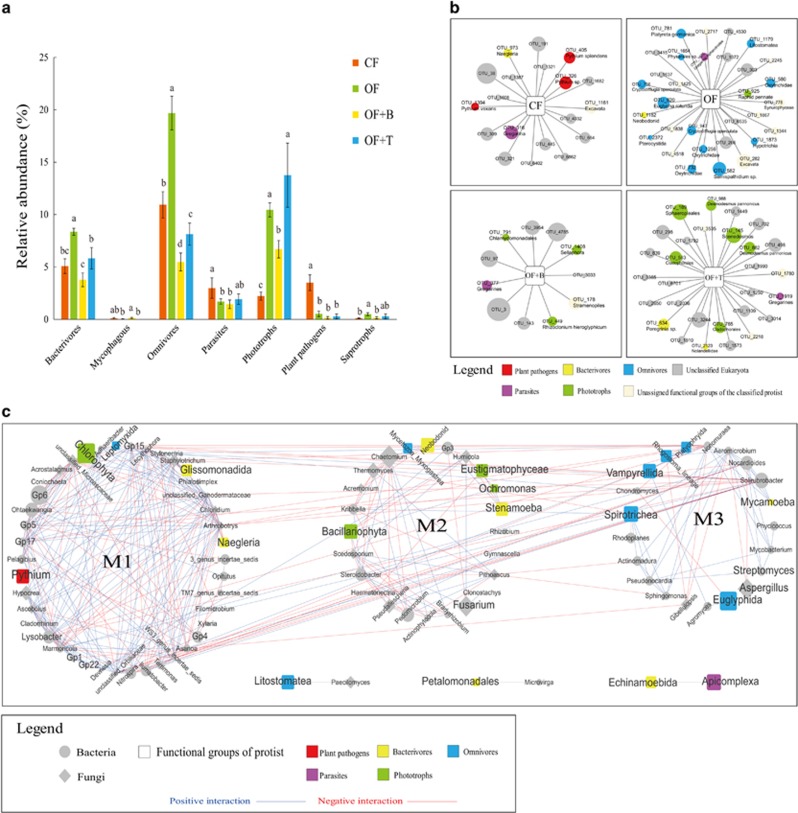
Changes in protist taxonomic community compositions induced by fertilizer regime subsequently resulted in shifts in the relative proportion of protist functional groups (Figure 2). Several potentially plant pathogenic Pythium spp., a group of widely distributed pathogens of thousands of plant species (Schroeder et al., 2012), were indicative for CF treatment (Figure 2b). In line, putative protist plant pathogens, including Pythium spp., were significantly reduced in all three organic fertilizer treatments (OF, OF+B and OF+T) compared with CF treatment (Figure 2a; Supplementary Table 2). This may be due either to direct inhibition by the introduced beneficial microbes, the stimulation of antagonistic microbes, or simply the promotion of a range of protists that consume or outcompete Pythium in organic matter. Apicomplexa, known as obligate parasites of vertebrates and invertebrate hosts (Kopečná et al., 2006), were lower in all organic fertilizer treatments as compared with the CF treatment (Supplementary Table 2). These results suggest a relatively ‘pathogen and parasite’ driven food-web in chemical fertilizer-treated soils. In contrast, soils treated with OF were not only higher in saprotrophs, but also in phototrophic algae (Figure 2a), indicative of less disturbed soils and increased soil carbon inputs (Zancan et al., 2006). The indicator taxa for OF were mainly omnivores consuming other eukaryotes (Figure 2b), suggesting a more complex food-web. The addition of Bacillus into OF, but not of Trichoderma, significantly reduced the saprotrophic and phototrophic protists. More targeted studies would be necessary to examine the functional changes in the protist communities after applying different biocontrol agents and their long-term impact on soil functioning. It has to be mentioned that we used a conservative approach to assign the classified protists into different functions, focusing merely on feeding mode. As any inference tool, our database approach should be interpreted with caution. It is necessarily reliant on the limited number of characterized reference species (Berney et al., 2017), and may therefore be biased toward specific functions as some elements of functional variation. Current efforts to expand and improve this taxonomic and functional database (Berney et al., 2017) will further improve this tool, and we anticipate that this approach will become more useful in the future to advance studies that seek to more meaningful integrate protists in soil microbiome research.
Figure 2.
Protists form a functional and dynamic hub in the soil microbiome induced by fertilizations. (a) Overview of the relative abundance of protist functional groups in the four fertilizer treatments. CF, chemical fertilizer; OF, organic matter fertilizer; OF+B, Bacillus enriched organic fertilizer; OF+T, Trichoderma enriched organic fertilizer. Different letters above the bars indicate a significant difference at the 0.05 probability level according to the Tukey’s test. (b) Protist indicator OTUs for the four fertilizer treatments. Circles represent protist OTUs and circle sizes correspond to their average relative abundance (log transformation) across all the samples. (c) Correlation-based network analysis showing potential interactions between abundant bacterial and fungal genera as well as protist functional groups. The node size is proportional to a taxon’s average relative abundance (log transformation) across all the samples. Lines connecting nodes (edges) represent positive (blue) or negative (red) co-occurrence relationships.
Finally, we examined the role of protists as an integral part of soil ecological networks. By constructing a combined co-occurrence network encompassing bacteria, fungi and functional groups of protists, we found that protists form distinct hubs in the soil network, linking a range of bacterial and fungal taxa (Supplementary Information; Figure 2c). Three main modules emerged in our constructed network, with protists present in all modules. Module 1 was dominated by bacteria such as Lysobacter and several Acidobacteria groups and contained five protist nodes from distinct taxonomic and functional groups (the mainly plant pathogenic Pythium, the bacterivorous Glissomonadida and Naegleria). Module 2 was phototroph-dominated (such as Bacillariophyta and Eustigmatophyceae). Module 3 was dominated by omnivorous protists (such as Spirotrichea, Vampyrellida and Euglyphida). Each module appeared to be generally associated with a specific range of functions (Zhou et al., 2011), suggesting interactions between similarly functioning microbes that provide either a stimulatory or inhibitory loop for soil functioning. Parasitic protist taxa, including the potential plant pathogenic Pythium and animal parasitic Apicomplexa, were present, but poorly connected to other microorganisms in the network. This disconnected position is likely related to the dependence of these organisms on plants (Xu et al., 2012) and soil animals (Geisen et al., 2015a) rather than other soil microbes.
Our study highlights the multi-trophic nature of the soil microbiome. This study is one of the first to link taxonomically assigned protist taxa to functional groups that are embedded within soil food webs. Soil amendments strongly impacted protist communities 1-year after application, with addition of organic material and beneficial microbes leading to profound changes of protist community composition, and eventually function. This study also serves as a plea to the scientific community to better integrate protists into microbiome studies. Given their large impacts on multiple soil functions, we propose that manipulation of soil protist communities offers new avenues to promote soil health, plant performance and other ecosystem services.
Acknowledgments
We thank Steven Lindow and four anonymous reviewers for their constructive comments on the manuscript. This research was supported by the National Key Basic Research Program of China (2015CB150506), the National Natural Science Foundation of China (31572212, 31672242 and 31501824), the Royal Netherlands Academy of Arts and Sciences (KNAW) joint network grant, the Natural Science Foundation of Jiangsu (BK20150059), National Key Scientific Research Project (2016YFD0800605, 2016YFD0200106 and 2016YFE0101100), the Priority Academic Program Development of the Jiangsu Higher Education Institutions (PAPD), and Qing Lan Project. Stefan Geisen was supported by the ERC advanced grant SPECIALS (ERC-Adv 260-55290) awarded to Wim H. van der Putten. Alexandre Jousset was supported by the Netherlands Organisation for Scientific Research (NWO) (ALW.870.15.050) and the Royal Netherlands Academy of Arts and Sciences (KNAW) (530-5CDP18). Wu Xiong was supported by the Sino-Dutch Bilateral Exchange Scholarship.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Alvarez F, Castro M, Príncipe A, Borioli G, Fischer S, Mori G et al. (2012). The plant-associated Bacillus amyloliquefaciens strains MEP218 and ARP23 capable of producing the cyclic lipopeptides iturin or surfactin and fengycin are effective in biocontrol of sclerotinia stem rot disease. J Appl Microbiol 112: 159–174. [DOI] [PubMed] [Google Scholar]
- Berney C, Ciuprina A, Bender S, Brodie J, Edgcomb V, Kim E et al. (2017). UniEuk: time to speak a common language in protistology!. J Eukaryot Microbiol 64: 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonkowski M. (2004). Protozoa and plant growth: the microbial loop in soil revisited. New Phytol 162: 617–631. [DOI] [PubMed] [Google Scholar]
- Cai F, Yu G, Wang P, Wei Z, Fu L, Shen Q et al. (2013). Harzianolide, a novel plant growth regulator and systemic resistance elicitor from Trichoderma harzianum. Plant Physiol Biochem 73: 106–113. [DOI] [PubMed] [Google Scholar]
- Expósito RG, Postma J, Raaijmakers JM, De Bruijn I. (2015). Diversity and activity of Lysobacter species from disease suppressive soils. Front Microbiol 6: 1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foissner W. (1997). Protozoa as bioindicators in agroecosystems, with emphasis on farming practices, biocides, and biodiversity. Agric Ecosyst Environ 62: 93–103. [Google Scholar]
- Geisen S. (2016. a). The bacterial-fungal energy channel concept challenged by enormous functional versatility of soil protists. Soil Biol Biochem 102: 22–25. [Google Scholar]
- Geisen S. (2016. b). Thorough high-throughput sequencing analyses unravels huge diversities of soil parasitic protists. Environ Microbiol 18: 1669–1672. [DOI] [PubMed] [Google Scholar]
- Geisen S, Laros I, Vizcaíno A, Bonkowski M, de Groot GA. (2015. a). Not all are free-living: high-throughput DNA metabarcoding reveals a diverse community of protists parasitizing soil metazoa. Mol Ecol 24: 4556–4569. [DOI] [PubMed] [Google Scholar]
- Geisen S, Tveit AT, Clark IM, Richter A, Svenning MM, Bonkowski M et al. (2015. b). Metatranscriptomic census of active protists in soils. ISME J 9: 2178–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heger TJ, Straub F, Mitchell EAD. (2012). Impact of farming practices on soil diatoms and testate amoebae: a pilot study in the DOK-trial at Therwil, Switzerland. Eur J Soil Biol 49: 31–36. [Google Scholar]
- Jousset A. (2012). Ecological and evolutive implications of bacterial defences against predators. Environ Microbiol 14: 1830–1843. [DOI] [PubMed] [Google Scholar]
- Jousset A, Lara E, Wall LG, Valverde C. (2006). Secondary metabolites help biocontrol strain Pseudomonas fluorescens CHA0 to escape protozoan grazing. Appl Environ Microbiol 72: 7083–7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopečná J, Jirků M, Oborník M, Tokarev YS, Lukeš J, Modrý D. (2006). Phylogenetic analysis of coccidian parasites from invertebrates: search for missing links. Protist 157: 173–183. [DOI] [PubMed] [Google Scholar]
- Mazzola M, de Bruijn I, Cohen MF, Raaijmakers JM. (2009). Protozoan-induced regulation of cyclic lipopeptide biosynthesis is an effective predation defense mechanism for Pseudomonas fluorescens. Appl Environ Microbiol 75: 6804–6811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murase J, Hida A, Ogawa K, Nonoyama T, Yoshikawa N, Imai K. (2015). Impact of long-term fertilizer treatment on the microeukaryotic community structure of a rice field soil. Soil Biol Biochem 80: 237–243. [Google Scholar]
- Pedersen AL, Winding A, Altenburger A, Ekelund F. (2011). Protozoan growth rates on secondary-metabolite-producing Pseudomonas spp. correlate with high-level protozoan taxonomy. FEMS Microbiol Lett 316: 16–22. [DOI] [PubMed] [Google Scholar]
- Rosenberg K, Bertaux J, Krome K, Hartmann A, Scheu S, Bonkowski M. (2009). Soil amoebae rapidly change bacterial community composition in the rhizosphere of Arabidopsis thaliana. ISME J 3: 675–684. [DOI] [PubMed] [Google Scholar]
- Schroeder KL, Martin FN, de Cock AWAM, Lévesque CA, Spies CFJ, Okubara PA et al. (2012). Molecular detection and quantification of pythium species: evolving taxonomy, new tools, and challenges. Plant Dis 97: 4–20. [DOI] [PubMed] [Google Scholar]
- Schulz-Bohm K, Geisen S, Wubs ERJ, Song C, de Boer W, Garbeva P. (2017). The prey’s scent – volatile organic compound mediated interactions between soil bacteria and their protist predators. ISME J 11: 817–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seppey CVW, Singer D, Dumack K, Fournier B, Belbahri L, Mitchell EAD et al. (2017). Distribution patterns of soil microbial eukaryotes suggests widespread algivory by phagotrophic protists as an alternative pathway for nutrient cycling. Soil Biol Biochem 112: 68–76. [Google Scholar]
- Wang B, Yuan J, Zhang J, Shen Z, Zhang M, Li R et al. (2013). Effects of novel bioorganic fertilizer produced by Bacillus amyloliquefaciens W19 on antagonism of Fusarium wilt of banana. Biol Fertil Soils 49: 435–446. [Google Scholar]
- Xiong W, Guo S, Jousset A, Zhao Q, Wu H, Li R et al. (2017). Bio-fertilizer application induces soil suppressiveness against Fusarium wilt disease by reshaping the soil microbiome. Soil Biol Biochem 114: 238–247. [Google Scholar]
- Xu L, Ravnskov S, Larsen J, Nilsson RH, Nicolaisen M. (2012). Soil fungal community structure along a soil health gradient in pea fields examined using deep amplicon sequencing. Soil Biol Biochem 46: 26–32. [Google Scholar]
- Zancan S, Trevisan R, Paoletti MG. (2006). Soil algae composition under different agro-ecosystems in North-Eastern Italy. Agric Ecosyst Environ 112: 1–12. [Google Scholar]
- Zhang J, Bayram Akcapinar G, Atanasova L, Rahimi MJ, Przylucka A, Yang D et al. (2016). The neutral metallopeptidase NMP1 of Trichoderma guizhouense is required for mycotrophy and self-defence. Environ Microbiol 18: 580–597. [DOI] [PubMed] [Google Scholar]
- Zhou J, Deng Y, Luo F, He Z, Yang Y. (2011). Phylogenetic molecular ecological network of soil microbial communities in response to elevated CO2. mBio 2: e00122–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.