Abstract
Plants alter chemical and physical properties of soil, and thereby influence rhizosphere microbial community structure. The structure of microbial communities may in turn affect plant performance. Yet, outside of simple systems with pairwise interacting partners, the plant genetic pathways that influence microbial community structure remain largely unknown, as are the performance feedbacks of microbial communities selected by the host plant genotype. We investigated the role of the plant circadian clock in shaping rhizosphere community structure and function. We performed 16S ribosomal RNA gene sequencing to characterize rhizosphere bacterial communities of Arabidopsis thaliana between day and night time points, and tested for differences in community structure between wild-type (Ws) vs clock mutant (toc1-21, ztl-30) genotypes. We then characterized microbial community function, by growing wild-type plants in soils with an overstory history of Ws, toc1-21 or ztl-30 and measuring plant performance. We observed that rhizosphere community structure varied between day and night time points, and clock misfunction significantly altered rhizosphere communities. Finally, wild-type plants germinated earlier and were larger when inoculated with soils having an overstory history of wild-type in comparison with clock mutant genotypes. Our findings suggest the circadian clock of the plant host influences rhizosphere community structure and function.
Introduction
In comparison with unvegetated soils, the presence of plants markedly affects the structure of soil microbial communities. Plant roots affect the physical, as well as chemical environment through the exudation of carbon into the rhizosphere zone, which immediately surrounds the roots (Bais et al., 2006; Jones et al., 2009; Dennis et al., 2010). Rhizosphere microbial community structure is dynamic and changes over the course of plant development (Lundberg et al., 2012), in part because of changes in exudation (Chaparro et al., 2014). Although much is known about rhizosphere assembly dynamics on longer time scales, there is currently little information regarding assembly dynamics on shorter, diurnal time scales. Further, although plant exudation may ‘feed-down’ and affect microbial community structure, rhizosphere communities can ‘feed-up’ and affect plant performance, by increasing plant access to nutrients (Çakmakçı et al., 2001; Chen et al., 2002; Richardson et al., 2009; Richardson and Simpson, 2011), relieving abiotic stress (Zolla et al., 2013), suppressing pathogens (Mendes et al., 2011, 2013), altering phenology (Wagner et al., 2014; Panke-Buisse et al., 2014) and promoting plant growth (Bashan, 1998; Lugtenberg and Kamilova, 2009; Henning et al., 2016). Some plant species, such as many legumes, have developmental genetic mechanisms that attract explicitly beneficial nitrogen-fixing rhizobia taxa (Bravo et al., 2016). The extent to which plants may attract complex beneficial communities remains largely unclear.
The use of experimental genetic lines available in plant model species may reveal specific genetic paths that affect microbial community structure. Comparing mutant vs wild-type plants of Arabidopsis thaliana, Lebeis et al. (2015) observed that salicylic acid, an immune signaling molecule, altered rhizosphere bacterial community structure. This finding suggests that genes regulating physiological traits, such as immune response, may have a role in shaping rhizosphere communities. Genes regulating additional physiological traits such as gas exchange and specifically carbon assimilation may also be worth examining, because of their effects on the photoassimilate pool available for allocation to root growth and for carbon exudation into the rhizosphere. More generally, the comparison of phenotypes between single-locus mutant genotypes with those expressed by wild-type genotypes removes potentially confounding effects of variation segregating elsewhere in the genome, and enables isolation of pathway-specific effects. Naturally occurring large-effect alleles at causal loci could in some cases have a role in shaping microbial community structure in natural plant populations or could be manipulated in crop species to improve plant growth.
Changes in the presence–absence (or abundance) of just a few microbial taxa can affect plant performance, because of the vast number of root-associated microbial cells and functions (Henning et al., 2016). Across diverse ecosystems, community structure and function are related (Tilman et al., 1997; Talbot et al., 2014), including in plant–rhizosphere associations and in cases where microbial community membership changes by one to few taxa. For instance, Henning et al. (2016) observed that the addition of single bacterial taxa to the rhizosphere of Populus trichocarpa led to drastic changes in plant growth traits. Similarly, Zolla et al. (2013) observed differences in drought response by Arabidopsis thaliana plants grown in soil that differed in community structure as a consequence of overstory history. Thus, differences in rhizosphere community structure can lead to differences in rhizosphere community function as estimated from plant performance.
In the current study, we tested the role of the plant circadian clock in determining rhizosphere community structure and function, where function was measured as plant performance. The circadian clock regulates up to 30% of the transcriptome, and affects diverse processes including patterned fluxes of carbon into (stomatal conductance, carbon assimilation) and out of (exudation) the plant on a diurnal scale (Watt and Evans, 1999; Harmer et al., 2000; McClung, 2006; Covington et al., 2008; Badri and Vivanco, 2009; Harmer, 2009; Greenham and McClung, 2015). We hypothesized that the circadian clock could shape rhizosphere community structure on a diurnal scale, if community structure responds to diurnally patterned fluxes of carbon into the rhizosphere, that is, we anticipated that microbial community structure might vary over the course of 24 h. We further hypothesized that rhizosphere communities of plant genotypes harboring clock mutations could differ from wild-type plants, because of differences in physiological phenotypes. Specifically, mutations in the clock genes TIMING OF CAB EXPRESSION 1 and ZEITLUPE lead plants to express altered clock period, or the duration of one circadian cycle (Millar et al., 1995; Kim et al., 2005). As a consequence of altered clock function, clock mutants express distinct physiological phenotypes, including reduced carbon assimilation, chlorophyll content and stomatal conductance (and thus root water uptake) relative to wild-type plants under 24 -h environmental cycles (Dodd et al., 2004, 2005). Clock misfunction may influence rhizosphere communities, if for instance the reduced flux of carbon into plants influences the flux of carbon exudation (Thornton et al., 2004) or if shifts in plant water use alter soil water potential and nutrient availability and hence the rhizosphere environment (Matimati et al., 2014). Finally, if rhizosphere community structure is altered by mutations in clock genes, then we hypothesize there may be differences in community function in the form of plant performance, in which microbial communities shaped by wild-type genotypes may lead to improved plant performance in comparison with microbial communities found in association with clock mutant genotypes.
Materials and methods
Plant material and growth conditions
To investigate the role of the circadian clock in shaping rhizosphere community structure and function, we used the Arabidopsis thaliana accession, Wassilewskija (Ws, CS2360), and two circadian clock period mutants in the Ws background, TIMING OF CAB EXPRESSION 1 (toc1-21) and ZEITLUPE (ztl-30). toc1-21 is a short-period mutant (~ 20 h), while ztl-30 is a long-period mutant (~28 h) in free-running conditions (Kevei et al., 2006; Fujiwara et al., 2008). Many prior studies have shown that the resonance between endogenous and environmental cycles affects plant phenotypes and performance (Dodd et al., 2005; Yerushalmi and Green, 2009; de Montaigu et al., 2015; Salmela et al., 2016); the current experiments extend prior research to test effects of the plant host clock on the rhizosphere microbiome.
For each experiment, seeds were surface sterilized using 15% bleach, 0.1% Tween and 84.9% RO H2O solution, cold stratified in the dark in 1 ml of RO H2O for 5 days at 4 °C, and placed in RO H2O to germinate in a Percival PGC-9/2 growth chamber (Percival Scientific, Perry, IN, USA) to ensure synchronous germination. Throughout this study, the growth chamber environment was set to 12/12 light–dark cycle (lights came on at 0700 hours and turned off at 1900 hours), 22 °C/18 °C day–night temperature cycles, 40% relative humidity, and photosynthetic photon flux density=350 μmol photons m−2 s−1. Upon the observation of root radicles, seedlings were aseptically transferred to 2 inch diameter pots filled with a mixture of sterilized potting media (N~400 p.p.m., P~90 p.p.m.) and microbial inoculate. To generate our sterilized media, Redi-Earth Potting Mix (Sungro Horticulture, Agawam, MA, USA) was autoclaved twice for 60 min. Next, 2 ml of microbial inoculate was added to each pot. The microbial inoculate was created by mixing 360 ml of RO H2O with 40 g of soil from the Catsburg region in Durham, North Carolina, USA (36.062294°N, –78.849644°W) and filtered through 1000 μm, 212 μm, 45 μm sieves, to remove soil nematodes that might negatively impact plant performance (van de Voorde et al., 2012). The Catsburg region has a well-documented history of A. thaliana occurrence, which has been naturalized in this region (Mauricio, 1998). Soil from the Catsburg region and our sterilized potting mix was characterized at the Colorado State Soil-Water-Plant Testing Lab (Fort Collins, CO, USA); of greatest relevance to microbial growth, the Catsburg and potting soils had similar pH values (5.4 vs 5.3, respectively). Following germination, seedlings were thinned to one plant per pot, and pots were watered at 0700 hours daily.
Experimental design
Experiment 1: temporal changes in rhizosphere community structure
To determine if rhizosphere bacterial communities are diurnally dynamic, replicates of wild-type Ws plants were grown for 4 weeks as described above. Starting at 0600 hours on 21 July and ending at 0600 hours on 22 July, 10 replicates were selected at random and harvested every 6 h for rhizosphere soil by separating the roots from the rosette (N=50), removing closely adhering soil particles from the roots as described in Bulgarelli et al. (2012), and storing the samples at –80 °C.
Experiment 2: candidate drivers of rhizosphere community structure
To characterize the effects of circadian period misfunction on rhizosphere bacterial community structure, 10 replicates of Ws, toc1-21 and ztl-30 genotypes were planted in a fully randomized design and grown for 4 weeks. Rhizosphere samples were collected as described above at 1800 hours on 21 July and stored at –80 °C. All samples were collected before visible signs of bolting, or the transition from a vegetative to a reproductive state, to avoid confounding effects of plant developmental stage (Lundberg et al., 2012; Chaparro et al., 2014). At the end of this experiment, we collected additional rhizosphere soil from four replicates of each of the three genotypes to generate the inoculum for Experiment 3.
Experiment 3: rhizosphere community feedbacks on plant performance
To test if rhizosphere microbiomes assembled by the three plant genotypes had differential effects on plant performance, we synchronously germinated seeds of the Ws genotype and planted these seeds in sterilized soil media inoculated with soil slurry generated by the Ws, toc1-21, or ztl-30 genotypes and collected at the end of Experiment 2 (N=60; 20 replicates × 3 inoculates). To determine the effects of the rhizosphere microbiome treatment on plant performance, rosette diameter was measured weekly for 3 weeks. In a second experiment, we allowed seeds to germinate naturally in sterilized soil media inoculated with the same soil slurries (N=60; 20 replicates x 3 inoculates). For this experiment, seeds were checked daily for germination as estimated from the first observation of cotyledons.
DNA extraction and amplicon sequencing
To extract microbial DNA, rhizosphere samples were placed into 15 ml Nunc Conical Centrifuge Tubes (Thermo Scientific, Waltham, MA, USA) containing 3 ml of phosphate-buffered saline, and then agitated for 15 min to separate soil particles from plant roots as described in Bulgarelli et al. (2012). Plant roots were then removed with sterilized forceps and the samples were centrifuged for 15 min at 3000 rcf. The supernatant was discarded, and 0.25 g of the pellet was put into bead tubes from the Mobio Power Soil DNA Isolation Kit (Mobio Laboratories, Carlsbad, CA, USA) using sterilized disposable spatulas. DNA was extracted from each sample following the manufacturer’s instructions. With each round of extractions, a soilless blank was included as a negative control. At the end of each round, PCR was performed to ensure sufficient DNA yields and reagent sterility.
DNA extracts were sent to the Marine Biological Laboratories (Woods Hole, MA, USA) for amplicon library preparation of the V4V5 region of the 16S ribosomal RNA gene using the 518 F and 926 R primers (Huse et al., 2014). Sequencing was performed on the Illumina MiSeq platform (Illumina, San Diego, CA, USA) as described in Nelson et al. (2014). Sequence reads were demultiplexed and quality filtered (Phred score⩾20, chimera removal by ChimeraSlayer) using QIIME 1.9.1, uclust was used to perform open reference operational taxonomic unit (OTU) picking at 97% sequence similarity using the Greengenes database (ver. 13.8), and all singletons were removed to avoid the possibility that a sequencing error was called as an OTU (Caporaso et al., 2010; Edgar, 2010; Haas et al., 2011; Bokulich et al., 2012; McDonald et al., 2012). We rarefied to 100 000 reads per sample to ensure common sampling effort. All sequences have been deposited into the Short Read Archive (SRA) under PRJNA391346.
Sequencing data analyses
To describe rhizosphere community structure, we generated Jaccard (presence–absence analysis) and Bray–Curtis (abundance analysis) dissimilarity matrices, and Shannon diversity estimates in QIIME (Caporaso et al., 2010). For Experiment 1, we used adonis to determine if rhizosphere community structure differed between day (1800 hours) and night (0600 hours) time points. To test if shifts in community structure were consistent between day and night time points, we used Pearson’s correlation coefficients to compare the percent change in OTU abundance between 0600 and 1800 hours on 21 July and the percent change in OTU abundance between 1800 hours on 21 July and 0600 hours on 22 July. We included only OTUs with >100 reads per sample to avoid potentially confounding effects of low-abundance taxa. For Experiments 2 and 3, we used one-way analysis of variance and Tukey’s honest significant differences post hoc comparison test using the car and agricolae R packages to characterize differences in principal coordinates between genotype along axis 1 and to determine differences in plant performance between soil treatments (Fox and Weisberg, 2011; de Mendiburu, 2016). Moreover, OTUs were split into common (>500 reads) or rare (<500 reads) categories, and presence–absence analyses and abundance analyses were performed again on the split data sets to determine if effects of clock genotype were detected using common or rare microbial taxa alone. Finally, sequence data were reanalyzed without rarefaction using the R package Phyloseq, to determine if results were consistent in the absence of rarefaction (McMurdie et al., 2014). Results were similar regardless of rarefaction, that is, the effect of host plant genotype was significant for both binary Jaccard (P=0.001) and Bray–Curtis dissimilarity (P=0.001) analyses with and without rarefaction; here, we present the results of analyses based on rarefaction. All plots were generated using the R package ggplot2 (Wickham, 2009).
To identify OTUs that explain observed differences in plant performance arising from soil overstory history in Experiment 3, we used the indicspecies package for indicator value analysis in R 3.0.3 and LefSe on the galaxy web platform (Dufrêne and Legendre, 1997; De Cáceres and Legendre, 2009; Segata et al., 2011; R Core Team, 2013). Indicator value analysis (IVA) has been used commonly in ecological studies to ascertain species that underlie treatment or site differences (Dufrêne and Legendre, 1997), and is used here to test which OTU(s) is(are) specific to a given level of a factor (for example, present/abundant in the rhizospheres of Ws replicates and absent from toc1-21 and ztl-30 rhizospheres). Notably, the calculation of IVA weights presence–absence and abundance, and as such may be sensitive to rare taxa. LefSe performs linear discriminant analysis on sequence data to identify marker taxa that underlie treatment differences, and is weighted preferentially by abundance differences of more common taxa. As rare OTUs contributed to microbiome differences between host plant genotypes, we used both IVA and Lefse. Finally, we coarsely estimated microbial community size by dividing the quantity of extracted DNA using a Qubit (ThermoFisher Scientific, Waltam, MA, USA) by the mass of soil used for each extraction to determine if microbial community size influenced plant performance in Experiment 3.
Results
Sequencing results
For Experiment 1, after quality filtering, chimera removal, OTU picking, outlier sample filtering and rarefaction to 100 000 reads per sample (Supplementary Figure 1a), there was a total of 3 700 000 high-quality reads out of 10 250 881 raw reads. For Experiment 2, after similar processing, but rarefaction to 116 000 reads per sample (Supplementary Figure 1b) there was a total 2 668 000 high-quality reads out of 6 487 790 raw reads. The number of reads after each processing step can be found in Supplementary Tables 1 and 2.
Experiment 1: temporal changes in rhizosphere community structure
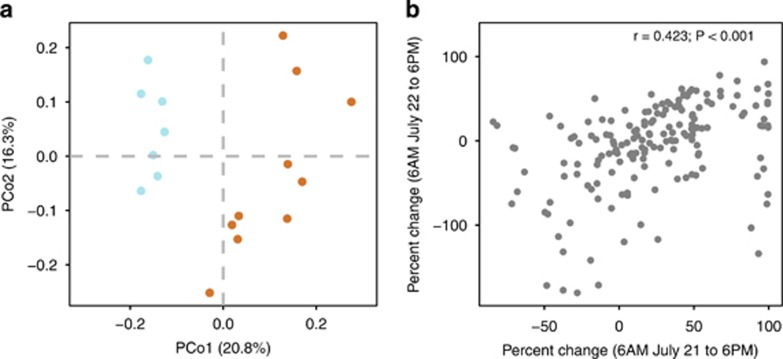
We observed significant differences in rhizosphere communities between day and night time points (P = 0.001; Figure 1a; Supplementary Table 3). The percent change in OTU abundance between 0600 hours on 21 July and 1800 hours was positively correlated with the percent change in OTU abundance between the 06 00 hours on 22 July and 1800 hours time points (r=0.423, P<0.001). This relationship suggests that the abundance of many common OTUs shift in a similar manner between day and night time points (Figure 1b).
Figure 1.
Day vs night timepoint influences rhizosphere community structure. (a) Principal coordinate analysis of Bray–Curtis dissimilarities (n=17). Differences between the 1800 hours (day, orange) and 06 00 hours (night, blue) time points were significant at P=0.001. (b) Bivariate relationship between percent change in OTU abundance from 0600 to 1800 hours 22 July and percent change in OTU abundance from 0600 hours on 21 July to 1800 hours (r=0.423; P<0.001).
Experiment 2: candidate drivers of rhizosphere community structure
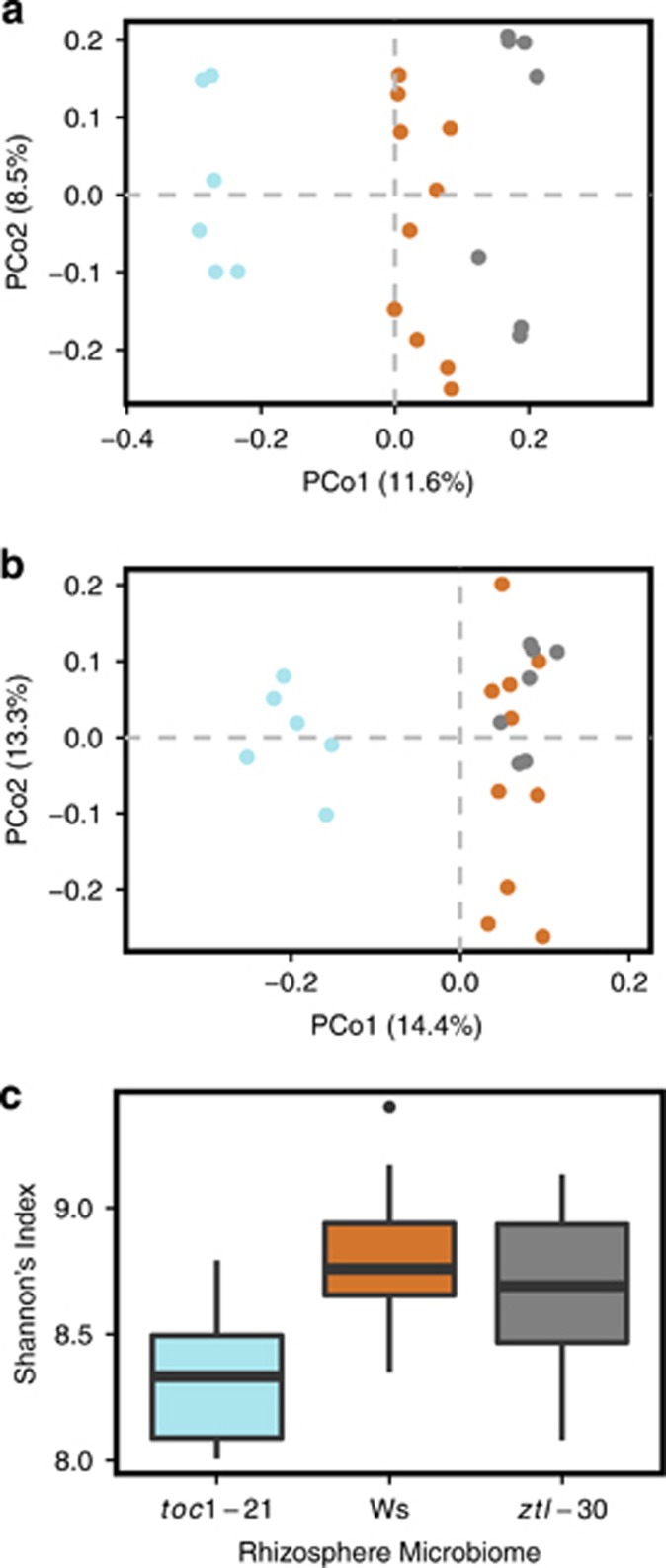
Rhizosphere community composition (presence vs absence of taxa), abundance, and diversity differed among genotypes (Figure 2). From Jaccard presence–absence analysis, Ws, toc1-21, and ztl-30 rhizosphere communities were significantly different from one another (P<0.001; Figure 2a; Supplementary Table 4). PCo1 describes the effect of progressive clock changes between short (lowest PCo values) vs wild-type (intermediate values) vs long (highest values) endogenous period lengths of the plants on rhizosphere community composition. Bray–Curtis relative abundance analysis revealed differences between the rhizosphere communities of the three clock genotypes (Supplementary Table 5), where toc1-21 communities were different from both Ws (P=0.02) and ztl-30 (P=0.04) communities, whereas Ws and ztl-30 communities were not significantly different from one another (P=0.39; Figure 2b). This result suggests that a period length shorter than 24 h specifically alters abundances of OTUs within the rhizosphere community. Similarly, the toc1-21 rhizosphere communities showed significantly reduced richness and evenness based on Shannon’s diversity index in comparison with Ws (P=0.03; Figure 2c).
Figure 2.
Clock function in A. thaliana alters rhizosphere community composition, abundances and diversity. (a) Principal coordinate analysis of Jaccard dissimilarities, where rhizosphere communities of Ws are represented by orange circles, toc1-21: blue circles, and ztl-30: gray circles (n=23). Rhizosphere community composition differs significantly between clock genotypes (P=0.001). (b) Principal coordinate analysis of Bray–Curtis dissimilarities (n=23). OTU abundances differ significantly between toc1-21 and both the Ws and ztl-30 genotypes (P=0.001). (c) Mean Shannon diversity index. The top and bottom of boxes represent the 75th and 25th percentiles, respectively. Whiskers represent 1.5 times the interquartile range. One-way analysis of variance and Tukey’s post hoc comparisons indicate significant differences between toc1-21 and both the Ws and ztl-30 genotypes (P=0.03).
To clarify the contributions of rare vs common OTUs to host plant genotype differences, we analyzed the data when culled to different minimum read numbers (Supplementary Table 6). When culling to a minimum read number > 500 for an OTU (or approximately 1% of the community), Jaccard and Bray–Curtis dissimilarities were significant, indicating that common taxa contribute at least partially to observed differences among the three host plant genotypes in the presence–absence of taxa (Figure 3a) and to differences in OTU abundance between toc1-21 and both Ws and ztl-30 (Figure 3b). Communities culled to OTUs with < 500 reads showed significant differences in both composition and abundance, indicating that rare microbial taxa respond to plant genotype (Figures 3c and d). In particular, when data for rare OTUs are analyzed, the distinction between Ws vs ztl-30 becomes significant (P=0.001) (cf Figure 2b vs 3d).
Figure 3.
Plant genotype influences both common (> 500 reads) and rare (< 500 reads) rhizosphere taxa. (a) Principal coordinate analysis of Jaccard dissimilarities of common taxa, where rhizosphere communities of Ws are represented by orange circles, toc1-21: blue circles and ztl-30: gray circles (n=23). Rhizosphere community composition differs significantly between all clock genotypes (P=0.001). (b) Principal coordinate analysis of Bray–Curtis dissimilarities of common taxa (n=23). OTU abundances differ significantly between toc1-21 and both the Ws and ztl-30 genotypes (P=0.001). (c) Principal coordinate analysis of Jaccard dissimilarities of rare taxa (n=23). Rhizosphere community composition differs significantly between all clock genotypes (P=0.001). (d) Principal coordinate analysis of Bray–Curtis dissimilarities of rare taxa (n=23). OTU abundances differ significantly between Ws and both toc1-21 and ztl-30 genotypes (P=0.001).
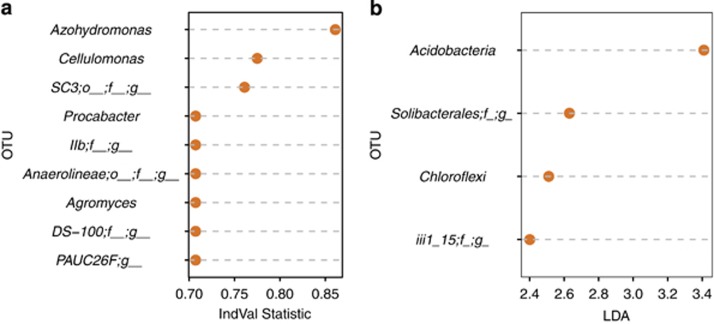
Combined, the IVA and Lefse analyses identified a total of 13 indicator OTUs associated with the Ws rhizosphere (Figure 4), 12 indicator OTUs associated with the toc1-21 rhizosphere (Supplementary Tables 7 and 9), and 12 indicator OTUs associated with the ztl-30 rhizosphere (Supplementary Table 8 and 9). As IVA is more sensitive to rare taxa, the two methods select somewhat different OTUs as biomarkers of host plant genotype. Notably, however, there is significant taxonomic overlap between the OTUs identified by IVA and Lefse. That is, taxa identified by IVA are phylogenetically related to those identified by Lefse, or vice versa. For instance, of the 13 indicator OTUs associated with the Ws rhizosphere, six taxa were members of the phylum Acidobacteria (IVA: DS-100;o_;f_;g_, llb;f_;g_, PAUC26;f_;g_; Lefse: Acidobacteria, Solibacterales;f_;g_, iii1_15;f_;g_) and two taxa were members of the Chloroflexi (indicator species analysis: Anaerolineae;o_;f_;g_, Lefse: Chloroflexi). From previous studies, some members of both phyla (Acidobacteria and Chloroflexi) and genera (Agromyces and Cellulomonas), have been described as growth promoting (Egamberdiyeva and Höflich, 2002; Kuffner et al., 2008; Chen et al., 2014; Kielak et al., 2016). Finally, community size as estimated from DNA per unit soil mass (P=0.11) and soil pH (P=0.53) did not significantly differ across clock genotypes (Supplementary Tables 10 and 11).
Figure 4.
Indicator taxa based on (a) IVA and (b) Lefse.
Experiment 3: rhizosphere community feedbacks on plant performance
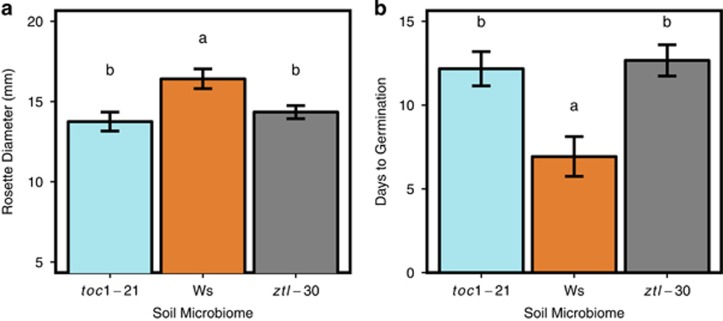
Soil overstory history had a significant influence on early plant performance (Figure 5). Wild-type plants grown in a soil with a history of Ws plants had significantly larger rosette diameters than plants grown in soils with a history of toc1-21 and ztl-30 after 1 week (19.4% and 14.4%, respectively; P=0.002) and 2 weeks of growth (10.8% and 8.3%, respectively; P=0.04). However, at the end of 3 weeks of growth, Ws plants grown in soils conditioned by each of the clock genotypes were only marginally different in size (P=0.11). In a germination experiment of similar design (in which seedlings were not transplanted but instead germinated directly on soil), Ws seeds in pots with Ws inoculum germinated an average of 5.2 days earlier than seeds planted into pots with toc1-21 (P=0.002) inoculum and 5.7 days earlier than those planted into ztl-30 (P=0.024) inoculated pots (P<0.001; Figure 5b).
Figure 5.
Effects of soil overstory history on plant growth. Letters denote significant differences between soil treatments based on Tukey’s Honest Significant Differences post hoc comparisons. (a) At week 1 (n=60), plants grown in a soil with a history of Ws had significantly larger rosette diameters than plants grown in soils with a history of toc1-21 or ztl-30 (P=0.002). (b) In a separate experiment where seeds were not germinated synchronously (n=35), seeds sowed onto a soil with a history of Ws occurrence germinated significantly earlier than seeds sown into soils with a history of toc1-21 or ztl-30 genotypes (P<0.001).
Discussion
The rhizosphere microbiome has been referred to as the ‘second genome’ of plants or the extended phenome (Berendsen et al., 2012). In part, these names reflect the role of the rhizosphere microbiome in determining plant performance. Empirical studies suggest complex feedbacks between plants and microbes, under which plant species may modulate rhizosphere community structure via carbon exudation and under which microbes may alter plant phenotypes directly or via ecosystem services such as nutrient accessibility (Bulgarelli et al., 2013). The mechanisms by which different plant genotypes may influence rhizosphere community structure remain largely unclear, as are the effects on plant performance of rhizosphere microbiomes selected by the plant host genotype (Heath and Tiffin, 2007; Panke-Buisse et al., 2014; Lebeis et al., 2015). Understanding plant–rhizosphere microbiome interactions is agroecologically relevant because rhizosphere communities can strongly influence plant fitness and biomass, which can in turn inform evolutionary studies of adaptation, conservation, and agronomic practices (Pérez-Jaramillo et al., 2016). In this study, we tested the role of the plant circadian clock as a mediator of plant–rhizosphere microbiome interactions. We hypothesized that (1) rhizosphere community structure may be temporally dynamic, if rhizosphere taxa respond to diurnally patterned fluxes of carbon, water, or nutrient availability into the rhizosphere (or other diurnally patterned plant phenotypes). (2) We further hypothesized that clock misfunction would have a role in shaping community structure, because differences in plant physiology attributable to genotype would lead to differences in rhizosphere community structure. (3) Finally, we hypothesized that differences in rhizosphere community structure attributable to plant genotype could lead to differences in community function with regards to plant performance.
The composition of plant-associated microbiomes is known to shift on long time scales, such as across seasons or across developmental stages of the plant host (Lundberg et al., 2012; Chaparro et al., 2014; Wagner et al., 2016). The short duration of many microbial life cycles means that microbial community composition may also respond to more rapid changes in the environment. Yet, it remains unclear if the community composition of microbes found in association with plants changes on short timeframes, such as across day-night transitions. We observed diurnally patterned shifts in rhizosphere community structure. That is, we observed consistent shifts in rhizosphere communities between day and night time points (Figure 1). Difference in community structure observed between the two time points may reflect the effects of day vs night conditions and carbon, water or nutrient availability in the rhizosphere. Several prior studies have shown that the concentration of certain exudates varies over the course of day (Watt and Evans, 1999; Badri and Vivanco, 2009). For instance, Iijima et al. (2003) observed higher rates of mucilage exudation at night, whereas other studies have observed higher prevalence of flavonoids and catechin during day conditions (Hughes et al., 1999; Iijima et al., 2003; Tharayil and Triebwasser, 2010). Further, rhizosphere water is depleted diurnally, depending on root and soil hydraulics (Sperry et al., 1998), and the transpiration stream increases nutrient flow (Matimati et al., 2014), potentially depleting soil nutrients in the rhizosphere zone. Therefore, rhizosphere taxa and populations may vary in abundance depending on soil resource availability, leading to our observed differences in community structure between day and night time points. Future experiments should be designed to tease apart the relative influence of root exudates, water dynamics, and nutrient uptake within the rhizosphere on microbes.
Specific plant genes have important roles in shaping rhizosphere communities (Bravo et al., 2016), and here we observed that circadian clock genes significantly influence rhizosphere community structure. In the current study, plant genotype explained 19.1% of the variation in community composition (presence vs absence of taxa), 21.7% of the variation in community relative abundances, and brought about differences in community diversity between short (toc1-21) vs longer (Ws, ztl-30) period genotypes (Figure 2). These differences in communities explained by clock genotype surpass variation explained by genotype in previous studies of the influence of plant genotype on rhizosphere community structure (Lundberg et al., 2012; Bulgarelli et al., 2012; Peiffer et al., 2013; Lebeis et al., 2015). Our results thus suggest that circadian clock misfunction has a strong influence on rhizosphere community structure. The large percent variance explained may arise from the pervasive transcriptomic and phenotypic effects of clock misfunction on the plant host, and potentially the microbial inoculant used here is one that amplifies the effect of host genotype, as demonstrated in other studies (Weinert et al., 2011; Peiffer et al., 2013).
TIMING OF CAB EXPRESSION 1 had a particularly pronounced impact on rhizosphere community structure, because strong mutant alleles in this gene led to changes in community composition, abundance and diversity (Figure 2). On the other hand, disruption of ZEITLUPE had less of an influence on rhizosphere community structure, with its effect limited to differences in OTU presence–absence relative to Ws (Figure 2). One possible explanation for the asymmetric effects of clock misfunction is that long period lengths theoretically enable better phase adjustment to dawn, such that period lengths shorter than 24 h may have more detrimental fitness consequences in nature (or in this case lead to greater deviations in rhizosphere microbial community structure) in comparison with period lengths greater than 24 h (Johnson and Kondo, 1992; McClung, 2006; Kevei et al., 2006; Hotta et al., 2007). Regardless of the exact mechanisms, clock misfunction and the mismatch between endogenous plant cycles and exogenous cycles affected aspects of microbial community structure.
As in any ecosystem, there is a link between community structure and function (Tilman et al., 1997). Several studies have illustrated this relationship in plant–rhizosphere microbiome interactions, where differences in plant performance can be attributed to differences in rhizosphere community structure (Mendes et al., 2011, 2013; Zolla et al., 2013; Wagner et al., 2014). Here, differences in rhizosphere communities (arising from mutations in host clock genes) affected performance of wild-type Ws plants grown in soils with differing overstory histories (Figure 5). Ws plants performed best when exposed to an inoculum from soils in which wild-type rather than clock mutant genotypes had been grown. We observed differences in the timing of germination, where Ws seeds sown in soils with a history of Ws occurrence germinated earlier. In comparison with untreated soil, autoclaved soil, such as that used here, differs in both chemical and physical properties and reflects a novel and possibly more stressful environment for plants (Trevors, 1996; Brulé et al., 2001; Lau and Lennon, 2011); differences in germination observed here may therefore reflect that the microbes from Ws-conditioned soil enable normal germination under the novel autoclaved soil conditions (rather than an acceleration of germination timing under natural conditions per se) (Lau and Lennon, 2011; Mahmood et al., 2014). Beyond germination timing, we observed that wild-type plants were also larger when grown in soils with a history of Ws rather than mutant genotype growth. These findings from two experiments in which soils were independently conditioned by Ws vs mutant genotypes suggest first that plants can select explicitly beneficial soil communities that improve initial offspring performance, and second that disruption of these communities by mutations in clock genes adversely affects initial offspring phenology and growth.
Although additional research is required to ascertain causality, community composition patterns and indicator analyses provide hypotheses as to which OTUs may lead to these differences in performance (De Cáceres and Legendre, 2009; DeAngelis et al., 2015). Here, we identified 13 indicator OTUs associated with Ws rhizosphere. Parallel to the differences in microbial community structure among host plant genotypes (Figures 2 and 3), one possibility is that rare OTUs underlie differences in plant performance observed between the Ws and ztl-30 rhizosphere microbiomes, whereas rare and common OTUs could contribute to plant performance differences observed between the Ws and toc1-21 microbiomes. Rare OTUs could affect plant performance via so-called indirect effects, such as facilitation of or competition with explicitly plant growth-promoting microbes, whereas common microbes could promote plant growth through indirect or direct interactions (Saleem et al., 2016). Specifically in regard to the indicator species analyses (IVA and Lefse), the presence and abundance of Acidobacteria (Kielak et al., 2016), Chloroflexi (Chen et al., 2014), Cellulomonas (Egamberdiyeva and Höflich, 2002) and Agromyces (Kuffner et al., 2008) in the Ws rhizosphere may explain the differences in plant size between rhizosphere treatments, as these OTUs have been previously associated with plant growth promotion.
In sum, we have shown that the plant circadian clock shapes rhizosphere community structure, particularly the presence of rare taxa. Further, this plant genetic driver of community assembly also influences community function, as estimated from plant performance. As community structure may shift in response to day and night conditions, future characterizations of the rhizosphere should account for differences in community structure attributable the timing of rhizosphere collection. Finally, additional research is needed into the role of host plant physiological loci in shaping rhizosphere communities.
Acknowledgments
This work was supported by the National Science Foundation grants IOS-1444571 to CW, BEE, LM and OIA-1655726 to CW and BEE, and a Wyoming INBRE sequencing and bioinformatics award to CH and CW. We thank the Powells’ for allowing us to collect soil from their property and Lindsay Leverett for collecting soil from the Catsburg site.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Badri DV, Vivanco JM. (2009). Regulation and function of root exudates. Plant Cell Environ 32: 666–681. [DOI] [PubMed] [Google Scholar]
- Bais HP, Weir TL, Perry LG, Gilroy S, Vivanco JM. (2006). The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57: 233–266. [DOI] [PubMed] [Google Scholar]
- Bashan Y. (1998). Inoculants of plant growth-promoting bacteria for use in agriculture. Biotechnol Adv 16: 729–770. [Google Scholar]
- Berendsen RL, Pieterse CMJ, Bakker PAHM. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci 17: 478–486. [DOI] [PubMed] [Google Scholar]
- Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R et al. (2012). Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10: 57–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo A, York T, Pumplin N, Mueller LA, Harrison MJ. (2016). Genes conserved for arbuscular mycorrhizal symbiosis identified through phylogenomics. Nat Plants 2: 15208. [DOI] [PubMed] [Google Scholar]
- Brulé C, Frey-Klett P, Pierrat J, Courrier S, Gérard F, Lemoine M et al. (2001). Survival in the soil of the ectomycorrhizal fungus Laccaria bicolor and the effects of a mycorrhiza helper Pseudomonas fluorescens. Soil Biol Biochem 33: 1683–1694. [Google Scholar]
- Bulgarelli D, Rott M, Schlaeppi K, Ver Loren van Themaat E, Ahmadinejad N, Assenza F et al. (2012). Revealing structure and assembly cues for Arabidopsis root-inhabiting bacterial microbiota. Nature 488: 91–95. [DOI] [PubMed] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S, van Themaat EVL, Schulze-Lefert P. (2013). Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 64: 807–838. [DOI] [PubMed] [Google Scholar]
- De Cáceres M, Legendre P. (2009). Associations between species and groups of sites: indices and statistical inference. Ecology 90: 3566–3574. [DOI] [PubMed] [Google Scholar]
- Çakmakçı R, Kantar F, Sahin F. (2001). Effect of N2-fixing bacterial inoculations on yield of sugar beet and barley. J Plant Nutr Soil Sci 164: 527. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7: 335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaparro JM, Badri DV, Vivanco JM. (2014). Rhizosphere microbiome assemblage is affected by plant development. ISME J 8: 790–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Condron L, Davis M, Sherlock R. (2002). Phosphorus dynamics in the rhizosphere of perennial ryegrass (Lolium perenne L.) and radiata pine (Pinus radiata D. Don.). Soil Biol Biochem 34: 487–499. [Google Scholar]
- Chen M, Li X, Yang Q, Chi X, Pan L, Chen N et al. (2014). Dynamic succession of soil bacterial community during continuous cropping of peanut (Arachis hypogaea L.). PLoS One 9: e101355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covington MF, Maloof JN, Straume M, Kay SA, Harmer SL. (2008). Global transcriptome analysis reveals circadian regulation of key pathways in plant growth and development. Genome Biol 9: R130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis KM, Pold G, Topcuoglu BD, van Diepen LTA, Varney RM, Blanchard JL et al. (2015). Long-term forest soil warming alters microbial communities in temperate forest soils. Front Microbiol 6: 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dennis PG, Miller AJ, Hirsch PR. (2010). Are root exudates more important than other sources of rhizodeposits in structuring rhizosphere bacterial communities? FEMS Microbiol Ecol 72: 313–327. [DOI] [PubMed] [Google Scholar]
- Dodd AN, Parkinson K, Webb AAR. (2004). Independent circadian regulation of assimilation and stomatal conductance in the ztl-1 mutant of Arabidopsis. New Phytol 162: 63–70. [Google Scholar]
- Dodd AN, Salathia N, Hall A, Kévei E, Tóth R, Nagy F et al. (2005). Plant circadian clocks increase photosynthesis, growth, survival, and competitive advantage. Science (80-) 309: 630–633. [DOI] [PubMed] [Google Scholar]
- Dufrêne M, Legendre P. (1997). Species assemblages and indicator species: the need for a flexible asymmetrical approach. Ecol Monogr 67: 345–366. [Google Scholar]
- Edgar RC. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26: 2460–2461. [DOI] [PubMed] [Google Scholar]
- Egamberdiyeva D, Höflich G. (2002). Root colonization and growth promotion of winter wheat and pea by Cellulomonas spp. at different temperatures. Plant Growth Regul 38: 219–224. [Google Scholar]
- Fox J, Weisberg S. (2011). An R companion to applied regression. Sage Publications: Thousand Oaks, CA, USA.
- Fujiwara S, Wang L, Han L, Suh S-S, Salome PA, McClung CR et al. (2008). Post-translational regulation of the Arabidopsis circadian clock through selective proteolysis and phosphorylation of pseudo-response regulator proteins. J Biol Chem 283: 23073–23083. [DOI] [PubMed] [Google Scholar]
- Greenham K, McClung CR. (2015). Integrating circadian dynamics with physiological processes in plants. Nat Rev Genet 16: 598–610. [DOI] [PubMed] [Google Scholar]
- Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G et al. (2011). Chimeric 16 S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21: 494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harmer SL. (2009). The circadian system in higher plants. Annu Rev Plant Biol 60: 357–377. [DOI] [PubMed] [Google Scholar]
- Harmer SL, Hogenesch JB, Straume M, Chang HS, Han B, Zhu T et al. (2000). Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 290: 2110–2113. [DOI] [PubMed] [Google Scholar]
- Heath KD, Tiffin P. (2007). Context dependence in the coevolution of plant and rhizobial mutualists. Proc Biol Sci 274: 1905–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henning JA, Weston DJ, Pelletier DA, Timm CM, Jawdy SS, Classen AT. (2016). Root bacterial endophytes alter plant phenotype, but not physiology. PeerJ 4: e2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotta CT, Gardner MJ, Hubbard KE, Baek SJ, Dalchau N, Suhita D et al. (2007). Modulation of environmental responses of plants by circadian clocks. Plant Cell Environ 30: 333–349. [DOI] [PubMed] [Google Scholar]
- Hughes M, Donnelly C, Crozier A, Wheeler CT. (1999). Effects of the exposure of roots of Alnus glutinosa to light on flavonoids and nodulation. Can J Bot 77: 1311–1315. [Google Scholar]
- Huse SM, Young VB, Morrison HG, Antonopoulos DA, Kwon J, Dalal S et al. (2014). Comparison of brush and biopsy sampling methods of the ileal pouch for assessment of mucosa-associated microbiota of human subjects. Microbiome 2: 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iijima M, Sako Y, Rao TP. (2003). A new approach for the quantification of root-cap mucilage exudation in the soil. Plant Soil 255: 399–407. [Google Scholar]
- Johnson CH, Kondo T. (1992). Light pulses induce ‘singular’ behavior and shorten the period of the circadian phototaxis rhythm in the CW15 strain of chlamydomonas. J Biol Rhythms 7: 313–327. [DOI] [PubMed] [Google Scholar]
- Jones DL, Nguyen C, Finlay RD. (2009). Carbon flow in the rhizosphere: carbon trading at the soil–root interface. Plant Soil 321: 5–33. [Google Scholar]
- Kevei E, Gyula P, Hall A, Kozma-Bognár L, Kim W-Y, Eriksson ME et al. (2006). Forward genetic analysis of the circadian clock separates the multiple functions of ZEITLUPE. Plant Physiol 140: 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kielak AM, Cipriano MAP, Kuramae EE. (2016). Acidobacteria strains from subdivision 1 act as plant growth-promoting bacteria. Arch Microbiol 198: 987–993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W-Y, Hicks KA, Somers DE. (2005). Independent roles for EARLY FLOWERING 3 and ZEITLUPE in the control of circadian timing, hypocotyl length, and flowering time. Plant Physiol 139: 1557–1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuffner M, Puschenreiter M, Wieshammer G, Gorfer M, Sessitsch A. (2008). Rhizosphere bacteria affect growth and metal uptake of heavy metal accumulating willows. Plant Soil 304: 35–44. [Google Scholar]
- Lau JA, Lennon JT. (2011). Evolutionary ecology of plant-microbe interactions: soil microbial structure alters selection on plant traits. New Phytol 192: 215–224. [DOI] [PubMed] [Google Scholar]
- Lebeis SL, Paredes SH, Lundberg DS, Breakfield N, Gehring J, McDonald M et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science (80-) 349: 860–864. [DOI] [PubMed] [Google Scholar]
- Lugtenberg B, Kamilova F. (2009). Plant-growth-promoting rhizobacteria. Annu Rev Microbiol 63: 541–556. [DOI] [PubMed] [Google Scholar]
- Lundberg DS, Lebeis SL, Paredes SH, Yourstone S, Gehring J, Malfatti S et al. (2012). Defining the core Arabidopsis thaliana root microbiome. Nature 488: 86–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahmood T, Mehnaz S, Fleischmann F, Ali R, Hashmi ZH, Iqbal Z. (2014). Soil sterilization effects on root growth and formation of rhizosheaths in wheat seedlings. Pedobiologia (Jena) 57: 123–130. [Google Scholar]
- Matimati I, Anthony Verboom G, Cramer MD. (2014). Do hydraulic redistribution and nocturnal transpiration facilitate nutrient acquisition in Aspalathus linearis? Oecologia 175: 1129–1142. [DOI] [PubMed] [Google Scholar]
- Mauricio R. (1998). Costs of resistance to natural enemies in field populations of the annual plant Arabidopsis thaliana. Am Nat 151: 20–28. [DOI] [PubMed] [Google Scholar]
- McClung CR. (2006). Plant circadian rhythms. Plant Cell 18: 792–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A et al. (2012). An improved green genes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J 6: 610–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, Holmes S, Hoffmann C, Bittinger K, Chen Y. (2014). Waste not, want not: why rarefying microbiome data is inadmissible McHardy AC (ed). PLoS Comput Biol 10: e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendes R, Garbeva P, Raaijmakers JM. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol Rev 37: 634–663. [DOI] [PubMed] [Google Scholar]
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JHM et al. (2011). Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science (80-), 332 1097–100. [DOI] [PubMed]
- de Mendiburu F. (2016). agricolae: Statistical Procedures for Agricultural Research https://cran.r-project.org/web/packages/agricolae/index.html.
- Millar AJ, Carré IA, Strayer CA, Chua NH, Kay SA. (1995). Circadian clock mutants in Arabidopsis identified by luciferase imaging. Science 267: 1161–1163. [DOI] [PubMed] [Google Scholar]
- de Montaigu A, Giakountis A, Rubin M, Tóth R, Cremer F, Sokolova V et al. (2015). Natural diversity in daily rhythms of gene expression contributes to phenotypic variation. Proc Natl Acad Sci USA 112: 905–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson MC, Morrison HG, Benjamino J, Grim SL, Graf J, Stahl D et al. (2014). Analysis, Optimization and Verification of Illumina-Generated 16S rRNA Gene Amplicon Surveys. PLoS One 9: e94249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panke-Buisse K, Poole AC, Goodrich JK, Ley RE, Kao-Kniffin J. (2014). Selection on soil microbiomes reveals reproducible impacts on plant function. ISME J 9: 980–989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peiffer JA, Spor A, Koren O, Jin Z, Tringe SG, Dangl JL et al. (2013). Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc Natl Acad Sci USA 110: 6548–6553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Jaramillo JE, Mendes R, Raaijmakers JM. (2016). Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol Biol 90: 635–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R Core Team. (2013) R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria. [Google Scholar]
- Richardson AE, Barea J-M, McNeill AM, Prigent-Combaret C. (2009). Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321: 305–339. [Google Scholar]
- Richardson AE, Simpson RJ. (2011). Soil microorganisms mediating phosphorus availability update on microbial phosphorus. Plant Physiol 156: 989–996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saleem M, Law AD, Moe LA. (2016). Nicotiana roots recruit rare rhizosphere taxa as major root-inhabiting microbes. Microb Ecol 71: 469–472. [DOI] [PubMed] [Google Scholar]
- Salmela MJ, Greenham K, Lou P, McClung CR, Ewers BE, Weinig C. (2016). Variation in circadian rhythms is maintained among and within populations in Boechera stricta. Plant Cell Environ 39: 1293–1303. [DOI] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L, Gevers D, Miropolsky L, Garrett WS et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol 12: R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sperry JS, Adler FR, Campbell GS, Comstock JP. (1998). Limitation of plant water use by rhizosphere and xylem conductance: results from a model. Plant, Cell Environ 21: 347–359. [Google Scholar]
- Talbot JM, Bruns TD, Taylor JW, Smith DP, Branco S, Glassman SI et al. (2014). Endemism and functional convergence across the North American soil mycobiome. Proc Natl Acad Sci USA 111: 6341–6346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tharayil N, Triebwasser DJ. (2010). Elucidation of a diurnal pattern of catechin exudation by centaurea stoebe. J Chem Ecol 36: 200–204. [DOI] [PubMed] [Google Scholar]
- Thornton B, Paterson E, Midwood AJ, Sim A, Pratt SM. (2004). Contribution of current carbon assimilation in supplying root exudates of Lolium perenne measured using steady-state 13C labelling. Physiol Plant 120: 434–441. [DOI] [PubMed] [Google Scholar]
- Tilman D, Knops J, Wedin D, Reich P, Ritchie M, Siemann E. (1997). The influence of functional diversity and composition on ecosystem processes. Science (80-) 277: 1300–1302. [Google Scholar]
- Trevors JT. (1996). Sterilization and inhibition of microbial activity in soil. J Microbiol Methods 26: 53–59. [Google Scholar]
- van de Voorde TFJ, van der Putten WH, Bezemer TM. (2012). Soil inoculation method determines the strength of plant–soil interactions. Soil Biol Biochem 55: 1–6. [Google Scholar]
- Wagner MR, Lundberg DS, Coleman-Derr D, Tringe SG, Dangl JL, Mitchell-Olds T. (2014). Natural soil microbes alter flowering phenology and the intensity of selection on flowering time in a wild Arabidopsis relative. Ecol Lett 17: 717–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner MR, Lundberg DS, del Rio TG, Tringe SG, Dangl JL, Mitchell-Olds T et al. (2016). Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat Commun 7: 12151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watt M, Evans JR. (1999). Linking development and determinacy with organic acid efflux from proteoid roots of white lupin grown with low phosphorus and ambient or elevated atmospheric CO2 concentration. Plant Physiol 120: 705–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert N, Piceno Y, Ding G-C, Meincke R, Heuer H, Berg G et al. (2011). PhyloChip hybridization uncovered an enormous bacterial diversity in the rhizosphere of different potato cultivars: many common and few cultivar-dependent taxa. FEMS Microbiol Ecol 75: 497–506. [DOI] [PubMed] [Google Scholar]
- Wickham H. (2009) ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag New York: New York. [Google Scholar]
- Yerushalmi S, Green RM. (2009). Evidence for the adaptive significance of circadian rhythms. Ecol Lett 12: 970–981. [DOI] [PubMed] [Google Scholar]
- Zolla G, Badri DV, Bakker MG, Manter DK, Vivanco JM. (2013). Soil microbiomes vary in their ability to confer drought tolerance to ArabidopsisAppl Soil Ecol 68: 1–9.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.