Abstract
High representation by ammonia-oxidizing archaea (AOA) in marine systems is consistent with their high affinity for ammonia, efficient carbon fixation, and copper (Cu)-centric respiratory system. However, little is known about their response to nutrient stress. We therefore used global transcriptional and proteomic analyses to characterize the response of a model AOA, Nitrosopumilus maritimus SCM1, to ammonia starvation, Cu limitation and Cu excess. Most predicted protein-coding genes were transcribed in exponentially growing cells, and of ~74% detected in the proteome, ~6% were modified by N-terminal acetylation. The general response to ammonia starvation and Cu stress was downregulation of genes for energy generation and biosynthesis. Cells rapidly depleted transcripts for the A and B subunits of ammonia monooxygenase (AMO) in response to ammonia starvation, yet retained relatively high levels of transcripts for the C subunit. Thus, similar to ammonia-oxidizing bacteria, selective retention of amoC transcripts during starvation appears important for subsequent recovery, and also suggests that AMO subunit transcript ratios could be used to assess the physiological status of marine populations. Unexpectedly, cobalamin biosynthesis was upregulated in response to both ammonia starvation and Cu stress, indicating the importance of this cofactor in retaining functional integrity during times of stress.
Introduction
The discovery of ammonia-oxidizing archaea (AOA) in marine and terrestrial environments has transformed our understanding of microbial controls on the global nitrogen cycle (Stahl and de la Torre, 2012). They comprise as much as 40% of the total microbial population and contribute a major fraction of transcripts in marine metatranscriptomes (Karner et al., 2001; Hollibaugh et al., 2011; Shi et al., 2011; Baker et al., 2012; Stewart et al., 2012), suggesting they have major roles in global biogeochemical cycles. Apart from serving a critical role in marine nitrification (Martens-Habbena et al., 2009, 2015; Santoro et al., 2010; Horak et al., 2013), AOA also contribute to carbon fixation (Könneke et al., 2014), production of the greenhouse gases nitrous oxide and methane (Santoro et al., 2011; Metcalf et al., 2012), and the provision of cobalamin (vitamin B12) to cobalamin-dependent populations in oceanic systems (Heal et al., 2017).
The ocean is a dynamic environment with often low and patchy distribution of nutrients (Guasto et al., 2012). The high abundance and activity of AOA throughout the marine environment suggest their resilience to such dynamic conditions. For example, AOA possess a high affinity ammonia uptake system that presumably helps sustain ammonia oxidation and growth despite low ammonium concentrations in large areas of the ocean (Martens-Habbena et al., 2009; Horak et al., 2013; Nakagawa and Stahl, 2013). In addition to ammonium, some marine AOA can utilize urea as a source of energy and nitrogen for growth (Qin et al., 2014; Bayer et al., 2016). Despite these adaptations, it is not clear how AOA respond to sporadic pulses of reduced nitrogen species over periods of time that span hours or days, and what molecular machinery facilitates such responses.
Similarly, the distinct trace element requirements of AOA may also contribute to their ecological success. Most notably, a large fraction of proteins in AOA genomes involved in electron transfer and ammonia oxidation are copper (Cu)-dependent enzymes (Walker et al., 2010). Such a reliance on copper rather than the typical iron-dependent proteins suggests that the AOA may be more competitive relative to other microbes in the open ocean where iron is often limiting and competition for bioavailable iron is high (Amin et al., 2013). However, laboratory and field studies also suggest that AOA may be limited by Cu concentrations prevalent in many regions of the marine environment (Amin et al., 2013; Jacquot et al., 2014).
In this study, we used transcription analyses to explore the whole cellular response of Nitrosopumilus maritimus strain SCM1 to varying ammonia and Cu concentrations, and relate those patterns to its global proteome. These analyses revealed that pathways for ammonia oxidation, electron transfer, and cobalamin biosynthesis are among the most responsive to changes in ammonia and Cu availability. This demonstration of clear transcriptional responses will allow the detection of ammonia and Cu stress among natural AOA populations.
Materials and methods
All materials and methods are described in detail in Supplementary Information.
Cell growth
N. maritimus strain SCM1 was cultured in synthetic seawater medium at 30 °C in the dark as described previously (Martens-Habbena et al., 2009; Amin et al., 2013; Qin et al., 2014).
Transcriptional response of N. maritimus to different ammonia and Cu levels
RNA was extracted from 0.22 μm Sterivex-GP filters (Millipore, Billerica, MA, USA) containing N. maritimus cells harvested following each of the described ammonia and Cu treatments. Microarray and RNA-seq were used to profile transcriptomic responses of N. maritimus induced by ammonia starvation and recovery and Cu limitation and toxicity, respectively. The microarray and RNA-sequencing data have been deposited in the NCBI GEO database under accession numbers GSE103848 and GSE103699, respectively. Transcript abundance of amo genes were determined by RT-qPCR on a Roche Light Cycler machine.
Protein analysis
Filters containing exponential phase N. maritimus cells were extracted using bead-beating to lyse cells. Proteins in the lysate underwent reduction of disulfides, alkylation of free cysteine residues, and in-solution protease digestion using either trypsin or GluC. Desalted samples were analyzed by a Waters nanoACQUITY liquid chromatography (LC) coupled to a Thermo QExactive HF Orbitrap high-resolution mass spectrometer (HRMS) equipped with a nano-electrospray ionization source. Data dependent acquisition was performed on the top 10 ions and data analysis was conducted using COMET (Eng et al., 2013) and software from the trans-proteomic pipeline (Nesvizhskii et al., 2007). Proteomics data have been deposited to the ProteomeXchange Consortium via the PRDE partner repository with the dataset identifier PXD007728.
Results and discussion
Global transcriptome and proteome of N. maritimus SCM1
A combined transcriptome and proteome analysis was used to identify the global gene expression profile of N. maritimus during exponential growth. Transcripts recovered from exponentially growing N. maritimus were quantified using both microarray and RNA-sequencing (RNA-seq) platforms with reference to the genome published in Walker et al. (2010). In sum, 1794 and 1759 transcripts were detected by microarray and RNA-seq, respectively, from a total of 1795 predicted N. maritimus protein-coding genes (Supplementary Dataset 1). The global proteome of exponential phase N. maritimus cells was analyzed by in-solution proteolytic digestion followed by LC-HRMS. MS-compatible surfactants were used to improve recovery of membrane-bound proteins, and two separate protease digestions, using either trypsin or GluC, which cleave proteins at different residues, were carried out to generate more comprehensive proteomic data sets (see Supplementary Information for experimental details).
Approximately 74% of the total predicted proteins were recovered by proteomics analysis (Supplementary Dataset 1), indicating that N. maritimus translates a large fraction of its genome during growth. The high level of proteome coverage was also observed for the oceanic species ‘Candidatus Nitrosopelagicus brevis’ (~70%) (Santoro et al., 2015), confirming the notion that the genomes of free-living marine AOA are streamlined with few redundant genes. Key enzymes for post-translational addition of an acetyl group to the N-terminal methionine or penultimate (subsequent to methionine cleavage) residue of a protein (Nt-acetylation) were detected in the proteome of N. maritimus. These consist of a putative methionine aminopeptidase (Nmar_0601), a putative N-terminal acetyltransferase (Nmar_0292), and members of the GCN5-related N-acetyltransferase (GNAT) superfamily (Nmar_0309, 0358, and 0540; Supplementary Dataset 1). We observed that ~6% of the N. maritimus proteome was modified by Nt-acetylation during exponential growth (Supplementary Dataset 2), which is a somewhat lower fraction than reported for other archaeal phyla characterized on a proteome-wide scale (~8–29% Nt-acetylated) (Eichler and Maupin-Furlow, 2013; Vorontsov et al., 2016). Within this set of proteins in N. maritimus, Nt-acetylation of ribosomal proteins, the α-subunit of proteasome, and the β-subunit of prefoldin (Supplementary Dataset 2) is also typical of other characterized archaea (Eichler and Maupin-Furlow, 2013).
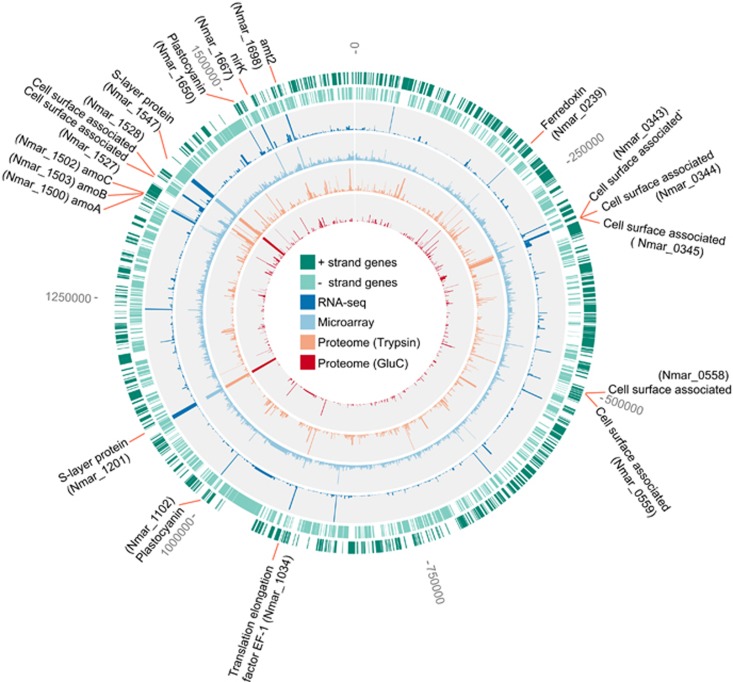
The most abundant transcripts and proteins of exponentially growing N. maritimus are mainly associated with cell surface, energy generation, carbon fixation and molecular processing (Figure 1; Supplementary Dataset 1). The global proteome showed that the two proteins with the highest spectral counts (Nmar_1547 and Nmar_1201) are predicted surface-layer (S-layer) proteins (Supplementary Figure 1), forming the prominent S-layer of N. maritimus (Urakawa et al., 2011) and many other archaea (Sara and Sleytr, 2000). Similarly, their homologs (T478_1299, T478_1300 and NVIE_016740) are among the most abundant proteins in the proteomes of N. brevis (Santoro et al., 2015) and the soil AOA Nitrososphaera viennensis (Kerou et al., 2016). A significant investment in S-layer protein biosynthesis is also mirrored by their high transcript abundance in both cultured (Figure 1) and environmental marine AOA populations (Hollibaugh et al., 2011). In addition, the high abundance of the ammonia monooxygenase (AMO) gene transcripts is consistent with the high demand on the ammonia oxidation pathway required for chemolithoautotrophic growth (Figure 1). Likewise, the B subunit of AMO (Nmar_1503) was among the top 5% most abundant proteins in the trypsin-based proteome, based on spectral counts (Supplementary Figure 1). In the trypsin digested samples, the apparent low spectral counts of the AmoA and AmoC (Nmar_1500 and Nmar_1502), which are expected to have comparable stoichiometry to AmoB, are consistent with other studies where low abundance (Santoro et al., 2015) or absence of AmoC was attributed to poor recovery of these presumptive membrane-associated proteins (Kerou et al., 2016). However, an in silico trypsin digestion of the AmoC sequence in N. maritimus and other AOA species revealed only a few tryptic cleavage sites that would generate peptides easily ionized and detected by HRMS (Supplementary Figure 2). Although surfactants were used to improve the recovery of AmoC from the membrane, trypsin digestion of AmoC in N. maritimus resulted in only two unique peptides capable of detection by HRMS and thus, likely contributed to its low apparent abundance. Additionally, one of these two unique peptides was derived from the N terminus of AmoC and was detected only if peptide identification took into account the Nt-acetylation (Supplementary Figure 3a; Supplementary Dataset 2).
Figure 1.
The global transcriptome and proteome of exponential phase N. maritimus. The first and second outermost rings (dark and light green) indicate the open reading frames (ORFs) on the positive and negative strands, respectively. The third and fourth rings (dark and light blue; histogram) are the relative abundance of transcripts mapped onto the genome based on the transcript read counts and hybridization florescence intensity, respectively. The fifth and sixth rings (orange and red; histogram) are the relative abundance of proteins based on the spectral counts detected in the trypsin and GluC based proteomes, respectively. The position and accession number of the most highly expressed genes in both protein and transcript levels are highlighted along the genome.
In contrast, a GluC digestion of N. maritimus proteins generated more AmoC peptides capable of HRMS detection (Supplementary Figures 2 and 4a), that consequently resulted in significantly higher apparent protein abundance of AmoC than in the trypsin digestion (Supplementary Figure 3b). The GluC digestion-based proteome of N. maritimus also revealed 12 proteins that were not observed in the trypsin digestion (Supplementary Dataset 1), including a plastocyanin-like protein (Nmar_1226) presumably functioning as a quinone reductase in the electron transfer pathway (Walker et al., 2010). Although the protein identifications from trypsin digestion showed greater proteome coverage compared to GluC digestion (c.f., 1313 proteins from trypsin and 480 proteins from GluC), it is clear that the exclusive use of trypsin digestion in previous (meta)proteomic analyses of AOA may have underrepresented the abundance of AmoC and other proteins (Williams et al., 2012; Hawley et al., 2014; Santoro et al., 2015; Kerou et al., 2016). These results highlight the value of using complementary proteases, such as trypsin and GluC, and assessing post-translational protein modifications for a more complete characterization of thaumarchaeotal (meta)proteomes.
Impact of changing ammonia and Cu concentration on activity and transcription
Marine AOA are known for their extremely high specific affinity for ammonia (Martens-Habbena et al., 2009). Yet, little is known about their response to periodic ammonia starvation, as frequently occurs in marine systems (Capone et al., 2008). We first evaluated the starvation and recovery behavior of N. maritimus by measuring changes in the rates of oxygen uptake following different periods of starvation and ammonia re-addition. The respiratory activity of N. maritimus as monitored by oxygen consumption immediately fell to near background levels after ammonia depletion (Supplementary Figure 4). When the period of starvation was short (<24 h), the initial activity was regained quickly after ammonia re-addition. However, when starved for significantly more than 24 h, recovery was impaired (Supplementary Figure 4). We therefore evaluated transcriptional changes at an intermediate period of starvation (24 h) and at 24 h following ammonia re-addition to starved cultures.
Replicate microarrays were used to evaluate the transcriptome of N. maritimus cells harvested at mid-exponential phase, after 24 h of ammonia starvation, and 24 h recovery. Hierarchical clustering of biological replicates of each treatment showed good reproducibility (Supplementary Figure 5a). Among the three physiological conditions examined, both the exponentially growing and recovery conditions had similar transcript profiles but were strikingly different from ammonia starvation (Supplementary Figure 5a). The transcript abundance changes in response to ammonia starvation and recovery displayed opposite global trends (Supplementary Figure 6a), suggesting N. maritimus recovers most metabolic activities in a relatively short time following ammonia availability. Relative to mid-log phase, 480 transcripts had significantly altered abundance in the ammonia-starved cells, 208 with reduced abundance and 272 with increased abundance (P<0.05) (Supplementary Dataset 3). In response to ammonia re-addition, significant changes in the abundance of 335 transcripts were observed, with 186 increased and 149 decreased relative to starved cells (Supplementary Dataset 3).
Based on earlier studies showing that free Cu concentrations control the growth of N. maritimus (Amin et al., 2013), RNA-seq was subsequently used to examine global changes in transcript abundance under Cu-limited (5 nm total Cu) and Cu-toxic (750 nm total Cu) conditions relative to Cu-replete conditions (50 nm total Cu). All Cu treatments concentrations are environmentally relevant (Moffett et al., 1997; Amin et al., 2013). Significant decreases in rates of ammonia oxidation and ultimately growth were observed at both Cu-limiting and Cu-toxic concentrations (Supplementary Figure 7). Of the 1759 transcripts detected by RNA-seq, 526 and 480 showed significant increases and decreases (P<0.05), respectively, in Cu-limited cells relative to control (Cu-replete) cells (Supplementary Dataset 3). In contrast, 460 and 344 showed significant increases and decreases, respectively, under the Cu-toxic condition relative to the control (Supplementary Dataset 3). Hierarchical clustering analysis showed a clear distinction between transcript profiles of control cultures and Cu-stressed treatments (Supplementary Figures 5b and 6b).
Transcriptional changes in pathways for ammonia oxidation and assimilation
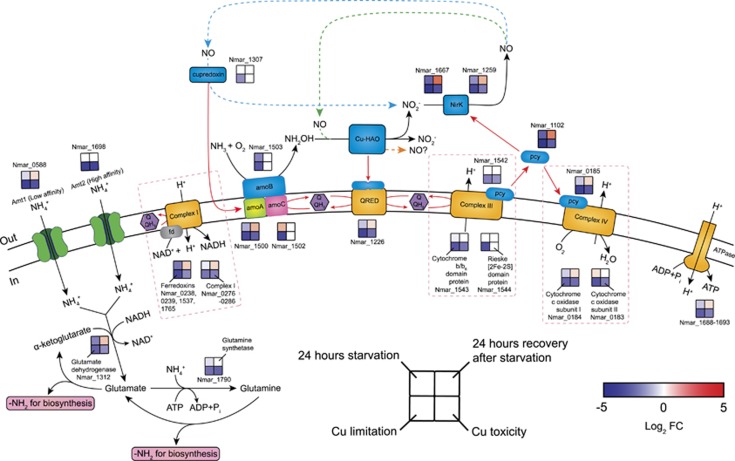
The genes encoding the three major subunits of the AMO (amoA, amoB, amoC) are co-localized in the N. maritimus genome but differ in organization and content relative to the bacterial amoCAB operon. In N. maritimus, amoC (Nmar_1502) and amoA (Nmar_1500) are separated by an ORF (Nmar_1501) of unknown function, and amoB (Nmar_1503) is located on the opposite strand from amoC and amoA (Figure 1; Walker et al., 2010), suggesting that the AOA amo genes may be under different regulatory control, which was supported by their divergent transcriptional responses to the three imposed stresses in this study. Transcription of all three amo genes was depressed with Cu limitation, whereas only amoA transcripts were reduced at toxic Cu concentrations (Figure 2). Loss of transcripts for amoA and amoB in response to ammonia starvation contrasted with retention of amoC transcripts (Figure 2). Thus, Cu and energy stress appear to be associated with distinct physiological states.
Figure 2.
Transcriptional changes for genes participating in proposed pathways of ammonia oxidation, electron transfer and ammonia assimilation in response to ammonia starvation, recovery from starvation, Cu limitation, and Cu toxicity conditions. Log2 fold changes in expression are shown for the 24 h starvation vs exponential growth, 24 h recovery after ammonia re-addition vs 24 h starvation, Cu limitation vs Cu-replete and Cu toxicity vs Cu-replete conditions. Upregulation is represented in red (P<0.05), downregulation in blue, and no significant differential expression in white. Alternative models for ammonia oxidation and electron transfer are represented by the blue, green, and orange dashed arrows, highlighting the as yet unresolved centrality of NO in AOA biochemistry. A possible function of NO as an electron shuttle was earlier posited based on energetic considerations, and may also suggest a role in electron transfer by an ORF (Nmar_1501) of unknown function in the archaeal AMO operon (Stahl and de la Torre, 2012). In this model, NO is produced by the reduction of nitrite by Cu-NirK (Nmar_1259 and Nmar_1667). Subsequently, NO is re-oxidized to nitrite by a purple cupredoxin (Nmar_1307) delivering electrons to AMO (Hosseinzadeh et al., 2016), transfer possibly facilitated by Nmar_1501 (blue dashed arrow). Alternatively, NO is a co-reactant with NH2OH to form nitrite facilitated by an unknown Cu-containing protein (green dashed arrow) (Kozlowski et al., 2016). Finally, the orange dashed arrow highlights the possibility, as recently shown in the AOB (Caranto and Lancaster, 2017), that NO is the primary product of NH2OH oxidation. Red arrows indicate possible electron flow. Pink dashed boxes contain the subunits and corresponding expression patterns of complexes I, III and IV. The expression patterns of complex I and ATPase were assigned as the averaged log2 fold changes in expression for their subunits. The log2 fold changes in the transcript abundances of four ferredoxins (Nmar_0238, 0239, 1537 and 1765) were averaged to represent the expression pattern of the putative complex I associated ferredoxins. No complex I and ATPase subunits and ferredoxin genes were regulated in the opposite directions (see Supplementary Dataset 3 for the fold-expression and statistical significance of each treatment).
Variation in amo gene transcript abundance over longer periods of starvation time (up to 200 h) was monitored by RT-qPCR. The per-cell copy numbers of amoA and amoB transcripts decreased by nearly 32-fold within 9 h of ammonia starvation and remained low for the remainder of the experiment (Supplementary Figure 8). In contrast, and consistent with the microarray analysis, amoC transcript abundance remained elevated during the initial 24 h of starvation, decreased by about 10-fold at 48 h, and remained more abundant than amoA and amoB transcripts over the entire 200-h experiment (Supplementary Figure 8). The persistence of the amoC mRNA, or continued transcription of the amoC, following depletion of ammonia has also been observed in ammonia-oxidizing bacteria (AOB) (Sayavedra-Soto et al., 1998; Hommes et al., 2001; Stein et al., 2013). Previous studies of β- and γ-AOB have shown that a monocistronic variant of bacterial AmoC has a chaperone-like function, likely acting to repair or stabilize the AMO holoenzyme (Berube et al., 2007; El Sheikh and Klotz, 2008; Berube and Stahl, 2012). The persistence of amoC transcripts in N. maritimus suggests that the archaeal AmoC may play a similar role in stabilizing ammonia oxidation function during periods of inactivity or energy stress. This hypothesis is also supported by Nt-acetylation of only the AmoC subunit of the holoenzyme (Supplementary Dataset 2), a modification controlling subunit stoichiometry and stability of multiprotein complexes in other organisms (Shemorry et al., 2013; Holmes et al., 2014).
These data also offer perspective for interpretation of environmental transcriptional data sets. For instance, the abundance of AOA amoC transcripts have been reported to be orders of magnitude greater than those of amoA and amoB in the oligotrophic open ocean (Shi et al., 2011). Analogous to the results of our study, these environmental results suggest that a considerable fraction of marine AOA in oligotrophic areas frequently face periods of energy starvation or limitation, which could result from variable remineralization rates or competition with ammonia-assimilating phytoplankton and heterotrophs.
Apart from the documented function of the AMO in the oxidation of ammonia to hydroxylamine (NH2OH) (Walker et al., 2010), the system of electron carriers linking oxidation of NH2OH to the terminal oxidase of N. maritimus is still unknown. It is hypothesized that this system involves some of the 17 small blue Cu-containing plastocyanins encoded by N. maritimus (Nmar_0004, 0185, 0815, 0918, 1102, 1129, 1142, 1161, 1226, 1273, 1307, 1443, 1542, 1637, 1650, 1665, 1678; Walker et al., 2010), all of which have no described functions apart from the nitric oxide (NO) oxidation activity of the biochemically characterized Nmar_1307 (Hosseinzadeh et al., 2016). Of these, Nmar_1102 was among the 5% most abundant proteins in the proteome (Figure 1; Supplementary Figure 1), and was also among the four plastocyanins (Nmar_0815, 1102, 1226, 1650) that showed reduced transcript abundances during ammonia starvation and Cu stress, and increased transcript abundances during recovery (Figure 2; Supplementary Dataset 4). Since Nmar_1102 has also been observed in metatranscriptomes of coastal waters (Hollibaugh et al., 2011), we hypothesize it functions in electron transfer with one, or both, of two putative nitrite reductases (NirK; Nmar_1667 and Nmar_1259; sharing ~91% amino acid identity) implicated in ammonia oxidation (Figure 2).
The high expression of the two NirK paralogs in the transcriptome and proteome of exponentially growing cells (Figure 1; Supplementary Dataset 1), their depression under ammonia limitation and Cu stress, and their increased transcription during recovery (Figure 2) together suggest they serve a central role in AOA ammonia catabolism. Recent studies have indicated that production of NO, possibly through NirK activity, is essential for ammonia oxidation by marine AOA pure cultures and natural populations (Martens-Habbena et al., 2015) and the soil AOA N. viennensis (Kozlowski et al., 2016). High expression of nirK-like genes has also been observed in marine metatranscriptomes and metaproteomes (Stewart et al., 2012; Hawley et al., 2014). Thus, similar to the observed rapid depletion of amoA transcripts in response to ammonia starvation, the expression of nirK may also be a useful molecular marker for monitoring the activity of natural marine AOA populations (Lund et al., 2012).
Recently, bacterial homologs of the AOA Cu-containing NirK present in AOB have been suggested to not function solely in nitrite reduction. Characterization of the Nitrosomonas europaea hydroxylamine oxidoreductase (HAO) showed that NO, not nitrite, is the major product of NH2OH oxidation (Caranto and Lancaster, 2017). Those authors further hypothesized that the bacterial NirK may not act as a nitrite reductase, but rather oxidizes NO to nitrite (Caranto and Lancaster, 2017). We previously showed nearly stoichiometric conversion of ammonia to NO when N. maritimus is exposed to very high ammonia levels (Martens-Habbena et al., 2015). The accumulation of NO at high ammonia concentrations suggests a kinetic limitation at one or more of the subsequent steps in the reaction pathway, and is consistent with NO production from NH2OH oxidation or by diversion of electrons to nitrite reduction (Figure 2). However, we also note that other copper proteins may function in NO oxidation, as recently reported for a cupredoxin (Nmar_1307) isolated from N. maritimus catalyzing oxidation of NO to nitrite (Hosseinzadeh et al., 2016).
Electrons transferred through a conventional complex III and IV would be used to generate a proton motive force, as well as possibly directed to the reduction of nitrite as suggested in one variant of current models (Figure 2). Transcripts of complex III genes (Nmar_1542-1544) showed no significant changes in response to ammonia starvation and recovery, but were significantly reduced under Cu stress (Figure 2). Transcripts of three complex IV genes (Nmar_0182, 0184, and 0185) were reduced during nutrient and Cu stress, and they were elevated during recovery (Figure 2). The lack of a gene encoding the NADH-binding subunit of NADH dehydrogenase (NuoF) in the genomes of N. maritimus and other AOA suggests that reduced cofactors (NAD(P)H) would be generated via ferredoxin or FAD-dependent oxidoreductases interacting with Complex I (NADH:ubiquinone oxidoreductase) (Nmar_0276-0286; Nuo ABCDHIJKMLN) (Urakawa et al., 2011). Ferredoxins, which are among the most highly expressed genes in the transcriptome and proteome (Figure 1; Supplementary Figure 1; Supplementary Dataset 1), showed the greatest variation in transcript abundance with changing nutrient status (Supplementary Datasets 3 and 4). Genes coding for both ferredoxins and Complex I were reduced during nutrient or Cu stress (Figure 2; Supplementary Dataset 4). High abundance of ferredoxin transcripts and proteins in coastal waters with relatively high iron bioavailability (Hollibaugh et al., 2011; Williams et al., 2012) further supports the importance of these electron carriers in marine AOA. In addition, as expected, genes for many ATP synthase subunits (Nmar_1688-1693) were among the most highly represented in the transcriptome and proteome of actively growing cells (Supplementary Figure 1; Supplementary Dataset 1) and showed significant reductions in stressed cells (Figure 2; Supplementary Dataset 4).
Apart from serving as an essential energy source, ammonia is also assimilated for biosynthesis by N. maritimus using one of two ammonia transporters (Amt1, Nmar_0588 and Amt2, Nmar_1698). Although amt1 was expressed at lower levels relative to amt2, they are both among the most abundant transcripts in the transcriptome of mid-log phase cells (Figure 1; Supplementary Dataset 1). A recent transcription study reported that actively growing N. maritimus cells sustain high levels of amt2 expression at environmentally relevant ammonium concentrations (low nanomolar range), whereas expression of amt1 was depressed at these concentrations (Nakagawa and Stahl, 2013). This is consistent with our observation of no significant change in amt2 transcript abundance after 24 h of ammonium starvation relative to mid-log control, and a significant decrease and increase (P<0.01) of amt1 transcripts in response to the short-term ammonium deprivation and replenishment, respectively (Figure 2). Since only Amt2 (Nmar_1698 and T478_1378) was abundant in the proteomes of exponential phase N. maritimus (Supplementary Dataset 4) and N. brevis cells (Santoro et al., 2015), the constitutive expression of amt2 suggests that N. maritimus and other AOA selectively retain the Amt2-like high affinity ammonium uptake system for reinitiating growth following ammonia limitation. After uptake, ammonia is assimilated into cellular nitrogen through the activities of glutamine synthetase (GS) and glutamate dehydrogenase (GDH). GDH (Nmar_1312 and T478_1059) was abundant in the proteomes of exponentially growing N. maritimus (Supplementary Dataset 4) and N. brevis (Santoro et al., 2015), and Nmar_1312 was upregulated during the recovery from ammonia starvation, together highlighting the primacy of GDH in driving ammonia assimilation (Figure 2).
Transcriptional changes in the pathway for carbon fixation
N. maritimus grows autotrophically by fixing inorganic carbon via a variant of the 3-hydroxypropionate/4-hydroxybutyrate (HP/HB) cycle, representing the most energy-efficient aerobic autotrophic pathway yet characterized (Könneke et al., 2014). All biochemically characterized thaumarchaeotal HP/HB cycle enzymes were identified in the proteome of exponentially growing cells, and many of them were among the most abundant proteins based on spectral counts, including acetyl-/propionyl-CoA carboxylase (Nmar_0273), malonic semialdehyde reductase (Nmar_1110), 3-hydroxypropionyl-CoA synthetase (Nmar_1309), 3-hydroxybutyryl-CoA dehydratase (Nmar_1308), 4-hydroxybutyryl-CoA dehydratase (Nmar_0207), and acetoacetyl-CoA β-ketothiolase (Nmar_1631) (Supplementary Figure 1; Supplementary Dataset 4). Their homologs have also been detected in marine metaproteomes (Williams et al., 2012; Hawley et al., 2014).
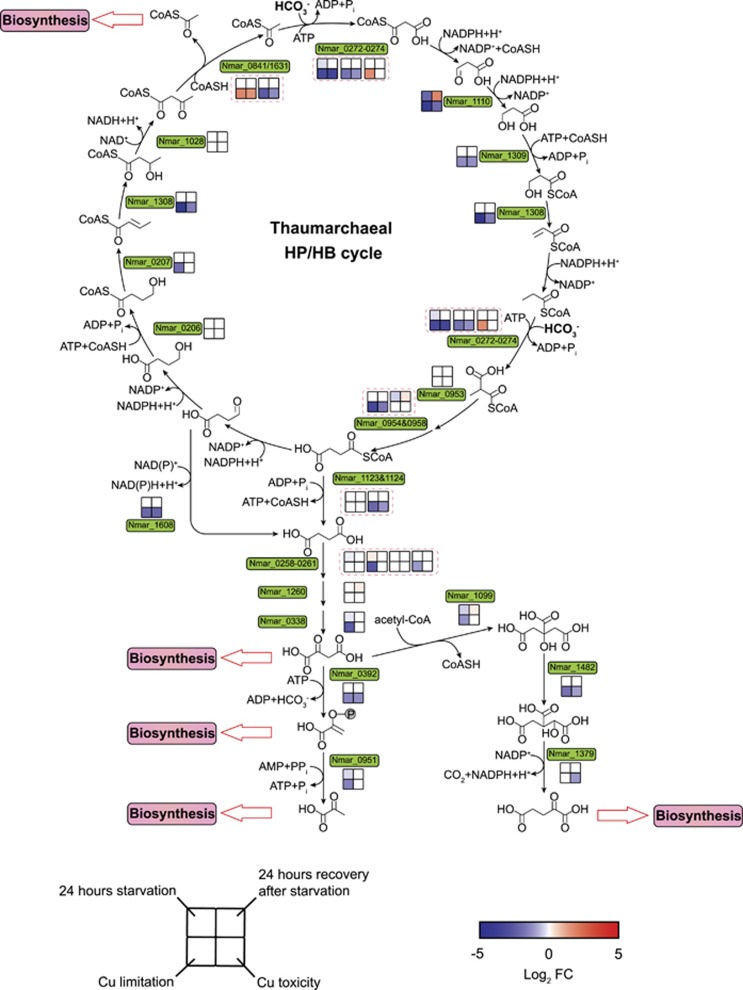
Under Cu-stressed conditions, N. maritimus transcripts for most of the HP/HB cycle genes were at significantly lower abundance (P<0.05) than under optimal growth conditions (Figure 3). In contrast, only a few transcripts in this pathway responded to changing ammonia availability. Among these, transcripts for malonic semialdehyde reductase (Nmar_1110), catalyzing the reduction of malonic semialdehyde to 3-hydroxypropionate, were most reduced in the absence of ammonia and the most elevated upon re-exposure to ammonia (Figure 3; Supplementary Dataset 4). This pattern appears to be an energy sparing response to ammonia limitation, since the conversion of 3-hydroxypropionate to 3-hydroxypropionyl-CoA by 3-hydroxypropionyl-CoA synthetase (Nmar_1309) is an ATP-consuming step. There would also be a kinetic advantage to regulatory control at this point of the HP/HB cycle. The Vmax of recombinant 3-hydroxypropionyl-CoA synthetase (0.59±0.03 μmol·min−1·mg−1 protein) is the lowest among the characterized enzymes of thaumarchaeotal HP/HB cycle, and its Km (1.2±0.2 mm) is the highest half-saturation constant of any characterized HP/HB pathway enzyme (Könneke et al., 2014; Otte et al., 2015). Since activation of 3-hydroxypropionate appears to be the major kinetic and energetic bottleneck in the thaumarchaeotal HP/HB cycle, regulation of 3-hydroxypropionate formation may serve to depress flux of carbon and energy through this pathway under conditions of energy limitation.
Figure 3.
Transcriptional changes for genes in the 3-HP/4-HB carbon fixation pathway in response to ammonia starvation, recovery from starvation, Cu limitation, and Cu toxicity conditions. The color scheme of expression and treatment comparison to controls are the same as that described in Figure 2. P-value cutoff of 0.05 was taken as the threshold of the significant alteration in expression relative to controls. Pink dashed boxes contain the expression patterns of multi-subunits or paralogs of carbon fixation enzymes.
Carbon fixed through the HP/HB cycle enters central metabolism as acetyl-CoA. Two acetyl-CoAs are generated in each turn of the pathway cycle by an acetoacetyl-CoA β-ketothiolase. N. maritimus and all characterized AOA encode two variants of acetoacetyl-CoA β-ketothiolase (Nmar_0841 and Nmar_1631) of low (~31.3%) amino acid identity to each other. Phylogenetic analysis revealed that these two β-ketothiolase variants affiliate with two distinct lineages (Supplementary Figure 9). Only Nmar_1631 was abundant in the proteome of N. maritimus under normal growth conditions (Supplementary Dataset 4) and is presumably the variant active in the HP/HB cycle. As expected, Nmar_1631 was transcribed at significantly lower levels (P<0.01) in the slowly metabolizing cells grown under Cu-stressed conditions compared to Cu-replete controls (Figure 3). In contrast, Nmar_0841 showed the opposite trend; its transcript increased about three-fold (P<0.01) under Cu-limited and Cu-toxic conditions relative to controls (Figure 3). Increased expression of this variant under stress indicates an alternative activity that may be related to the Nt-acetylation observed throughout the N. maritimus proteome. This potential role of Nmar_0841 in Nt-acetylation is also supported by a concurrent increase in the transcript abundance of an Nt-acetylating GNAT protein gene (Nmar_0309) (Vetting et al., 2005) under ammonia and Cu stress conditions (Supplementary Dataset 4). It has been shown in other organisms that a response to varying environmental and growth conditions is modulation of protein stability and activity by differential Nt-acetylation, and that the extent of Nt-acetylation is determined in part by the availability of acetyl-CoA (Yi et al., 2011; Hoshiyasu et al., 2013). Thus, the conservation of Nmar_0841-like variants in different lineages of AOA suggests it plays a similar role in regulating stress-related Nt-acetylation.
Transcriptional changes in the pathway for cobalamin biosynthesis
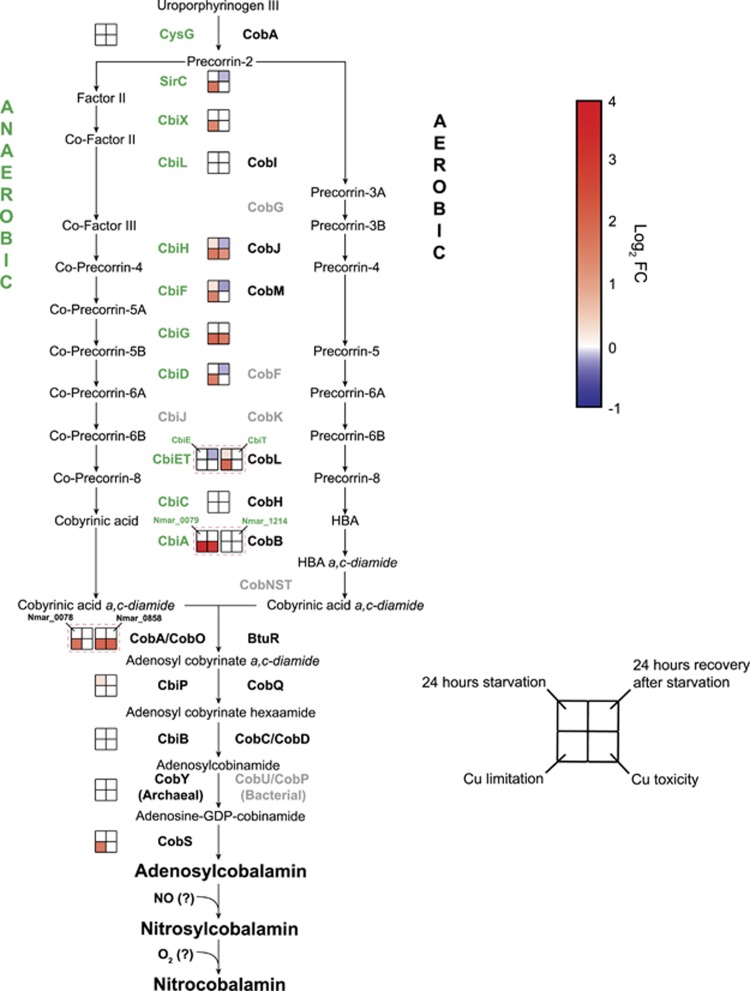
The putative thaumarchaeotal cobalamin biosynthetic pathway has been previously identified in marine AOA genomes and metagenomes (Walker et al., 2010; Doxey et al., 2015; Santoro et al., 2015). Many cobalamin biosynthetic genes were found transcribed and translated in the marine environment (Hollibaugh et al., 2011; Williams et al., 2012). Critical cobalamin-dependent enzymes include the different isomerase and methyltransferase activities of methylmalonyl-CoA mutase (MCM), methionine synthase (MetH), and ribonucleotide reductase (Walker et al., 2010; Doxey et al., 2015). The production of cobalamin by marine AOA was recently confirmed in several pure cultures (Heal et al., 2017), and the enzymes diagnostic for catalyzing its multi-step biosynthesis were identified here in N. maritimus (Supplementary Dataset S4). The culture studies also revealed remarkably high carbon-specific cobalamin cell quotas (2800–11 600 nmol cobalamin per mol C), suggesting that marine AOA are a major source of cobalamin in seawater, particularly below the euphotic zone (Heal et al., 2017).
Surprisingly, transcripts associated with N. maritimus genes in the cobalamin biosynthetic pathway increased under ammonia-depleted and Cu-stressed conditions, and their transcripts decreased during recovery from ammonia starvation (Figure 4). A possible explanation for increased biosynthetic demand for cobalamin was suggested by the earlier observation that AOA produce NO as an intermediate in the ammonia oxidation pathway (Martens-Habbena et al., 2015). Nitric oxide can rapidly bind and react with the reduced forms of cobalamin to produce nitrosylcobalamin (NOCbl) (Danishpajooh et al., 2001; Kambo et al., 2005), which is subsequently oxidized to nitrocobalamin (NO2Cbl) in the presence of oxygen (Wolak et al., 2006). Notably, a recent analysis by our group confirmed that under Cu stress and ammonia starvation, NO2Cbl was a significant fraction of the cobalamin pool of N. maritimus cells (Heal, Qin, et al., unpublished results). Nitrosylated cobalamin has diminished ability to serve as a cofactor in some organisms’ MCM and MetH (Brouwer et al., 1996; Danishpajooh et al., 2001; Kambo et al., 2005). Thus, during unbalanced growth of AOA associated with stress, the possible accumulation of NO may act to deplete the biologically active pool of cobalamin and necessitate upregulation of its biosynthetic pathway to meet cellular demand.
Figure 4.
Transcriptional changes for genes in the pathway for cobalamin synthesis in response to ammonia starvation, recovery from starvation, Cu limitation, and Cu toxicity conditions. Genes and gene clusters are shown as upregulated (red) (P<0.05), downregulated (blue), or not differentially regulated (white) in stressed or recovery conditions relative to reference cultures as described in Figure 2. Gene names in gray correspond to enzymes not yet identified in thaumarchaeotal genomes.
Conclusions
Overall, the transcriptome of N. maritimus revealed a reduction in ammonia oxidation, ammonia assimilation and carbon fixation during ammonia starvation and Cu-stressed conditions, and an activation of major energy production and biosynthesis pathways during ammonia-addition recovery. As previously suggested for the AOB, AmoC appears to serve an additional function in the starvation stress response of AOA. Notably, in consideration of the high demand for cobalamin indicated by our earlier report of high cellular quotas of cobalamin in AOA likely due to its role in the carbon fixation pathway (Heal et al., 2017), upregulation of the biosynthetic pathway under ammonia starvation and Cu stress may be a response to nitrosative stress associated with unbalanced growth, compensating for the loss of functional cobalamin. The global responses documented here represent diagnostic features of physiological state that should be of value in assessing the activity status of environmental populations. In comparison to existing environmental transcriptomic data sets our results suggest that despite the extremely high ammonia affinity, a large proportion of marine AOA experience extended periods of starvation in oligotrophic oceans.
Acknowledgments
This study was supported by National Science Foundation Grants MCB-0604448 and MCB-0920741 (to DAS), Dimensions of Biodiversity Program OCE-1046017 (to DAS, AEI, EVA, AHD and JWM), Swiss National Science Foundation (P2EZP2_155522 to RAL), and NSF GRFP (to KRH). Supplementary Information is available at ISME Journal’s website.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Amin SA, Moffett JW, Martens-Habbena W, Jacquot JE, Han Y, Devol A et al. (2013). Copper requirements of the ammonia-oxidizing archaeon Nitrosopumilus maritimus SCM1 and implications for nitrification in the marine environment. Limnol Oceanogr 58: 2037–2045. [Google Scholar]
- Baker BJ, Lesniewski RA, Dick GJ. (2012). Genome-enabled transcriptomics reveals archaeal populations that drive nitrification in a deep-sea hydrothermal plume. ISME J 6: 2269–2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayer B, Vojvoda J, Offre P, Alves RJE, Elisabeth NH, Garcia JAL et al. (2016). Physiological and genomic characterization of two novel marine thaumarchaeal strains indicates niche differentiation. ISME J 10: 1051–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube PM, Samudrala R, Stahl DA. (2007). Transcription of all amoC copies is associated with recovery of Nitrosomonas europaea from ammonia starvation. J Bacteriol 189: 3935–3944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berube PM, Stahl DA. (2012). The divergent AmoC3 subunit of ammonia monooxygenase functions as part of a stress response system in Nitrosomonas europaea. J Bacteriol 194: 3448–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouwer M, Chamulitrat W, Ferruzzi G, Sauls DL, Weinberg JB. (1996). Nitric oxide interactions with cobalamins: biochemical and functional consequences. Blood 88: 1857–1864. [PubMed] [Google Scholar]
- Capone DG, Bronk DA, Mulholland MR, Carpenter EJ. (2008) Nitrogen in the Marine Environment. Academic Press: Burlington, MA, USA. [Google Scholar]
- Caranto JD, Lancaster KM. (2017). Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proc Natl Acad Sci USA 114: 8217–8222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danishpajooh IO, Gudi T, Chen YC, Kharitonov VG, Sharma VS, Boss GR. (2001). Nitric oxide inhibits methionine synthase activity in vivo and disrupts carbon flow through the folate pathway. J Biol Chem 276: 27296–27303. [DOI] [PubMed] [Google Scholar]
- Doxey AC, Kurtz DA, Lynch MDJ, Sauder LA, Neufeld JD. (2015). Aquatic metagenomes implicate Thaumarchaeota in global cobalamin production. ISME J 9: 461–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichler J, Maupin-Furlow J. (2013). Post-translation modification in Archaea: lessons from Haloferax volcanii and other haloarchaea. FEMS Microbiol Rev 37: 583–606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Sheikh AF, Klotz MG. (2008). Ammonia-dependent differential regulation of the gene cluster that encodes ammonia monooxygenase in Nitrosococcus oceani ATCC 19707. Environ Microbiol 10: 3026–3035. [DOI] [PubMed] [Google Scholar]
- Eng JK, Jahan TA, Hoopmann MR. (2013). Comet: an open-source MS/MS sequence database search tool. Proteomics 13: 22–24. [DOI] [PubMed] [Google Scholar]
- Guasto JS, Rusconi R, Stocker R. (2012). Fluid mechanics of planktonic microorganisms. Annu Rev Fluid Mech 44: 373–400. [Google Scholar]
- Hawley AK, Brewer HM, Norbeck AD, Pasa-Tolic L, Hallam SJ. (2014). Metaproteomics reveals differential modes of metabolic coupling among ubiquitous oxygen minimum zone microbes. Proc Natl Acad Sci USA 111: 11395–11400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heal KR, Qin W, Ribalet F, Bertagnolli AD, Coyote-Maestas W, Hmelo LR et al. (2017). Two distinct pools of B12 analogs reveal community interdependencies in the ocean. Proc Natl Acad Sci USA 114: 364–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollibaugh JT, Gifford S, Sharma S, Bano N, Moran MA. (2011). Metatranscriptomic analysis of ammonia-oxidizing organisms in an estuarine bacterioplankton assemblage. ISME J 5: 866–878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes WM, Mannakee BK, Gutenkunst RN, Serio TR. (2014). Loss of amino-terminal acetylation suppresses a prion phenotype by modulating global protein folding. Nat Commun 5: 4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hommes NG, Sayavedra-Soto LA, Arp DJ. (2001). Transcript analysis of multiple copies of amo (encoding ammonia monooxygenase) and hao (encoding hydroxylamine oxidoreductase) in Nitrosomonas europaea. J Bacteriol 183: 1096–1100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horak REA, Qin W, Schauer AJ, Armbrust EV, Ingalls AE, Moffett JW et al. (2013). Ammonia oxidation kinetics and temperature sensitivity of a natural marine community dominated by Archaea. ISME J 7: 2023–2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshiyasu S, Kohzuma K, Yoshida K, Fujiwara M, Fukao Y, Yokota A et al. (2013). Potential involvement of N-terminal acetylation in the quantitative regulation of the epsilon subunit of chloroplast ATP synthase under drought stress. Biosci Biotechnol Biochem 77: 998–1007. [DOI] [PubMed] [Google Scholar]
- Hosseinzadeh P, Tian SL, Marshall NM, Hemp J, Mullen T, Nilges MJ et al. (2016). A purple cupredoxin from Nitrosopumilus maritimus containing a mononuclear type 1 copper center with an open binding site. J Am Chem Soc 138: 6324–6327. [DOI] [PubMed] [Google Scholar]
- Jacquot JE, Horak REA, Amin SA, Devol AH, Ingalls AE, Armbrust EV et al. (2014). Assessment of the potential for copper limitation of ammonia oxidation by Archaea in a dynamic estuary. Mar Chem 162: 37–49. [Google Scholar]
- Kambo A, Sharma VS, Casteel DE, Woods VL, Pilz RB, Boss GR. (2005). Nitric oxide inhibits mammalian methylmalonyl-CoA mutase. J Biol Chem 280: 10073–10082. [DOI] [PubMed] [Google Scholar]
- Karner MB, DeLong EF, Karl DM. (2001). Archaeal dominance in the mesopelagic zone of the Pacific Ocean. Nature 409: 507–510. [DOI] [PubMed] [Google Scholar]
- Kerou M, Offre P, Valledor L, Abby SS, Melcher M, Nagler M et al. (2016). Proteomics and comparative genomics of Nitrososphaera viennensis reveal the core genome and adaptations of archaeal ammonia oxidizers. Proc Natl Acad Sci USA 113: E7937–E7946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Könneke M, Schubert DM, Brown PC, Hugler M, Standfest S, Schwander T et al. (2014). Ammonia-oxidizing archaea use the most energy-efficient aerobic pathway for CO2 fixation. Proc Natl Acad Sci USA 111: 8239–8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozlowski JA, Stieglmeier M, Schleper C, Klotz MG, Stein LY. (2016). Pathways and key intermediates required for obligate aerobic ammonia-dependent chemolithotrophy in bacteria and Thaumarchaeota. ISME J 10: 1836–1845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund MB, Smith JM, Francis CA. (2012). Diversity, abundance and expression of nitrite reductase (nirK-like genes in marine thaumarchaea. ISME J 6: 1966–1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens-Habbena W, Berube PM, Urakawa H, de la Torre JR, Stahl DA. (2009). Ammonia oxidation kinetics determine niche separation of nitrifying Archaea and Bacteria. Nature 461: 976–979. [DOI] [PubMed] [Google Scholar]
- Martens-Habbena W, Qin W, Horak REA, Urakawa H, Schauer AJ, Moffett JW et al. (2015). The production of nitric oxide by marine ammonia-oxidizing archaea and inhibition of archaeal ammonia oxidation by a nitric oxide scavenger. Environ Microbiol 17: 2261–2274. [DOI] [PubMed] [Google Scholar]
- Metcalf WW, Griffin BM, Cicchillo RM, Gao JT, Janga SC, Cooke HA et al. (2012). Synthesis of methylphosphonic acid by marine microbes: a source for methane in the aerobic ocean. Science 337: 1104–1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moffett JW, Brand LE, Croot PL, Barbeau KA. (1997). Cu speciation and cyanobacterial distribution in harbors subject to anthropogenic Cu inputs. Limnol Oceanogr 42: 789–799. [Google Scholar]
- Nakagawa T, Stahl DA. (2013). Transcriptional response of the archaeal ammonia oxidizer Nitrosopumilus maritimus to low and environmentally relevant ammonia concentrations. Appl Environ Microbiol 79: 6911–6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nesvizhskii AI, Vitek O, Aebersold R. (2007). Analysis and validation of proteomic data generated by tandem mass spectrometry. Nat Methods 4: 787–797. [DOI] [PubMed] [Google Scholar]
- Otte J, Mall A, Schubert DM, Könneke M, Berg IA. (2015). Malonic semialdehyde reductase from the archaeon Nitrosopumilus maritimus is involved in the autotrophic 3-hydroxypropionate/4-hydroxybutyrate cycle. Appl Environ Microbiol 81: 1700–1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin W, Amin SA, Martens-Habbena W, Walker CB, Urakawa H, Devol AH et al. (2014). Marine ammonia-oxidizing archaeal isolates display obligate mixotrophy and wide ecotypic variation. Proc Natl Acad Sci USA 111: 12504–12509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro AE, Casciotti KL, Francis CA. (2010). Activity, abundance and diversity of nitrifying archaea and bacteria in the central California Current. Environ Microbiol 12: 1989–2006. [DOI] [PubMed] [Google Scholar]
- Santoro AE, Buchwald C, McIlvin MR, Casciotti KL. (2011). Isotopic signature of N2O produced by marine ammonia-oxidizing archaea. Science 333: 1282–1285. [DOI] [PubMed] [Google Scholar]
- Santoro AE, Dupont CL, Richter RA, Craig MT, Carini P, McIlvin MR et al. (2015). Genomic and proteomic characterization of ‘Candidatus Nitrosopelagicus brevis’: An ammonia-oxidizing archaeon from the open ocean. Proc Natl Acad Sci USA 112: 1173–1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sara M, Sleytr UB. (2000). S-layer proteins. J Bacteriol 182: 859–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayavedra-Soto LA, Hommes NG, Alzerreca JJ, Arp DJ, Norton JM, Klotz MG. (1998). Transcription of the amoCamoA and amoB genes in Nitrosomonas europaea and Nitrosospira sp, NpAV. FEMS Microbiol Lett 167: 81–88. [DOI] [PubMed] [Google Scholar]
- Shemorry A, Hwang CS, Varshavsky A. (2013). Control of protein quality and stoichiometries by N-terminal acetylation and the N-end rule pathway. Mol Cell 50: 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi YM, Tyson GW, Eppley JM, DeLong EF. (2011). Integrated metatranscriptomic and metagenomic analyses of stratified microbial assemblages in the open ocean. ISME J 5: 999–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl DA, de la Torre JR. (2012). Physiology and diversity of ammonia-oxidizing archaea. Annu Rev Microbiol 66: 83–101. [DOI] [PubMed] [Google Scholar]
- Stein LY, Campbell MA, Klotz MG. (2013). Energy-mediated vs. ammonium-regulated gene expression in the obligate arnmonia-oxidizing bacterium, Nitrosococcus oceani. Front Microbiol 4: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart FJ, Ulloa O, DeLong EF. (2012). Microbial metatranscriptomics in a permanent marine oxygen minimum zone. Environ Microbiol 14: 23–40. [DOI] [PubMed] [Google Scholar]
- Urakawa H, Martens-Habbena W, Stahl DA. (2011) Physiology and genomics of ammonia-oxidizing archaea. In: Ward MK BB, Arp DJ (ed). Nitrification. ASM Press: Washington, DC, pp 117–155. [Google Scholar]
- Vetting MW, de Carvalho LPS, Yu M, Hegde SS, Magnet S, Roderick SL et al. (2005). Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys 433: 212–226. [DOI] [PubMed] [Google Scholar]
- Vorontsov EA, Rensen E, Prangishvili D, Krupovic M, Chamot-Rooke J. (2016). Abundant lysine methylation and N-terminal acetylation in Sulfolobus islandicus revealed by bottom-up and top-down proteomics. Mol Cell Proteomics 15: 3388–3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker CB, de la Torre JR, Klotz MG, Urakawa H, Pinel N, Arp DJ et al. (2010). Nitrosopumilus maritimus genome reveals unique mechanisms for nitrification and autotrophy in globally distributed marine crenarchaea. Proc Natl Acad Sci USA 107: 8818–8823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams TJ, Long E, Evans F, DeMaere MZ, Lauro FM, Raftery MJ et al. (2012). A metaproteomic assessment of winter and summer bacterioplankton from Antarctic Peninsula coastal surface waters. ISME J 6: 1883–1900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolak M, Stochel G, van Eldik R. (2006). Reactivity of aquacobalamin and reduced cobalamin toward S-nitrosoglutathione and S-nitroso-N-acetylpenicillamine. Inorg Chem 45: 1367–1379. [DOI] [PubMed] [Google Scholar]
- Yi CH, Pan HL, Seebacher J, Jang IH, Hyberts SG, Heffron GJ et al. (2011). Metabolic regulation of protein N-alpha-acetylation by Bcl-xL promotes cell survival. Cell 146: 607–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.