Abstract
The tropical ascidian Lissoclinum patella hosts two enigmatic cyanobacteria: (1) the photoendosymbiont Prochloron spp., a producer of valuable bioactive compounds and (2) the chlorophyll-d containing Acaryochloris spp., residing in the near-infrared enriched underside of the animal. Despite numerous efforts, Prochloron remains uncultivable, restricting the investigation of its biochemical potential to cultivation-independent techniques. Likewise, in both cyanobacteria, universally important parameters on light-niche adaptation and in situ photosynthetic regulation are unknown. Here we used genome sequencing, transcriptomics and metabolomics to investigate the symbiotic linkage between host and photoendosymbiont and simultaneously probed the transcriptional response of Acaryochloris in situ. During high light, both cyanobacteria downregulate CO2 fixing pathways, likely a result of O2 photorespiration on the functioning of RuBisCO, and employ a variety of stress-quenching mechanisms, even under less stressful far-red light (Acaryochloris). Metabolomics reveals a distinct biochemical modulation between Prochloron and L. patella, including noon/midnight-dependent signatures of amino acids, nitrogenous waste products and primary photosynthates. Surprisingly, Prochloron constitutively expressed genes coding for patellamides, that is, cyclic peptides of great pharmaceutical value, with yet unknown ecological significance. Together these findings shed further light on far-red-driven photosynthesis in natural consortia, the interplay of Prochloron and its ascidian partner in a model chordate photosymbiosis and the uncultivability of Prochloron.
Introduction
Colonial ascidians (family didemnidae) are the only chordates known to form an obligate photosymbiosis (Hirose et al., 2009) with the chorophyll (Chl) b-containing cyanobacterium Prochloron spp. (Lewin and Withers, 1975). Prochloron is found at high cell densities in the cloacal cavity (=CC) of some didemnid species, or more rarely, as epibionts (Hirose et al., 2009; Nielsen et al., 2015). Certain aspects of this obligate symbiosis have been resolved, for example, it is known that Prochloron contributes to carbon fixation/nutrient translocation within its host (Pardy and Lewin, 1981; Kremer et al., 1982; Griffiths and Thinh, 1983) and produces bioactive secondary metabolites such as patellamides (Schmidt et al., 2005). Other physiological aspects, such as N2 fixation (Kline and Lewin, 1999), reactive-oxygen species detoxification (Lesser and Stochaj, 1990), host interaction and the diel regulation of metabolic pathways have remained largely unstudied, mostly due to the inability to maintain cultures of Prochloron. Culture-independent methods have elucidated certain aspects of Prochloron biology, and the publication of a draft-genome (Donia et al., 2011) allowed insights into the functional ecology of this symbiont. This first genomic analysis (Donia et al., 2011) demonstrated that Prochloron carries all primary metabolic genes required for survival outside of its host, along with a large amount of paralogous high-light inducible genes (Hli). Additionally, Prochloron has genes coding for the nitrate reduction pathway but appears to lack the capability to fix dinitrogen (Donia et al., 2011), contrasting previous reports that demonstrated active N2-fixation (Kline and Lewin, 1999).
Prochloron is the most abundant bacterium within the didemnid ascidian Lissoclinum patella (Behrendt et al., 2012) but other bacteria co-occur in the CC, the surface and the underside (US) of the animal (Behrendt et al., 2012). One of these bacteria is Acaryochloris marina, a Chl d-containing cyanobacterium residing below Prochloron on the US of the host (Kühl et al., 2005; Behrendt et al., 2012). In this particular microenvironment, Acaryochloris uses Chl d to sustain oxygenic photosynthesis under near-infrared radiation, that propagates through the overlying ascidian tissue and Prochloron cells which selectively filter out visible light (Kühl et al., 2005; Behrendt et al., 2012). Acaryochloris is a significant member of the microbial community found on the US of L. patella, accounting for up to 14% of all sequences (Behrendt et al., 2012). Because it contains Chl d and is easily cultivated, Acaryochloris has been studied extensively; yet little is known about its in situ ecology and functional niche adaptation mechanisms.
Most information about the diversity and physiology of Prochloron and Acaryochloris has been obtained through metagenomics and amplicon sequencing (Donia et al., 2011; López-Legentil et al., 2011; Behrendt et al., 2012), and a few studies on in vitro gene expression of Acaryochloris (Pfreundt et al., 2012; Yoneda et al., 2016). Here we used transcriptomics on in vivo microbial communities within the CC and the US of L. patella to obtain insights to the regulatory pathways employed by Prochloron and Acaryochloris during the noon and midnight. For the assessment of in situ transcripts, we used the published A. marina MBIC11017 genome (Swingley et al., 2008) and generated a new draft genome of Prochloron (=P5), using refined assembly methods (Albertsen et al., 2013). Moreover, metabolomics was used on the CC to assess the diel biochemical modulation between Prochloron and its host. The results are discussed against the background of the microenvironment of Prochloron and Acaryochloris (Behrendt et al., 2012; Kühl et al., 2012) and in the light of +40 years of unsuccessful attempts to cultivate Prochloron.
Materials and methods
Sampling
Intact specimens (5–15 cm2) of 5–10 mm thick L. patella were collected on the outer reef flat off Heron Island (S23°26′055, E151°55′850) at ca. 30 cm water depth during low tide (~2.8 m tidal range). Collected specimens were kept in an outdoor aquarium (max. ~200 μmol photons m−2 s−1) with a continuous supply of fresh seawater (26–28 °C). Sampling of L. patella-associated microbial biofilms for transcriptomics was performed during night-time (0000–0300 hours, hereafter referred to as ‘midnight’) or during midday (1200–1400 hours, hereafter referred to as ‘noon’) over 2 consecutive days (14–16.02.2011). During noon, photon irradiances at the sampling site routinely reached >1500 μmol photons m2 s−1. Sampling for metabolomic analysis was done for a single diel cycle during a different field campaign (07–18.02.2017) using L. patella specimens from the same sampling site and water depth. Sampling was done with a sharpened cork-borer (10 mm diameter), inserted into live L. patella specimen. The resulting core was subsampled into two microbial consortia: the CC, containing the internal symbiont Prochloron and the biofilm associated with the US of L. patella (Figure 1a and Behrendt et al., 2012). For transcriptomics, three subsamples were pooled and flash-frozen in liquid N2. Three biological replicates were sampled for each time point and day. For genomic sequencing, Prochloron cells were collected via a Pasteur pipette inserted through the siphon into the CC of L. patella. The resulting cells were filtered through meshes (100 μm and 50 μm) and concentrated using centrifugation (2000 g for 2 min). The supernatant was removed and Prochloron cells were flash frozen in liquid N2. For metabolomics, 10 biological replicates were sampled in a randomized fashion from four individual animals and the CC cores were snap frozen in liquid N2 during the day and night. All samples were transported back to the laboratory on dry ice and stored at −80 °C upon arrival.
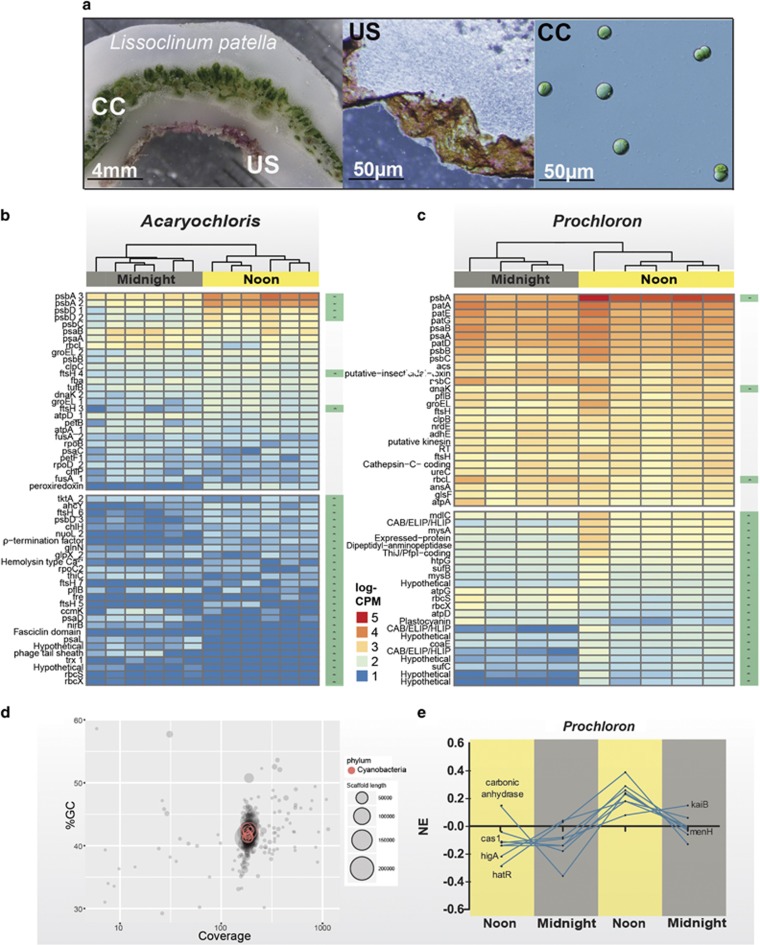
Figure 1.
The in situ transcriptome of Acaryochloris marina MBIC11017 and Prochloron sp. associated with their ascidian host Lissoclinum patella. (a) Cross-section of the L. patella holobiont with the Prochloron-containing cloacal cavity (CC) and the underside (US) harboring the Chl d-containing cyanobacterium Acaryochloris sp. (b and c) Heatmaps displaying the 25 most expressed and differentially expressed genes ranked by the average expression during the noon (top-heatmap) for Acaryochloris (b) and Prochloron (c). The remaining statistically significant differentially expressed genes for Acaryochloris and Prochloron are found in the bottom heatmaps. Within heatmaps, the displayed values correspond to log10-transformed gene expression data normalized to counts per million (CPM) within each sample. Top dendrograms are based on Euclidean distances of values displayed and color-coded according to time of sampling (noon=yellow vs midnight=grey). Stars besides rows within the bottom heatmaps indicate genes (B=30, C=26) that were differentially expressed with statistical significance (adjusted P-values <0.05). (d) Percent GC vs sequence coverage of assembled scaffolds from extracted Prochloron cells. Taxonomic affiliation of the scaffolds (represented as circles) was based on the comparison with selected marker genes (where red indicates their affiliation to the phylum cyanobacteria). The size distribution of scaffolds is given by the relative size of radii. Note the absence of contamination, most likely attributable to the clean extraction of Prochloron cells from its host followed by a set of filtration steps. (e) K-medoid clustering of normalized gene expression (NE) over two diurnal cycles for Prochloron sp. The eight selected genes from the cluster were chosen based on their similarity in diurnal expression.
Prochloron genome sequencing
Frozen Prochloron cells were homogenized in liquid N2 in a bleach-cleaned mortar. The resulting powder was processed using the standard FastDNA for soil kit (MP Biomedicals, Illkirch Cedex, France) with two additional bead-beating cycles. The resulting DNA was eluted in TAE buffer and quantified using a Qubit-fluorometer (Invitrogen, San diego, CA, USA), checked for integrity on a 0.8% agarose gel and stored at −20 °C. Sequencing was performed using Illumina technology (Illumina Inc., San diego, CA, USA) while assembly and binning procedures were done according to Albertsen et al. (2013). The resulting Prochloron genome consisted of 395 contigs with a combined genome size of 5 699 313 bp (=the P5 genome) and was submitted for Rapid Annotation using Subsystems Technology (RAST) (Aziz et al., 2008). The previously described patellamide gene cluster (Schmidt et al., 2005) was manually identified in the draft genome using BLAST.
Transcriptome analysis—experimental overview
Three reference genomes were used in the transcriptome analysis: (i) the Prochloron (P5) genome to which reads from the CC were mapped (Figure 1d); (ii) the A. marina genome MBIC11017 (Swingley et al., 2008) to which reads from the US were mapped; and (iii) the Ciona intestinalis genome (Dehal et al., 2002) to which all unmapped reads from (i) and (ii) were mapped (Ciona intestinalis is the most comprehensively annotated member of the ascidian family, phylogenetically belonging to a sister clade of didemnid-containing Aplousobranchia Tsagkogeorga et al., 2009). For the metatranscriptomic analysis, all reads not aligning against (i), (ii) or (iii) were mapped against contigs (n=5769) originating from a de novo assembly of the original reads. For details on transcriptomic analysis, please refer to Supplementary Materials.
Metabolomics analysis
Metabolite extraction and quantitation followed established protocols (see Supplementary Materials) and was conducted by Metabolomics Australia, the University of Melbourne. Sugars, sugar alcohols and organic acids were quantified on dried aliquots using gas chromatography-mass spectrometry as described in Dias et al. (2015). Amino acids and biogenic amines were quantified using liquid chromatography-mass spectrometry as described by Boughton et al. (2011). Targeted metabolites were identified and quantified using corresponding calibration series of authentic standards using the Agilent MassHunter Quantitative and Quantitative Analysis software packages (B.07.00).
Results
The Prochloron (P5) draft genome and comparative genomics
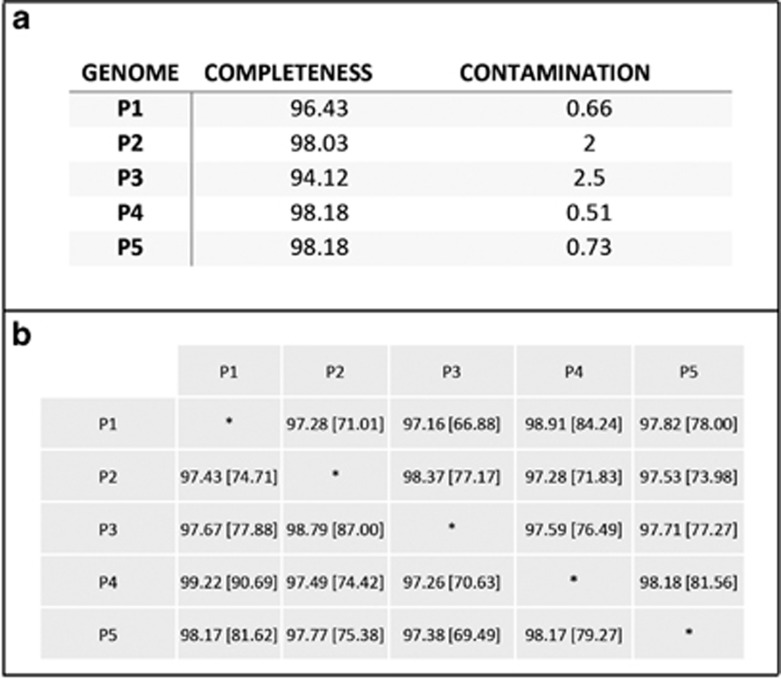
Binning of metagenome reads, originating from extracted Prochloron cells (Figure 1a, CC), was done based on GC-content, taxonomic affiliation and read coverage and resulted in a total of 395 contigs hereafter referred to as P5 (Figure 1d). The complexity of the metagenome was low, resulting in good binning resolution and minimal contamination in the binned population genomes (Figure 2). A comparison of P5 with four published Prochloron draft-genome assemblies revealed G+C% contents ranging from 41.8% (P2) to 42% (P1). The quality of all five genomes was assessed using CheckM and demonstrated little contamination (<1%) and high completeness (>98%) in the P5 genome (Figure 2a). The average nucleotide identities based on BLAST (ANIb) in the Prochloron genomes were high and all tested pairs had ANIb values above the suggested species level cutoff (>95%, Figure 2b).
Figure 2.
Overall statistics and comparative genomics of the P5-Prochloron genome. (a) Genome quality parameters for four previously published Prochloron genomes (P1–P4) and the genome generated in this study (P5), all determined via the CheckM pipeline. The genomes were truncated to 1 kb scaffold sizes before analysis and the ‘cyanobacterial marker lineage’ used for quality assessment. Genome completeness and contamination was assessed by use of broad marker genes placed within a reference genome tree also taking into account co-location of genes. (b) Average nucleotide identity (ANIb) of all five sequenced Prochloron genomes (P1–P5) and their aligned nucleotide fraction (% nucleotides). All comparisons were done in JSpeciesWS and the P2–P4 genomes were truncated to contigs of >1 kb before analysis. Note the very high nucleotide conservation between all analyzed Prochloron genomes.
Sequence overview
Despite an rRNA removal step, reads originating from the CC consisted of ~40±12% (mean±s.d.; n=10) rRNA (mostly eukaryotic 18S/28S, Supplementary Table S1) and ~73±2% in the US (mean±s.d.; n=12), again mostly 18S/28S, estimated using the SortMeRNA pipeline (Kopylova and Noe, 2012). A total of ~ 8.4 × 106 (n=10) of reads derived from the CC mapped against the P5 draft-genome (Supplementary Table S2). The mapping frequencies were low for Acaryochloris and a total of only ~3.5 × 105 (n=12) of the US reads mapped to this genome (Supplementary Table S2). Reads from the CC and US, not mapping against either of the two cyanobacterial genomes, were mapped against the Ciona intestinalis genome. Here a total of ~2.2 × 106 (n=22) of the reads mapped to the genome (Supplementary Table S2). Lastly, reads from the US and CC not mapping against any of the above reference genomes were mapped against de novo assembled transcriptome contigs, where a total of ~2.1 × 107 (CC, n=10) and ~1.2 × 107 (US, n=12) of reads mapped against 5.769 contigs above 1 kb.
Highly expressed genes in Acaryochloris and Prochloron
The 25 most transcribed genes for Acaryochloris and Prochloron were clustered according to their relative expression levels (Figures 1b and c, respectively). For P5, the majority of transcripts were at an abundance that allowed for a detailed transcriptomic investigation (Supplementary Figure S1A), whereas the low read-density for Acaryochloris demands careful interpretation of the differential expression patterns (Supplementary Figure S1B). Genes involved in the general maintenance of photosynthesis were constitutively expressed in both cyanobacteria. For Acaryochloris, this included genes coding for the photosystem (PS) I subunits (psaA, psaB, psaC), PSII CP47/43 (psbB, psbC), CO2 fixation (rbcL) and stress mitigation clpC) (Lehel et al., 1992). For Prochloron, photosynthesis-related genes were among the top 25 transcripts and encompassed genes coding for PSII CP47/43 (psbB, psbC), PSI subunits (psaA, psaB) and ATP synthase coding genes (Acaryochloris: atpA, atpD; Prochloron: atpA) (Alam et al., 1986). Other highly expressed genes include those functionally related to nitrogen metabolism (Prochloron: glsF, ureC) and osmoregulation/maintenance of photosynthesis (both Prochloron and Acaryochloris: ftsH) (Bailey et al., 2001; Stirnberg et al., 2007). For Prochloron, genes coding for cyclic peptides, patellamides, were transcribed at levels similar to regular photosynthesis housekeeping genes. Notably, all members of the patellamide gene cluster were transcribed, with patA and patE (the patellamide precursor molecule) (cf. Schmidt et al., 2005) reaching the highest expression levels.
Differentially expressed genes in Acaryochloris and Prochloron
Differential regulation of global gene expression was found in Acaryochloris (n=30; Figure 1b) and Prochloron (n=26; Figure 1c). Global gene expression patterns in both Prochloron and Acaryochloris were influenced by time of sampling, that is, noon vs midnight (PCoA plots in Supplementary Figures S2A and B, respectively). Acaryochloris displayed skewed expression patterns as determined via MA-plot characterization (Supplementary Figure S1B), demanding careful interpretation of the results. In Acaryochloris, differential regulation was found in 30 genes (Figure 1b), most of which were upregulated during noon and included photosynthesis-related genes, psbA, psbD and psaD, coding for the basic building blocks of PSII and PSI, and chlH a gene taking part in chlorophyll biosynthesis (Addlesee et al., 1996; Jensen et al., 1996). During midnight, several genes were upregulated: (i) rbcX and rbcS (coding for the assembly chaperone and small subunit of RuBisCO) (Saschenbrecker et al., 2007), (ii) nirB (involved in nitrite respiration) (Zumft, 1997), (iii) psaL (coding for the PSI reaction center subunit XI). During noon, significant upregulation of photosynthesis-related genes occurred in Prochloron (Figure 1c), where psbA, three members of the CAB/ELIP/HLIP superfamily (Kilian et al., 2008) and two genes coding for UV-protecting mycosporine molecules (mysA, mysB) (Gao and Garcia-pichel, 2011a,2011b) were upregulated. In Prochloron, significant upregulation during midnight occurred in three RuBisCO coding genes (rbcL, rbcS, rbcX) (Spreitzer and Salvucci, 2002; Emlyn-Jones et al., 2006) and a plastocyanin coding gene (petE) (Hervas et al., 1993). Diel expression patterns in Prochloron were further investigated via k-medoid clustering (Figure 1e). Genes involved in circadian rhythmicity (kaiB) (Johnson et al., 2011), CO2 interconversion (carbonic anhydrase) (Badger and Price, 1994) and prokaryotic immune response (CRISPR cas1) (Makarova, 2011) were found to follow time-dependent expression patterns.
The metabolome of the L. patella cloacal cavity
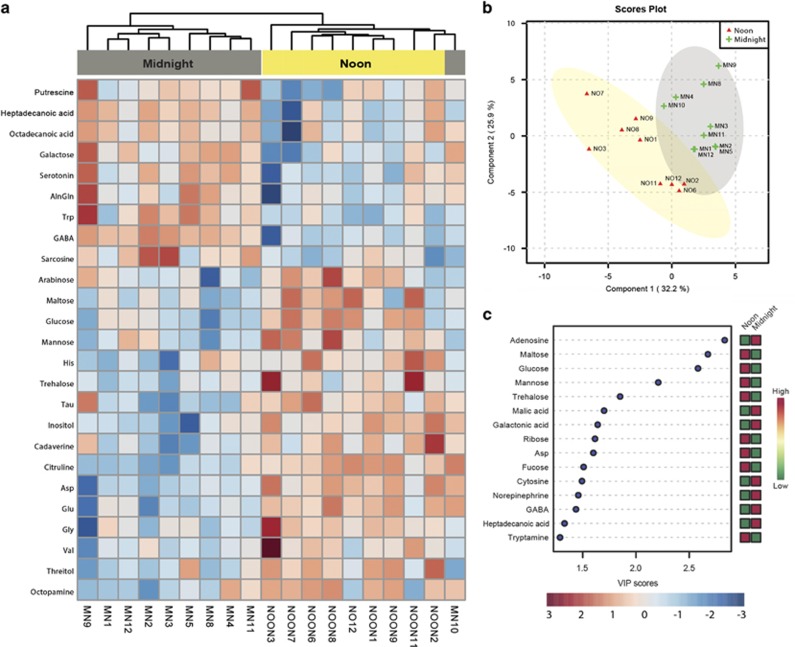
Metabolomic investigation of the CC resulted in a total of 100 detected compounds, which were normalized to mg of fresh tissue weight and are reported in Supplementary Table S3. Distinctly regulated metabolite concentrations are evident by hierarchical clustering analysis (Figure 3a). Amino-acids were the most abundant metabolites with concentrations ranging from 71±31 (mean±s.d., Asp) to 575±199 (Glu) pmol mg−1. The microtubule-stabilizing Tau-protein reached a concentration of 2600±520 pmol mg−1 and the neurotransmitter 4-amino butyric acid (GABA) a concentration of ~1600±560 pmol mg−1, while other vertebrate neurotransmitters were lower in concentration (that is, phenethylamine: 1.3±0.3, octopamine: 0.38±0.15, norepinephrine: 1.2±0.9 pmol mg−1). Saturated fatty acids occurred in low concentrations (octadecanoic acid: 2.9±1.1, heptadecanoic acid: 0.1±0.04 pmol mg−1). Various sugars (that is, galactose, trehalose, arabinose, maltose, mannose, glucose, fucose) and their derivatives (that is, inositol, arabitol) had concentrations ranging from 0.39±0.12 (galactose), 13±13 (maltose) to 38±15 (inositol) pmol mg−1. Other notable metabolites include citrulline (3.1±0.9 pmol mg−1), an amino-acid and key intermediate in the urea cycle and L-alanyl-L-glutamine (AlnGln, 0.67±0.14 pmol mg−1) a soluble dipeptide of alanine and glutamine. Partial Least Square Discriminant Analysis (PLS-DA) clustered samples from noon/midnight according to the first (32.2%) and second component (25.9% Figure 3b). Pairwise statistical comparison of metabolite concentrations between midnight and noon (P<0.05, FDR=0.1, fold-change >1.5) identified one upregulated metabolite during the night (GABA) and four upregulated metabolites during noon (glucose, Asp, mannose, maltose, trehalose) (Supplementary Table S4). Variables of importance projection (VIP) identified the metabolites that best explained the observed variance within PLS-DA, and included metabolites driving the separation at midnight (for example, adenosine, malic and galactonic acid and cytosine) and noon (for example, maltose, ribose, Asp and fucose) (Figure 3c).
Figure 3.
The in situ metabolome of the Prochloron-containing cloacal cavity of Lissoclinum patella. (a) Heatmap displaying the top 25 normalized and ranked metabolites resulting from GC-MS analysis of a total of 9 × CC samples taken during noon (yellow) and 10 × CC samples taken during midnight (gray). Ranking was performed via univariate t-testing and values within heatmaps correspond to the log2-transformed and sum-normalized values of pmol per mg−1 tissue fresh weight (scale in the bottom right). A single outlier was removed from the analysis due to missing metabolite information (=NOON10). Top dendograms are based on Euclidean clustering using Ward’s method. (b) 2D score ordination plot of all 19 samples from the noon (‘NO’) or midnight (‘MN’) with displayed 95% confidence intervals (noon=yellow, midnight=gray). Data clustering was performed using Partial Least Square Discriminant Analysis (PLS-DA) after component optimization. (c) The most important metabolites ranked by their variable importance in projection (VIP) for PLS-DA. Higher VIP scores denote metabolites that are better at explaining the observed variance within PLS. The heatmap on the right indicates the detected concentration variations during noon or midnight. All data analysis and visualization was performed in MetaboAnalyst 3.0.
Gene expression in L. patella
The Ciona intestinalis genome was used as a proxy for the investigation of gene expression in L. patella. Using a proxy genome is challenging, reflected in the overall low read mapping (see MA plot, Supplementary Figure S1C) and the absence of statistically differentially expressed genes. Given these obstacles, we found expressed genes that coded for translational products typically associated with general housekeeping; actin isoforms, histones (H3, H2B), tubulins and actin filament assembly (talin2). More specific for ascidians, expression of genes involved in the formation of metalloproteins, vanabins was observed in the CC (vanadium binding protein 1, Supplementary Figure S3) (Ueki et al., 2015).
The metatranscriptome of the cloacal cavity and underside of L. patella
To investigate the transcription of other L. patella-associated organisms all remaining reads (=not mapping to any reference genome) were mapped onto contig-assemblies generated via de novo assembly. Quality control via MA-plot characterization revealed disproportionate expression patterns (Supplementary Figures S1D and E), possibly affecting subsequent normalization and testing for differential expression. Owing to this bias, we refrain from inferring statistical significance and will only provide expression values. Reads re-mapped from both the US (Supplementary Figure S4A) and CC (Supplementary Figure S4B) displayed high expression of genes involved in sulfate metabolism (cysD) (Malo and Loughlin, 1990), L-lysine transport (lysE) (Vrljic et al., 1996) and the entry of bacterial pathogens into eukaryotic cells (Invasin) (Palumbo and Wang, 2006). Within the US, expression of genes related to the production of bioactive compounds (TOMM C/D)(Melby et al., 2011) and heat-shock proteins/chaperones (groEL, dnaK, clpB) (Eriksson and Clarke, 1996; Török et al., 1997; Calloni et al., 2012) was observed.
Discussion
We present the first in situ metabolomic- and transcriptomic investigation of Prochloron and Acaryochloris in association with L. patella.
Photosynthesis: light utilization
Accurate timing of gene expression is crucial for cyanobacteria, enabling appropriate responses to anticipated daily fluctuations and allowing for, for example, temporally decoupled N2 fixation and modulated metabolic responses to changes in extracellular pH, temperature and O2 (Steunou et al., 2008; Jensen et al., 2011). Circadian rhythmicity is entrained by a light-induced phosphorylation cycle between kaiA/B/C proteins, a mechanism leading to sustained and anticipatory gene-expression in cyanobacteria (Liu et al., 1995; Johnson et al., 2011). In Prochloron, the gene coding for one of the clock proteins (kaiB) is expressed in a cyclic fashion, suggesting light as a primary stimulus for the activation of oxygenic photosynthesis during the noon and respiration during the night (Kim et al., 2012). As expected for such phototrophs, both Prochloron and Acaryochloris express the D1, D2 and CP43/CP47 coding genes during noon. To sustain photosynthesis under high-light regimes (=noon), appropriate stress quenching and repair mechanisms are required. For chloroplasts, the repair mechanisms of the reaction center proteins D1 and D2 are well understood and characterized by a high turnover rate of these proteins during high irradiance (Melis, 1999). Metalloproteases such as FtsH can expedite the rate limiting step of D1 protein degradation (Bailey et al., 2001) and high expression of the ftsH gene in Acaryochloris and Prochloron supports FtsH-facilitated D1-protein degradation during noon in order to maintain maximum photosynthetic quantum yield. In Prochloron, photoprotection is likely provided by translational products of mysA/mysB (producing UV-screening mycosporine amino-acids) and CAB/ELIP/HLIP genes, known to maintain photosynthesis under high-light conditions (HLIP), protect against oxidative damage (ELIP), and extend light harvesting capabilities by binding to Chl a/b (CAB) (Kilian et al., 2008). Earlier metagenomic analysis of Prochloron revealed such genes (Donia et al., 2011) and our expression data substantiate their use as photoprotectants. Acaryochloris resides in a near-infrared radiation-enriched microniche below Prochloron, relatively shielded from shorter, more damaging wavelengths (Kühl et al., 2005). Yet, the expression of peroxiredoxin-like genes in Acaryochloris (involved in oxidative stress mitigation) advocates the occurrence of light generated reactive-oxygen species (Dietz, 2011). A study on reactive oxygen species production under near-infrared radiation demonstrated that Acaryochloris does encounter light-stress, albeit at a lower level than under white light (Behrendt et al., 2013). Although light stress must be reduced in Acaryochloris, because of its cryptic location, the expression of clpC, groEL and dnaK genes (all expressed during noon and involved in protein stabilization under stress/heat-shock conditions) (Török et al., 1997; Charpentier et al., 2000; Horváth et al., 2008) still suggests a requirement for reactive oxygen species quenching.
Photosynthesis: CO2 fixation
Photosynthesis drives carbon fixation, a process linked to RuBisCO, the carboxylase enzyme which facilitates chemical incorporation of CO2 into organic molecules (Spreitzer and Salvucci, 2002). Early studies on ascidians used 14CO2 labeling to demonstrate that glycolic acid is produced under light and accounts for up to 7% of isotopes fixed by Prochloron (Fisher and Trench, 1980). Illumination of isolated Prochloron cells triggered the accumulation of amino-acids (Glu, Gly, Ala and Asp) (Kremer et al., 1982). Our sampling procedure cannot distinguish between host- and Prochloron-derived metabolites, yet, amino acids and primary photosynthates (that is, maltose, glucose, fructose, mannose and sucrose) were elevated during noon, and so was the expression of an ATP-synthase-coding gene in Prochloron. This suggests the active production (and possible translocation) of Prochloron-derived photosynthates in the CC. In Prochloron and Acaryochloris, RuBisCO-associated genes (rbcL, rbcS, rbcX) and ccmK (a gene coding for the basic building block in carboxysome formation, Kerfeld et al., 2005) were elevated during the night. While this was unexpected—given the high demand for CO2 in photosynthetic carbon fixation during the day—it has been previously reported for other cyanobacteria (Huang and McCluskey, 2002). This trend could relate to photorespiration, that is, the displacement of CO2 in RuBisCO by O2, which induces its oxygenase function and production of phosphoglycolate (2PG), acting as a sensor molecule for the regulation of carbon concentration mechanisms (Haimovich-Dayan et al., 2015). At a photon irradiance of 250 μmol photons m−2 s−1, the CC and US of L. patella reach hyperoxic O2 and very high pH levels (Kühl et al., 2012), a microenvironment where O2 could outcompete CO2 in RuBisCO. Within the metabolomics data set, we find no direct evidence of 2PG accumulation, yet in other cyanobacteria 2PG can re-enter the Calvin-Benson-Bassham cycle through a metabolic conversion via several intermediary products and pathways (Eisenhut, 2006; Eisenhut et al., 2008). One of these intermediary species is glycine, the transamination product of glyoxylate (Maurino and Peterhansel, 2010), which was found to occur predominantly during noon in the metabolome. The utilization of the C2 pathway in Prochloron is supported by the transcription (data not shown) of most components of the glycine cleavage system (the H, T and P protein coding genes are present, L appears missing), suggesting that 2PG accumulates and is biochemically converted to glycine during high-light. Yet, we cannot exclude that glycine is associated with host-specific behavior, for example, ascidian locomotion via glycine-specific receptors (Nishino et al., 2010) or the biosynthesis of UV-protective mycosporine-glycine (Dunlap and Yamamoto, 1995; Maruyama et al., 2003). However, previous microenvironmental measurements, current expression data on RuBisCO genes and metabolomic signatures give strong indications for high photorespiration during noon. We hypothesize that more efficient CO2 fixation occurs pre- and post-noon under less hyperoxic conditions.
Nitrogen metabolism
The uptake and utilization of nitrogen in Prochloron was suggested to be driven by nitrogen fixation (Paerl, 1984; Kline and Lewin, 1999). Contrary to these findings, no essential dinitrogen fixation (nif) genes were found within the P1–P4- (Donia et al., 2011) or the P5-Prochloron genome. In P5, the occurrence of a full ferredoxin-dependent glutamate synthase pathway (Fd-GOGAT) and its partial expression (glsF) suggests that Prochloron instead utilizes host-excretion products as its primary nitrogen source, a mechanism proposed earlier (Parry, 1985; Donia et al., 2011). In addition to transcriptional evidence for ammonium utilization, we find that the CC contains chemical signatures of citrulline, a chemical intermediate in the urea cycle. Citruline can react with aspartate and via additional steps form urea (Herman and Shambaugh, 1977); a compound which ascidians excrete (Markus and Lambert, 1983). Urea can be decomposed, via the urease enzyme, into ammonia and carbonic acid (Solomon et al., 2010) and be used as a nitrogen source. In Prochloron, urea decomposition is corroborated by the expression of the urease subunit-α coding gene (ureC), additional β/γ subunits (data not shown) and earlier suggestions on the genetic potential of Prochloron to utilize urea (Donia et al., 2011). These results substantiate that Prochloron relies on host-associated nitrogen recycling and is not capable of fixing dinitrogen.
For Acaryochloris, N2 fixation has only been described in strain HICR111A (Pfreundt et al., 2012) and the remaining Acaryochloris genomes, MBIC11017 (Swingley et al., 2008) and CCMEE5415 (Miller et al., 2005) contain no N2 fixation genes. In this study most reads mapped to strain MBIC11017, suggesting that the Acaryochloris ecotypes found on L. patella are most closely related to this non-N2 fixing strain (or, alternatively, that the genome coverage is better). The absence of N2 fixation is (non-statistically) supported by metatranscriptomic analysis; only two solitary nif-related genes (=nifU) were detected within 9.744 coding sequences, and both demonstrated only weak expression. In an in vitro nitrogen starvation experiment Acaryochloris CCMEE5410 upregulated glnN, a gene coding for a type III glutamine synthetase (Yoneda et al., 2016) which provides an advantage during periods of nitrogen starvation (Sauer et al., 2000). We find glnN expressed in Acaryochloris during noon, suggesting a low availability of nitrogen and a fine-tuned adaptation to conserve limited nitrogen resources during times of oxygenic photosynthesis.
Why does Prochloron resist cultivation?
The presence of all primary metabolic genes and little genome modifications (P1–P5 are >95% identical across a >8000 km geographic transect), suggested that Prochloron sp. is associating with its host in a facultative manner (Donia et al., 2011). This hypothesis is corroborated by the finding of surface-associated Prochloron cells (for example, Nielsen et al., 2015). However, Prochloron was successfully cultivated in only one (unreplicated) occasion (Patterson et al., 1982); here the addition of tryptophan or serine + indole initiated growth, particularly at low pH (~5.5), a proton concentration almost two orders of magnitude lower than measured in the CC of intact L. patella specimens (Kühl et al., 2012). The authors attributed the observed growth to a biochemical deficit in the first steps of tryptophan biosynthesis (the shikimate pathway). An inspection of the P5 genome revealed all genes involved in the shikimate pathway and Prochloron appears fully equipped to biosynthesize tryptophan, confirming earlier reports (Donia et al., 2011). Within the metabolome, amino-acids (including Trp and Ser) are abundant and the expression of amino-acid transporters such as TRAP (in Prochloron, Mulligan et al., 2011) underscores that translocation could occur across the membrane of Prochloron and its host. In our experience, adding Trp/Ser/Ind and lowering the pH does not sustain cell divisions in Prochloron (Behrendt et al., unpubl), questioning the amino acid auxotrophy of this cyanobacterium.
Other clues to the uncultivability of Prochloron might lie hidden in its microenvironment. Most ascidians, including L. patella, accumulate trace metals at high concentrations, often 2–4 orders of magnitude higher than in surrounding waters (Krupp et al., 2012). Specifically for vanadium, concentration mechanisms involve the chemical reduction by microbial communities and transport within blood plasma using vanabins (Ueki et al., 2015), specialized carrier proteins for which we find expression signatures in L. patella. Whether Prochloron benefits, suffers or is involved in facilitating such metal concentration is unknown, but we can reason that unspecific uptake would cause an influx of metals at high concentrations, instigating the disruption of regular physiological functions (Baptista and Vasconcelos, 2006). To this end, Prochloron appears well equipped to maintain metal homeostasis, and the P5 genome contains signatures of various metal influx/efflux transporters most of which are transcribed (data not shown), for example, for Cu2+ (P-type ATPases), Zn2+ (ZnuABC, Patzer and Hantke, 1998), Mn2+ (MgtE, Hattori et al., 2009) and Fe3+ (futA1/futB/futC, Morrissey and Bowler, 2012). Besides these metal influx and efflux mechanisms, Prochloron produces a whole array of cyanobactins, prominently patellamides, microcin-like cyclic peptides (Long et al., 2005; Schmidt et al., 2005). Prochloron expresses the patellamide gene cluster with levels of patE (the precursor molecule) on-par with levels for psbA, the major D1 protein for PSII. This suggests an important role of patellamides for the metabolism and fitness of Prochloron. We speculate that patellamides might have the potential to scavenge metal cations, a theory put forward earlier (Bertram and Pattenden, 2007; Hirose et al., 2009), but never confirmed experimentally. Patellamide-like peptides comfortably bind Cu2+ and Zn2+ (Comba and Eisenschmidt, 2017), and do so even within Prochloron cells (Comba et al., 2017), yet show no affinity for binding to Mg2+ and Ca2+ (Bertram and Pattenden, 2007). Intriguingly, Cu (II)-complexed patellamide pseudo-peptides exhibit a wide spectrum of pH-dependent hydrolytic activities, in vitro these copper-bound analogs function as carbonic anhydrase or phosphatases at pH 7–8 (Comba et al., 2013, 2012), as glycosidases at pH ~10, and as lactamases at pH ~11.5 (Comba et al., 2016), evocative of a ‘chemical Swiss-army knife’. We speculate that Patellamides might thus ‘be imbued with biological activity’ upon metal complexation (Krupp et al., 2012), opening a functional parameter space that adjusts with the above indicated influx/efflux mechanisms. Given the strong diel pH fluctuations in L. patella (Kühl et al., 2012), we hypothesize that this spectrum of catalytic patellamide functions could be accessed by Prochloron and/or L. patella and might present an essential component of their host–microbe interaction.
We anticipate that future experiments with metabolite-augmented media will enable the establishment of stable Prochloron cultures. Further, we foresee that such an approach will highlight the value of metabolomics-assisted cultivation efforts and facilitate the artificial reconstruction of other symbiotic relationships in vitro.
Acknowledgments
This study was supported by a Sapere Aude starting postdoc grant (LB, DFF-1323-00747), a Sapere-Aude advanced grant (MK, DFF-1323-00065B), a research project grant from the Danish Council for Independent Research (MA, PH-N, DFF-4005-00369) and a research grant from VILLUM FONDEN (MA, 15510). We thank the staff at Heron Island Research Station for assistance (conducted under permit no. G12/35118.1 and G16/38423.1). Metabolite analysis was conducted at Metabolomics Australia (The University of Melbourne, Australia), a NCRIS initiative under Bioplatforms Australia Pty Ltd.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Addlesee HA, Gibson LCD, Jensen PE, Hunter CN. (1996). Cloning, sequencing and functional assignment of the chlorophyll biosynthesis gene, chZP, of Synechocystis sp. PCC 6803. FEBS Lett 389: 2–6. [DOI] [PubMed] [Google Scholar]
- Alam J, Whitaker RA, Krogmann DW, Curtis SE. (1986). Isolation and sequence of the gene for ferredoxin I from the cyanobacterium Anabaena sp. strain PCC 7120. J Bacteriol 168: 1265–1271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertsen M, Hugenholtz P, Skarshewski A, Nielsen KL, Tyson GW, Nielsen PH. (2013). Genome sequences of rare, uncultured bacteria obtained by differential coverage binning of multiple metagenomes. Nat Biotechnol 31: 533–538. [DOI] [PubMed] [Google Scholar]
- Aziz R, Bartels D, Best A, DeJongh M, Disz T, Edwards R et al. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9: 1–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badger MR, Price GD. (1994). The role of carbonic anhydrase in photosynthesis. Annu Rev Plant Physiol Plant Mol Biol 45: 369–392. [Google Scholar]
- Bailey S, Silva P, Nixon P, Mullineaux C, Robinson C, Mann N. (2001). Auxiliary functions in photosynthesis: the role of the FtsH protease. Biochem Soc Trans 29: 455–459. [DOI] [PubMed] [Google Scholar]
- Baptista MS, Vasconcelos MT. (2006). Cyanobacteria metal interactions: requirements, toxicity, and ecological implications. Crit Rev Microbiol 32: 127–137. [DOI] [PubMed] [Google Scholar]
- Behrendt L, Larkum A, Trampe E, Norman A, Sørensen S, Kühl M. (2012). Microbial diversity of biofilm communities in microniches associated with the didemnid ascidian Lissoclinum patella. ISME J 6: 1222–1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrendt L, Staal M, Cristescu SM, Harren FJ, Schliep M, Larkum AW et al. (2013). Reactive oxygen production induced by near-infrared radiation in three strains of the Chl d-containing cyanobacterium Acaryochloris marina. F1000Res 2: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertram A, Pattenden G. (2007). Marine metabolites: metal binding and metal complexes of azole-based cyclic peptides of marine origin. Nat Prod Rep 24: 18–30. [DOI] [PubMed] [Google Scholar]
- Boughton BA, Callahan DL, Silva C, Bowne J, Nahid A, Rupasinghe T et al. (2011). Comprehensive profiling and quantitation of amine group containing metabolites. Anal Chem 83: 7523–7530. [DOI] [PubMed] [Google Scholar]
- Calloni G, Chen T, Schermann SM, Chang HC, Genevaux P, Agostini F et al. (2012). DnaK functions as a central hub in the E. coli chaperone network. Cell Rep 1: 251–264. [DOI] [PubMed] [Google Scholar]
- Charpentier E, Novak R, Tuomanen E. (2000). Regulation of growth inhibition at high temperature, autolysis, transformation and adherence in Streptococcus pneumoniae by ClpC. Mol Microbiol 37: 717–726. [DOI] [PubMed] [Google Scholar]
- Comba P, Eisenschmidt A. (2017), Structures, electronics and reactivity of copper(II) complexes of the cyclic pseudo-peptides of the ascidians. In: Hanson G, Berliner L (eds). Future Directions in Metalloprotein and Metalloenzyme Research. Springer International Publishing: Cham, pp 13–32.
- Comba P, Eisenschmidt A, Gahan LR, Herten D-P, Nette G, Schenk G et al. (2017). Is Cu II coordinated to patellamides inside Prochloron cells? Chem—A Eur J 23: 12264–12274. [DOI] [PubMed] [Google Scholar]
- Comba P, Eisenschmidt A, Kipper N, Schießl J. (2016). Glycosidase- and β-lactamase-like activity of dinuclear copper(II) patellamide complexes. J Inorg Biochem 159: 70–75. [DOI] [PubMed] [Google Scholar]
- Comba P, Gahan LR, Hanson GR, Maeder M, Westphal M. (2013). Carbonic anhydrase activity of dinuclear Cu(II) complexes with patellamide model ligands. Dalt Trans 43: 3144–3152. [DOI] [PubMed] [Google Scholar]
- Comba P, Gahan LR, Hanson GR, Westphal M. (2012). Phosphatase reactivity of a dicopper(ii) complex of a patellamide derivative—possible biological functions of cyclic pseudopeptides. Chem Commun 48: 9364. [DOI] [PubMed] [Google Scholar]
- Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A et al. (2002). The draft genome of Ciona intestinalis : insights into chordate and vertebrate origins. Science 298: 2157–2167. [DOI] [PubMed] [Google Scholar]
- Dias DA, Hill CB, Jayasinghe NS, Atieno J, Sutton T, Roessner U. (2015). Quantitative profiling of polar primary metabolites of two chickpea cultivars with contrasting responses to salinity. J Chromatogr B 1000: 1–13. [DOI] [PubMed] [Google Scholar]
- Dietz K-J. (2011). Peroxiredoxins in plants and cyanobacteria. Antioxid Redox Signal 15: 1129–1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donia MS, Fricke WF, Partensky F, Cox J, Elshahawi SI, White JR et al. (2011). Complex microbiome underlying secondary and primary metabolism in the tunicate-Prochloron symbiosis. Proc Natl Acad Sci USA 108: 1423–1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunlap WC, Yamamoto Y. (1995). Small-molecule antioxidants in marine organisms: antioxidant activity of mycosporine-glycine. Comp Biochem Physiol Part B Biochem Mol Biol 112: 105–114. [Google Scholar]
- Eisenhut M. (2006). The plant-Like C2 glycolate cycle and the bacterial-like glycerate pathway cooperate in phosphoglycolate metabolism in cyanobacteria. Plant Physiol 142: 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenhut M, Ruth W, Haimovich M, Bauwe H, Kaplan A, Hagemann M. (2008). The photorespiratory glycolate metabolism is essential for cyanobacteria and might have been conveyed endosymbiontically to plants. Proc Natl Acad Sci USA 105: 17199–17204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emlyn-Jones D, Woodger FJ, Price GD, Whitney SM. (2006). RbcX can function as a rubisco chaperonin, but is non-essential in Synechococcus PCC7942. Plant Cell Physiol 47: 1630–1640. [DOI] [PubMed] [Google Scholar]
- Eriksson M-J, Clarke AK. (1996). The heat shock protein ClpB mediates the development of thermotolerance in the Cyanobacterium Synechococcus sp. Strain PCC 7942. J Bacteriol 178: 4839–4846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher CRJ, Trench RK. (1980). In vitro carbon fixation by Prochloron sp. isolated from Diplosoma virens. Biol Bull 159: 636–648. [Google Scholar]
- Gao Q, Garcia-pichel F. (2011. a). Microbial ultraviolet sunscreens. Nat Rev Microbiol 9: 791–802. [DOI] [PubMed] [Google Scholar]
- Gao Q, Garcia-Pichel F. (2011. b). An ATP-grasp ligase involved in the last biosynthetic step of the iminomycosporine shinorine in Nostoc punctiforme ATCC 29133. J Bacteriol 193: 5923–5928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths D, Thinh L. (1983). Transfer of photosynthetically fixed carbon between the prokaryotic green alga Prochloron and its ascidian host. Aust J Mar Freshw Res 34: 431–440. [Google Scholar]
- Haimovich-Dayan M, Lieman-Hurwitz J, Orf I, Hagemann M, Kaplan A. (2015). Does 2-phosphoglycolate serve as an internal signal molecule of inorganic carbon deprivation in the cyanobacterium Synechocystis sp. PCC 6803? Environ Microbiol 17: 1794–1804. [DOI] [PubMed] [Google Scholar]
- Hattori M, Iwase N, Furuya N, Tanaka Y, Tsukazaki T, Ishitani R et al. (2009). Mg2+-dependent gating of bacterial MgtE channel underlies Mg2+ homeostasis. EMBO J 28: 3602–3612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herman H, Shambaugh E. (1977). Urea biosynthesis I. The urea cycle and relationships acid cycle. Am J Clin Nutr 30: 2083–2087. [DOI] [PubMed] [Google Scholar]
- Hervas M, Navarro F, Navarro JA, Chavez S, Diaz A, Florencio FJ et al. (1993). Synechocystis 6803 plastocyanin isolated from both the cyanobacterium and E. coli transformed cells are identical. FEBS Lett 319: 257–260. [DOI] [PubMed] [Google Scholar]
- Hirose E, Neilan B, Schmidt E, Murakami A. (2009), Enigmatic life and evolution of Prochloron and related cyanobacteria inhabiting colonial ascidians. In: Gault PM, Marler HJ (eds). Handbook on cyanobacteria: biochemistry, biotechnology and applications. Nova Science Publishers: Hauppauge, NY, USA, pp 161–189.
- Horváth I, Multhoff G, Sonnleitner A, Vígh L. (2008). Membrane-associated stress proteins: more than simply chaperones. Biochim Biophys Acta 1778: 1653–1664. [DOI] [PubMed] [Google Scholar]
- Huang L, McCluskey MP. (2002). Global gene expression profiles of the cyanobacterium Synechocystis sp. strain PCC 6803 in response to irradiation with UV-B and white light. J Bacteriol 184: 6845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen PE, Gibson LCD, Henningsen KW, Hunter CN. (1996). Expression of the chlIchlD, and chlH genes from the Cyanobacterium Synechocystis PCC6803 in Escherichia coli and demonstration that the three cognate proteins are required for magnesium-protoporphyrin chelatase activity. J Biol Chem 271: 16662–16667. [DOI] [PubMed] [Google Scholar]
- Jensen SI, Steunou A-S, Bhaya D, Kühl M, Grossman AR. (2011). In situ dynamics of O2, pH and cyanobacterial transcripts associated with CCM, photosynthesis and detoxification of ROS. ISME J 5: 317–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson CH, Stewart PL, Egli M. (2011). The cyanobacterial circadian system: from biophysics to bioevolution. Annu Rev Biophys 40: 143–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerfeld CA, Sawaya MR, Tanaka S, Nguyen CV, Phillips M, Beeby M et al. (2005). Protein structures forming the shell of primitive bacterial organelles. Science 309: 936–938. [DOI] [PubMed] [Google Scholar]
- Kilian O, Steunou AS, Grossman AR, Bhaya D. (2008). A novel two domain-fusion protein in cyanobacteria with similarity to the CAB/ELIP/HLIP superfamily: evolutionary implications and regulation. Mol Plant 1: 155–166. [DOI] [PubMed] [Google Scholar]
- Kim Y-I, Vinyard DJ, Ananyev GM, Dismukes GC, Golden SS. (2012). Oxidized quinones signal onset of darkness directly to the cyanobacterial circadian oscillator. Proc Natl Acad Sci USA 109: 17765–17769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kline TJ, Lewin R. (1999). Natural 15N/14N abundance as evidence for N2 fixation by Prochloron (Prochlorophyta) endosymbiotic with didemnid ascidians. Symbiosis 26: 193–198. [Google Scholar]
- Kopylova E, Noe L. (2012). Sequence analysis SortMeRNA : fast and accurate filtering of ribosomal RNAs in metatranscriptomic data. Bioinformatics 28: 3211–3217. [DOI] [PubMed] [Google Scholar]
- Kremer B, Pardy R, Lewin R. (1982). Carbon fixation and photosynthates of Prochloron, a green alga symbiotic with an ascidian, Lissoclinum patella. Phycologia 21: 258–263. [Google Scholar]
- Krupp E, Wright SH, Raab A. (2012), Marine metabolites and metal ion chelation. In: Fattorusso E, Gerwick WH, Taglialatela-Scafati O (eds). Handbook of Marine Natural Products. Springer Netherlands: Dordrecht, pp 861–892.
- Kühl M, Behrendt L, Trampe E, Qvortrup K, Schreiber U, Borisov SM et al. (2012). Microenvironmental ecology of the chlorophyll b-containing symbiotic Cyanobacterium Prochloron in the didemnid ascidian Lissoclinum patella. Front Microbiol 3: 402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kühl M, Chen M, Ralph P, Schreiber U, Larkum AWD. (2005). A niche for cyanobacteria containing chlorophyll d. Nature 433: 820. [DOI] [PubMed] [Google Scholar]
- Lehel C, Wada H, Kovács E, Török Z, Gombos Z, Horváth I et al. (1992). Heat shock protein synthesis of the cyanobacterium Synechocystis PCC 6803: purification of the GroEL-related chaperonin. Plant Mol Biol 18: 327–336. [DOI] [PubMed] [Google Scholar]
- Lesser MP, Stochaj WR. (1990). Photoadaptation and protection against active forms of oxygen in the symbiotic procaryote Prochloron sp. and its ascidian host. Appl Environ Microbiol 56: 1530–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewin R, Withers N. (1975). Extraordinary pigment composition of a prokaryotic alga. Nature 256: 735–737. [Google Scholar]
- Liu Y, Tsinoremas NF, Johnson CH, Lebedeva NV, Golden SS, Ishiura M et al. (1995). Circadian orchestration of gene expression in cyanobacteria. Genes Dev 9: 1469–1478. [DOI] [PubMed] [Google Scholar]
- Long PF, Dunlap WC, Battershill CN, Jaspars M. (2005). Shotgun cloning and heterologous expression of the patellamide gene cluster as a strategy to achieving sustained metabolite production. Chembiochem 6: 1760–1765. [DOI] [PubMed] [Google Scholar]
- López-Legentil S, Song B, Bosch M, Pawlik J, Turon X. (2011). Cyanobacterial diversity and a new Acaryochloris-like symbiont from Bahamian sea-squirts. PLoS One 6: e23938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makarova KS. (2011). Evolution and classification of the CRISPR–Cas systems. Nat Rev Microbiol 9: 467–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malo MS, Loughlin RE. (1990). Promoter elements and regulation of expression of the cysD gene of Escherichia coli K-12. Gene 87: 127–131. [DOI] [PubMed] [Google Scholar]
- Markus JA, Lambert CC. (1983). Urea and ammonia excretion by solitary ascidians. J Exp Mar Bio Ecol 66: 1–10. [Google Scholar]
- Maruyama T, Hirose E, Ishikura M. (2003). Ultraviolet-light-absorbing tunic cells in didemnid ascidians hosting a symbiotic photo-oxygenic prokaryote. Prochloron Biol Bull 204: 109–113. [DOI] [PubMed] [Google Scholar]
- Maurino VG, Peterhansel C. (2010). Photorespiration: current status and approaches for metabolic engineering. Curr Opin Plant Biol 13: 248–255. [DOI] [PubMed] [Google Scholar]
- Melby JO, Nard NJ, Mitchell DA. (2011). Thiazole/oxazole-modified microcins: complex natural products from ribosomal templates. Curr Opin Chem Biol 15: 369–378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melis A. (1999). Photosystem-II damage and repair cycle in chloroplasts: what modulates the rate of photodamage in vivo? Trends Plant Sci 4: 130–135. [DOI] [PubMed] [Google Scholar]
- Miller S, Augustine S, Olson T, Le, Blankenship R, Selker J, Wood M. (2005). Discovery of a free-living chlorophyll d-producing cyanobacterium with a hybrid proteobacterial/cyanobacterial small-subunit rRNA gene. Proc Natl Acad Sci USA 102: 850–855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J, Bowler C. (2012). Iron utilization in marine cyanobacteria and eukaryotic algae. Front Microbiol 3: 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan C, Fischer M, Thomas GH. (2011). Tripartite ATP-independent periplasmic (TRAP) transporters in bacteria and archaea. FEMS Microbiol Rev 35: 68–86. [DOI] [PubMed] [Google Scholar]
- Nielsen DA, Pernice M, Schliep M, Sablok G, Jeffries TC, Kühl M et al. (2015). Microenvironment and phylogenetic diversity of Prochloron inhabiting the surface of crustose didemnid ascidians. Environ Microbiol 17: 4121–4132. [DOI] [PubMed] [Google Scholar]
- Nishino A, Okamura Y, Piscopo S, Brown ER. (2010). A glycine receptor is involved in the organization of swimming movements in an invertebrate chordate. BMC Neurosci 11: 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paerl HW. (1984). N2 fixation (nitrogenase activity) attributable to a specific Prochloron (Prochlorophyta)-ascidian association in Palau, Micronesia. Mar Biol 81: 251–254. [Google Scholar]
- Palumbo RN, Wang C. (2006). Bacterial invasin: structure, function, and implication for targeted oral gene delivery. Curr Drug Deliv 3: 47–53. [DOI] [PubMed] [Google Scholar]
- Pardy RL, Lewin RA. (1981). Colonial ascidians with Prochlorophyte symbionts: evidence for translocation of metabolites from alga to host. Bull Mar Sci 31: 817–823. [Google Scholar]
- Parry D. (1985). Nitrogen assimilation in the symbiotic marine algae Prochloron spp. Mar Biol 222: 219–222. [Google Scholar]
- Patterson GML, Withers NW, Url S. (1982). Laboratory cultivation of Prochloron, a tryptophan auxotroph. Science 217: 1034–1035. [DOI] [PubMed] [Google Scholar]
- Patzer SI, Hantke K. (1998). The ZnuABC high-affinity zinc uptake system and its regulator Zur in Escherichia coli. Mol Microbiol 28: 1199–1210. [DOI] [PubMed] [Google Scholar]
- Pfreundt U, Stal LJ, Voß B, Hess WR. (2012). Dinitrogen fixation in a unicellular chlorophyll d-containing cyanobacterium. ISME J 2: 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saschenbrecker S, Bracher A, Rao KV, Rao BV, Hartl FU, Hayer-Hartl M. (2007). Structure and function of RbcX, an assembly chaperone for hexadecameric rubisco. Cell 129: 1189–1200. [DOI] [PubMed] [Google Scholar]
- Sauer J, Dirmeier U, Forchhammer K. (2000). The Synechococcus strain PCC 7942 glnN product (Glutamine Synthetase III) helps recovery from prolonged nitrogen chlorosis. J Bacteriol 182: 5615–5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt E, Nelson J, Rasko D, Sudek S, Eisen J, Haygood M et al. (2005). Patellamide A and C biosynthesis by a microcin-like pathway in Prochloron didemni, the cyanobacterial symbiont of Lissoclinum patella. Proc Natl Acad Sci USA 102: 7315–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon C, Collier J, Berg G, Glibert P. (2010). Role of urea in microbial metabolism in aquatic systems: a biochemical and molecular review. Aquat Microb Ecol 59: 67–88. [Google Scholar]
- Spreitzer RJ, Salvucci ME. (2002). Rubisco: structure, regulatory interactions, and possibilities for a better enzyme. Annu Rev Plant Biol 53: 449–475. [DOI] [PubMed] [Google Scholar]
- Steunou A-S, Jensen SI, Brecht E, Becraft ED, Bateson MM, Kilian O et al. (2008). Regulation of nif gene expression and the energetics of N2 fixation over the diel cycle in a hot spring microbial mat. ISME J 2: 364–378. [DOI] [PubMed] [Google Scholar]
- Stirnberg M, Fulda S, Huckauf J, Hagemann M, Krämer R, Marin K. (2007). A membrane-bound FtsH protease is involved in osmoregulation in Synechocystis sp. PCC 6803 : the compatible solute synthesizing enzyme GgpS is one of the targets for proteolysis. Mol Microbiol 63: 86–102. [DOI] [PubMed] [Google Scholar]
- Swingley W, Chen M, Cheung P, Conrad A, Dejesa L, Hao J et al. (2008). Niche adaptation and genome expansion in the chlorophyll d-producing cyanobacterium Acaryochloris marina. Proc Natl Acad Sci USA 105: 2005–2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Török Z, Horváth I, Goloubinoff P, Kovács E, Glatz A, Balogh G et al. (1997). Evidence for a lipochaperonin: association of active protein-folding GroESL oligomers with lipids can stabilize membranes under heat shock conditions. Proc Natl Acad Sci USA 94: 2192–2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsagkogeorga G, Turon X, Hopcroft RR, Tilak K, Feldstein T, Shenkar N et al. (2009). An updated 18S rRNA phylogeny of tunicates based on mixture and secondary structure models. BMC Evol Biol 9: 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki T, Yamaguchi N, Isago Y, Tanahashi H. (2015). Vanadium accumulation in ascidians: a system overview. Coord Chem Rev 301–302: 301–308. [Google Scholar]
- Vrljic M, Sahm H, Eggeling L. (1996). A new type of transporter with a new type of cellular function: L-lysine export from Corynebacterium glutamicum. Mol Microbiol 22: 815–826. [DOI] [PubMed] [Google Scholar]
- Yoneda A, Wittmann BJ, King JD, Blankenship RE, Dantas G. (2016). Transcriptomic analysis illuminates genes involved in chlorophyll synthesis after nitrogen starvation in Acaryochloris sp. CCMEE 5410. Photosynth Res 129: 171–182. [DOI] [PubMed] [Google Scholar]
- Zumft WG. (1997). Cell biology and molecular basis of denitrification. Microbiol Mol Biol Rev 61: 533–616. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.