Abstract
The diet provides carbohydrates that are non-digestible in the upper gut and are major carbon and energy sources for the microbial community in the lower intestine, supporting a complex metabolic network. Fermentation produces the short-chain fatty acids (SCFAs) acetate, propionate and butyrate, which have health-promoting effects for the human host. Here we investigated microbial community changes and SCFA production during in vitro batch incubations of 15 different non-digestible carbohydrates, at two initial pH values with faecal microbiota from three different human donors. To investigate temporal stability and reproducibility, a further experiment was performed 1 year later with four of the carbohydrates. The lower pH (5.5) led to higher butyrate and the higher pH (6.5) to more propionate production. The strongest propionigenic effect was found with rhamnose, followed by galactomannans, whereas fructans and several α- and β-glucans led to higher butyrate production. 16S ribosomal RNA gene-based quantitative PCR analysis of 22 different microbial groups together with 454 sequencing revealed significant stimulation of specific bacteria in response to particular carbohydrates. Some changes were ascribed to metabolite cross-feeding, for example, utilisation by Eubacterium hallii of 1,2-propanediol produced from fermentation of rhamnose by Blautia spp. Despite marked inter-individual differences in microbiota composition, SCFA production was surprisingly reproducible for different carbohydrates, indicating a level of functional redundancy. Interestingly, butyrate formation was influenced not only by the overall % butyrate-producing bacteria in the community but also by the initial pH, consistent with a pH-dependent shift in the stoichiometry of butyrate production.
Introduction
The relationship between the gut microbiota and host health is well established. The highest concentration and diversity of gut microbes is found in the colon, which acts as a fermentor system for dietary compounds that escape the digestive system of the host. Quantitatively, non-digestible dietary carbohydrates (NDCs) are the main energy sources for bacterial growth in the colon. It is estimated that between 20 and 60 g of NDCs, including plant cell wall polysaccharides, resistant starches (RSs), oligosaccharides and sugar alcohols, escape the digestive enzymatic breakdown and reach the human colon each day (Cummings and Macfarlane, 1991). Over the last years, research has established that gut bacteria possess an enormous variety of carbohydrate-degrading enzyme activities, which allow them to access NDCs (Flint et al., 2012). Microbial fermentation of NDCs mainly leads to the production of the short-chain fatty acids (SCFAs) acetate, propionate and butyrate, and of lactate, succinate, ethanol, methane, carbon dioxide and hydrogen (Cummings and Macfarlane, 1991). SCFAs are of particular interest for maintaining host health as they are known not only to contribute directly to energy metabolism, but also have positive effects on the host’s physiology. Butyrate is mainly metabolised by colonic cells (Hamer et al., 2008), whereas acetate and propionate are absorbed and metabolised by the liver and peripheral organs (Den Besten et al., 2013). Besides serving as an energy source, SCFA are associated with a number of health benefits for the host. Whereas butyrate and propionate have been shown to modulate cell differentiation and to exert anti-carcinogenic and anti-inflammatory effects (Hamer et al., 2008; Louis et al., 2014), acetate and propionate are of interest because of their potential to enhance satiety and suppress appetite either through receptor-mediated or other central mechanisms (Frost et al., 2014; Arora et al., 2011).
The intake of NDCs can have direct (primary) and secondary effects on the microbial community in the large intestine, and therefore on the host’s physiology. Particular NDCs can lead to the stimulation of specialised groups of microorganisms that possess the carbohydrate active enzymes necessary for their utilisation (Flint et al., 2012). Through cross-feeding, NDC breakdown intermediates or fermentation products from primary degraders can serve as substrates for secondary degraders, which are not directly capable of degrading a certain carbohydrate. This has been demonstrated in vitro (Belenguer et al., 2006; Rogowski et al., 2015) and reflects the complex nature of the intestinal ecosystem. The decrease of pH in the colon because of the production of SCFAs can also lead to selective effects on the microbial community as has been demonstrated in vitro (Walker et al., 2005; Duncan et al., 2009).
The human gut microbiota is composed of several phyla, with the Firmicutes and Bacteroidetes being the most abundant. Firmicutes not only include the major butyrate-producing species (Louis et al., 2010), but also include propionate producers and acetogens (Louis et al., 2014; Reichardt et al., 2014). Bacteroidetes possess genes encoding for the succinate pathway and therefore represent the main propionate producers in the gut (Reichardt et al., 2014). Many Bacteroides species are able to degrade a wide range of soluble plant cell wall polysaccharides (Flint et al., 2012; Martens et al., 2014). Firmicutes, on the other hand, tend to have fewer genes involved in carbohydrate breakdown (Flint et al., 2012), but specific members appear to have key roles in insoluble polysaccharide degradation (Ze et al., 2013). For example, Ruminococcus bromii is of key importance for the degradation of RS (Ze et al., 2015).
It is important to obtain a good understanding of how different NDCs are degraded and how this affects the gut microbiota and its fermentation products in order to reach conclusions on their effects upon the host’s health. This study investigated the degradation of 15 different NDCs by human faecal bacteria during in vitro fermentations. They included α- and β-glucans, pectins, galactomannans, arabinoxylan and fructans to achieve a good representation of different dietary NDCs. Fermentations were run at two different initial pH values to simulate proximal and distal colon conditions. The aim was to gain a comprehensive overview of the microbial changes and SCFA production in a complex community of human faecal microbiota in response to NDC breakdown.
Materials and methods
In vitro fermentations
Anaerobic in vitro incubations were carried out in a total volume of 10 ml in triplicate in Hungate tubes sealed with butyl rubber stoppers and screw caps (Bellco Glass, Shrewsbury, UK). The medium (details provided in Supplementary Methods) contained minerals, bile salts, volatile fatty acids, vitamins and 0.2% (wt/vol) of the test NDC. Cysteine was added to the medium following boiling and dispensed into Hungate tubes while they were flushed with CO2. The vitamin solution and the NDCs were added from stock solutions after autoclaving of the medium, directly before inoculation with the faecal suspension. NDC stock solutions were prepared anaerobically by flushing with CO2 at 1% in water and boiled for 1 min. Fifteen different NDCs (Table 1) were used in fermentation 1 and four in fermentation 2 to assess reproducibility. The supplier of one of the NDCs used in fermentation 2 was changed (Table 1), as this study formed part of a larger project that investigated carbohydrate effects in vivo (to be reported elsewhere). Ethical approval for the study was granted by the Rowett Institute Ethical review panel (number 09/005).
Table 1. NDCs used for anaerobic in vitro incubations with human faecal samples in fermentation (f) 1 and 2 and their suppliers.
| Class | NDC | Commercial name and supplier | f1 | f2 |
|---|---|---|---|---|
| α-Glucans | Pyrodextrin | Fibersol-2; gifted by Matsutani, Itami-City, Japan | x | |
| Pullulan | Megazyme, Bray, Ireland (cat no. P-PULLN) | x | ||
| Resistant starch type II | Hylon VII, National Starch and Chemical Comp., Bridgewater, USA | x | ||
| Resistant starch type III | Novelose330, National Starch and Chemical Comp., Bridgewater, USA | x | ||
| β-Glucans | β-Glucan from barley | Megazyme, Bray, Ireland (cat no. P-BGBL) | x | |
| β-Glucan from barley | Glucagel, PolyCell Technologies, Crookston, USA | x | ||
| Laminarin | Sigma Aldrich, UK (cat no. L9634) | x | ||
| Deoxy-hexose | Rhamnose | Sigma Aldrich, UK (cat no. W373011) | x | x |
| Pectins | Rhamnogalacturonan from potato | Megazyme, Bray, Ireland (cat no. P-RHAM1) | x | |
| Apple pectin | Sigma Aldrich, UK (cat no. 76282) | x | x | |
| Galactomannans | Carob galactomannan | Megazyme, Bray, Ireland (cat no. P-GALML) | x | |
| Guar galactomannan | Megazyme, Bray, Ireland (cat no. P-GGMMV) | x | ||
| Hemicellulose | Arabinoxylan | Megazyme, Bray, Ireland (cat no. P-WAXYL) | x | |
| Inulin-type fructans | Oligofructose, DP=2–8 | Orafti P95, gifted by Beneo, Tienen, Belgium | x | |
| Medium-chain inulin, average DP⩾10 | Orafti GR, Beneo, Tienen, Belgium | x | ||
| Long-chain inulin, average DP⩾23 | Orafti HP, gifted by Beneo, Tienen, Belgium | x | x |
Abbreviations: DP, degree of polymerisation; NDC, non-digestible dietary carbohydrate.
Fresh faecal samples were obtained from four different donors (fermentation 1, donors 1, 2 and 3; fermentation 2, approximately 12 months later, donors 2b, 3b and 4) with no history of gastrointestinal disorders or antibiotic treatment for at least 3 months before the study. Faecal samples were processed within 2 h after defecation. Eight ml of pre-reduced phosphate-buffered saline were added to 2 g of faecal sample and then homogenised in a Dispomix Drive (Medic Tools, Lussiwag, Switzerland) and 0.5 ml of the homogenised faecal suspension was used as an inoculum for the fermentation tubes (final faecal concentration: 1%). Incubations for each NDC were carried out in triplicate at pH 5.5, and 6.5, respectively, on a rotator (Stuart SB3, Bibby Scientific, Stone, UK) at 25 r.p.m. for 24 h at 37 °C. A no-NDC control was run in triplicate with every fermentation experiment. At 0, 6 and 24 h, an aliquot of 2 ml was taken from the fermentation vessels and centrifuged at 10 000 × g for 10 min at 4 °C. The supernatant was stored at −20 °C for analysis of SCFA. The cell pellet was resuspended in 800 μl of sodium phosphate buffer and 122 μl of MT buffer, transferred to a Lysing Matrix E tube (all part of the FastDNA spin kit for soil, MP Biomedicals, Illkirch, France) and stored at −70 °C until DNA extraction. Six-hour samples were processed if the 24-h sample was not available, as growth had taken place by then. Only primary data of those samples were included (Supplementary Tables S1a and S2b, c, shown in grey font), and they were excluded for any statistical analyses.
DNA extractions
DNA from the faecal inoculates and the cell pellets from the fermentation experiments (resuspended in buffer and stored as described above) was extracted using the FastDNA spin kit for soil (MP Biomedicals). For the DNA extraction of the faecal inoculates, an aliquot of 500 μl of the homogenised faecal suspension was transferred to a Lysing Matrix E tube and 300 μl of sodium phosphate buffer and 122 μl of MT buffer was added. The samples were stored at −70 °C until DNA extraction.
SCFA analysis
SCFA concentrations were measured in culture supernatants (0.5 ml) using gas chromatography as described previously (Richardson et al., 1989). After derivatisation, 1 μl of sample was analysed using a Hewlett-Packard (Palo Alto, CA, USA) gas chromatograph fitted with a fused silica capillary column with helium as a carrier gas. The SCFA concentrations were calculated from the relative response factor with respect to the internal standard two-ethylbutyrate.
Quantitative PCR
The pooled triplicate DNA samples from the in vitro fermentations 1 and 2 were analysed by quantitative real-time PCR as described previously (Fuller et al., 2007; Ramirez-Farias et al., 2009) with the following modifications. Reactions were performed in duplicate with iTaq Universal SYBR Green Supermix (Bio-Rad, Hemel Hempstead, UK) in a total volume of 10 μl with primers at 500 nM and 5 ng of DNA in optical-grade 384-well plates sealed with optical sealing tape in the presence of 1 μg ml–1 herring sperm DNA (Promega, Madison, WI, USA). Amplification was performed with a CFX384 Real-Time System (Bio-Rad, Hemel Hempstead, UK) with the following protocol: 1 cycle of 95 °C for 3 min, 40 cycles of 95 °C for 5 s and annealing temperature as per Supplementary Table S3 for 30 s, 1 cycle of 95 °C for 10 s and a stepwise increase of the temperature from 65 °C to 95 °C (at 5 s per 0.5 °C) to obtain melt curve data. Primers used for the quantification of the specific bacterial groups are given in Supplementary Table S3. Standard curves consisted of dilution series of amplified bacterial 16S ribosomal RNA (rRNA) genes from reference strains. The abundance of 16S rRNA genes was determined from standard curves and bacterial groups were either expressed as a percentage of total bacteria determined by universal primers or as 16S rRNA gene copies per ml culture. The detection limit was determined with negative controls containing only herring sperm DNA.
454 Sequencing
Amplicon sequencing of the V1–V3 region of the 16S rRNA genes was performed on GS FLX 454 platform by the Centre of Genomic Research of the University of Liverpool and Bioinformatics were conducted in-house using Mothur v. 1.34.4. software platform (Schloss et al., 2009) on the University of Aberdeen’s HPC cluster (Maxwell). Full details are given in Supplementary Methods. Reads per sample varied from 426 to 82 791 (average 9069.1±11 044.5). Good’s coverage was over 95% for all but one sample (Supplementary Table S2b) and rarefaction and collectors curves (Supplementary Figure S1) showed that a good coverage was achieved for all samples. Exclusion of samples with Good’s coverage of <97% resulted in very similar results (Table 2) and subsampling to 426 sequence reads per sample revealed that the relative abundance of the top 50 operational taxonomic units (OTUs; representing >88% of all reads) was very similar to the full data set (Supplementary Figure S2). It was therefore decided to work with the full data set to preserve as much of the data as possible. OTUs were generated at ⩾97% sequence identity, which resulted in 1552 OTUs (Supplementary Table S2c), and the relative abundance was calculated. OTUs with an overall abundance of >100 reads (201 OTUs, 95.6–99.8% of sequence reads per sample) were analysed using the BLAST algorithm (Altschul et al., 1990) and compared with the taxonomy from the SILVA database (Quast et al., 2013). OTUs were then assigned to their corresponding quantitative PCR (qPCR) assays if possible as detailed in Supplementary Methods and Supplementary Table S2a.
Table 2. OTUs from 454 sequencing analysis of fermentation 1 exhibiting a significant increase on specific NDCs and pH values (P<0.001, see Supplementary Table S2a).
| OTU no. | Closest relative bacterial species (BLAST) | Identity |
Non-digestible carbohydrate
|
|
|---|---|---|---|---|
| pH 5.5 | pH 6.5 | |||
| Otu0002 | Faecalibacterium prausnitzii | 99% | Apple pectina, b | Apple pectina, b |
| Carob galactomannana, b | ||||
| Otu0003 | Clostridium spiroforme | 93% | Laminarina, b | |
| Otu0005 | Bacteroides uniformis | 100% | Pyrodextrina, b | |
| Laminarina, b | ||||
| Guar galactomannana, b | ||||
| Otu0006 | Blautia faecis | 99% | Rhamnosea, b | Rhamnosea, b |
| Otu0010 | Fusicatenibacter saccharivorans | 99% | Laminarina | |
| Carob galactomannana, b | ||||
| Otu0013 | Subdoligranulum variabile | 99% | Arabinoxylana, b | |
| Otu0017 | Oscillibacter ruminantium | 96% | Rhamnogalacturonana | |
| Otu0018 | Dorea longicatena | 99% | Pullulan | |
| Guar galactomannan | ||||
| Medium-chain inulina, b | ||||
| Otu0024 | Lactobacillus rogosae | 96% | Rhamnogalacturonana | |
| Guar galactomannan | ||||
| Otu0026 | Parabacteroides merdae | 98% | Guar galactomannana | |
| Otu0027 | Bacteroides thetaiotaomicron | 99% | Barley β-glucana | |
| Otu0031 | Unclassified | Carob galactomannana | ||
| Guar galactomannan | Guar galactomannana | |||
| Otu0037 | Unclassified | Rhamnogalacturonana | ||
| Otu0041 | Flavonifractor plautii | 96% | Pullulana | Rhamnogalacturonana |
| Otu0043 | Eubacterium ventriosum | 98% | Pyrodextrin | |
| Pullulana | ||||
| Laminarina | ||||
| Otu0045 | Bacteroides ovatus | 100% | Barley β-glucana | |
| Otu0055 | Clostridium bolteae | 97% | Rhamnogalacturonan | |
Abbreviations: NDC, non-digestible dietary carbohydrate; OTU, operational taxonomic unit.
Also significant after removal of samples with <97% Good’s coverage.
Also significant after subsampling to lowest coverage (426 sequence reads).
Polysaccharide analysis
All analyses were performed in duplicate. To evaluate authenticity and purity of the NDCs used in this study, their monosaccharide composition was analysed by high performance anion exchange chromatography with pulsed amperometric detection after acid hydrolysis as described previously (Wefers and Bunzel, 2015). Details of the hydrolysis conditions for the different NDCs are given in Supplementary Methods.
Statistical analysis
Data from each fermentation study were analysed by analysis of variance with random effect for donor and fixed effects for NDC, pH and their interaction, followed by post-hoc t-test. Principal components analysis and partial least squares were used to investigate associations between bacteria and SCFA production. Associations of interest were quantified by random effects regression with donor as random effect and with pH, microbial abundance and their interaction as fixed effects. The agreement between the 454 sequencing and qPCR methods between corresponding bacterial groups (expressed as percentage of total bacteria) was investigated by linear regression. Full details are given in Supplementary Methods.
For the regression analyses, P<0.05 was regarded significant. For the analysis of variance analyses and subsequent post-hoc comparisons, however, to reduce the reporting of false-positives because of the large number of comparisons, an effect was considered significant only when P<0.001.
Results
SCFA from in vitro batch culture incubations
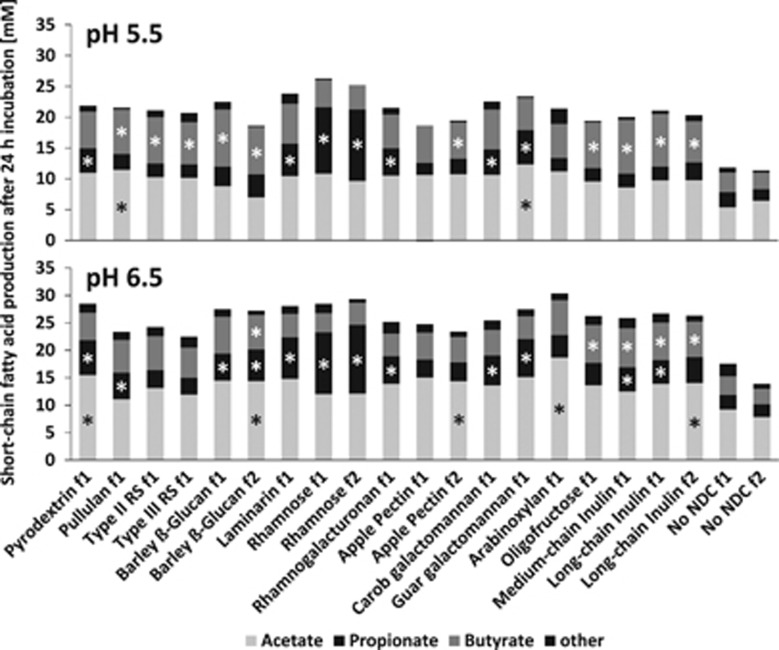
Anaerobic incubations were conducted with 15 different NDCs as energy sources (Table 1; 0.2% w/v) in the presence of human faecal slurries from three healthy donors (fermentation 1) at initial pH values of 5.5 and 6.5. For four NDCs, another fermentation was carried out approximately 1 year later to assess reproducibility of the responses (fermentation 2, two of the three donors were the same as in fermentation 1). Profiles of net SCFA production after 24 h of incubation differed between NDCs, especially for those from different NDC classes. Analysis of the chemical composition of NDCs used (shown in Supplementary Table S4) showed that their composition was consistent with their description. For NDCs used in both fermentations, the SCFA profiles were very reproducible (Figure 1).
Figure 1.
Net SCFA production after 24-h incubation of human faecal samples with different NDCs. Average of three donors in two fermentations (f1 and f2) at pH 5.5 and 6.5 (t=24 h minus t=0, standard error of the difference and percentages given in Supplementary Table S5). Faecal donors were donors 1, 2 and 3 in f1; and 2, 3 and 4 in f2. Analysed by analysis of variance (ANOVA) with donor as random effect and with NDC, pH and their interaction as fixed effects. NDCs that differ (P<0.001) from the no-NDC control for each of the two pH levels are indicated by *.
After 24 h, the cumulative amount of fermentation acids produced was significantly higher at pH 6.5 compared with pH 5.5 in both fermentations (Supplementary Table S5, P<0.001). The main SCFAs produced were acetate, propionate and butyrate, with pH 6.5 leading to higher acetate and propionate and pH 5.5 to higher butyrate formation for most NDCs (Figure 1, P<0.001). The branched-chain fatty acids iso-butyrate and iso-valerate, as well as formate, valerate and lactate were only detected in minor amounts (<1.13 mM each), whereas succinate was not detected in any of the incubations.
When individual NDCs were compared with the no-NDC control, starch, β-glucan and inulin/oligofructose gave rise to significantly (P<0.001) increased butyrate concentrations when the initial pH was 5.5. At an initial pH of 6.5, this butyrogenic effect was seen for inulin/oligofructose but not for the starches (Figure 1). Pyrodextrin, laminarin, rhamnose, rhamnogalacturonan and the two galactomannans all gave rise to increased propionate at initial pH 5.5. These same NDCs increased propionate when the initial pH was 6.5, but in addition pullulan, β-glucan and some of the inulin-type NDCs also promoted propionate significantly at the higher pH (all P<0.001). The NDC that stands out with regard to propionate production, however, is rhamnose. The absolute amount produced was between 1.5- and 5.5-fold higher than in the presence of the other NDCs, and the molar ratio (40–42% of total SCFA) was similar to the percentage of acetate in both fermentations and independent of the pH (Figure 1, Supplementary Table S5).
Microbial population changes detected by qPCR
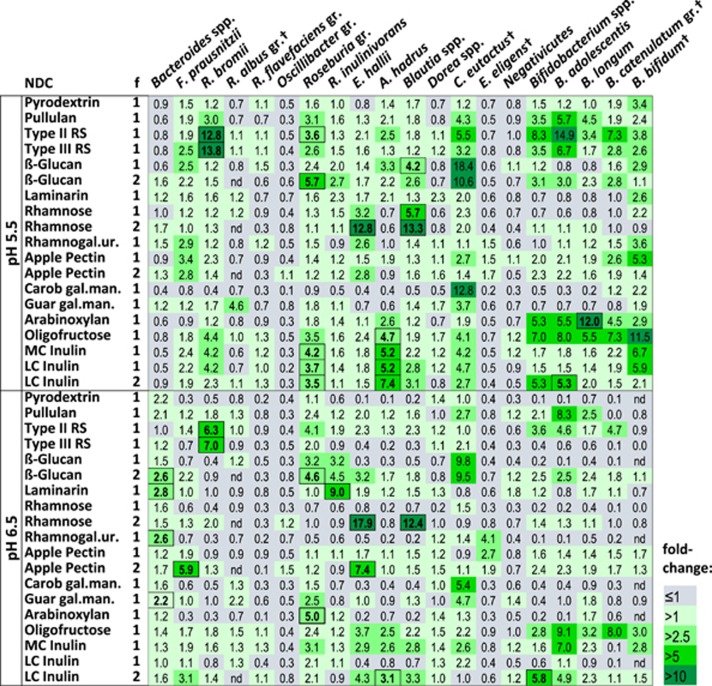
The microbial composition of the in vitro incubations from both fermentation experiments was analysed using qPCR against 21 different bacterial species and groups, in addition to total bacteria and methanogenic Archaea (Supplementary Table S3). Analysis of the inocula revealed high inter-individual variability of bacterial composition, as well as intra-individual differences for the donors used in both fermentations (donors 2 and 3). In addition to quantitative differences, certain microbial groups were only found in some faecal samples (some ruminococci, Coprococcus eutactus, Eubacterium eligens, certain bifidobacteria and methanogenic Archaea), and none of the donors had detectable levels of Prevotella spp. (Supplementary Table S1a). After 24 h of incubation, the total amount of bacteria had increased in all incubations, including the no-NDC control (average fold change of total 16S rRNA gene copies 8.0±2.5). In order to investigate bacterial changes specific to the different NDCs, the data were expressed as the ratio of the absolute 16S rRNA gene copies per ml culture between each carbohydrate incubation and the no-NDC control after 24 h of incubation (Figure 2,Supplementary Table S1b). For several bacterial groups and NDCs, responses were similar in the different donors. Thus, R. bromii significantly (P<0.001) increased on both types of RS at both pH values, with highest levels reached at pH 5.5 (Figure 2). Bifidobacteria and several other Firmicutes, especially the Roseburia group, also increased on RSs and pullulan, in particular at the lower pH value, but due to inter-individual variability this mostly did not reach significance. Barley β-glucan resulted in significant increases in the Roseburia group at both pH values in fermentation 2, whereas R. inulinivorans showed a significant increase on laminarin only at pH 6.5 (Figure 2, all P<0.001). At the lower pH value, Blautia spp. increased significantly on barley β-glucan in fermentation 1, whereas at the higher pH value, Bacteroides spp. increased on two of the three β-glucan-type incubations (Figure 2). Coprococcus eutactus, which was only detected in one donor, increased markedly on barley β-glucan in both fermentations at both pH tested, but not with laminarin (Figure 2).
Figure 2.
Increase of bacterial groups analysed by qPCR after 24-h incubation of human faecal samples with different NDCs. Average of three donors in two fermentations (f1 and f2) at pH 5.5 and pH 6.5 in relation to the increases with no NDC (given as relative fold change). Faecal donors were donors 1, 2 and 3 in f1 (for pH 5.5 only donors 1 and 3 were included, as no-NDC control was not available at 24 h for donor 2; data for all donors are shown in Supplementary Table S1) and 2, 3 and 4 in f2. Bacterial 16S rRNA gene copies ml–1 culture were expressed relative to the no-NDC control for each donor and pH. The log-transformed ratios were analysed by analysis of variance (ANOVA) with donor as random effect and with NDC, pH and their interaction as fixed effects. Presented here are the back-transformed mean log ratios. Test NDCs that differ (P<0.001) from the no-NDC control for each of the two pH levels are shown in bold and with a border. Rhamnogal.ur., rhamnogalacturonan; gal.man., galactomannan; MC, medium-chain; LC, long-chain; gr., group; †, bacterial group not detected in all donors.
Rhamnose led to a significant (P<0.001) increase of both Eubacterium hallii and Blautia spp. at both pH values in fermentation 2, whereas this response was weaker and only observed for Blautia spp. at pH 5.5 in fermentation 1 (Figure 2). NDCs of the pectin class resulted in the highest fold changes relative to the no-NDC control for F. prausnitzii, E. hallii and B. bifidum at pH 5.5, but this did not reach significance. At pH 6.5, F. prausnitzii and E. hallii showed a significant response on apple pectin during fermentation 2, whereas Bacteroides spp. increased significantly (P<0.001) on rhamnogalacturonan. Eubacterium eligens, which was not detected in all donors (Supplementary Table S1a), competed poorly on most of the NDCs tested (fold changes relative to no-NDC control <1; Figure 2), but showed a numerical increase for the pectin-type NDCs (Figure 2), especially at the higher pH value. For the two galactomannans, C. eutactus (present in only one donor, Supplementary Table S1a) increased relative to no-NDC control, and Bacteroides spp. had significantly (P<0.001) higher levels on guar galactomannan at pH 6.5. Bifidobacteria showed the strongest response to arabinoxylan, at pH 5.5, which reached significance for B. longum, whereas Roseburia spp. increased significantly on arabinoxylan at pH 6.5 only (Figure 2).
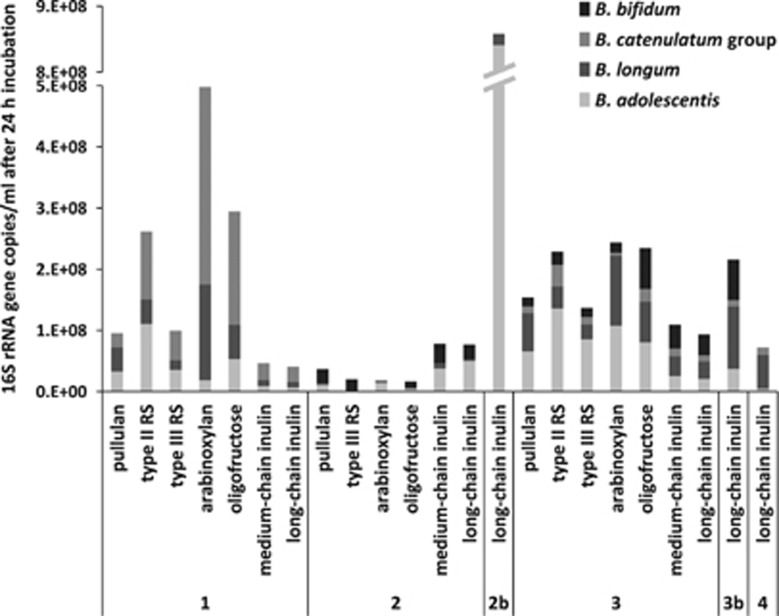
The inulin-type fructans tested resulted in significant (P<0.001) increases of the Roseburia group and A. hadrus at pH 5.5. Increases were also seen for several other groups, including bifidobacteria, R. bromii (pH 5.5 only) and Blautia spp., but were mostly not significant (P>0.001, Figure 2,Supplementary Table S1b). Interestingly, Bifidobacterium spp. showed mostly higher increases on oligofructose than on medium- or long-chain inulin (Figure 2). Individual Bifidobacterium species were subject to large inter-individual differences (Figure 3). For example, B. adolescentis and B. longum, detected in all donors, responded with higher increases on oligofructose in donors 1 and 3, whereas for donor 2 stronger responses were seen with the longer chain inulin-type fructans. B. adolescentis showed a much stronger stimulation in donor 2 compared with B. longum, regardless of the type of fructan, whereas B. longum achieved high levels of stimulation on various NDCs in the other donors. The B. catenulatum group responded strongly to several NDCs in donor 1, whereas B. bifidum showed the strongest response to fructan-type NDCs in donor 3 (Figure 3).
Figure 3.
Growth response of individual Bifidobacterium species on α-glucans, arabinoxylan and fructans. Data shown are from individual faecal incubations (fermentation 1 donors 1, 2 and 3; fermentation 2 donors 2b, 3b and 4) analysed by qPCR after 24-h incubation at pH 5.5. At 0 h, Bifidobacterium species levels in all donors ranged from 6.1 × 104 to 9.7 × 106 ml–1 (see Supplementary Table S1a).
Interestingly, the R. flavefaciens group, Oscillibacter group, Dorea spp. and Negativicutes group were not significantly stimulated by any of the NDCs tested (P>0.001, Figure 2). Methanogenic archaea did not exhibit big increases compared with no-NDC control for most incubations they were detected in (Supplementary Table S1). Regression analysis of SCFA and bacterial groups showed a significant (P<0.05) positive correlation of Bacteroides spp. and propionate production and of A. hadrus, F. prausnitzii and Roseburia group, respectively, and butyrate production (Supplementary Figure S3).
Relationship between microbiota composition and SCFA formation
In addition to qPCR, the influence of the different NDCs on the microbiota of fermentation 1 was analysed using 454 sequencing. Like qPCR, OTU analysis revealed large inter-individual differences in the inocula. In the sample of donor 1 and donor 3, we detected 116 and 114 different OTUs, respectively, with an abundance of >100 reads, whereas the inoculum of donor 2 contained 162. OTU 1 (Blautia obeum) was the most abundant OTU in donors 2 and 3 and the second most abundant in donor 1 after OTU 8 (A. hadrus) (Supplementary Table S2). Statistical analysis of all OTUs that were detected in at least two-thirds of all samples after 24 h of incubation revealed that 17 OTUs, covering a range of different Bacteroidetes and Firmicutes species, were significantly (P<0.001) increased compared with the inoculum on at least one NDC (Table 2).
In order to compare the qPCR results with sequencing results, the OTUs were assigned to groups that would be targeted by the qPCR primers used (Supplementary Figure S4, Supplementary Table S2a). This led to an assignment rate of 29.2–83.2% per sample (average 59.1%) of all sequences. The results in Supplementary Figure S4 show that the changes detected by 454 sequencing agree well with those detected by qPCR. This applies, for example, to the increase in Blautia spp. with rhamnose, in R. bromii with RS and in A. hadrus with fructans. Correlations were calculated between relative data from qPCR and the sum of OTUs assigned to the corresponding primer set. Significant (P<0.05) correlations were found for all OTU groups, which could be assigned to corresponding qPCR primers, except for the Oscillibacter group (Supplementary Figure S5). Weaker correlations likely reflect technical differences arising from either qPCR or sequencing methodology, or a limited understanding of the groups under study, which may affect the accuracy of assigning sequence OTUs to the corresponding qPCR group. Bifidobacteria were not included in this comparison as they are underestimated by 454 sequencing with the primers used here (Walker et al., 2015).
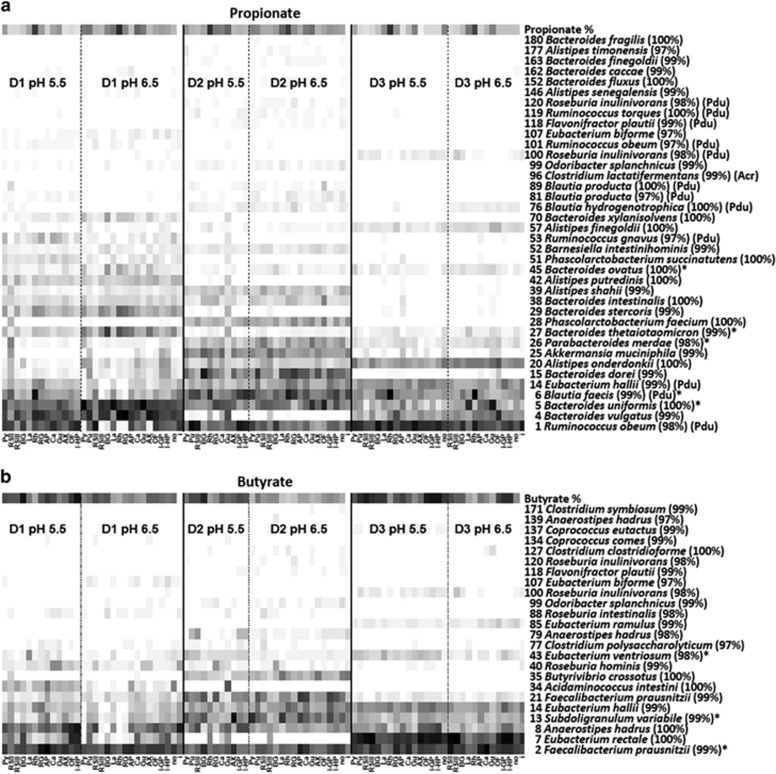
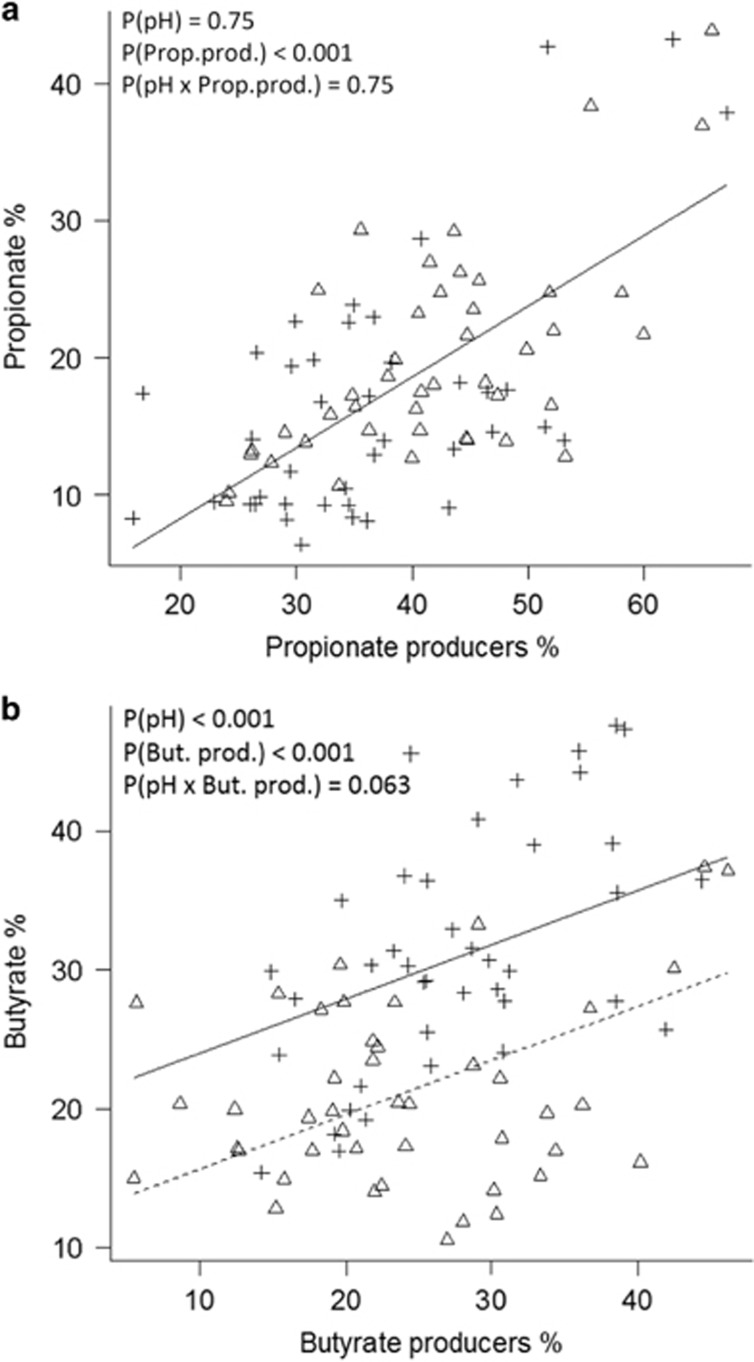
Heat map analysis of relative OTU abundance revealed high inter-individual variation (Supplementary Figure S6 most abundant OTUs). Propionate- and butyrate-producing status was assigned to all classified OTUs (Supplementary Table S2a; 39–87% of sequence data per sample assigned to fermentation product formation based on at least 97% sequence identity to known species) and heat maps for propionate- and butyrate-producing OTUs, respectively, were generated (Figure 4). This shows that different OTUs contribute to SCFA production in different donors. However, regression analyses of the sum of all propionate- or butyrate-producing bacteria (as percentage of total sequences) to percentage propionate or butyrate produced over 24 h of incubation showed a strong correlation (Figure 5, P<0.001 for both SCFAs). For propionate, the initial pH of the incubations had no effect on this relationship, but for butyrate a strong effect of initial pH was found (Figure 5, P<0.001). Partial least squares regression was carried out on all classified OTUs to reveal any associations with acetate, propionate or butyrate, which revealed some strong associations that mostly were individual-specific (Supplementary Table S2a). These may reflect not just a direct conversion of NDCs to SCFA, but could also include cross-feeding effects.
Figure 4.
Relative abundance of propionate- and butyrate-producing OTUs with at least 97% sequence identity to known bacterial species after 24- h incubation of human faecal samples from fermentation 1 with different NDCs (white–black: a, Propionate-producing OTUs 0–34% b, butyrate-producing OTUs 0–24%). Rationale for assignment of SCFA production capacity is given in Supplementary Table S2a. Relative production of the corresponding SCFA is given at the top of each heat map (white–black: a, Propionate % 0-43% b, butyrate % 0–47%). OTUs showing a significant (P<0.001) increase under certain conditions, compared with inoculum, are indicated by * (for details see Table 2). AP, apple pectin; AX, arabinoxylan; BG, β-glucan; Ca, carob galactomannan; Gu, guar galactomannan; i, inoculum; I-GP, medium-chain inulin; I-HP, long-chain inulin; La, laminarin; no, no-NDC; OF, oligofructose; Pu, pullulan; Py, pyrodextrin; RG, rhamnogalacturonan; Rh, rhamnose; RSII, type II RS; RSIII, type III RS. a: Acr, acrylate pathway; Pdu, propanediol pathway.
Figure 5.
Relationship between sum of all propionate- (a) or butyrate-producing (b) OTUs with at least 97% sequence identity to known bacterial species after 24 h of incubation of human faecal samples from fermentation 1, expressed as percentage of total sequences, and SCFA production. Relative propionate or butyrate production (percentage of total SCFA produced) was regressed on percentage of propionate or butyrate producers, using mixed effect models with donor as random effect and with fixed effects for pH, percentage producers, and their interaction. pH 5.5, crosses; pH 6.5, triangles. Lines correspond to the fit for each pH where a significant effect of pH was observed (b, solid line pH 5.5, dashed line pH 6.5). But.prod., butyrate production; Prop.prod., propionate production.
Relative qPCR and SCFA data obtained from NDCs that were examined in years 1 and 2 after 24 h of incubation were further analysed by principal component analysis. This revealed some clustering by donor, but the samples originating from the same donor in different years showed little overlap, showing a relatively large intra-individual variation (Supplementary Figure S7a). Rhamnose incubations in particular clustered separately and were associated with propionate production, Blautia spp. and E. hallii (Supplementary Figures S7b and d). Long-chain inulin also tended to result in a bigger difference to no-NDC control than the other NDCs examined (Supplementary Figure S7b). A separation by pH could be observed especially for the NDCs other than rhamnose, which was associated with butyrate formation at pH 5.5 and propionate formation at pH 6.5 (Supplementary Figures S7c and d).
Discussion
This study investigated the impact of 15 different NDCs upon microbiota composition in anaerobic batch cultures inoculated with human faecal samples. The in vitro batch culture system provided a fast and cost effective way to study the effects of an extensive set of NDCs on the microbial community from four donors. As the pH of batch cultures cannot be controlled precisely as acids are produced during incubation, we set the initial pH at two different values (5.5 and 6.5) to simulate prevailing conditions in the healthy proximal and distal colon. Using 0.2% carbohydrate, we anticipate that acid production during incubation will have reduced the pH further by 0.5–1 unit by 24 h so that growth will have occurred largely under mildly acidic conditions. When pH is controlled at 6.5 using a continuous flow fermentor system, we have shown that Bacteroides spp. outcompete Firmicutes and Actinobacteria within the human colonic microbiota for soluble carbohydrate substrates (Walker et al., 2005; Duncan et al., 2003; Chung et al., 2016). By contrast in the present study, this dominance of Bacteroides spp. was curtailed by the lower pH conditions, and probably also by the reduction in the peptide content of the medium (to 0.1% casitone and 0.1% yeast extract). This has helped to reveal the response of Firmicutes and Actinobacteria to different carbohydrates. Lowering of gut pH because of increased fermentation may also contribute to the reduced abundance of Bacteroides species often observed in human dietary trials with NDC (Martinez et al., 2013; Duncan et al., 2009).
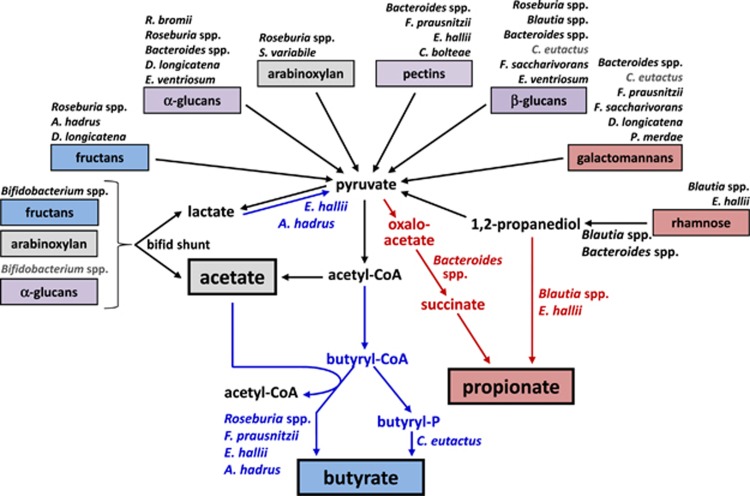
An overview of the major microbiota responses on the different carbohydrate classes and corresponding pathways for SCFA formation is provided in Figure 6. The NDC that promoted by far the highest SCFA proportion of propionate was rhamnose. This can be explained by the fact that rhamnose is fermented via the propanediol pathway in some anaerobic bacteria, yielding propionate and sometimes also propanol (Reichardt et al., 2014; Scott et al., 2006; Louis and Flint, 2017). The distribution of the propanediol pathway of propionate formation from deoxy sugars is, however, quite limited, being found so far in Roseburia inulinivorans and in Blautia spp. (Reichardt et al., 2014). This fits very well with the observed highly specific enrichment of Blautia spp. on rhamnose that was detected both by 16S rRNA-based qPCR and sequence analysis. In other bacteria that can utilise deoxy sugars, such as Bacteroides spp. (Rodionova et al., 2013) and Anaerostipes rhamnosivorans (Bui et al., 2014), propionate is not formed and 1,2-propanediol is an end product. E. hallii also showed a high qPCR ratio for rhamnose relative to the no-NDC control. Existing E. hallii strains are not known to grow on rhamnose (Holdeman and Moore, 1974), but its stimulation is likely to be indirect, because of cross-feeding of 1,2-propanediol formed from rhamnose by Bacteroides spp. and A. rhamnosivorans, as a recent study demonstrated the ability of E. hallii to metabolise 1,2-propanediol (Engels et al., 2016). E. hallii also has the ability to utilise lactate (Duncan et al., 2004), which is a major fermentation product of Blautia faecis (Park et al., 2013), the Blautia species that was most strongly stimulated by rhamnose in these experiments. The propanediol pathway may also contribute significantly to propionate formation from rhamnose residues on rhamnogalacturonan and pectin (25.3 and 4.8% of monosaccharide composition, Supplementary Table S4), but for the remaining polysaccharides it is expected that propionate will originate mainly via the succinate pathway found in the Bacteroidetes (Reichardt et al., 2014). Consistent with this, the level of propionate produced showed a significant correlation with the abundance of Bacteroides spp. based on qPCR results (Supplementary Figure S3). Based on sequencing data, relative propionate production correlated more strongly with the sum of all propionate producers (Figure 5) than with propionate producers that employ either the succinate or propanediol pathway (data not shown), confirming that both pathways contribute to propionate formation. The percentage of butyrate among SCFA was highest for fructans at both initial pHs, and for pullulan, RS type II and III and β-glucan at pH 5.5. This appeared to reflect the stimulation of known butyrate-producing species, in particular the Roseburia group, F. prausnitzii, A. hadrus, and C. eutactus, depending on the NDC.
Figure 6.
Overview of known major fermentation pathways for SCFA formation in human gut bacteria. NDC class colour is based on whether they mainly stimulated propionate (red) or butyrate (blue) production or both (purple) (see Figure 1). Responses of gut bacteria to different NDCs detected in this study are shown above each NDC class; significant responses detected by qPCR (see Figure 2, compared with no-NDC) and 454 sequencing (see Table 2, compared with inoculum, only OTUs with >97% identity to known species given) are shown in black, nonsignificant qPCR-based responses with a fold change >10 are shown in grey.
A number of responses to particular NDCs agreed well with previous reports from in vivo and in vitro studies. They include stimulation of R. bromii on RS (Abell et al., 2008; Martinez et al., 2010; Walker et al., 2011; Ze et al., 2013, 2015), of bifidobacteria (Ramirez-Farias et al., 2009; Scott et al., 2014; Selak et al., 2016; McLaughlin et al., 2015), butyrate-producing Roseburia/Eubacterium rectale group and A. hadrus (Louis et al., 2010; van den Abbeele et al., 2011; Scott et al., 2014) and Dorea longicatena (Taras et al., 2002) on inulin-type fructans, and of Bacteroides spp., F. prausnitzii and E. eligens on pectin-type NDCs (Salyers et al., 1977; Lopez-Siles et al., 2012; Chung et al., 2016). Arabinoxylan increased bifidobacteria at the lower pH, which reached significance for B. longum based on qPCR results. This is in agreement with another in vitro study that investigated pure culture growth of different Bifidobacterium species and found good growth on arabinoxylan only for strains belonging to B. longum (McLaughlin et al., 2015).
SCFA production was surprisingly reproducible for the different NDCs investigated here compared with the high microbiota variation between donors, which indicated that different OTUs contributed to NDC breakdown and SCFA formation in the different donors. For example, OTU 11 (closest relative R. bromii, 94% identity) responded strongly to RS in donor one, whereas it was low in donor 3 and OTU 9 (R. bromii, 99%) responded strongly to RS in this donor (Supplementary Figure S6). When looking specifically at propionate- or butyrate-producing bacteria, it becomes clear that they show a heterogeneous response to different carbohydrates (Figure 4), but their combined response correlates very strongly with the corresponding SCFA output (Figure 5), revealing functional redundancy in the microbiota. The activities of each microbiota member will be dependent not only on their genetic potential to degrade certain NDCs and produce certain SCFA, but also on their interaction with other microbes and their competitive fitness. This likely underlies the different response seen for some OTUs in different donors (for example, OTU 8, A. hadrus (100%) showing an increase on arabinoxylan in donor 1 and 3, but not donor 2). The poor response of R. inulinivorans on inulin and rhamnose likely also reflects its poor ability to compete effectively in the complete microbiota, despite the fact that it can grow on those NDCs in pure culture (Duncan et al., 2006; Reichardt et al., 2014; Scott et al., 2014). This agrees with a human intervention study, which also failed to see an increase in this species after inulin supplementation in subjects with high baseline levels of this species (Louis et al., 2010).
Our data also reveal that while the % butyrate among SCFA products was related to the proportion of butyrate-producing bacteria, the initial pH altered this relationship considerably (Figure 5). It is known that species such as F. prausnitzii and Roseburia spp. that use the butyryl-CoA:acetate CoA-transferase route for butyrate formation, exhibit a shift in fermentation stoichiometry in pure culture at lower pH (5.5) in favour of greater butyrate production and greater acetate consumption per mol of carbohydrate consumed (Kettle et al., 2015; Louis and Flint, 2017). The relationships seen in Figure 5 indicate that this shift in stoichiometry applies also to butyrate production by the mixed community, whereas propionate production was simply related to the % propionate-producing bacteria regardless of the initial pH.
In conclusion, the work presented here is one of few in vitro studies that compares the impact of a large variety of NDCs on the composition and metabolic activity of the human faecal microbiota. Some of the NDCs investigated here are currently classed as prebiotics, but this study does not reveal a clear distinction between those and NDCs currently classed as dietary fibre, in terms of a selective stimulation of specific bacteria. Prebiotic NDCs are considered to have consequences for health mainly through their impact upon the gut microbiota. These impacts can be ascribed to two types of mechanism. First, as shown here, prebiotics can promote the growth of a limited number of bacterial species, boosting their populations and their representation within the gut microbiota, although the pattern of stimulation can vary between microbiota from different individuals. Some of these bacteria may interact with the host’s immune system, but we can expect that inter-individual variability in the microbiota and the selective effects of different NDCs may result in wide variation in health consequences. Second, we know that the metabolites produced by the microbial community have important consequences for health. Here our results suggest, at least for SCFAs, that the consequences of NDC fermentation are likely to be more consistent between individuals. This is explained by the large number of gut anaerobes capable of producing the major SCFA, resulting in functional redundancy that tends to mask inter-individual variation at the species level. Thus, we have shown that, in spite of inter-individual differences in microbiota composition, SCFA profiles were very similar for each individual and for a given NDC. We should also note, however, that for metabolites whose production is limited to a smaller number of species, individual variability is likely to be correspondingly greater. In addition, if keystone species are absent, the capacity of the microbiota to ferment NDC can be greatly reduced, as shown for individuals lacking R. bromii on diets high in RS (Walker et al., 2011). There is currently much debate on the prebiotic concept, and the stipulation that they have to selectively stimulate certain microbes is increasingly challenged (Bindels et al., 2015; Louis et al., 2016; Steinert et al., 2016). The data presented here are in support of a more general definition with regard to the modulation of the gut microbiota in order to achieve a beneficial effect on the host.
Acknowledgments
The Rowett Institute and Biomathematics and Statistics Scotland receive financial support from the Scottish Government Rural and Environmental Sciences and Analytical Services. Nicole Reichardt was funded by a Scottish Government Strategic Partnership on Food and Drink Science. We would like to acknowledge the support of the Maxwell compute cluster funded by the University of Aberdeen. We would like to thank Donna Henderson for carrying out GC analysis and Alan Walker for help and advice with bioinformatic sequence analysis.
Footnotes
Supplementary Information accompanies this paper on The ISME Journal website (http://www.nature.com/ismej)
The authors declare no conflict of interest.
Supplementary Material
References
- Abell GCJ, Cooke CM, Bennett CN, Conlon MA, McOrist AL. (2008). Phylotypes related to Ruminococcus bromii are abundant in the large bowel of humans and increase in response to a diet high in resistant starch. FEMS Microbiol Ecol 66: 505–515. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. (1990). Basic local alignment search tool. J Mol Biol 215: 403–410. [DOI] [PubMed] [Google Scholar]
- Arora T, Sharma R, Frost G. (2011). Propionate. Anti-obesity and satiety enhancing factor? Appetite 56: 511–515. [DOI] [PubMed] [Google Scholar]
- Belenguer A, Duncan SH, Calder AG, Holtrop G, Louis P, Lobley GE et al. (2006). Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol 72: 3593–3599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindels LB, Delzenne NM, Cani PD, Walter J. (2015). Opinion: towards a more comprehensive concept for prebiotics. Nat Rev Gastroenterol Hepatol 12: 303–310. [DOI] [PubMed] [Google Scholar]
- Bui TPN, de Vos WM, Plugge CM. (2014). Anaerostipes rhamnosivorans sp. nov., a human intestinal, butyrate-forming bacterium. Int J Syst Evol Microbiol 64: 787–793. [DOI] [PubMed] [Google Scholar]
- Chung WSF, Walker AW, Louis P, Parkhill J, Vermeiren J, Bosscher D et al. (2016). Modulation of the human gut microbiota by dietary fibres occurs at the species level. BMC Biol 14: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JH, Macfarlane GT. (1991). The control and consequences of bacterial fermentation in the human colon. J Appl Bacteriol 70: 443–459. [DOI] [PubMed] [Google Scholar]
- Den Besten G, Van Eunen K, Groen AK, Venema K, Reijngoud DJ, Bakker BM. (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54: 2325–2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Thomson JM, Flint HJ. (2009). The role of pH in determining the species composition of the human colonic microbiota. Environ Microbiol 11: 2112–2122. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Aminov RI, Scott KP, Louis P, Stanton TB, Flint HJ. (2006). Proposal of Roseburia faecis sp. nov., Roseburia hominis sp. nov. and Roseburia inulinivorans sp. nov., based on isolates from human faeces. Int J Syst Evol Microbiol 56: 2437–2441. [DOI] [PubMed] [Google Scholar]
- Duncan SH, Louis P, Flint HJ. (2004). Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl Environ Microbiol 70: 5810–5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan SH, Scott KP, Ramsay AG, Harmsen HJM, Welling GW, Stewart CS et al. (2003). Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Appl Environ Microbiol 69: 1136–1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engels C, Ruscheweyh HJ, Beerenwinkel N, Lacroix C, Schwab C. (2016). The common gut microbe Eubacterium hallii also contributes to intestinal propionate formation. Front Microbiol 7: 713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. (2012). Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3: 289–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frost G, Sleeth ML, Sahuri-Arisoylu M, Lizarbe B, Cerdan S, Brody L et al. (2014). The short-chain fatty acid acetate reduces appetite via a central homeostatic mechanism. Nat Commun 5: 3611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuller Z, Louis P, Mihajlovski A, Rungapamestry V, Ratcliffe B, Duncan AJ. (2007). Influence of cabbage processing methods and prebiotic manipulation of colonic microflora on glucosinolate breakdown in man. Br J Nutr 98: 364–372. [DOI] [PubMed] [Google Scholar]
- Hamer HM, Jonkers D, Venema K, Vanhoutvin S, Troost FJ, Brummer R-J. (2008). Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther 27: 104–119. [DOI] [PubMed] [Google Scholar]
- Holdeman LV, Moore WEC. (1974). New genus, Coprococcus, twelve new species, and emended descriptions of four previously described species of bacteria from human feces. Int J Syst Bacteriol 24: 260–277. [Google Scholar]
- Kettle H, Louis P, Holtrop G, Duncan SH, Flint HJ. (2015). Modelling the emergent dynamics and major metabolites of the human colonic microbiota. Environ Microbiol 17: 1615–1630. [DOI] [PubMed] [Google Scholar]
- Lopez-Siles M, Khan TM, Duncan SH, Harmsen HJM, Garcia-Gil LJ, Flint HJ. (2012). Cultured representatives of two major phylogroups of human colonic Faecalibacterium prausnitzii can utilize pectin, uronic acids, and host-derived substrates for growth. Appl Environ Microbiol 78: 420–428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louis P, Flint HJ. (2017). Formation of propionate and butyrate by the human colonic microbiota. Environ Microbiol 19: 29–41. [DOI] [PubMed] [Google Scholar]
- Louis P, Flint HJ, Michel C. (2016). How to manipulate the microbiota: prebiotics. Adv Exp Med Biol 902: 119–142. [DOI] [PubMed] [Google Scholar]
- Louis P, Hold GL, Flint HJ. (2014). The gut microbiota, bacterial metabolites and colorectal cancer. Nat Rev Microbiol 12: 661–672. [DOI] [PubMed] [Google Scholar]
- Louis P, Young P, Holtrop G, Flint HJ. (2010). Diversity of human colonic butyrate-producing bacteria revealed by analysis of the butyryl-CoA:acetate CoA-transferase gene. Environ Microbiol 12: 304–314. [DOI] [PubMed] [Google Scholar]
- Martens EC, Kelly AG, Tauzin AS, Brumer H. (2014). The devil lies in the details: how variations in polysaccharide fine-structure impact the physiology and evolution of gut microbes. J Mol Biol 426: 3851–3865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I, Kim J, Duffy PR, Schlegel VL, Walter J. (2010). Resistant starches types 2 and 4 have differential effects on the composition of the fecal microbiota in human subjects. PLoS One 5: e15046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez I, Lattimer JM, Hubach KL, Case JA, Yang J, Weber CG et al. (2013). Gut microbiome composition is linked to whole grain-induced immunological improvements. ISME J 7: 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin HP, Motherway MO, Lakshminarayanan B, Stanton C, Paul Ross R, Brulc J et al. (2015). Carbohydrate catabolic diversity of bifidobacteria and lactobacilli of human origin. Int J Food Microbiol 203: 109–121. [DOI] [PubMed] [Google Scholar]
- Park SK, Kim MS, Bae JW. (2013). Blautia faecis sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 63: 599–603. [DOI] [PubMed] [Google Scholar]
- Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al. (2013). The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res 41: D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Farias C, Slezak K, Fuller Z, Duncan A, Holtrop G, Louis P. (2009). Effect of inulin on the human gut microbiota: stimulation of Bifidobacterium adolescentis and Faecalibacterium prausnitzii. Br J Nutr 101: 541–550. [DOI] [PubMed] [Google Scholar]
- Reichardt N, Duncan SH, Young P, Belenguer A, McWilliam Leitch C, Scott KP et al. (2014). Phylogenetic distribution of three pathways for propionate production within the human gut microbiota. ISME J 8: 1323–1335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson AJ, Calder AG, Stewart CS, Smith A. (1989). Simultaneous determination of volatile and non-volatile acidic fermentation products of anaerobes by capillary gas chromatography. Lett Appl Microbiol 9: 5–8. [Google Scholar]
- Rodionova IA, Li X, Thiel V, Stolyar S, Stanton K, Fredrickson JK et al. (2013). Comparative genomics and functional analysis of rhamnose catabolic pathways and regulons in bacteria. Front Microbiol 4: 407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogowski A, Briggs JA, Mortimer JC, Tryfona T, Terrapon N, Lowe EC et al. (2015). Glycan complexity dictates microbial resource allocation in the large intestine. Nat Commun 6: 7481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers AA, Vercellotti JR, West SEH, Wilkins TD. (1977). Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl Environ Microbiol 33: 319–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB et al. (2009). Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75: 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott KP, Martin JC, Duncan SH, Flint HJ. (2014). Prebiotic stimulation of human colonic butyrate-producing bacteria and bifidobacteria in vitro. FEMS Microbiol Ecol 87: 30–40. [DOI] [PubMed] [Google Scholar]
- Scott KP, Martin JC, Campbell G, Mayer CD, Flint HJ. (2006). Whole-genome transcription profiling reveals genes up-regulated by growth on fucose in the human gut bacterium 'Roseburia inulinivorans'. J Bacteriol 188: 4340–4349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selak M, Rivière A, Moens F, van Den Abbeele P, Geirnaert A, Rogelj I et al. (2016). Inulin-type fructan fermentation by bifidobacteria depends on the strain rather than the species and region in the human intestine. Appl Microbiol Biotechnol, 1–11. [DOI] [PubMed]
- Steinert RE, Sadaghian Sadabad M, Harmsen HJM, Weber P. (2016). The prebiotic concept and human health: a changing landscape with riboflavin as a novel prebiotic candidate. Eur J Clin Nutr 70: 1348–1353. [DOI] [PubMed] [Google Scholar]
- Taras D, Simmering R, Collins MD, Lawson PA, Blaut M. (2002). Reclassification of Eubacterium formicigenerans Holdeman and Moore 1974 as Dorea formicigenerans gen. nov., comb. nov., and description of Dorea longicatena sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 52: 423–428. [DOI] [PubMed] [Google Scholar]
- van den Abbeele P, Gérard P, Rabot S, Bruneau A, El Aidy S, Derrien M et al. (2011). Arabinoxylans and inulin differentially modulate the mucosal and luminal gut microbiota and mucin-degradation in humanized rats. Environ Microbiol 13: 2667–2680. [DOI] [PubMed] [Google Scholar]
- Walker AW, Martin JC, Scott P, Parkhill J, Flint HJ, Scott KP. (2015). 16 S rRNA gene-based profiling of the human infant gut microbiota is strongly influenced by sample processing and PCR primer choice. Microbiome 3: 87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AW, Ince J, Duncan SH, Webster LM, Holtrop G, Ze X et al. (2011). Dominant and diet-responsive groups of bacteria within the human colonic microbiota. ISME J 5: 220–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker AW, Duncan SH, McWilliam Leitch EC, Child MW, Flint HJ. (2005). pH and peptide supply can radically alter bacterial populations and short-chain fatty acid ratios within microbial communities from the human colon. Appl Environ Microbiol 71: 3692–3700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wefers D, Bunzel M. (2015). Characterization of dietary fiber polysaccharides from dehulled common buckwheat (Fagopyrum esculentum) seeds. Cereal Chem 92: 598–603. [Google Scholar]
- Ze X, Ben David Y, Laverde-Gomez JA, Dassa B, Sheridan PO, Duncan SH et al. (2015). Unique organization of extracellular amylases into amylosomes in the resistant starch-utilizing human colonic Firmicutes bacterium Ruminococcusbromii. mBio 6: e01058–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ze X, Le Mougen F, Duncan SH, Louis P, Flint HJ. (2013). Some are more equal than others: the role of 'keystone' species in the degradation of recalcitrant substrates. Gut Microbes 4: 236–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.