Abstract
Gastric cancer (GC) is one of the leading types of cancer in terms of mortality cases worldwide. Doxorubicin (Dox), a common chemotherapy drug, is frequently used to treat GC; however, acquired resistance to Dox hinders the chemotherapeutic outcome and causes shorter survival in GC patients. Several Dox-resistant GC cell lines, including SGC7901, SNU-1 and SNU-5 were generated to investigate the mechanism of Dox resistance in GC. Various methods were used to test the response of Dox-resistant GC cells and parental cells, including flow cytometry, Cell Counting kit-8 assay, reverse transcription polymerase chain reaction and western blot analysis. In the present study, various Dox-resistant cells presented reduced apoptosis and cell cycle arrest in response to Dox treatment. Western blot results revealed that cyclin D1 was upregulated in Dox-resistant cells, whereas inhibition or depletion of cyclin D1 re-sensitized the resistant cells to Dox treatment, which indicated that the induction of cyclin D1 expression was a result of the Dox resistance in GC cells. Furthermore, it was observed that a transcription activated form of p73 (TAp73), is the upstream modulator of cyclin D1, manipulating the cyclin D1 transcription with the assistance of activator protein 1 (AP-1). Overall, the present study data provided a rational strategy to overcome the Dox resistance in GC treatment by inhibiting cyclin D1 expression.
Keywords: cyclin D1, transcription activated p73, cyclin-dependent kinase 4 inhibitor, gastric cancer
Introduction
As one of the most prevalent types of cancer, gastric cancer (GC) accounted for almost 9% of all mortalities caused by cancer worldwide in 2012 (1). Chemotherapy has been recognized as an effective and frequently used therapeutic method for advanced GC with or without metastasis (2). Doxorubicin (Dox) is a member of the anthracycline family of drugs and, along with other chemotherapy agents, such as mitomycin and 5-fluorouracil, constitutes the gold standard treatment in advanced GC patients (3). However, treatment based on Dox has a number of adverse effects, which lead to poor survival of GC patients (4,5). Chemotherapy drug resistance serves as the main contributor to treatment failure, bringing about tumor relapse and metastasis (6). The underlying genetic mechanism of chemotherapy resistance is complicated and linked with multiple processes, including the repair of DNA damage, cell death, and transport and metabolism of medicine (6).
Cyclin D1 serves an essential role in tumorigenesis and disease progression of various types of cancer, including lung, esophagus, breast and bladder cancer (7,8). Cyclin D1 is proto-oncogenic since it serves as a cell cycle regulator and is frequently involved in G1/S transition (9). Once cyclin D1 binds to cyclin-dependent kinase 4 (CDK4) or CDK6, phosphorylation of retinoblastoma protein (Rb) is triggered at the early stage of G1 phase, causing the release of E2F factors, which serve as transcription factors of the genes pushing the cell cycle from G1 phase to S phase (10,11). Therefore, overexpression of cyclin D1 tends to cause a rapid transition from G1 phase to S phase in fibroblasts. In addition, cyclin D1 serves an important but complicated role in the promotion or inhibition of apoptosis based on the cell status and cell type (12). In particular, elevated level of endogenous cyclin D1 hinders the apoptosis in hepatocellular carcinoma (13), while overexpression of cyclin D1 attenuates apoptosis triggered by drugs in rat embryonic fibroblasts (14). According to these previous findings, it is suggested that cyclin D1 promotes survival in cancer cells.
It has previously been suggested that a large number of GC patients are accompanied with overexpression of cyclin D1 (15). Furthermore, enhanced expression of cyclin D1 is associated with worse prognosis and shorter survival in GC patients (7,16). Although overexpression of cyclin D1 has been associated with a poor clinical outcome, the association between elevated cyclin D1 and chemoresistance in GC cells has not been extensively studied.
In order to identify the mechanism underlying the cyclin D1-mediated chemoresistance in gastric carcinoma and to assist the development of an innovative strategy to overcome drug resistance, the present study attempted to examine agents sensitizing Dox in GC treatment and its underlying mechanism. Several Dox-resistant human GC cell lines, SGC7901, SNU-1 and SNU-5 were generated and investigated. The results indicated that cyclin D1 expression was induced in Dox-resistant cells, while knockdown of cyclin D1 by small interfering RNA (siRNA) re-sensitized the resistant cells to Dox. As to the mechanism of cyclin D1 induction, the current study observed that transcription activated (TA)p73 is the upstream regulator of cyclin D1, which further confirmed the tumor pro-survival function of TAp73.
Materials and methods
Reagents and cell culture
Dox was obtained from Sigma-Aldrich (Merck KGaA, Darmstadt, Germany) and diluted in phosphate-buffered saline (PBS). The CDK4 inhibitors (CDK4i), PD-0332991 (PD; Pfizer, Inc., New York, NY, USA) and LEE011 (Selleck Chemicals, Houston, TX, USA), were diluted in dimethyl sulfoxide (DMSO). TAp73 (cat. no. SC-7238; 1:1,000), p53 (cat. no. SC-126; 1:1,000), ΔΝp73 (cat. no. SC-70966; 1:1,000), cyclin D1 (cat. no. SC-4074; 1:1,000), cleaved caspase-3 (cat. no. SC-113,427; 1:1,000), activator protein 1 (AP-1; cat. no. SC-8047; 1:1,000) and β-actin (cat. no. SC-58673, 1:1,000) primary antibodies were purchased from Santa Cruz Biotechnology, Inc. (Dallas, TX, USA). The HRP conjugated mouse (ab6789) and rabbit (ab6728) secondary antibodies were purchased from Abcam (Cambridge, USA). The remaining chemicals conformed to the highest standard of quality that can be purchased commercially.
Human GC cell line SGC-7901 was provided by the Cell Bank of Shanghai Institute of Cell Biology, Chinese Academy of Sciences (Shanghai, China). The GC cell lines, SNU-1 (cat. no. CRL-5971) and SNU-5 (cat. no. CRL-5973) were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA). RPMI-1640 (Hyclone; GE Healthcare Life Sciences, Little Chalfont, UK) with added antibiotics (100 mg/ml streptomycin and 100 U/ml penicillin) and 10% fetal bovine serum (FBS; Hangzhou Sijiqing Biotech Co., Ltd., Hangzhou, China) was used for cell culture under humidified conditions in 5% CO2 at 37.8°C in an incubator (Thermo Fisher Scientific, Inc., Waltham, MA, USA).
Establishment of Dox-resistant GC cell lines
SGC7901, SNU-1 and SNU-5 cells resistant to Dox were generated by incubation in gradually elevated level of Dox. Briefly, cells (1×105/ml) were initially inoculated in Dox-free media for 24 h at 37.8°C. Subsequently, the medium was substituted by medium containing a low Dox concentration of 2 nM and incubated at 37.8°C for the next 48 h. The surviving cells were collected and transferred into Dox-free medium prior to the next Dox treatment. Following the adaption of cells, the concentration of Dox was increased, and the Dox and Dox-free treatment was repeated in turns until survival was maintained when cells were incubated with 10 nM Dox. These Dox-resistant SGC7901, SNU-1 and SNU-5 cells were recognized as SGC7901_R, SNU-1_R and SNU-5_R, respectively. The cells were cultured at 37.8°C under Dox-free circumstances in medium without medicine for 2 days prior to the initiation of the study.
Cytotoxicity assay
Cell Counting kit-8 assay (KeyGen Biotech Co., Ltd., Nanjing, China) was applied to determine the cytotoxicity of Dox. Briefly, 96-well plates were used for cell seeding with 100 µl RPMI-1640 medium supplemented with 10% FBS at a density of 5×104 cells/well. Following incubation at 37.8°C for 24 h, 10 µl Dox at different concentrations was supplemented into the medium, and cultured for 24 h under a humidified atmosphere of 5% CO2 at 37.8°C. A microplate reader (Thermo Fisher Scientific, Inc.) was used to detect the absorbance at 450 nm. Cells without drug treatment were used as the control group. The relative drug resistance was evaluated by a comparison of the half maximal inhibitory concentration (IC50) as previously described (17).
Western blot analysis
The GC cells with or without the indicated treatment were homogenized in a lysis buffer [100 mmol/l NaCl, 10 mmol/l Tris-HCl (pH 7.6), 0.1% NP-40, 0.1% protease inhibitor, 0.5% 100 mM phenylmethane sulfonyl fluoride and 1% phosphatase inhibitor] and centrifuged at 10,000 × g for 15 min at 4°C. The supernatant was collected and the protein concentration was determined by BCA assay according to the manufacturer's protocol. A total of 40 µg/lane of protein were loaded onto 10% SDS-PAGE and then transferred to 0.22 µm polyvinylidene difluoride membranes. The membranes were blocked with 5% fat-free milk [5% skimmed milk powder in PBS with 0.1% Tween-20 (PBST)] and incubated with primary antibodies overnight at 4°C. Following three washes with PBST, each for 5 min, the membranes were incubated with secondary antibodies (1:4,000) for 1 h at room temperature. Following washing with PBST, specific antibody binding was detected using a chemiluminescence detection system (Pierce; Thermo Fisher Scientific, Inc.), according to the manufacturer's protocol. The western blot films were captured using a scanner (Canon, Inc., Tokyo, Japan). All bands were normalized to β-actin.
Crystal violet staining
The GC cells were seeded in 12-well plates with 1 ml RPMI-1640 medium supplemented with 10% FBS at a density of 5×104 cells/well. Following incubation at 37.8°C for 24 h, the cells were treated with 20 nM Dox and/or 40 nM PD-0332991, and cultured for 24 h under a humidified atmosphere of 5% CO2 at 37.8°C. DMSO (0.1%) was used for the control group. The attached cells in 12 wells were stained with a crystal violet staining buffer [0.5% crystal violet powder (Sigma-Aldrich; Merck KGaA), 79.5% distilled H2O, 20% methanol], and incubated at room temperature for 1 h. The plates were subsequently washed with H2O three times and visualized using a digital camera (Canon, Inc.).
Apoptosis and cell cycle analysis
GC cells (1×106) were washed twice with ice-cold PBS, treated with trypsin and fixed in 70% ethanol at 4°C for 30 min. The cell pellet was then incubated in a solution containing 50 ng/ml propidium iodide, 0.2 mg/ml RNase and 0.1% Triton X-100 at room temperature for 30 min. Subsequently, the cells were analyzed by flow cytometry using an EPICS XL-MCL FACScan device (Beckman Coulter, Inc., Brea, CA, USA). The data were analyzed with MultiCycle software, version 306 for Windows (Phoenix Flow Systems, San Diego, CA, USA).
SGC7901, SNU-1, SNU-5 parental or resistant cells were treated with 20 nM Dox, 40 nM PD and 20 nM LEE011 alone or in combination at 37.8°C for 24 h. Cells were treated with 0.1% DMSO as the control. The apoptosis of GC cells was evaluated by Hoechst 33258 staining. The cells were stained with 10 µg/ml Hoechst 33258 (Invitrogen; Thermo Fisher Scientific, Inc.), in 0.5% Nonidet P-40 and 3.7% formaldehyde at room temperature for 30 min. Subsequently, a microscope was used to observe the cell apoptosis, which was characterized by micronucleation and condensed chromatin in the cells (18).
Transfections of DNA plasmids
To overexpress cyclin D1 cells were transfected with RC/CMV cyclin D1 plasmids and to knockdown TAp73, AP-1 and cyclin D1, cells were transfected with siRNA specific for TAp73, AP-1 and cyclin D1. This was performed using Lipofectamine 2000® (Invitrogen; Thermo Fisher Scientific, Inc.). The RC/CMV plasmid was used as the control for transfection with the cyclin D1 plasmid, and control siRNA was used for TAp73, AP-1 and cyclin D1 siRNA transfection. The RC/CMV cyclin D1 and control RC/CMV plasmid was purchased from Addgene, Inc. (Cambridge, MA, USA). Transfection was performed with the TAp73 siRNA (5′-GGATTCCAGCATGGACGTCTT-3′) (19), AP-1 siRNA (cat. no. SC-97430), control siRNA (cat. no. SC-37007) and cyclin D1 siRNA (cat. no. SC-29286) (all from Santa Cruz Biotechnology, Inc.). Briefly, 0.4 µg plasmid or 200 pMol siRNA were mixed with 2 µl Lipofectamine 2000 in 100 µl serum-free cell culture medium. After 30 min, the mixture was dropped into targeted cells, and incubated at 37.8°C overnight. The efficiency of transfection was tested by western blot analysis at 24 h following transfection.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The AxyPrep™ Purification kit (Axygen Scientific, Inc., Union City, CA, USA) was utilized to obtain the total RNA from SGC7901 cells. Next, the quality and quantity of total RNA were examined by a Nanodrop 2000 micro-volume spectrophotometer (Thermo Fisher Scientific, Inc.) through evaluation of absorbance. For cDNA synthesis, 1 µg of RNA was mixed with Oligo(dT) primers, dNTP mix, 5X first-strand buffer, Ditiotreitol, RNAseOUT and SuperScript II RT, all according to the manufacturer's protocol (Invitrogen; Thermo Fisher Scientific, Inc.). The PCR reaction mixture contained 5 µl cDNA, 1 µl TaqMan Master mix (Thermo Fisher Scientific, Inc.), 400 nM of primer for the targeted gene, 400 nM Primer mix for the examined and reference genes (Gene Tech Co., Ltd., Hong Kong, China), 200 nM of each hydrolysis TaqMan probe (for the examined and reference genes) and PCR-grade H2O to adjust to the final 20 µl reaction volume. The thermocycling conditions were 10 min of initial denaturation at 95°C, followed by 45 cycles of denaturation at 95°C for 10 sec, annealing at 60°C for 30 sec and extension at 72°C for 30 sec with a single fluorescence acquisition step at the end of extension, followed by final cooling at 40°C for 30 sec. PCR analysis was then conducted using the Mx3000P qPCR system (Stratagene; Agilent Technologies, Inc., Santa Clara, CA, USA). GAPDH served as an internal control. The results were calculated by the 2−ΔΔCq method as previously described (20). The primers used were as follows: Cyclin D1, 5′-CCTCGGTGTCCTACTTCAAA-3′ (sense) and 5′-GGGATGGTCTCCTTCATCTT-3′ (antisense); GAPDH, 5′-CTCCCCACACACATGCACTTA-3′ (sense) and 5′-CCTAGTCCCAGGGCTTTGATT-3′ (antisense).
Luciferase reporter assay
To analyze the cyclin D1 promoter activity, the cyclin D1 promoter with a 12-O-tetradecanoylphorbol-13-acetate-responsive element (TRE/AP-1 site), along with the control β-galactosidase reporter pCMVβ (Promega Corp., Madison, WI, USA), was transfected into SGC7901 cells using Lipofectamine Plus reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. Luciferase activities were then measured and normalized to those of β-galactosidase. Experiments were performed in triplicate and all measurements were repeated at least three times.
Chromatin immunoprecipitation (ChIP)
For the AP-1 binding assay, the ChIP Assay kit (Upstate Biotechnology, Inc., Lake Placid, NY, USA) was used, as previously described (21) on SGC7901 cells that were transfected with control or TAp73 siRNA and treated with Dox as previous described. A total of 2 µg antibodies against AP-1 (cat. no. SC-8047; Santa Cruz Biotechnology, Inc.) and isotype matched immunoglobulin G (R&D System, Inc., Minneapolis, MN, USA) were applied to examine the immunoprecipitation.
Statistical analysis
Data are presented in the terms of the mean ± standard error. One-way analysis of variance and student's t-test were applied to evaluate the statistical significance with the SPSS version 13.0 software (SPSS, Inc., Chicago, IL, USA). A value of P<0.05 was considered to indicate a statistically significant difference.
Results
Dox-resistant GC cells present reduced apoptosis and cell cycle arrest
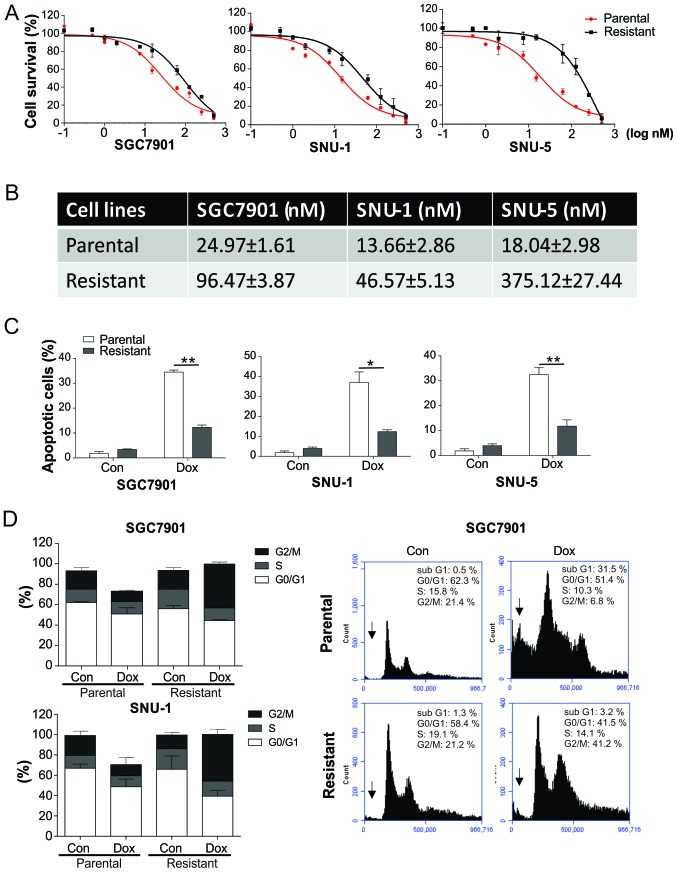
In order to study the mechanism of drug resistance in GC, Dox-resistant GC cell lines were generated through processing the SGC7901, SNU-1 and SNU-5 cells with four successive rounds of Dox treatment. As tested by a Cell Counting kit-8 assay, the resistant cells (denoted by _R) were accompanied with higher IC50 values in comparison with the parent cells (denoted by _P; Fig. 1A and B). The cell lines resistant to Dox were reported to present significantly impaired apoptosis in comparison with parent cell lines (Fig. 1C). Notably, the Dox-resistant SGC7901_R and SNU-1_R cells demonstrated significantly increased cell cycle arrest in the G2/M phase in response to Dox treatment, compared with the parental cells (Fig. 1D), indicating that the dysfunction of cell cycle may contribute to the Dox resistance in GC cells.
Figure 1.
Dox-resistant GC cells presented reduced apoptosis and cell cycle arrest. (A) SGC7901, SNU-1 and SNU-5 parental and resistant cells underwent treatment with Dox for 24 h, and were subsequently analyzed by Cell Counting kit-8 assay. (B) Half maximal inhibitory concentration values of SGC7901, SNU-1 and SNU-5 parental and resistant cells for Dox are presented. (C) Apoptosis rate of SGC7901, SNU-1 and SNU-5 parental and resistant cells treated with 20 nM Dox, as determined by Hoechst 33258 staining. Cells with condensed and fragmented nuclei were recognized as apoptotic cells. (D) Cell cycle progression was investigated by propidium iodide staining and flow cytometry in SGC7901 and SNU-1 parental and resistant cells treated with 20 nM. The percentage of cells at the different phases were calculated and plotted. Representative graphs for each group in SGC7901 cells are displayed, with the arrows indicating the apoptotic cells. Error bars indicate the standard deviation of triplicate detection. *P<0.05 and **P<0.01. GC, gastric cancer; Dox, doxorubicin; Con, control.
Upregulation of cyclin D1 is associated with drug resistance in GC
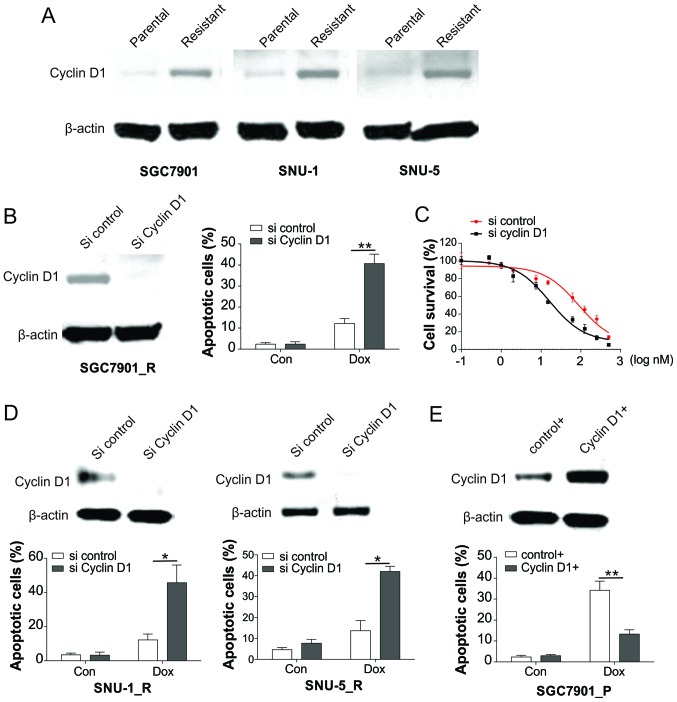
As cyclin D1 is a vital gene in modulating the cell cycle, its expression was investigated in resistant and parental cells. As demonstrated in Fig. 2A, the expression level of cyclin D1 was elevated in resistant cells in comparison with that in parent cells, indicating that cyclin D1 upregulation may be the cause of Dox resistance. Furthermore, knockdown of cyclin D1 in resistant cells by siRNA transfection sensitized the SGC7901_R cells to Dox by inducing increased apoptosis (Fig. 2B). Accordingly, the survival of SGC7901-R cells was markedly suppressed by cyclin D1 knockdown in response to Dox treatment (Fig. 2C). Similarly, knockdown of cyclin D1 also recovered the apoptotic effect of Dox in SNU-1_R and SNU-5_R cells (Fig. 2D). By contrast, overexpression of cyclin D1 in SGC7901_P cells significantly reduced the apoptosis in response to Dox treatment (Fig. 2E). These results indicated that GC cells gained resistance to Dox by upregulation of cyclin D1.
Figure 2.
Induction of cyclin D1 results in Dox resistance in GC cells. (A) Expression of cyclin D1 in SGC7901, SNU-1 and SNU-5 parental and resistant cells was analyzed by western blot analysis. (B) SGC7901 resistant cells were transfected with cyclin D1 siRNA, and then the expression of cyclin D1 was analyzed by western blot analysis (left), while the apoptotic cells following Dox treatment were analyzed by Hoechst 33258 staining (right). (C) SGC7901 resistant cells underwent transfection with cyclin D1 siRNA, followed by treatment with Dox, and then the viability of cells was examined using Cell Counting kit-8 assay. (D) SNU-1 and SNU-5 resistant cells were transfected with cyclin D1 siRNA. The expression of cyclin D1 was analyzed by western blot assay (upper), and the apoptotic cells following Dox treatment were analyzed by Hoechst 33258 staining (lower). (E) SGC7901 parental cells were transfected with cyclin D1 plasmid, and the expression of cyclin D1 was analyzed by western blot assay (upper), while the apoptotic cells after Dox treatment were analyzed by Hoechst 33258 staining (lower). Error bars represent the standard deviation of triplicate detection. *P<0.05 and **P<0.01. GC, gastric cancer; Dox, doxorubicin; siRNA, small interfering RNA; Con, control.
CDK4 inhibitors (CDK4i) and Dox therapy triggers apoptosis
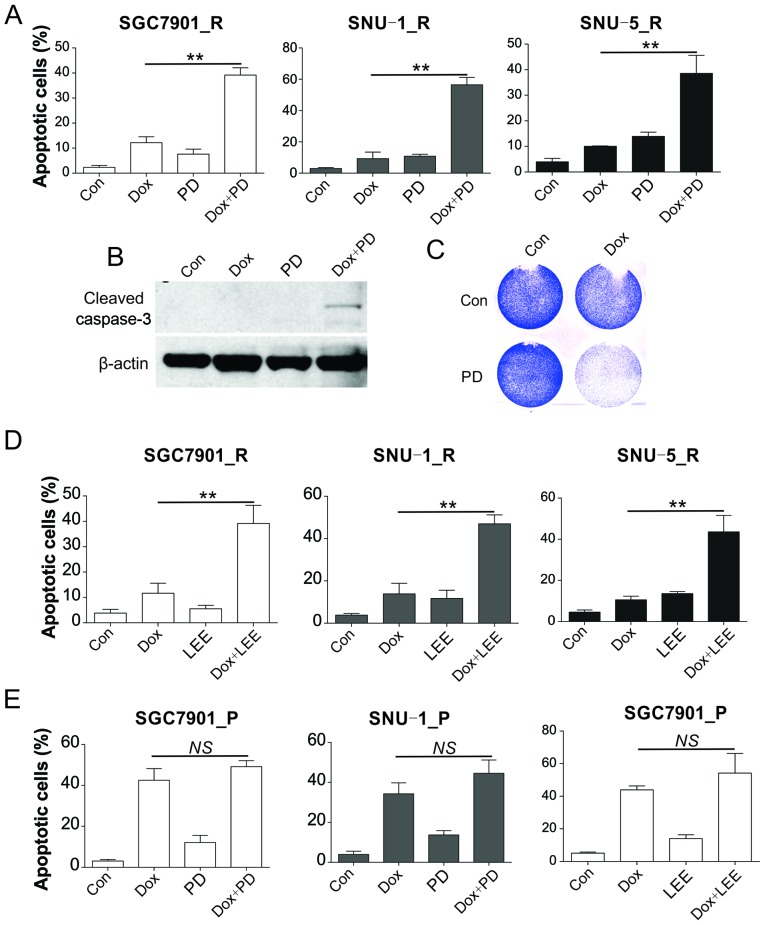
The present study also evaluated whether combination treatment with CDK4 inhibitors was able to re-sensitize the resistant cells to Dox treatment. The Dox-resistant SGC7901_R, SNU-1_R cells and SNU-5_R were supplemented with control DMSO, PD, Dox or combination therapy for 48 h, and then Hoechst 33258 staining was applied for the determination of apoptosis. In comparison with the control group, it was observed that SGC7901_R and SNU-1_R cells presented a slight elevation in apoptosis subsequent to single Dox or PD treatment. Combination treatment displayed the maximum apoptosis in reaction to therapy (Fig. 3A). The combination treatment also markedly induced a higher level of caspase-3 and reduced the cell viability in SGC7901_R cells (Fig. 3B and C). Furthermore, another CDK4 inhibitor, LEE011, enhanced the apoptosis when combined with Dox treatment in SGC7901_R and SNU-1_R cells (Fig. 3D). However, the combination of PD and Dox did not significantly increase the apoptosis in SGC7901_P and SNU-1_P cells (Fig. 3E). Similarly, the effect of LEE011, also had no significant effects on Dox induced apoptosis in SGC7901_P cells. According to these findings, upregulation of cyclin D1 may be the cause for Dox resistance in GC cells.
Figure 3.
CDK inhibitors sensitized the resistant GC cells to Dox. (A) SGC7901_R, SNU-1_R and SNU-5_R cells were treated with 20 nM Dox and/or 40 nM PD-0332991 for 24 h. The apoptotic cells were analyzed by Hoechst 33258 staining. (B) Caspase-3 expression was analyzed by western blot analysis, and (C) cell viability was examined by crystal violet staining in SGC7901_R cells treated with 20 nM Dox and/or 40 nM PD-0332991 for 24 h. (D) Apoptosis was analyzed by Hoechst staining in SGC7901_R, SNU-1_R, and SNU-5_R cells treated with 20 nM Dox and/or 20 nM LEE011 for 24 h. (E) Apoptosis was analyzed by Hoechst staining in SGC7901_P and SNU-1_P cells treated with 20 nM Dox and/or 40 nM PD-0332991 or 20 nM LEE011 for 24 h. Error bars indicate the standard deviation of triplicate detection. **P<0.01. NS, non-significant; GC, gastric cancer; Dox, doxorubicin; _P, parental; _R, resistant; Con, control.
TAp73 modulates cyclin D1 expression
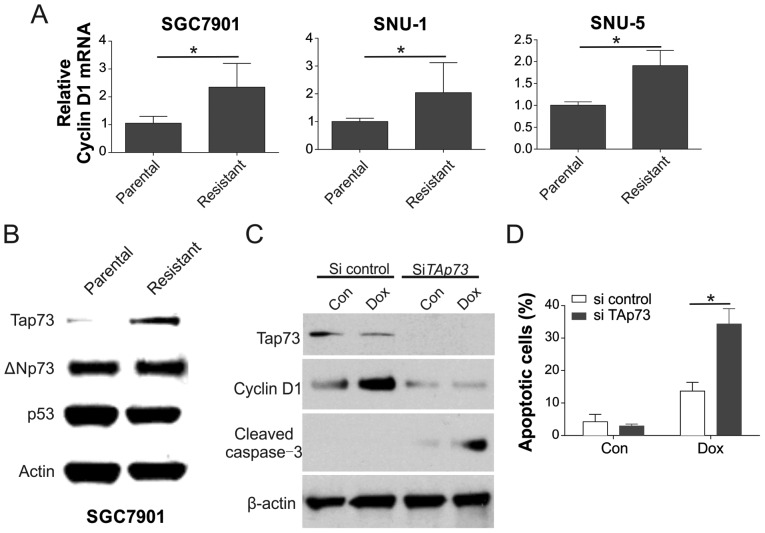
The mechanism underlying the cyclin D1 induction in the resistant cells was then examined. It was observed that the concentration of cyclin D1 mRNA was elevated in the SGC7901, SNU-1, and SNU-5 resistant cells, suggesting that cyclin D1 expression was modulated by its upstream regulator (Fig. 4A). As cyclin D1 transcription is modulated by p53, TAp73 and ΔΝp73 (22,23), the expression levels of these transcription factors were investigated. As exhibited in Fig 4B, only TAp73 was notably upregulated in resistant cells, p53 and ΔΝp73 were not. Accordingly, cyclin D1 expression was continuously upregulated in SGC7901_R cells upon Dox treatment, but reduced when TAp73 was knocked down by siRNA (Fig. 4C). Furthermore, depletion of TAp73 by siRNA also sensitized the SGC7901_R cells to Dox treatment by increasing apoptosis (Fig. 4D).
Figure 4.
TAp73 modulated cyclin D1 regulation. (A) mRNA level of cyclin D1 in SGC7901, SNU-1 and SNU-5 parental and resistant cells was analyzed by reverse transcription-quantitative polymerase chain reaction. (B) The expression levels of the indicated proteins in SGC7901 parental and resistant cells were analyzed by western blot analysis. (C) Expression levels of the indicated proteins were analyzed by western blot analysis, and (D) apoptosis was evaluated by Hoechst 33258 staining in SGC7901 resistant cells transfected with TAp73 siRNA, followed by 20 nM Dox treatment. Error bars represent the standard deviation of triplicate detection. *P<0.05. Dox, doxorubicin; siRNA, small interfering RNA; Con, control.
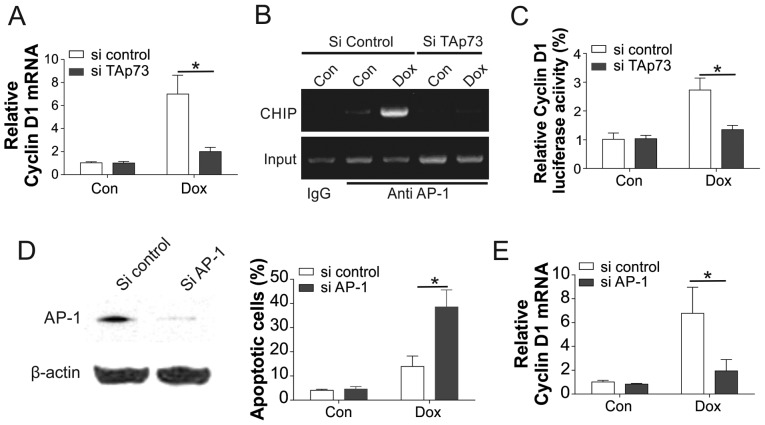
To further understand how TAp73 modulates cyclin D1 regulation, RT-PCR was used to investigate the cyclin D1 mRNA expression. The results indicated that the upregulation of cyclin D1 mRNA by Dox in the control knockdown cells was also reduced by the TAp73 knockdown (Fig. 5A). As TAp73 coordinates with the AP-1 to control the transcription of cyclin D1, the present study further evaluated the influence of TAp73 on the AP-1 transcription activity on cyclin D1. Upon Dox treatment, the binding capability of AP-1 in the cyclin D1 promoter was enhanced, which was reduced following TAp73 knockdown (Fig. 5B). Similarly, knockdown of TAp73 also abrogated the luciferase activity of the cyclin D1 promoter with the AP-1 binding site (Fig. 5C). Furthermore, depletion of AP-1 also sensitized the SGC7901_R cells to Dox treatment (Fig. 5D), and suppressed the mRNA level of cyclin D1 (Fig. 5E). Collectively, the aforementioned findings suggested that TAp73 modulated the transcription of cyclin D1 to Dox treatment by mediating the binding of AP-1 to the cyclin D1 promoter.
Figure 5.
TAp73 modulates the transcription of AP-1, a promoter of cyclin D1. (A) mRNA level of cyclin D1 analyzed by qPCR and (B) ChIP assay using anti-AP-1, in SGC7901 resistant cells transfected with TAp73 siRNA, followed by 20 nM Dox treatment. Immunoprecipitated products and input were amplified by PCR with primer annealing in the cyclin D1 promoter with the AP-1 binding site, and IgG served as a negative control. (C) SGC7901 resistant cells were transfected with cyclin D1 luciferase reporter and then transfected with TAp73 siRNA, followed by 20 nM Dox treatment. The luciferase activity were analyzed and plotted. (D) SGC7901 resistant cells were transfected with AP-1 siRNA. The expression of AP-1 was analyzed by western blot analysis (left), while the apoptotic cells following DOX treatment were analyzed by Hoechst 33258 staining (right). (E) SGC7901 resistant cells were transfected with AP-1 siRNA and the mRNA level of cyclin D1 was examined using qPCR. Error bars represent the standard deviation of triplicate detection. *P<0.05. AP-1, activator protein 1; Dox, doxorubicin; Con, control; siRNA, small interfering RNA; qPCR, quantitative polymerase chain reaction.
Discussion
As one of the most prevalent types of cancer, GC has a high incidence rate in China, (22.0 per 100,000 men and 10.4 per 100,000 women in 2002) (24). Chemotherapy is a commonly used treatment for GC; however, its effects are hindered by simultaneous resistance of cancer cells to chemotherapy drugs. Although Dox serves as an essential drug against GC (3), its chemotherapeutic effect is hampered by the acquired resistance, resulting in a poor clinical outcome (17).
It has been reported that enhanced cyclin D1 expression is associated with the early stage of tumorigenesis in a range of tumors, including non-small cell lung cancer, metachronous lung cancer, breast cancer, esophageal cancer, and carcinoma of the neck and head (15). It has been reported that cyclin D1 controls cell adhesion and the migration of various cell types and tumors, consequently affecting their metastatic potential (25). In the present study, three Dox-resistant cell lines, SGC7901, SNU-1 and SNU-5 were generated by long-term and incremental Dox treatment, resulting in drug resistance. The findings of current study are in accordance with previous observations, suggesting that Dox-resistant GC cells present a higher expression of cyclin D1 (25,26). Inhibition or depletion of cyclin D1 re-sensitized the resistant GC cells to Dox treatment. Thus, these results provided novel evidence to enhance the killing effect of Dox on GC cells, which may assist in the development of an effective strategy for the clinical therapy of GC.
Inhibitors of pan-CDK4 have been applied in phase I clinical trials of solid cancer; however, the effects were not satisfactory, possibly due to nonspecificity of CDK4 suppression and insufficiency of cyclin D1 analysis in the trials (27,28). A more thorough understanding of the role of specific cyclins and their CDK4 complements, in addition to the direct study of CDK4i in specific types of cancer that overexpress cyclin, is necessary to reveal the therapeutic potential of these agents. Currently, only a limited number of clinical trials focusing on the combination treatment of CDKi and chemotherapy have demonstrated a partial response for patients with terminal solid cancer (27). In the present study, it was observed that inhibition of cyclin D1 by a CDK4 inhibitor (PD-0332991) was able to significant overcome the Dox resistance. These results provide support for the implementation of CDKi therapy, combined with cytotoxic chemotherapy, which may result in improved cytotoxic efficacy and improved patient outcomes.
Regarding the mechanism underlying the cyclin D1 induction in Dox-resistant GC cells, the present study observed that TAp73 regulates cyclin D1 expression via AP-1. TAp73 is known to share a similar structure with p53 (29). The majority of previous studies have demonstrated that TAp73 acts as an apoptosis stimulator (30,31), however, a number of types of cancer display overexpression of TAp73 (29,32). It has been previously reported that TAp73 promotes cell growth by activating targeted genes of AP-1, such as cyclin D1, in association with c-Jun, which forms a complex wit AP-1 (23,33). Furthermore, the promotion of cellular growth by TAp73 was also verified by the reported TAp73 activation of pentose phosphate pathway, as well as cell proliferation (32). Accordingly, the results of the current study demonstrate that TAp73 modulated the upregulation of cyclin D1 in Dox-resistant GC cells, indicating that the expression of TAp73 may also contribute toward Dox resistance in the GC cells. Consequently, these data suggest that inhibition of TAp73 and AP-1 may be an avenue to inhibit tumor cell growth, particularly in types of cancer that overexpress TAp73.
In conclusion, the current study demonstrated that upregulation of cyclin D1 significantly inhibited the sensitivity of GC cells to the anticancer effect of Dox, and decreased the percentage of apoptotic cells. Upregulation of cyclin D1 in GC cells induced cell cycle arrest. Furthermore, the combination of a CDK inhibitor and Dox was able to re-sensitize the Dox-resistant cells and induce apoptosis, which may be a potential strategy to overcome drug resistance in GC therapy. Thus, it is speculated that targeting cyclin D1 may aid in the treatment of GC in the future.
References
- 1.Giordano AI, Domènech I, Torres A, Skufca J, Callejo A, Palomino L, Aparicio A, Junyent J, Mañós M. Results in the surgical treatment of giant acoustic neuromas. Acta Otorrinolaringol Esp. 2012;63:194–199. doi: 10.1016/j.otorri.2011.11.003. (In Spanish) [DOI] [PubMed] [Google Scholar]
- 2.Glimelius B, Hoffman K, Haglund U, Nyrén O, Sjödén PO. Initial or delayed chemotherapy with best supportive care in advanced gastric cancer. Ann Oncol. 1994;5:189–190. doi: 10.1093/oxfordjournals.annonc.a058778. [DOI] [PubMed] [Google Scholar]
- 3.MacDonald JS, Schein PS, Woolley PV, Smythe T, Ueno W, Hoth D, Smith F, Boiron M, Gisselbrecht C, Brunet R, Lagarde C. 5-Fluorouracil, doxorubicin and mitomycin (FAM) combination chemotherapy for advanced gastric cancer. Ann Intern Med. 1980;93:533–536. doi: 10.7326/0003-4819-93-4-533. [DOI] [PubMed] [Google Scholar]
- 4.Lise M, Nitti D, Marchet A, Sahmoud T, Buyse M, Duez N, Fiorentino M, Dos Santos JG, Labianca R, Rougier P, et al. Final results of a phase III clinical trial of adjuvant chemotherapy with the modified fluorouracil, doxorubicin, and mitomycin regimen in resectable gastric cancer. J Clin Oncol. 1995;13:2757–2763. doi: 10.1200/JCO.1995.13.11.2757. [DOI] [PubMed] [Google Scholar]
- 5.Zhang YW, Zhang YL, Pan H, Wei FX, Zhang YC, Shao Y, Han W, Liu HP, Wang ZY, Yang SH. Chemotherapy for patients with gastric cancer after complete resection: A network meta-analysis. World J Gastroenterol. 2014;20:584–592. doi: 10.3748/wjg.v20.i2.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dai Z, Huang Y, Sadée W. Growth factor signaling and resistance to cancer chemotherapy. Curr Top Med Chem. 2004;4:1347–1356. doi: 10.2174/1568026043387746. [DOI] [PubMed] [Google Scholar]
- 7.Aaltonen K, Amini RM, Landberg G, Eerola H, Aittomäki K, Heikkilä P, Nevanlinna H, Blomqvist C. Cyclin D1 expression is associated with poor prognostic features in estrogen receptor positive breast cancer. Breast Cancer Res Treat. 2009;113:75–82. doi: 10.1007/s10549-008-9908-5. [DOI] [PubMed] [Google Scholar]
- 8.Knudsen KE, Diehl JA, Haiman CA, Knudsen ES. Cyclin D1: Polymorphism, aberrant splicing and cancer risk. Oncogene. 2006;25:1620–1628. doi: 10.1038/sj.onc.1209371. [DOI] [PubMed] [Google Scholar]
- 9.Pardee AB. G1 events and regulation of cell proliferation. Science. 1989;246:603–608. doi: 10.1126/science.2683075. [DOI] [PubMed] [Google Scholar]
- 10.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 11.Lundberg AS, Weinberg RA. Functional inactivation of the retinoblastoma protein requires sequential modification by at least two distinct cyclin-cdk complexes. Mol Cell Biol. 1998;18:753–761. doi: 10.1128/MCB.18.2.753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Han EK, Ng SC, Arber N, Begemann M, Weinstein IB. Roles of cyclin D1 and related genes in growth inhibition, senescence and apoptosis. Apoptosis. 1999;4:213–219. doi: 10.1023/A:1009618824145. [DOI] [PubMed] [Google Scholar]
- 13.Albanese C, D'Amico M, Reutens AT, Fu M, Watanabe G, Lee RJ, Kitsis RN, Henglein B, Avantaggiati M, Somasundaram K, et al. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J Biol Chem. 1999;274:34186–34195. doi: 10.1074/jbc.274.48.34186. [DOI] [PubMed] [Google Scholar]
- 14.Tan A, Bitterman P, Sonenberg N, Peterson M, Polunovsky V. Inhibition of Myc-dependent apoptosis by eukaryotic translation initiation factor 4E requires cyclin D1. Oncogene. 2000;19:1437–1447. doi: 10.1038/sj.onc.1203446. [DOI] [PubMed] [Google Scholar]
- 15.Kumari S, Prasad Puneet SB, Yadav SS, Kumar M, Khanna A, Dixit VK, Nath G, Singh S, Narayan G. Cyclin D1 and cyclin E2 are differentially expressed in gastric cancer. Med Oncol. 2016;33:40. doi: 10.1007/s12032-016-0754-8. [DOI] [PubMed] [Google Scholar]
- 16.Kornmann M, Ishiwata T, Itakura J, Tangvoranuntakul P, Beger HG, Korc M. Increased cyclin D1 in human pancreatic cancer is associated with decreased postoperative survival. Oncology. 1998;55:363–369. doi: 10.1159/000011879. [DOI] [PubMed] [Google Scholar]
- 17.Yan LH, Wei WY, Cao WL, Zhang XS, Xie YB, Xiao Q. Overexpression of E2F1 in human gastric carcinoma is involved in anti-cancer drug resistance. BMC Cancer. 2014;14:904. doi: 10.1186/1471-2407-14-904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen D, Ming L, Zou F, Peng Y, Van Houten B, Yu J, Zhang L. TAp73 promotes cell survival upon genotoxic stress by inhibiting p53 activity. Oncotarget. 2014;5:8107–8122. doi: 10.18632/oncotarget.2440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rocco JW, Leong CO, Kuperwasser N, DeYoung MP, Ellisen LW. p63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9:45–56. doi: 10.1016/j.ccr.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Wang P, Yu J, Zhang L. The nuclear function of p53 is required for PUMA-mediated apoptosis induced by DNA damage. Proc Natl Acad Sci USA. 2007;104:4054–4059. doi: 10.1073/pnas.0700020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen X, Bargonetti J, Prives C. p53, through p21 (WAF1/CIP1), induces cyclin D1 synthesis. Cancer Res. 1995;55:4257–4263. [PubMed] [Google Scholar]
- 23.Subramanian D, Bunjobpol W, Sabapathy K. Interplay between TAp73 protein and selected activator protein-1 (AP-1) family members promotes AP-1 target gene activation and cellular growth. J Biol Chem. 2015;290:18636–18649. doi: 10.1074/jbc.M115.636548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang L. Incidence and mortality of gastric cancer in China. World J Gastroenterol. 2006;12:17–20. doi: 10.3748/wjg.v12.i1.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bustany S, Bourgeais J, Tchakarska G, Body S, Hérault O, Gouilleux F, Sola B. Cyclin D1 unbalances the redox status controlling cell adhesion, migration, and drug resistance in myeloma cells. Oncotarget. 2016;7:45214–45224. doi: 10.18632/oncotarget.9901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan LH, Wang XT, Yang J, Kong FB, Lian C, Wei WY, Luo W, Xie YB, Xiao Q. Reversal of multidrug resistance in gastric cancer cells by E2F-1 downregulation in vitro and in vivo. J Cell Biochem. 2014;115:34–41. doi: 10.1002/jcb.24652. [DOI] [PubMed] [Google Scholar]
- 27.Dickson MA, Schwartz GK. Development of cell-cycle inhibitors for cancer therapy. Curr Oncol. 2009;16:36–43. doi: 10.3747/co.v16i2.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Witkiewicz AK, Cox D, Knudsen ES. CDK4/6 inhibition provides a potent adjunct to Her2-targeted therapies in preclinical breast cancer models. Genes Cancer. 2014;5:261–272. doi: 10.18632/genesandcancer.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yang A, Walker N, Bronson R, Kaghad M, Oosterwegel M, Bonnin J, Vagner C, Bonnet H, Dikkes P, Sharpe A, et al. p73-deficient mice have neurological, pheromonal and inflammatory defects but lack spontaneous tumours. Nature. 2000;404:99–103. doi: 10.1038/35003607. [DOI] [PubMed] [Google Scholar]
- 30.Dabiri Y, Kalman S, Gürth CM, Kim JY, Mayer V, Cheng X. The essential role of TAp73 in bortezomib-induced apoptosis in p53-deficient colorectal cancer cells. Sci Rep. 2017;7:5423. doi: 10.1038/s41598-017-05813-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, Jacks T. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–564. doi: 10.1038/416560a. [DOI] [PubMed] [Google Scholar]
- 32.Du W, Jiang P, Mancuso A, Stonestrom A, Brewer MD, Minn AJ, Mak TW, Wu M, Yang X. TAp73 enhances the pentose phosphate pathway and supports cell proliferation. Nat Cell Biol. 2013;15:991–1000. doi: 10.1038/ncb2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dulloo I, Phang BH, Othman R, Tan SY, Vijayaraghavan A, Goh LK, Martin-Lopez M, Marques MM, Li CW, de Wang Y, et al. Hypoxia-inducible TAp73 supports tumorigenesis by regulating the angiogenic transcriptome. Nat Cell Biol. 2015;17:511–523. doi: 10.1038/ncb3130. [DOI] [PubMed] [Google Scholar]