Abstract
The present study aimed to design, synthesize and screen specifically targeted antimicrobial peptides (STAMPs) that can selectively kill Streptococcus mutans (S. mutans) in the biofilm, and to detect protein metabolism, in order to investigate the mechanism of the antibacterial functions of STAMPs against S. mutans. A series of STAMPs were synthesized, and their effects on the selective antibacterial activity of S. mutans on single species and multi-species biofilms under the condition of the planktonic state were studied. The total protein of S. mutans was extracted before and after C11H, and matrix-assisted laser adsorption ionization-time of flight mass spectrometry identification was performed. The antibacterial activity on planktonic S. mutans was increased 3- to 4-fold via C8H, C11H, C12H, C13H, and C14H compared with hLF1-11 (H) alone, and there was no difference between Streptococcus gordonii (S. gordonii) and Streptococcus sanguis (S. sanguis). C8H, C11H, C12H, C13H, and C14H had significant inhibitory effects on the growth of S. mutans biofilm, but there were no significant effects on S. gordonii and S. sanguis biofilms. The number of S. mutans in biofilm decreased at 4 h after C8H, C11H, C12H, C13H and C14H and C8, C11, C12, C13 and C14 had no effect on the growth of planktonic and biofilm states of S. mutans, S. gordonii and S. sanguis species. C11H and C12H exhibited the most obvious effects, followed by C13H and C14H, and then C8H. A total of 21 protein spots with a mean change ratio of 1.5 were identified, all of which were downregulated after C11H. A total of 19 proteins were successfully identified, including cell cycle-relative proteins, nucleic acid metabolism-related enzymes and proteins, virulence factors, protein biosynthesis and regulation, proteins involved in energy metabolism, and proteins with unknown function. In the present study, STAMPs with selective antibacterial activity against S. mutans grown in planktonic or biofilm states but without obvious effects on oral Streptococci and multi-species biofilm were successfully designed and synthesized. Differential protein expression before and after C11H was identified. The mechanism of the antibacterial function was also discussed. Results of the present study laid the foundation for application of STAMPs in the prevention and treatment of dental caries.
Keywords: proteomics, Streptococcus mutans, dental caries, specifically targeted antimicrobial peptides
Introduction
Dental caries affect a large proportion of the world population. S. mutans is considered a major etiologic agent involved in human dental caries. In the past several decades, efforts have been made to remove S. mutans from the biofilm community via replacement therapy, passive or active immunization attempts, and targeted antimicrobial agents. However, most antimicrobial agents have broad spectra of activity, which can usually kill benign and pathogenic organisms indiscriminately. When the normal dental flora is disturbed by any number of factors, microbial infections at the mucosal surface may happen. Candida albicans and Staphylococcus aureus are two examples of classical opportunistic pathogens that take advantage of increased niche size after antibiotic treatment (1,2). The problems resulting from wide-spectrum antibiotic use highlight the fundamental need for new ‘targeted’ antibiotic therapies to combat mucosal pathogens with a minimal impact on normal microflora. In 2006, Eckert et al suggested the concept of specifically targeted antimicrobial peptides (STAMPs) (3). STAMPs containing the target peptide, antibacterial peptide and/or connecting regions can selectively target and kill bacteria. STAMPs independently play roles in different regions to greatly increase anti-bactericidal capacity.
At present, bacterial pheromone or signal peptide is usually used as the targeting region for STAMPs (3–7). Pheromone or signal peptide is a kind of special signaling molecule that mediates the exchange of cellular signals between gram-positive bacteria and fungi. The signal peptide-mediated quorum-sensing (QS) system indicates that S. mutans is a type of cariogenic bacteria in dental plaque biofilm. Deformation of the S. mutans QS system mainly regulates the gene expression state, bacteriocin production and biological behavior through the functions of competence-stimulating peptide (CSP) and the dual signal transduction system. CSP and its fragments are the targets of the antibacterial functions of STAMPs against S. mutans, and the region from 6 th threonine to 13 th arginine is sufficient for targeting (8). Seven allele genes from 36 clinical isolates of S. mutans were isolated from three different mature CSPs, and four kinds of different amino acids were the same with 12 different CSP sequences (9). CSP had two functional regions: Τhe C-terminal region of the CSP and the structure of the α helix core, which is located between the fourth leucine and the twentieth glycine, and constitutes the essential region of the receptor binding (10). Previous findings showed that the hydrophobic property of CSP was very important for the recognition of CSP, and that the activity of CSP was not affected by the hydrophobic leucine or phenylalanine substitution. The 9th, 12th and 13th hydrophilic residues were not affected, and 18th alanine was also important for the function of CSP (11). Thus, CSP of S. mutans may be used as the targeting peptide domain. However, the functional fragment of CSP and the receptor-binding region remain to be determined. Furthermore, the molecular mechanism of STAMPs against S. mutans has yet to be elucidated.
Thus, in the present study, a series of STAMPs were designed and chemically synthesized. The aim was to examine the possibility of different fragments of CSP to serve as the STAMP-targeting domain. In addition, the proteomic analysis with two-dimensional differential in gel electrophoresis and matrix-assisted laser adsorption ionization-time of flight mass spectrometry (MALDI-TOF-MS) were performed to investigate the molecular mechanism of the function of STAMPs against S. mutans.
Materials and methods
Strains and peptides
Strains in the study are shown in Table I. In this experiment, peptide synthesis was performed by Invitrogen; Thermo Fisher Scientific (Shanghai) Co., Ltd. (Shanghai, China) and all sequences are listed in Table II.
Table I.
Strains in the study.
| Strains | Source |
|---|---|
| S. mutans UA 159 | Institute of Stomatological Research, Sun Yat-sen University |
| S. mutans GS-5 | Institute of Stomatological Research, Sun Yat-sen University |
| luc-ldh reporter strain of S. mutans | The present study, and refs, 12 and 13 |
| S. gordonii ATCC 10558 | China General Microbiological Culture Collection Management Center |
| S. sanguis ATCC 49295 | China General Microbiological Culture Collection Management Center |
Table II.
Peptides in the study.
| Peptides | Amino acid sequence | Molecular weight |
|---|---|---|
| CSPC8 (C8) | TFFRLFNR | 1,100.6 |
| CSPC11 (C11) | TFFRLFNRSFT | 1,435.66 |
| CSPC12 (C12) | LSTFFRLFN | 1,144.35 |
| CSPC13 (C13) | STFFRLFNRSFTQ | 1,650.87 |
| CSPC14 (C14) | LSTFFRLFNRSFTQ | 1,764.03 |
| hLF1-11 (H) | GRRRRSVQWCA | 1,493.6 |
| CSPC8hLF1-11 (C8H) | TFFRLFNRGRRRRSVQWCA | 2,575.88 |
| CSPC11hLF1-11 (C11H) | TFFRLFNRSFTGRRRRSVQWCA | 2,912.1 |
| CSPC12hLF1-11 (C12H) | LSTFFRLFNGRRRRSVQWCA | 2,619.93 |
| CSPC13hLF1-11 (C13H) | STFFRLFNRSFTQGRRRRSVQWCA | 3,126.45 |
| CSPC14hLF1-11 (C14H) | LSTFFRLFNRSFTQGRRRRSVQWCA | 3,239.61 |
STAMPs MICs of S. mutans, S. gordonii, and S. sanguis
The antimicrobial activities of the peptides against oral Streptococci strains were investigated by serial microbroth dilution. Minimal inhibitory concentrations (MICs) of the peptides were determined using the broth microdilution method. S. mutans. UA 159, S. mutans GS-5, S. gordonii ATCC-10558 and S. sanguis ATCC-49295 were cultured anaerobically overnight in brain heart infusion (BHI) broth (BD Biosciences, San Diego, CA, USA) at a temperature of 37°C. The medium was diluted (1:20) in fresh BHI and cultured anaerobically for 2.5 h to reach the mid-logarithmic growth phase (diluted to 1×105-106 CFU/ml). Briefly, 2-fold serial dilutions of each peptide were prepared in BHI medium at a volume of 200 µl per well in 96-well round-bottom microtitre plates (Costar, Cambridge, MA, USA). Then, 20 µl per well of bacterial cell suspension with a final concentration of 1×105 CFU/ml for all bacterial species was transferred to the microtitre plates for cell culture. After anaerobic incubation at 37°C for 24 h, the MIC endpoint was defined as the lowest concentration of the test agents that completely inhibited cell growth compared with the drug-free control. The MIC values were expressed as the median of at least three independent experiments.
Effect of STAMPs on the biofilm of a single species
The S. mutans UA 159, S. gordonii ATCC-10558 and S. sanguis ATCC-49295 were cultured anaerobically overnight in BHI liquid medium. Then culture medium was diluted (1:20) in fresh BHI and cultured anaerobically for 2.5 h to reach the mid-logarithmic growth phase. The concentration was then adjusted to 107 CFU/ml. S. mutans UA 159 BHI medium contained 0.5% sucrose, S. gordonii ATCC-10558 and S. sanguis ATCC-49295 BHI medium contained 0.5% glucose and mannose, respectively. Bacteria were transferred to 96-well flat bottom plate with 200 µl for each well. After centrifugation for 10 min at 4,390 × g, the plates were incubated at 37°C for 4 h to form a biofilm. The supernatant was discarded and 100 µl phosphate buffered saline (PBS) containing diluted peptide was added to reach the final concentration of 128 µg/ml, followed by incubation for 5 min with PBS as the control. The supernatant was discarded, and 100 µl fresh BHI culture medium containing diluted residue peptide was added, followed by centrifugation for 10 min at 4,390 × g. Subsequently, fresh medium containing sugar was added, followed by incubation at 37°C. An automatic enzyme standard instrument was used to measure OD600, once every 30 min for 5 h. Three technical and biological repeats were used for each experiment. Statistical analysis software SPSS 13.0 (SPSS, Inc., Chicago, IL, USA) was used, single-factor analysis of variance was performed to analyze the differences in OD600 values among S. mutans UA 159, S. gordonii ATCC-10558, and S. sanguis ATCC-49295. P<0.05 was considered to indicate a statistically significant analysis.
Effect of STAMPs on the biofilm of multi species
The luc-ldh reporter strain of S. mutans, S. gordonii ATCC-10558, and S. sanguis ATCC-49295 were cultured anaerobically overnight in BHI liquid medium. After that the medium was diluted (1:20) in fresh BHI and cultured anaerobically for 2.5 h to reach the mid-logarithmic growth phase, and then the concentration was adjusted to 1×107 CFU/ml. BHI medium for the luc-ldh reporter strain of S. mutans contained 0.5% sucrose, BHI medium for S. gordonii ATCC-10558 and S. sanguis ATCC-49295 contained 0.5% glucose and mannose, respectively. Bacteria were transferred to a 96-well plate with 200 µl for each well, followed by centrifugation for 2 min at 4,000 × g. Biofilm was formed after anaerobic culture was carried out for 4 h at 37°C. The supernatant was discarded, and 100 µl PBS containing diluted peptide was added to the final concentration of 128 µg/ml. The supernatant was then removed, and 200 µl fresh BHI culture medium containing diluted residue peptide was added, followed by centrifugation for 10 min at 4,390 × g. Subsequently, 200 µl fresh BHI medium containing sugar was added, followed by anaerobic cultivation at 37°C for 5,10,60 and 120 min, respectively, to measure OD600 values to detect biofilm growth. Then 50 µl of 1 mM D-luciferin (Promega Corp., Madison, WI, USA) was added, and the growth of S. mutans was detected using a luminous RLU detection analyzer. Technical and biological repeats were performed for each experiment. Statistical analysis software SPSS 13.0 (SPSS, Inc., Chicago, IL, USA) was used, and single-factor analysis of variance was performed to analyze the difference in OD600 values. P<0.05 was considered to indicate a statistically significant analysis.
Bacterial total protein extraction
The S. mutans UA 159 were cultured anaerobically overnight in BHI liquid medium, and the medium was diluted (1:20) in fresh BHI and cultured anaerobically for 2.5 h to reach the mid-logarithmic growth phase. The concentration was adjusted to 1×106 CFU/ml. C11H was added to the experimental group to reach 20 µg/ml, followed by anaerobic culture at 37°C for 1 h, and the same volume of sterile ultrapure water was added to the control group. Centrifugation for 15 min at 10,000 × g at 4°C was performed to collect bacteria in the experimental and control groups. The supernatant was discarded, followed by an ice bath. Then, 500 µl of lysate containing protease inhibitors (1 mm PMSF, 1 µg/ml pepstatin, 5 µg/ml aprotinin) and 1 mg/ml lysozyme were added, followed by an ice bath for 15 min. Ultrasound (5 sec on/off, with an intensity of 37% for 10 min) was performed, followed by centrifugation (DHS 4 C; 10,000 × g for 50 min) to isolate protein. The extracted protein was purified using an Amersham clean-up kit (GE Healthcare Life Sciences, Uppsala, Sweden), quantified with a GE 2-D Quant kit (Amersham Biosciences, Sweden) and fluorescence labeling was performed with CyDye DIGE Flour (minimal Dye) Labelling kit (GE Healthcare, Arlington, TX, USA).
MALDI-TOF-MS identification of protein spots
Targeted points were placed into the MALDI-TOF-MS, and the parameter was set to reflection mode, ion source accelerating voltage of 24 kV, 1 and 2 for 22 kV acceleration voltage, ion delay extract 0,000 ns, vacuum 1.4×10-7 torr, mass spectrum signal accumulation 300 times in a single scan, with standard maker peak as external standard calibration mass spectrum peak, positive ion spectrum measurement, measurement range control in 700–4,000. Peptide mass fingerprint (PMF) of the sample was obtained. Using LIFT software, four peaks with main strength of PMF were subjected to cascade mass spectrum analysis. The obtained maps were retrieved using Biotools software, with Mascot (Matrix Science, Ltd., London, UK) as the search engine. Search parameter settings were as follows: Database for NCBInr, key species of bacteria, data retrieval method for combined, maximum allowable leakage cutting locus was 1, and enzyme trypsin. The quality error range was calculated as: PMF 50 ppm, MS/MS 0.5 Da.
Results
Antimicrobial activities of STAMPs and its components against planktonic S. mutans, S. gordonii and S. sanguis
Antimicrobial activities of STAMPs and its components against S. mutans, S. gordonii and S. sanguis are shown in Table III. MICs of C8H, C11H, C12H, C13H and C14H were used against the S. mutans strains at a dose of 10.67–13.33 µg/ml, and hLF1-11 was used against the S. mutans strains of MICs at a dose of 37.33–48.00 µg/ml. C8H, C11H, C12H, C13H and C14H increased antibacterial activity against S. mutans by 3–4 times compared with hLF1-11 alone. There were no obvious differences on the effects of C8H, C11H, C12H, C13H, C14H and hLF1-11 on antimicrobial activity against S. gordonii and S. sanguis. The MICs of C8, C11, C12, and C13 and C14 against S. mutans, S. gordonii and S. sanguis were >256.00 g/ml. The results suggested that binding to the targeting peptide domain improved the selective antibacterial effect of hLF1-11 on S. mutans without affecting S. gordonii and S. sanguis.
Table III.
Effects of STAMPs and its components on S. mutans, S. gordonii and S. sanguis MICs.
| IMC ± SD (µg/ml) | ||||
|---|---|---|---|---|
| Peptides | S. mutans UA159 | S. mutans GS-5 | S. gordonii ATCC-10558 | S. sanguis ATCC-49295 |
| hLF 1–11 | 37.33±13.06 | 48.00±17.53 | 64.00±13.06 | 138.67±62.92 |
| C8 | >256.00 | >256.00 | >256.00 | >256.00 |
| C8H | 10.67±4.00 | 12.44±4.22 | 49.78±16.87 | 99.56±33.73 |
| C11 | >256.00 | >256.00 | >256.00 | >256.00 |
| C11H | 10.67±4.00 | 12.44±4.22 | 49.78±16.87 | 99.56±33.73 |
| C12 | >256.00 | >256.00 | >256.00 | >256.00 |
| C12H | 10.67±4.00 | 13.33±4.00 | 49.78±16.87 | 99.56±33.73 |
| C13 | >256.00 | >256.00 | >256.00 | >256.00 |
| C13H | 10.67±4.00 | 13.33±4.00 | 49.78±16.87 | 99.56±33.73 |
| C14 | >256.00 | >256.00 | >256.00 | >256.00 |
| C14H | 10.67±4.00 | 13.33±4.00 | 49.78±16.87 | 99.56±33.73 |
Effects of STAMPs and its components on the single bacteria biofilm
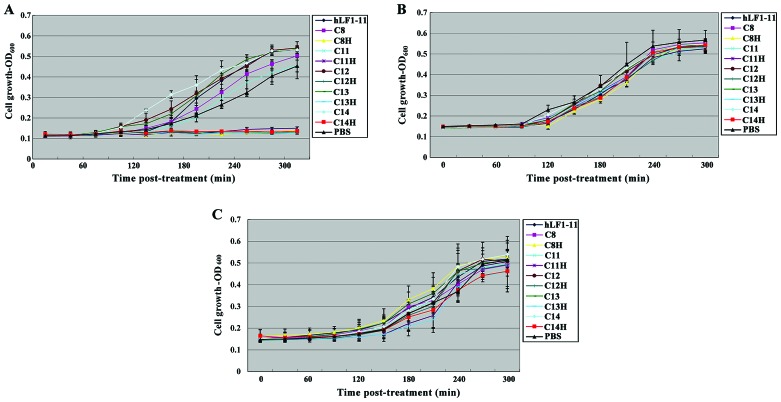
S. mutans in the body usually occurs in the form of dental plaque biofilm. In order to detect the roles of STAMPs on S. mutans in biofilm, biofilms formed by S. mutans, S. gordonii and S. sanguis and after treatment with STAMPS for 5 min were used. Biofilm growth is evident in Fig. 1. After S. mutans was treated with C8H, C11H, C12H, C13H and C14H for 5 min, the growth of biofilm was significantly suppressed, whereas hLF1-11, C8, C11, C12, C13 and C14 did not affect S. mutans biofilm growth (Fig. 1A). Biofilm growth of S. gordonii and S. sanguis was not affected by C8H, C11H, C12H, C13H, C14H, hLF1-11, C8, C11, C12, and C13 and C14 (Fig. 1B and C). The results suggested specific targeting of antimicrobial peptides C8H, C11H, C12H, C13H and C14H in a short period of time can specifically increase the antibacterial effects on bacterial biofilm of S. mutans.
Figure 1.
Effects of STAMPs and its composition on the growth of biofilm of single species. (A) S. mutans, (B) S. gordonii, and (C) S. sanguis. STAMPs, specifically targeted antimicrobial peptides.
Effects of STAMPs and its components on bacterial multi-species biofilms
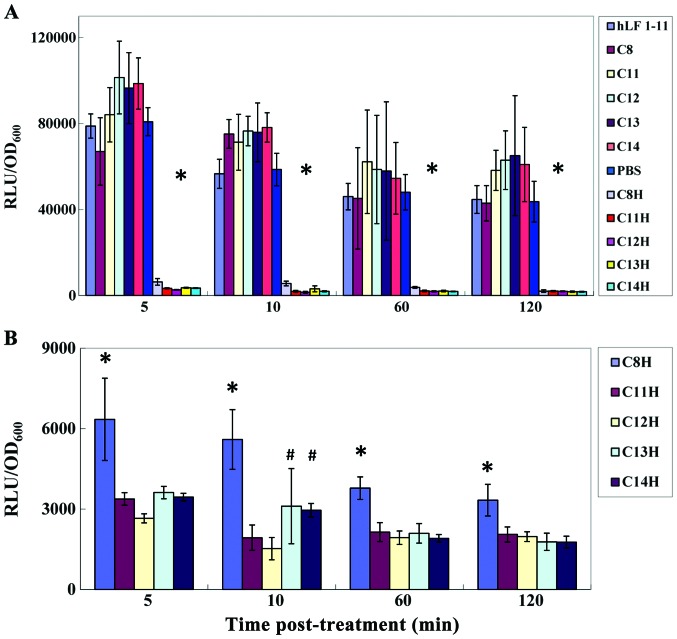
The dental plaque biofilm is a complex structure comprising a variety of bacteria that simulate the S. mutans growth environment in vivo. Bacterial biofilms formed by luc-ldh reporter strains of S. mutans, S. gordonii and S. sanguis were used, and the effects of STAMPs and its components are shown in Fig. 2. RLU values indicated the number of living bacteria in the biofilm of S. mutans. After treatment with C8H, C11H, C12H, C13H and C14H for 5 min, RLU/OD600 was significantly decreased, resulting in the number of living bacterium decreasing significantly. RLU/OD600 remained at a low level at 5,10,60 and 120 min following removal of C8H, C11H, C12H, C13H and C14H, indicating long-term effects. hLF1-11, C8, C11, C12, and C13 and C14 showed no significant effects on growth, and RLU/OD600 values of bacterial biofilm formed by S. mutans (Fig. 2A). Statistical analysis revealed statistically significant effects of those factors on bacterial biofilm formed by S. mutans (P<0.05). As shown in Fig. 2B, statistical analysis revealed that C8H and C11H, C8H and C12H, C8H and C13H, and C8H and C14H showed statistically significant effects at 5,60 and 120 min after fresh BHI culture medium was added. C8H and C11H, C8H and C12H, C8H and C13H, C8H and C14H, C11H and C13H, C12H and C13H, C11H and C14H, and C12H and C14H showed statistically significant differences at 10 min after fresh BHI culture was added. No significant difference was found between C11H and C12H, and C13H and C14H. Thus, C11H and C12H have the strongest effects, followed by C13H and C14H, and then C8H.
Figure 2.
Effects of STAMPs and its composition on the growth of Biofilm of multi species. (A) Effect of STAMPs and different components on the Streptococcus species in the multi species biofilm. *C8H, C11H, C12H, C13H and C14H were significantly different from hLF1-11, C8, C11, C12, C13 and C14. (B) Effect of STAMPs and different components on the growth of S. mutans in the multi-species biofilm. *C8H was significantly different from C11H, C12H, C13H and C14H; #difference of C13H, C14H and C12H, C11H was statistically significant.
Mass spectrum identification
A total of 21 protein spots with a mean change ratio of 1.5 were identified, all of which were downregulated after C11H treatment, and 19 proteins were successfully identified. The proteins included cell cycle relative proteins, nucleic acid metabolism-related enzymes and proteins, virulence factors, protein biosynthesis and regulation, proteins involved in energy metabolism and proteins with unknown function (Table IV).
Table IV.
Differential protein expression profiles for C11H.
| Cellular role | Master no. | Average ration | Protein name | Accession no. | Coverage (%) | Molecular mass Kda (theoretical) | PI (theoretical) |
|---|---|---|---|---|---|---|---|
| Cell cycle relative proteins | 654 | −4.75 | Putative 40K cell wall protein precursor | gi|24379086 | 16 | 67.474 | 5.66 |
| 1327 | −11.09 | Putative cell division protein DivIVA | gi|24379038 | 34 | 30.803 | 4.48 | |
| 1341 | −5.58 | Putative cell division protein DivIVA | gi|24379038 | 34 | 30.803 | 4.48 | |
| 1355 | −8 | Putative cell division A protein DivIV | gi|24379038 | 34 | 30.803 | 4.48 | |
| Nucleotide metabolism | 824 | −12.84 | Inositol-5 monophosphate dehydrogenase | gi|24380485 | 18 | 53.118 | 5.66 |
| 2068 | −5.54 | Putative dTDP-4-keto-L-rhamnose reductase | gi|24379857 | 34 | 22.407 | 4.98 | |
| 2141 | −4.63 | Orotate phosphoribo-syltransferase | gi|24379645 | 27 | 22.802 | 5.37 | |
| 2334 | −5.37 | Single-strand DNA-binding protein | gi|24380219 | 40 | 18.321 | 4.98 | |
| Virulence factors | 261 | −17.52 | Hypothetical protein SMU.689 | gi|24379161 | 14 | 107.128 | 5.22 |
| Protein biosynthesis and regulation | 2035 | −4.19 | Elongation factor P | gi|24380208 | 24 | 20.768 | 4.89 |
| 2072 | −4.7 | Ribosome recycling factor | gi|24380004 | 30 | 20.63 | 5. 58 | |
| 1781 | −4.2 | Heat shock protein GrpE (HSP-70 cofactor) | gi|24378605 | 35 | 20.676 | 4.6 | |
| Energy metabolism amino acid metabolism | 1241 | −5.4 | Branch ed-chain amino acid aminotransferase | gi|24379628 | 32 | 37.822 | 5.03 |
| 1431 | −5.09 | Putative cystein e sythetase A; O-acetylserine lyase | gi|24378980 | 34 | 32.417 | 5.82 | |
| 1805 | −4.51 | Putative amino acid ABC transporter, ATP-binding protein | gi|24379908 | 21 | 28.934 | 5.85 | |
| Fatty acid metabolism | 1551 | −4.34 | Putative trans-2-enoyl-ACP reductase | gi|24380114 | 44 | 33.571 | 5.74 |
| Nitrogen metabolism | 1628 | −4.12 | NAD synthetase | gi|24378953 | 28 | 30.156 | 5.16 |
| Glucometabolism | 2008 | −6.09 | Putative N-acetyglucosamine-6-phosphate isomerase | gi|24379109 | 42 | 25.601 | 5.34 |
| Unknown function | 1506 | −5.93 | Hypothetical protein SMU.1760c | gi|24380128 | 30 | 33.119 | 5.12 |
| 1576 | −6.04 | Hypothetical protein SMU.1789c | gi|24380155 | 31 | 25.725 | 4.51 | |
| 2188 | −6.14 | Hypothetical protein SMU.1716c | gi|24380090 | 55 | 19.895 | 5.13 |
Discussion
In the present study, CSP of S. mutans is employed as the targeting peptide domain. However, the functional fragment of CSP and receptor binding has not been clearly determined. Therefore, five targeting peptide domains were designed: i) CSPC8 (C8)8, 6–13 amino acid of CSP, TFFRLFNR; ii) CSPC11 (C11), contained the 7th, 11st, and 15th amino acid of CSP, TFFRLFNRSFT; iii) CSPC12 (C12), 4–12 amino acid of CSP, LSTFFRLFN; iv) CSPC13 (C13), four amino acids of the amino terminal and four amino acids of the carboxyl terminal of CSP were removed, STFFRLFNRSFTQ; v) CSPC14 (C14), three amino acids of the amino terminal and four amino acids of the carboxyl terminal of CSP were removed, LSTFFRLFNRSFTQ. Antibacterial peptides were selected at an early stage by the research group to demonstrate the promising antimicrobial activity of hLF1-11 (12–14). STAMPs can be used to connect the targeting peptide domain to the antimicrobial peptide via a short fragment amino acid sequence, but can also be directly connected. In addition, the use of the connector is determined by use of the targeting peptide and antimicrobial peptide domains (3–5,15). He et al have found that, for antimicrobial peptides rich in arginine and tryptophan, the connector did not improve the targeting of STAMPs (16). hLF1-11 contains five arginine, which may reduce the number of amino acids in STAMPs, and it was easier to be synthesized. Therefore, there was no connecting domain in the STAMPs in the present study.
To determine the specific-targeted antimicrobial activities of STAMPs against the S.mutans, the antimicrobial activities of the peptides against oral Streptococci strains were investigated by serial microbroth dilution. MICs of the peptides showed that C8H, C11H, C12H, C13H and C14H increased antimicrobial activity specifically against planktonic S. mutans by 3- to 4-fold compared with hLF1-11 alone. In addition, there was no significant difference of antimicrobial activities of C8H, C11H, C12H, C13H, C14H and hLF1-11 against S. gordonii and S. sanguis. C8, C11, C12, C13 and C14 did not show antimicrobial activities against S. mutans, S. gordonii and S. sanguis. The results suggest that antimicrobial activities and specificity against S. mutans of hLF1-11 were enhanced after the connection to targeted domains, but other tested oral Streptococci were not affected. S. mutans is present in oral biofilms (17). Additionally, the antimicrobial sensitivity of drugs is reduced 100–1,000 times in biofilm bacteria compared with bacteria in the planktonic state (18). Thus, the effects of STAMPs and its components against S. mutans, S. gordonii and S. sanguis in single species biofilm were tested. It was found that, 5 min after treatment with C8H, C11H, C12H, C13H and C14H, respectively, the number of S. mutans live bacteria in a biofilm were significantly reduced. However, 5 min after C8H, C11H, C12H, C13H, C14H, hLF1-11, C8, C11, C12, and C13 and C14, respectively, the growth of S. gordonii and S. sanguis biofilms was not affected. The results showed the specific antimicrobial activity of C8H, C11H, C12H, C13H and C14H against S. mutans in single species biofilm was significant even after treatment over a short period of time (5 min). S. mutans coexists with many bacteria in the dental plaque biofilm, a complex structure comprising a variety of bacteria. In order to mimic the growing environment of S. mutans, this experiment observed the roles of the STAMPs and its components in multispecies bacterial biofilms. However, 5 min after treatment with C8H, C11H, C12H, C13H and C14H, respectively, the number of S. mutans live bacteria in a biofilm was significantly reduced. hLF1-11, C8, C11, C12, and C13 and C14 had no effect on the growth of S. mutans. However, 5 min after the treatment of C8H, C11H, C12H, C13H, C14H, hLF1-11, C8, C11, C12, and C13 and C14, respectively, the growth of S. gordonii and S. sanguis in biofilm was not affected. These data suggested that STAMPs can improve the selective inhibition on the growth of S. mutans without affecting other bacteria in the biofilm.
Previous findings showed that hLF1-11 can penetrate the cell wall and the cell membrane without destroying the integrity of the cell membrane, and hLF1-11 has DNA binding force (14). Thus, we speculated that DNA may be the target. In order to study the mechanism underlying the effects of STAMPs on S. mutans, changes in the protein expression of S. mutans treated with C11H was examined. Additionally, 19 proteins were successfully identified, including cell cycle relative proteins, nucleic acid metabolism-related enzymes and proteins, virulence factors, protein biosynthesis and regulation, proteins involved in energy metabolism and proteins with unknown function. We found that C11H inhibited inositol-5-monophosphate dehydrogenase (IMPDH), dTDP-4-keto-L-rhamnose reductase and orotate phosphoribosyl transferase expression. These enzymes participate in nucleic acid metabolism. IMPDH is a key enzyme involved in purine metabolism. Through the oxidation reduction reaction and hydrolysis catalyzed by phosphoric acid cyclohexyl 6, IMPDH plays an important role in controlling the number of guanine nucleotides (19). Deoxidation thymidine diphosphate-4-keto l is a kind of rhamnose reductase belonging to the oxidoreductase family, which was mainly involved in nucleotide sugar metabolism, streptomycin synthesis and polyketide synthesis of sugar and other metabolic reactions (20). Oral acid phosphoryl glycosyltransferase is one of the members of the phosphoribosyl transferase family and is involved in the synthesis and the recovery of the synthesis of purine nucleotides and pyrimidine nucleotides. Orotate phosphoribosyl transferase catalytic orotic acid and phosphoribosyl pyrophosphate generate inorganic pyrophosphate and 5′-phosphate acid whey acid riboside (OMP) in the presence of Mg2+, while OMP generates 5′-uridine with the function of decarboxylase, and the former is the key factor in the synthesis of pyrimidine nucleotide (21,22). Single-stranded DNA binding protein (SSBP) is a single-stranded region along the replication fork that prevents the formation of new single-strand DNA to form double-stranded DNA or nucleic acid enzyme degradation of protein. In cells, a large number of SSBPs bind to single-strand DNA to stretch the DNA without causing nodules and bending to facilitate the role of single DNA to serve as a template. Thus, SSBP does not possess enzyme activity nor binds to ATP, which is an essential ingredient for DNA replication, recombination and repair (23,24). Proteomic analysis confirmed that C11H inhibited DNA-related enzymes and proteins, which is one of the mechanisms of its antibacterial effect on S. mutans. It also indicates that the antibacterial effect of C11H was achieved by the hLF1-11.
In conclusion, STAMPs were successfully designed and screened and the overall level of protein metabolism was explored to investigate the mechanism of antibacterial function. The major findings of this study are i) STAMPs had selective antibacterial activity against S. mutans grown in liquid or biofilm states but did not affect other oral Streptococci tested. ii) C11H and C12H exhibited the strongest effects, followed by C13H and C14H, and C8H. iii) C11H inhibited the expression of nucleic acid metabolism-related enzymes, which was one of the mechanisms of the inhibitory effects on S. mutans. Therefore, STAMPs can be used to improve antimicrobial therapies in the treatment of dental caries.
References
- 1.Mitchell TJ. The pathogenesis of streptococcal infections: From tooth decay to meningitis. NAT REV MICROBIOL. 2003;1:219–230. doi: 10.1038/nrmicro771. [DOI] [PubMed] [Google Scholar]
- 2.Sansonetti PJ. War and peace at mucosal surfaces. Nat Rev Immunol. 2004;4:953–964. doi: 10.1038/nri1499. [DOI] [PubMed] [Google Scholar]
- 3.Eckert R, Qi F, Yarbrough DK, He J, Anderson MH, Shi W. Adding selectivity to antimicrobial peptides: Rational design of a multidomain peptide against Pseudomonas spp. Antimicrob Agents Chemother. 2006;50:1480–1488. doi: 10.1128/AAC.50.4.1480-1488.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qiu XQ, Wang H, Lu XF, Zhang J, Li SF, Cheng G, Wan L, Yang L, Zuo JY, Zhou YQ, et al. An engineered multidomain bactericidal peptide as a model for targeted antibiotics against specific bacteria. Nat Biotechnol. 2003;21:1480–1485. doi: 10.1038/nbt913. [DOI] [PubMed] [Google Scholar]
- 5.Qiu XQ, Zhang J, Wang H, Wu GY. A novel engineered peptide, a narrow-spectrum antibiotic, is effective against vancomycin-resistant Enterococcus faecalis. Antimicrob Agents Chemother. 2005;49:1184–1189. doi: 10.1128/AAC.49.3.1184-1189.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl HG, de Kruijff B. Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science. 1999;286:2361–2364. doi: 10.1126/science.286.5448.2361. [DOI] [PubMed] [Google Scholar]
- 7.Kristiansen PE, Fimland G, Mantzilas D, Nissen-Meyer J. Structure and mode of action of the membrane-permeabilizing antimicrobial peptide pheromone plantaricin A. J Biol Chem. 2005;280:22945–22950. doi: 10.1074/jbc.M501620200. [DOI] [PubMed] [Google Scholar]
- 8.Eckert R, He J, Yarbrough DK, Qi F, Anderson MH, Shi W. Targeted killing of Streptococcus mutans by a pheromone-guided ‘smart’ antimicrobial peptide. Antimicrob Agents Chemother. 2006;50:3651–3657. doi: 10.1128/AAC.00622-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allan E, Hussain HA, Crawford KR, Miah S, Ascott ZK, Khwaja MH, Hosie AH. Genetic variation in comC, the gene encoding competence-stimulating peptide (CSP) in Streptococcus mutans. FEMS Microbiol Lett. 2007;268:47–51. doi: 10.1111/j.1574-6968.2006.00593.x. [DOI] [PubMed] [Google Scholar]
- 10.Syvitski RT, Tian XL, Sampara K, Salman A, Lee SF, Jakeman DL, Li YH. Structure-activity analysis of quorum-sensing signaling peptides from Streptococcus mutans. J Bacteriol. 2007;189:1441–1450. doi: 10.1128/JB.00832-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tian X, Syvitski RT, Liu T, Livingstone N, Jakeman DL, Li YH. A method for structure-activity analysis of quorum-sensing signaling peptides from naturally transformable streptococci. Biol Proced Online. 2009;11:207–226. doi: 10.1007/s12575-009-9009-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Does AMVD, Bogaards SJP, Ravensbergen B, Beekhuizen H, Dissel JTV, Nibbering PH. Antimicrobial peptide hLF1-11 directs granulocyte-macrophage colony-stimulating factor-driven monocyte differentiation toward macrophages with enhanced recognition and clearance of pathogens. Antimicrob Agents Chemother. 2010;54:811. doi: 10.1128/AAC.00652-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stallmann HP, Faber C, Bronckers AL, Amerongen Nieuw AV, Wuisman PI. Osteomyelitis prevention in rabbits using antimicrobial peptide hLF1-11- or gentamicin-containing calcium phosphate cement. J Antimicrob Chemother. 2004;54:472–476. doi: 10.1093/jac/dkh346. [DOI] [PubMed] [Google Scholar]
- 14.Huo L, Zhang K, Ling J, Peng Z, Huang X, Liu H, Gu L. Antimicrobial and DNA-binding activities of the peptide fragments of human lactoferrin and histatin 5 against Streptococcus mutans. Arch Oral Biol. 2011;56:869–876. doi: 10.1016/j.archoralbio.2011.02.004. [DOI] [PubMed] [Google Scholar]
- 15.Lu X, Wan L, Yang H, Zhang J, Li S, Kang M, Li Y, Cheng J. Fusion of fungicidal peptide dhvar4 to enterococcal peptide pheromone increases its bactericidal activity against Enterococcus faecalis. Chem Biol Drug Des. 2006;68:220–224. doi: 10.1111/j.1747-0285.2006.00434.x. [DOI] [PubMed] [Google Scholar]
- 16.He J, Eckert R, Pharm T, Simanian MD, Hu C, Yarbrough DK, Qi F, Anderson MH, Shi W. Novel synthetic antimicrobial peptides against Streptococcus mutans. Antimicrob Agents Chemother. 2007;51:1351–1358. doi: 10.1128/AAC.01270-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Banas J. Virulence properties of Streptococcus mutans. Front Biosci. 2004;9:1267–1277. doi: 10.2741/1305. [DOI] [PubMed] [Google Scholar]
- 18.Lutkenhaus J. FtsZ ring in bacterial cytokinesis. Mol Microbiol. 1993;9:403–409. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- 19.Marston AL, Thomaides HB, Edwards DH, Sharpe ME, Errington J. Polar localization of the MinD protein of Bacillus subtilis and its role in selection of the mid-cell division site. Genes Dev. 1998;12:3419–3430. doi: 10.1101/gad.12.21.3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hedstrom L, Gan L. IMP dehydrogenase: Structural schizophrenia and an unusual base. Curr Opin Chem Biol. 2006;10:520–525. doi: 10.1016/j.cbpa.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 21.Shibaev VN. Biosynthesis of bacterial polysaccharide chains composed of repeating units. Adv Carbohydr Chem Biochem. 1986;44:277–339. doi: 10.1016/S0065-2318(08)60080-3. [DOI] [PubMed] [Google Scholar]
- 22.Gonzalez-Segura L, Witte JF, McClard RW, Hurley TD. Ternary complex formation and induced asymmetry in orotate phosphoribosyltransferase. Biochemistry. 2007;46:14075–14086. doi: 10.1021/bi701023z. [DOI] [PubMed] [Google Scholar]
- 23.Yablonski MJ, Pasek DA, Han BD, Jones ME, Traut TW. Intrinsic activity and stability of bifunctional human UMP synthase and its two separate catalytic domains, orotate phosphoribosyltransferase and orotidine-5′-phosphate decarboxylase. J Biol Chem. 1996;271:10704–10708. doi: 10.1074/jbc.271.18.10704. [DOI] [PubMed] [Google Scholar]
- 24.Bochkarev A, Pfuetzner RA, Edwards AM, Frappier L. Structure of the single-stranded-DNA-binding domain of replication protein A bound to DNA. Nature. 1997;385:176–181. doi: 10.1038/385176a0. [DOI] [PubMed] [Google Scholar]