Abstract
The present study aimed to measure the expression of microRNA (miRNA/miR) −429 in gastric cancer and investigate the associated mechanism of action. A total of 30 patients with gastric cancer who received radical or palliative resection at Jining No. 1 People's Hospital between January-October 2016 were included in the present study. Resected gastric cancer and tumor-adjacent tissues were resected. Gastric cancer BGC-823 cells were transfected with miR-429 mimics to induce the overexpression of miR-429, or transfected with small interfering (si)RNA of heparanase (HPSE) for the silencing of HPSE. Reverse transcription-quantitative polymerase chain reaction was used to measure the expression of miR-429 or HPSE mRNA. Western blotting was employed to determine the protein expression of HPSE. Cell-Counting Kit-8 assay was carried out to test cell proliferation and a Transwell assay was used to determine cell invasion ability. Expression of HPSE mRNA and protein in gastric cancer tissues was increased compared with tumor-adjacent tissues. Reduced expression of miR-429 in gastric cancer tissues may be associated with the targeting of HPSE mRNA by miR-429. Overexpression of miR-429 inhibited the transcription and translation of the HPSE gene. However, overexpression of miR-429 did not affect the proliferation of gastric cancer cells. Notably, overexpression of miR-429 reduced the invasion ability of gastric cancer cells. Transfection with HPSE siRNA decreased the expression of the HPSE protein in BGC-823 cells and inhibited the occurrence and development of gastric cancer by reducing the invasion ability of the cells. The present study demonstrated that expression of miR-429 in gastric cancer tissues was significantly reduced compared with tumor-adjacent tissues. As a tumor-suppressor gene, miR-429 decreases the invasion ability of gastric cancer cells by downregulating the expression of HSPE.
Keywords: microRNA-429, heparanase, invasion, gastric cancer
Introduction
Gastric cancer is a type of tumor with a high incidence, and the metastasis of gastric cancer is the main reason for the failure of surgical treatment (1). The first process in the invasion and metastasis of tumor cells is the breakthrough of a tissue barrier formed by basement membrane and extracellular matrix (2). Studies show that heparanase (HPSE) is a β-D-glucuronate endonuclease of the major component of the tissue barrier, heparan sulfate proteoglycan (HSPG) (3,4). HPSE degrades HSPG by cleaving heparan sulfate glycosaminoglycan chain (HS-GAG), and directly and indirectly promotes the invasion and metastasis of tumors in the cooperation of multiple factors (3,4). Nobuhisa et al report that the expression of HPSE in colorectal cancer tissues is high (69%), and is especially high in patients with higher stages of colorectal cancer or lymphatic vessel metastasis (5). In addition, the invasion ability of tumor cells transfected with HPSE is enhanced, while HPSE inhibitor can reduce the invasion ability of tumor cells (5).
MicroRNA (miRNA or miR) is small non-coding RNA molecules with 18–25 nucleotides, which can regulate mRNA translation by binding with its 3′-untranslated region (3′-UTR) (6). Functional analysis shows that upregulated or downregulated expression of miRNA affects the proliferation, differentiation, and apoptosis of cells, as well as the occurrence and metastasis of tumors (7). Interactions between miRNA and tumor-associated proteins may promote or inhibit the biological function of tumors (8). The coding gene of miR-429 is located on human chromosome 1 (1p36.33), which is related with some kinds of tumors (9). It is shown that inhibition of miR-429 expression enhances the cytotoxic activity of cisplatin on endometrial endometrioid carcinoma (10). In addition, miR-429 acts as a tumor-suppressor gene for breast cancer (11). In the present study, we measure the expression of miR-429 in gastric cancer and investigate its mechanism of action.
Materials and methods
Patients
A total of 30 patients with gastric cancer who received radical or palliative resection at Jining No. 1 People's Hospital between January 2016 and October 2016 were included in the present study. Resected gastric cancer tissues were diagnosed and classified by two individual pathologists following the tumor classification standards published by World Health Organization in 2003. Tumor-adjacent tissues more than 5 cm away from tumor tissues were also resected as control. The clinical information and pathological data of all subjects were collected. All procedures were approved by the Ethics Committee of Jining No. 1 People's Hospital. Written informed consents were obtained from all patients or their families.
Cells
Gastric cancer BGC-823 cells (Cell Bank, Chinese Academy of Sciences, Shanghai, China) were seeded into 24-well plates and divided into negative control and miR-429 mimics groups. The cells were cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum. When reaching 70–90% confluency, 1.25 µl miR-429 mimics (20 µM) and 2 µl liposome (Lipofectamine® 2000; Thermo Fisher Scientific, Inc., Waltham, MA, USA) were mixed with 50 µl Opti Memi medium, respectively, in individual Eppendorf tubes. After standing still for 5 min, the mixtures in the two Eppendorf tubes were mixed and kept under room temperature for 15 min, followed by addition into each culture well. Six h later, the medium was replaced by fresh RPMI-1640 medium supplemented with 10% fetal bovine serum, and the cells were cultured under normal condition for 48 h before use.
For the silencing of human HPSE mRNA (GenBank ID: 10855), BGC-823 cells were transfected with small interfering RNA (siRNA) of HPSE (siRNA sequences: sense, 5′-GGAUAUUUGCAAAUAUGGATT-3′; anti-sense, 5′-UCCAUAUUUGCAAAUAUCCTG-3′) or negative control (NC) siRNA (NC siRNA sequences: 5′-UUCUCCGAACGUGUCACGUTT-3′; 5′-ACGUGACACGUUCGGAGAATT-3′) (GenePharma, Shanghai, China). BGC-823 cells were seeded onto 6-well plates and cultured at 37°C and 5% CO2. When the cells reached 70% confluency, the medium was discarded and washed with phosphate-buffered saline (PBS) for three times, before addition of 2 ml Opti-Memi medium into each well. Then, siRNA and 5 µl liposome were dissolved in 250 µl Opti-MEM in two individual Eppendorf tubes, respectively. After standing still at room temperature for 5 min, the two tubes were combined before incubation at room temperature for another 20 min. Subsequently, the mixture was added into each well. After incubation at 37°C and 5% CO2 for 6 h, the medium was changed to normal medium.
Bioinformatics
Bioinformatics prediction is a powerful tool for the study of the functions of miRNAs. To understand whether HPSE was a target gene of miR-429, we used TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org/microrna/home.do) for analysis.
Quantitative polymerase chain reaction (qPCR)
Gastric cancer and tumor-adjacent tissues (100 mg) were ground into powder in liquid nitrogen and mixed with 1 ml Trizol (Thermo Fisher Scientific, Inc.) for lysis. For cells, 2×105 cells were thoroughly mixed with 1 ml Trizol for lysis. Then, total RNA was extracted using phenol chloroform method. The purity of RNA was determined by A260/A280 using ultraviolet spectrophotometry (Nanodrop ND2000; Thermo Fisher Scientific, Inc.). Then, cDNA was obtained by reverse transcription using Reverse Transcription System (Takara Bio, Dalian, China) from 1 µg RNA and stored at −20°C. Reverse transcription of miRNA was carried out using SYBR PrimeScript miRNA RT-PCR Kit (Takara Bio).
The expression of miR-429 was determined by SYBR-Green qPCR kit (Kapa Biosystems, Wilmington, MA, USA), using U6 as internal reference. The reaction system (25 µl) contained 12.5 µl SYBR Premix Ex Taq, 1 µl PCR forward primer (miR-429, 5′-UAAUACUGUCUGGUAAAACCGU-3′), 1 µl PCR reverse primer (5′-UUCUCCGAACGUGUCACGUTT-3′), 2 µl template and 8.5 µl ddH2O. The reaction protocol was: initial denaturation at 95°C for 30 sec; 40 cycles of 95°C for 5 sec and 60°C for 20 sec (iQ5; Bio-Rad, Hercules, CA, USA). The 2−ΔΔCq method was used to calculate the relative expression of miR-429 against U6. Each sample was tested in triplicate.
SYBR-Green qPCR kit (Kapa Biosystems,) was used to detect mRNA expression of HSPE, using GADPH as internal reference. The reaction system (20 µl) was composed of 10 µl SYBR Ex Taq-Mix, 0.5 µl upstream primer (HSPE, 5′-ACTGGCAATCTCAAGTCAAC-3′; GADPH, 5′-AAGGTGAAGGTCGGAGTCA-3′), 0.5 µl downstream primer (HSPE, 5′-CTTAGCCGTCTTTCTTCG-3′; GADPH, 5′-GGAAGATGGTGATGGGATTT-3′), 1 µl cDNA and 8 µl ddH2O. PCR condition was: Iinitial denaturation at 95°C for 10 min; 40 cycles of denaturation at 95°C for 1 min, annealing at 60°C for 40 sec and elongation at 72°C for 30 sec; final elongation at 72°C for 1 min (iQ5; Bio-Rad). The 2−ΔΔCq method was used to calculate the relative expression of HSPE mRNA against GADPH. Each sample was tested in triplicate.
Western blotting
Cells were seeded into 6-well plates at a density of 1×106/well. At 48 h after transfection, the cells were collected and mixed with 100 µl precooled Radio-Immunoprecipitation assay (RIPA) lysis buffer containing 1 mM phenylmethylsulfonyl fluoride for lysis of 15 min at 4°C. Then, the mixture was centrifuged at 12,000 × g and 4°C for 5 min. The supernatant was used to determine protein concentration by bicinchoninic acid (BCA) protein concentration determination kit (RTP7102; Real-Times Biotechnology Co., Ltd., Beijing, China). Protein samples (50 µg) were then mixed with 2X sodium dodecyl sulfate loading buffer before denaturation in boiling water bath for 5 min. Afterwards, the samples (10 µl) were subjected to 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis at 100 V. The resolved proteins were transferred to polyvinylidene difluoride membranes on ice (300 mA, 1.5 h) and blocked with 5 g/l skimmed milk at room temperature for 1 h. Then, the membranes were incubated with rabbit anti-human HPSE polyclonal primary antibody (1:1,000; Abcam, Cambridge, UK) at 4°C overnight. After extensive washing with phosphate-buffered saline with Tween 20 for 3 times of 15 min, the membranes were incubated with polyclonal goat anti-rabbit horseradish peroxidase-conjugated secondary antibody (1:1,000; Abcam) for 1 h at room temperature before washing with phosphate-buffered saline with Tween-20 for 3 times of 15 min. Then, the membrane was developed with enhanced chemiluminescence detection kit (Sigma-Aldrich, St. Louis, MO, USA) for imaging. Image lab v3.0 software (Bio-Rad) was used to acquire and analyze imaging signals. The relative expression of HPSE protein was calculated against β-actin.
Cell-counting Kit 8 (CCK-8) assay
Cells were seeded at 2,000/well in 96-well plates for transfection. At 48 h after transfection, the cells were subjected to CCK-8 assay for the detection of proliferation. At 24, 48 and 72 h, the medium was discarded, and the cells were washed with phosphate-buffered saline twice, followed by addition of 10% CCK-8 reaction reagent diluted in DMEM medium. After incubation at 37°C for 30 min, the absorbance of each well was measured at 450 nm for plotting cell proliferation curves. Each group was tested in 3 replicate wells and the values were averaged.
Transwell assay
Growth-factor depleted Matrigel invasion chambers (BD Biosciences, Franklin Lakes, NJ, USA) were used to determine the invasion ability of cells. In Matrigel chambers, 500 µl serum-free DMEM medium was added and kept for 1 h at room temperature. In the lower chamber, 750 µl DMEM medium supplemented with 20% serum was added. After trypsinization, the transfected cells were resuspended to a density of 4×105 cells/ml using DMEM containing 0.1% bovine serum albumin. After adding 500 µl cell suspension into the invasion chamber, the cells were incubated at 37°C and 5% CO2 for 18 h. Then, the cells in invasion chamber were wiped by cotton swab. Cells that moved to the other side of the chamber were fixed with 100% methanol for 10 min. After being stained with 0.1% crystal violet, the number of cells was counted under a microscope.
Statistical analysis
Statistical analysis was performed using SPSS16.0 (IBM, Armonk, NY, USA). Measurement data were expressed as means ± standard deviations. Two groups of data were compared using t-test. Differences with P<0.01 were considered statistically significant.
Results
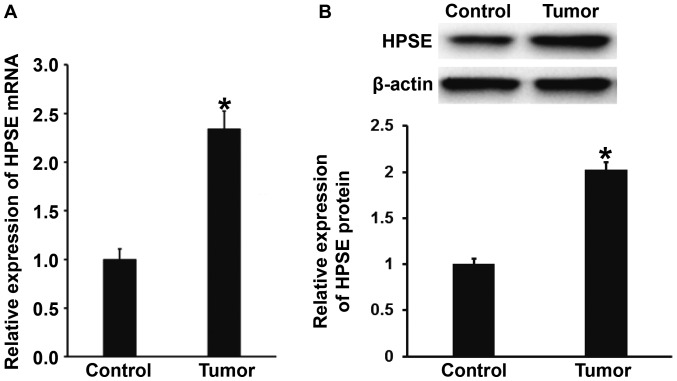
Expression of HPSE mRNA and protein in gastric cancer tissues is enhanced compared with that in tumor-adjacent tissues
To measure the expression of HPSE mRNA and protein in gastric cancer and tumor-adjacent tissues, qPCR and western blotting were employed, respectively. The qPCR data showed that the levels of HPSE mRNA in gastric cancer tissues were significantly higher than those in tumor-adjacent tissues (P<0.01) (Fig. 1A). Consistently, western blotting showed that the protein expression of HPSE in gastric cancer tissues was significantly elevated compared with that in tumor-adjacent tissues (P<0.01) (Fig. 1B). The results suggest that the expression of HPSE mRNA and protein in gastric cancer tissues is enhanced compared with that in tumor-adjacent tissues.
Figure 1.
Expression of (A) mRNA and (B) protein of HPSE in tumor-adjacent tissues and gastric cancer tissues. Quantitative polymerase chain reaction was used to determine the expression of mRNA, and western blotting was employed to measure protein expression. *P<0.01 compared with control. HPSE, heparanase.
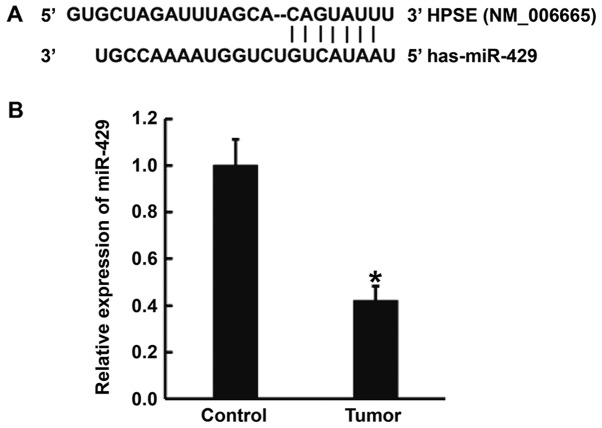
Reduced expression of miR-429 in gastric cancer tissues may be related to the targeting of HPSE mRNA by miR-429
To predict whether miR-429 can target HPSE, TargetScan and miRanda were used. The data showed that miR-429 was able to bind with the 3′-untranslated region (3′-UTR) of HPSE mRNA (Fig. 2A). To determine the expression of miR-429 in tissues, qPCR was carried out. The data showed that the level of miR-429 in gastric cancer tissues was significantly lower than that in tumor-adjacent tissues (P<0.01) (Fig. 2B). The result indicates that reduced expression of miR-429 in gastric cancer tissues may be related to the targeting of HPSE mRNA by miR-429.
Figure 2.
Prediction of target gene and determination of miR-429 expression in gastric cancer tissues. (A) Bioinformatics prediction of direct interactions between miR-429 and HPSE. Bioinformatics prediction is a powerful tool for the study of the functions of miRNAs. To understand the regulatory mechanism of HPSE in gastric cancer, TargetScan (http://www.targetscan.org) and miRanda (http://www.microrna.org/microrna/home.do) were used to predict the interaction between miR-429 and HPSE mRNA. (B) Expression of miR-429 in tumor-adjacent tissues and gastric cancer tissues. Expression of miR-429 was determined by quantitative polymerase chain reaction. *P<0.01 compared with control. miR, microRNA; HPSE, heparanase.
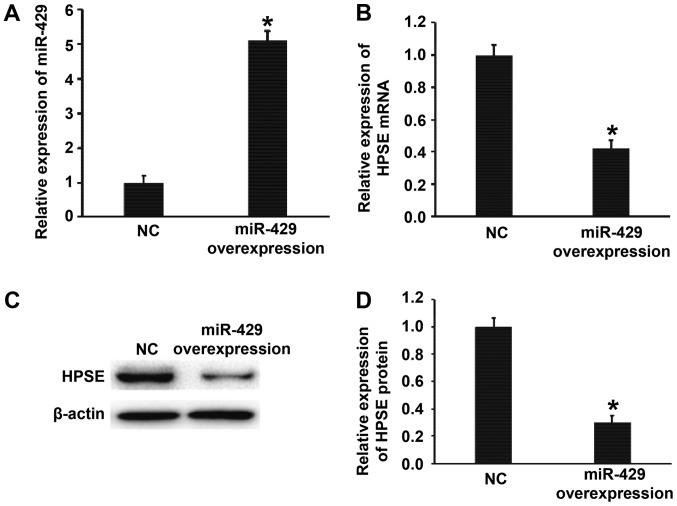
Overexpression of miR-429 inhibits the transcription and translation of HPSE gene
To test whether miR-429 regulates the expression of HPSE, BGC-823 cells were transfected with miR-429 mimics. The data showed that BGC-823 cells transfected with miR-429 mimics had significantly higher miR-429 level than NC group (P<0.01) (Fig. 3A). In addition, the expression of HPSE mRNA and protein in BGC-823 cells transfected with miR-429 mimics was significantly lower than that in NC group (P<0.01) (Fig. 3B-D). The results suggest that overexpression of miR-429 inhibits the transcription and translation of HPSE gene.
Figure 3.
Effect of miR-429 on the expression of HPSE in BGC-823 cells. (A) Relative expression of miR-429 in BGC-823 cells of NC and miR-429 overexpression groups. (B) Relative expression of HPSE mRNA in BGC-823 cells of NC and miR-429 mimics groups. (C and D) Relative expression of HPSE protein in BGC-823 cells of NC and miR-429 mimics groups. Quantitative polymerase chain reaction was used to determine the expression of miR-429 and HPSE mRNA, and western blotting was employed to measure HPSE protein expression. *P<0.01 compared with NC group. miR, microRNA; HPSE, heparanase; NC, negative control.
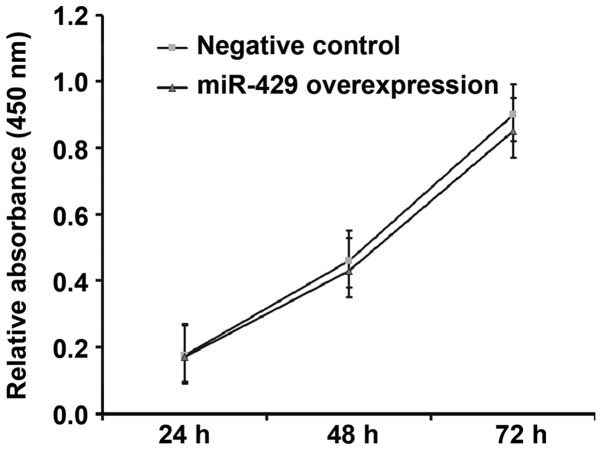
Overexpression of miR-429 does not affect the proliferation of gastric cancer cells
To determine how miR-429 affects the proliferation of BGC-823 cells, CCK-8 assay was used. The data showed that the absorbance of BGC-823 cells transfected with miR-429 mimics was not significantly different from that in NC group at 48 h or 72 h (P>0.05) (Fig. 4). The result indicates that overexpression of miR-429 does not affect the proliferation of gastric cancer cells.
Figure 4.
Proliferation of BGC-823 cells at 24, 48 and 72 h after transfection with NC or miR-429 overexpression. Cell counting kit-8 assay was used to determine the proliferation of the cells. Absorbance of each well was measured at 450 nm with a microplate reader and cell proliferation curves were plotted. miR, microRNA; NC, negative control.
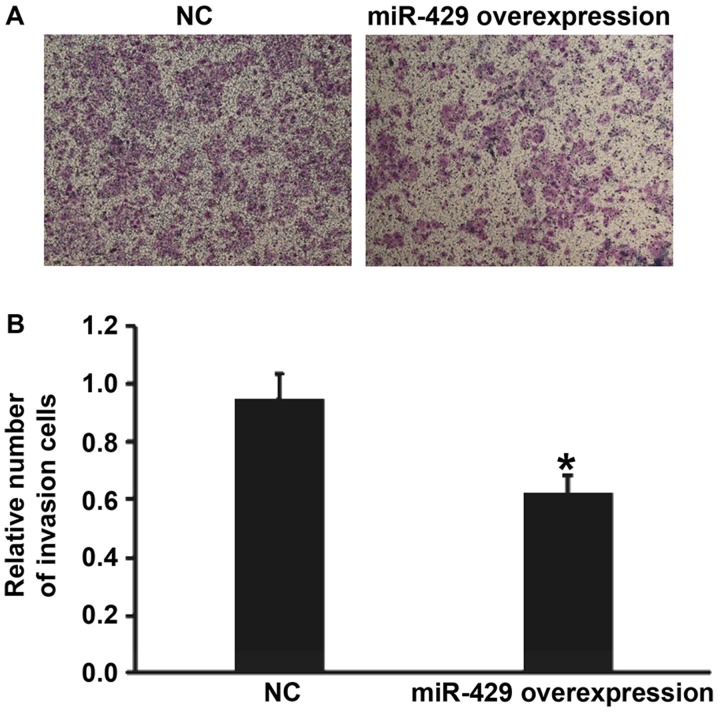
Overexpression of miR-429 reduces the invasion ability of gastric cancer cells
To examine the effect of miR-429 on the invasion ability of BGC-823 cells, transwell assay was used. The data showed that the number of cells that crossed the chamber membrane in miR-429 mimics group was significantly lower than that in NC group (P<0.01) (Fig. 5). The result suggests that overexpression of miR-429 reduces the invasion ability of gastric cancer cells.
Figure 5.
Effect of miR-429 on the invasion ability of BGC-823 cells transfected with NC or miR-429 overexpression. (A) Images of invasion cells that crossed chamber membrane. (B) Relative number of invasion cells. Transwell assay was used to determine the invasion ability of the cells. *P<0.01 compared with NC group. miR, microRNA; NC, negative control.
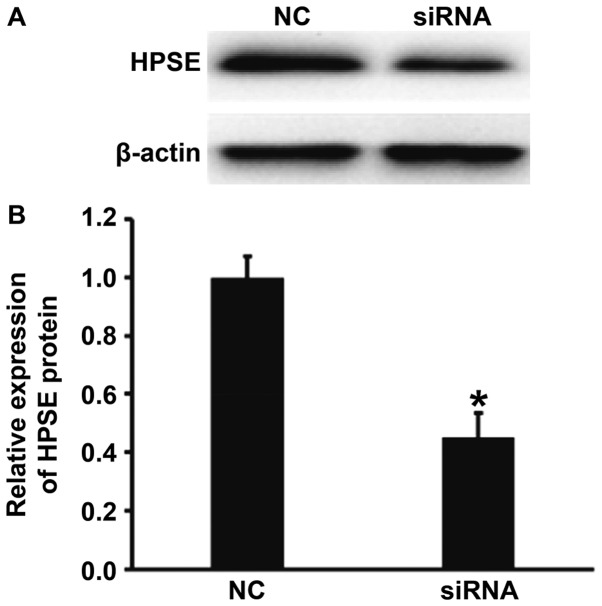
Transfection with HPSE siRNA decreases the expression of HPSE protein in BGC-823 cells
To measure the expression of HPSE in BGC-823 cells transfected with the siRNA of HPSE, western blotting was carried out. The data showed that HPSE protein expression in BGC-823 cells transfected with HPSE siRNA was significantly lower than that in NC group (P<0.01) (Fig. 6). The result indicates that transfection with HPSE siRNA decreases the expression of HPSE protein in BGC-823 cells.
Figure 6.
Effect of siRNA of HPSE on the protein expression of HPSE in BGC-823 cells. (A) Western blots of HPSE protein in cells transfected with NC or siRNA of HPSE. (B) Relative expression of HPSE protein in BGC-823 cells transfected with NC or siRNA of HPSE. Western blotting was employed to measure HPSE protein expression. *P<0.01 compared with NC group. HPSE, heparanase; NC, negative control; si, small interferring.
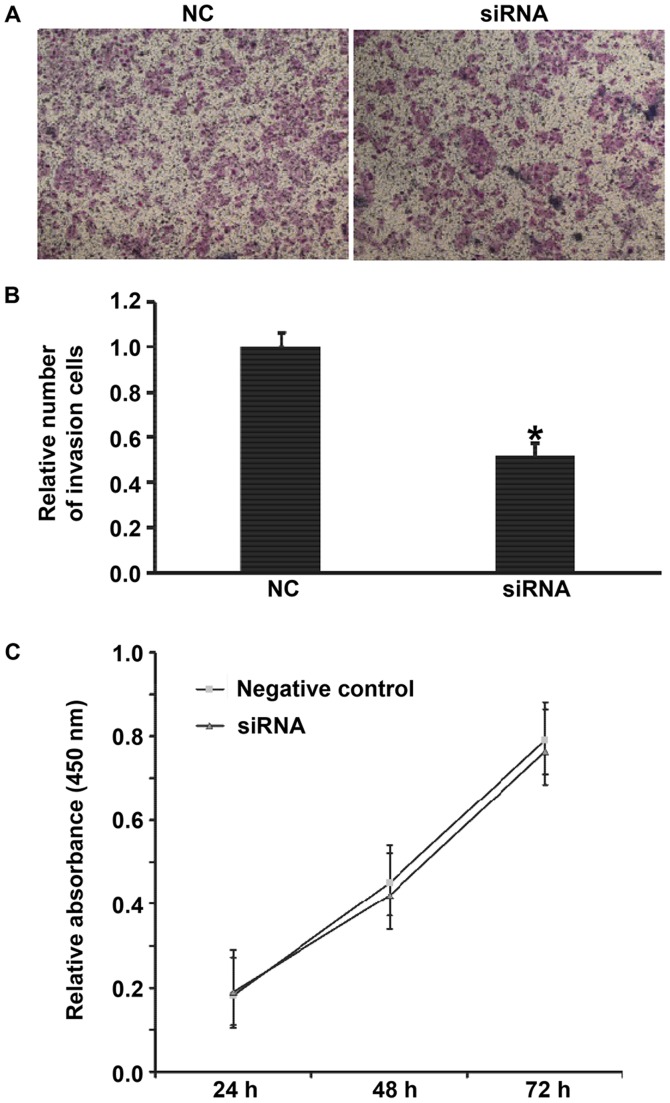
Downregulated HPSE expression inhibits the occurrence and development of gastric cancer by reducing the invasion ability of the cells. To determine the effect of silencing of HPSE on the biological functions of BGC-823 cells, Transwell assay and CCK-8 assay were carried out. The data showed that reduced HPSE expression decreased the invasion ability of BGC-823 cells (Fig. 7A and B), but had no effect on the proliferation of the cells (Fig. 7C). The results suggest that downregulated HPSE expression inhibits the occurrence and development of gastric cancer by reducing the invasion ability of the cells.
Figure 7.
Effect of siRNA of HPSE on the biological functions of BGC-823 cells. (A) Images of invasion cells that crossed chamber membrane. (B) Relative number of invasion cells transfected with NC or siRNA of HPSE. Transwell assay was used to determine the invasion ability of the cells. *P<0.01 compared with NC group. (C) Proliferation of BGC-823 cells at 24, 48 and 72 h after transfection with negative control (NC) or siRNA of HPSE. CCK-8 assay was used to determine the proliferation of the cells. Absorbance of each well was measured at 450 nm with a microplate reader and cell proliferation curves were plotted. *P<0.01 compared with NC group. si, small interferring; HPSE, heparanase; NC, negative control.
Discussion
miR-429 is an important member of miR-200 family that includes miR-200a, miR-200b, miR-200c, miR-141 and miR-429. miR-200 family plays important roles in the growth, development and apoptosis of tumor cells (12,13). Studies show that miR-200 family members are key regulators of epithelial mesenchymal transition in tumor cells (13,14). Of note, low expression of miR-200 induces the occurrence of epithelial to mesenchymal transition (15,16), and overexpression of miR-429 reverses epithelial to mesenchymal transition and induces mesenchymal to epithelial transition in ovarian cancer cells (17). In most types of tumors, miR-429 acts as a tumor-suppressor gene. For example, miR-429 is downregulated in non-small-cell lung cancer, colorectal cancer, renal cell carcinoma and esophageal cancer tissues, and upregulation of miR-429 inhibits the proliferation, migration and invasion of tumor cells by targeting Onecut2, bcl-2 and SP-1 genes (18–21). The results of the present study show that the expression of miR-429 in gastric cancer tissues is lower than that in tumor-adjacent normal tissues, suggesting that miR-429 participates in the occurrence and development of gastric cancer as a tumor-suppressor miRNA. In addition, low expression of miR-429 may become an important marker for the clinical progression of patients with gastric cancer. In vitro study shows that miR-429 promotes the invasion ability of gastric cancer cells by downregulating the expression of HPSE gene.
HPSE is the only β-D-glucuronate endonuclease that degrades HSPG. It damages tissue barriers composed of extracellular matrix and basement membrane, and promotes the invasion and metastasis of tumors (22). For example, Murry et al discover that the expression of HPSE in metastasized melanoma is significantly higher than that in primary melanoma (22). Uno et al show that HPSE facilitates the invasion and metastasis of esophageal cancer, and inhibition of HPSE effectively prevents the pleural metastasis of esophageal cancer (23). Parish et al report that competitive inhibition of HPSE activity by sulfate oligosaccharide compounds reduces the invasion and metastasis of tumors (24). Edovitsky et al find that mice inoculated with tumor cells with HPSE gene knockout have significantly decreased tumor invasion and metastasis rates, as well as prolonged survival time (25). These studies further strengthen the fact that high expression of HPSE facilitates the invasion and metastasis of tumor cells, and reduces survival time of patients. In the present study, bioinformatics shows that binding sites of miR-429 exist in the 3′-UTR of HPSE mRNA. In addition, the invasion ability of BGC-823 cells is significantly lowered after silencing HPSE expression. In conclusion, the present study demonstrates that reduced expression of miR-429 in gastric cancer tissues leads to enhanced HPSE expression, as well as further activated invasion of gastric cancer. Therefore, miR-429 can be a potential diagnostic marker and therapeutic target in the treatment of gastric cancer.
Acknowledgements
This study was supported by Jining No. 1 People's Hospital (Jining, China). We would also like to thank Dr Shuyin Sun, the Director of Jining No. 1 People's Hospital, for his valuable instructions on experimental designs. In addition, we would like to express our sincere gratitude towards Dr Shusheng Ji for his instructions on the writing-up.
References
- 1.Yoo CH, Noh SH, Shin DW, Choi SH, Min JS. Recurrence following curative resection for gastric carcinoma. Br J Surg. 2000;87:236–242. doi: 10.1046/j.1365-2168.2000.01360.x. [DOI] [PubMed] [Google Scholar]
- 2.Cairns RA, Khokha R, Hill RP. Molecular mechanisms of tumor invasion and metastasis: An integrated view. Curr Mol Med. 2003;3:659–671. doi: 10.2174/1566524033479447. [DOI] [PubMed] [Google Scholar]
- 3.Reiland J, Sanderson RD, Waguespack M, Barker SA, Long R, Carson DD, Marchetti D. Heparanase degrades syndecan-1 and perlecan heparan sulfate: Functional implications for tumor cell invasion. J Biol Chem. 2004;279:8047–8055. doi: 10.1074/jbc.M304872200. [DOI] [PubMed] [Google Scholar]
- 4.Chen XP, Luo JS, Tian Y, Nie CL, Cui W, Zhang WD. Downregulation of heparanase expression results in suppression of invasion, migration, and adhesion abilities of hepatocellular carcinoma cells. Biomed Res Int. 2015;2015:241983. doi: 10.1155/2015/241983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nobuhisa T, Naomoto Y, Ohkawa T, Takaoka M, Ono R, Murata T, Gunduz M, Shirakawa Y, Yamatsuji T, Haisa M, et al. Heparanase expression correlates with malignant potential in human colon cancer. J Cancer Res Clin Oncol. 2005;131:229–237. doi: 10.1007/s00432-004-0644-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Akao Y, Nakagawa Y, Naoe T. let-7 microRNA functions as a potential growth suppressor in human colon cancer cells. Biol Pharm Bull. 2006;29:903–906. doi: 10.1248/bpb.29.903. [DOI] [PubMed] [Google Scholar]
- 7.Bao B, Wang Z, Ali S, Ahmad A, Azmi AS, Sarkar SH, Banerjee S, Kong D, Li Y, Thakur S, Sarkar FH. Metformin inhibits cell proliferation, migration and invasion by attenuating CSC function mediated by deregulating miRNAs in pancreatic cancer cells. Cancer Prev Res (Phila) 2012;5:355–364. doi: 10.1158/1940-6207.CAPR-11-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gregory RI, Shiekhattar R. MicroRNA biogenesis and cancer. Cancer Res. 2005;65:3509–3512. doi: 10.1158/0008-5472.CAN-05-0298. [DOI] [PubMed] [Google Scholar]
- 9.Uhlmann S, Zhang JD, Schwäger A, Mannsperger H, Riazalhosseini Y, Burmester S, Ward A, Korf U, Wiemann S, Sahin O. miR-200bc/429 cluster targets PLCgamma1 and differentially regulates proliferation and EGF-driven invasion than miR-200a/141 in breast cancer. Oncogene. 2010;29:4297–4306. doi: 10.1038/onc.2010.201. [DOI] [PubMed] [Google Scholar]
- 10.Lee JW, Park YA, Choi JJ, Lee YY, Kim CJ, Choi C, Kim TJ, Lee NW, Kim BG, Bae DS. The expression of the miRNA-200 family in endometrial endometrioid carcinoma. Gynecol Oncol. 2011;120:56–62. doi: 10.1016/j.ygyno.2010.09.022. [DOI] [PubMed] [Google Scholar]
- 11.Bockmeyer CL, Christgen M, Muller M, Fischer S, Ahrens P, Länger F, Kreipe H, Lehmann U. MicroRNA profiles of healthy basal and luminal mammary epithelial cells are distinct and reflected in different breast cancer subtypes. Breast Cancer Res Treat. 2011;130:735–745. doi: 10.1007/s10549-010-1303-3. [DOI] [PubMed] [Google Scholar]
- 12.Korpal M, Lee ES, Hu G, Kang Y. The miR-200 family inhibits epithelial-mesenchymal transition and cancer cell migration by direct targeting of E-cadherin transcriptional repressors ZEB1 and ZEB2. J Biol Chem. 2008;283:14910–14914. doi: 10.1074/jbc.C800074200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, Calin G, Wang H, Siefker-Radtke A, McConkey D, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–5072. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, Vadas MA, Khew-Goodall Y, Goodall GJ. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 16.Chen J, Wang L, Matyunina LV, Hill CG, McDonald JF. Overexpression of miR-429 induces mesenchymal-to-epithelial transition (MET) in metastatic ovarian cancer cells. Gynecol Oncol. 2011;121:200–205. doi: 10.1016/j.ygyno.2010.12.339. [DOI] [PubMed] [Google Scholar]
- 17.Yi C, Wang Q, Wang L, Huang Y, Li L, Liu L, Zhou X, Xie G, Kang T, Wang H, et al. MiR-663, a microRNA targeting p21(WAF1/CIP1), promotes the proliferation and tumorigenesis of nasopharyngeal carcinoma. Oncogene. 2012;31:4421–4433. doi: 10.1038/onc.2011.629. [DOI] [PubMed] [Google Scholar]
- 18.Cheng T, Wang L, Li Y, Huang C, Zeng L, Yang J. Differential microRNA expression in renal cell carcinoma. Oncol Lett. 2013;6:769–776. doi: 10.3892/ol.2013.1460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Y, Li M, Zang W, Ma Y, Wang N, Li P, Wang T, Zhao G. MiR-429 up-regulation induces apoptosis and suppresses invasion by targeting Bcl-2 and SP-1 in esophageal carcinoma. Cell Oncol (Dordr) 2013;36:385–394. doi: 10.1007/s13402-013-0144-6. [DOI] [PubMed] [Google Scholar]
- 20.Liu D, Xia P, Diao D, Cheng Y, Zhang H, Yuan D, Huang C, Dang C. MiRNA-429 suppresses the growth of gastric cancer cells in vitro. J Biomed Res. 2012;26:389–393. doi: 10.7555/JBR.26.20120029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Teoh-Fitzgerald ML, Fitzgerald MP, Jensen TJ, Futscher BW, Domann FE. Genetic and epigenetic inactivation of extracellular superoxide dismutase promotes an invasive phenotype in human lung cancer by disrupting ECM homeostasis. Mol Cancer Res. 2012;10:40–51. doi: 10.1158/1541-7786.MCR-11-0501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Murry BP, Greiter-Wilke A, Paulsen DP, Hiatt KM, Beltrami CA, Marchetti D. Selective heparanase localization in malignant melanoma. Int J Oncol. 2005;26:345–352. [PubMed] [Google Scholar]
- 23.Uno F, Fujiwara T, Takata Y, Ohtani S, Katsuda K, Takaoka M, Ohkawa T, Naomoto Y, Nakajima M, Tanaka N. Antisense-mediated suppression of human heparanase gene expression inhibits pleural dissemination of human cancer cells. Cancer Res. 2001;61:7855–7860. [PubMed] [Google Scholar]
- 24.Parish CR, Freeman C, Brown KJ, Francis DJ, Cowden WB. Identification of sulfated oligosaccharide-based inhibitors of tumor growth and metastasis using novel in vitro assays for angiogenesis and heparanase activity. Cancer Res. 1999;59:3433–3441. [PubMed] [Google Scholar]
- 25.Edovitsky E, Elkin M, Zcharia E, Peretz T, Vlodavsky I. Heparanase gene silencing, tumor invasiveness, angiogenesis, and metastasis. J Natl Cancer Inst. 2004;96:1219–1230. doi: 10.1093/jnci/djh230. [DOI] [PubMed] [Google Scholar]