Abstract
Over 30 years have passed since co-infection with human immunodeficiency virus (HIV)/hepatitis C virus (HCV) was first documented in hemophilia patients in Japan. In such cases, the leading cause of mortality is reportedly HCV-associated end-stage liver disease. However, the current characteristics of hemophilia patients co-infected with HIV/HCV are unknown. The aim of the present study was to reveal the current characteristics, notably HCV geno-prevalence and liver function, among hemophilia patients co-infected with HIV/HCV in Japan. Current characteristics were evaluated using cross-sectional retrospective data of 44 hemophilia patients positive for anti-HCV and anti-HIV antibodies who underwent screening of liver dysfunction. A total of 56.8% of hemophilia patients co-infected with HIV/HCV were positive for HCV RNA. The most common HCV genotypes were 1a, 1b and 3a. Liver cirrhosis was diagnosed in 26.3% patients negative for HCV RNA and 60.0% patients positive for HCV RNA. Decompensated liver cirrhosis was diagnosed in 33.3% HCV RNA-positive patients and none of the HCV RNA-negative patients. The rate of liver cirrhosis was greater for HCV genotype 3a compared with other genotypes. Overall, the current primary HCV RNA genotypes among hemophilia patients co-infected with HIV/HCV are 1a, 1b and 3a. Over 50% of HIV/HCV co-infected hemophilia patients positive for HCV RNA were diagnosed with liver cirrhosis and some were diagnosed with decompensated liver cirrhosis.
Keywords: human immunodeficiency virus, hepatitis C virus, hemophilia, genotype, liver cirrhosis
Introduction
Hemophilia increases the risk of co-infection with human immunodeficiency virus (HIV) and hepatitis C virus (HCV) due to contaminated or unheated blood products (1,2). According to a survey by the National Institute of Infectious Diseases, 1,431 hemophilia patients in Japan were infected with HIV due to contaminated or unheated blood products and 95% were co-infected with HCV (HIV/HCV co-infected).
Over 30 years have passed since co-infection with HIV/HCV was first documented in a hemophilia patient in Japan. It is well known that co-infection with HIV/HCV is associated with a greater rate of rapid progression to liver cirrhosis and end-stage liver disease (2–5). Additionally, several studies have reported that the prognosis of hemophilia patients co-infected with HIV/HCV has continued to improve along with advancements in highly active anti-retroviral therapy for HIV (6–8), resulting in a shift in the leading cause of death to HCV-associated end-stage liver disease (1,2,9,10). Therefore, the degree of liver dysfunction mainly affects the prognosis of co-infected hemophilia patients. However, there is currently limited data of HCV genoprevalence and liver function in co-infected hemophilia patients in Japan. Hence, it is important to examine the current characteristics of these patients.
Since 2009, our hospital routinely screens for liver dysfunction among hemophilia patients who are positive for both anti-HIV and anti-HCV antibodies. The aim of this study was to reveal the current characteristics of hemophilia patients co-infected with HIV/HCV.
Materials and methods
Study design and participants
The study protocol was approved by the Ethics Committee of Nagasaki University Graduate School of Biomedical Sciences (approval no. 1502271); all participants gave their informed consent prior to participation. We included 44 hemophilia patients who were positive for both anti-HCV and anti-HIV antibodies and admitted to Nagasaki University Hospital for screening of liver dysfunction from October 2009 to February 2014.
Hematochemical and serological tests were performed on admission. Hepatitis B virus (HBV) antigens and antibodies (HBsAg, HBsAb, HBcAb, and HBeAb) and HCV antibodies were tested using commercial immunoassays (Lumipulse, Fujirebio Diagnostics, Inc., Tokyo, Japan). HCV and HIV RNA levels were determined using polymerase chain reaction (PCR), and the presence of IL28B single-nucleotide polymorphisms (rs8099917) was detected using pyrosequencing analysis (11). In addition, blood samples were collected from patients admitted between June 2012 and March 2014, and sera were extracted and frozen for future analysis. A diagnosis of underlying liver disease, such as liver cirrhosis, was based on blood parameters and findings of ultrasonography, dynamic computed tomography, and upper gastrointestinal endoscopy. History of previous anti-HCV and anti-HIV treatment were obtained from patient interview. The regimen of anti-HIV therapy on admission were categorized to protease inhibitors (PI)-unboosted combination with two nucleoside/nucleotide reverse transcriptase inhibitor (NRTI) (PI plus 2-NRTI), PI-boosted with two NRTI (Boosted PI plus 2-NRTI), PI-boosted with integrase strand transfer inhibitor (INSTI) (Boosted PI plus INSTI), PI-boosted with two NRTI and INSTI (Boosted PI plus 2-NRTI and INSTI), INSTI with two NRTI (INSTI plus 2-NRTI), non-nucleoside reverse transcriptase inhibitor (NNRTI) with two NRTI (NNRTI plus 2-NRTI), and others.
HCV genotyping
HCV genotyping was conducted on admission for a few patients positive for HCV RNA (HCV core genotype; SRL, Inc., Tokyo, Japan), whereas most were determined by phylogenic analysis of the HCV core region using sera that had been extracted and stored after admission. In brief, HCV RNA was extracted from serum using the QIAamp MinElute Virus Spin kit (Qiagen, Hilden, Germany), and reverse transcription was performed with random primers using the SuperScript III First-Strand Synthesis System (Invitrogen; Thermo Fisher Scientific, Inc., Waltham, MA, USA). The HCV core region was amplified by nested PCR with specific primers (1st forward: 5′-GGGAGGTCTCGTAGACCGTGCACCATG-3′, 1st reverse: 5′-GAGMGGKATRTACCCCATGAGRTCGGC-3′; 2nd forward: 5′-AGACCGTGCACCATGAGCAC-3′, 2nd reverse: 5′-TACGCCGGGGGTCAKTRGGGCCCCA-3′). After purification with the QIAquick PCR Purification Kit (Qiagen), amplified PCR products were sequenced using the 3130 Genetic Analyzer (Applied Biosystems; Thermo Fisher Scientific, Inc.) with the 2nd forward primer. To determine the HCV genotype, nucleotide sequence alignment and phylogenic analysis were performed using GENETYX ver.13 (Genetyx Corporation, Tokyo, Japan) with 278 bp of the HCV core region.
Statistical analysis
The Mann-Whitney U test was used for the analysis of continuous ordinal data, and the Yates' corrected Chi-square test was used to identify associations between qualitative variables. StatFlex ver. 6.0 software was used for all statistical analyses. P<0.05 was considered to indicate a statistically significant difference.
Results
Clinical characteristics of hemophilia patients co-infected with HIV/HCV
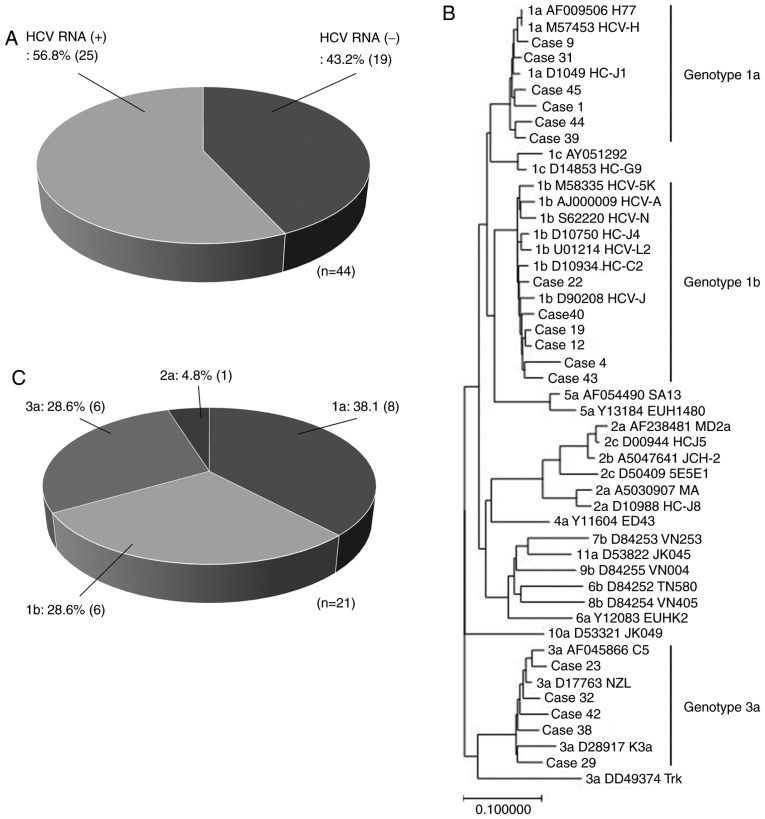
All 44 patients in this study were males with a median age of 43.5 years (age range, 30–68 years), of which 39 were diagnosed with hemophilia A and 5 with hemophilia B. Of these, 19 patients (43.2%) were negative for HCV RNA and 25 (56.8%) were positive (Fig. 1A). The median level of HCV RNA among the positive patients was 6.3 log IU/ml. After anti-retroviral therapy, 37 patients (84.1%) were negative for HIV RNA, whereas 7 (15.9%) remained positive.
Figure 1.
Current genoprevalence in hemophilia patients co-infected with HIV/HCV. (A) The proportion of HCV RNA-positive and -negative hemophilia patients co-infected with HIV/HCV. A phylogenetic tree of the HCV core region from 17 hemophilia patients co-infected with HIV/HCV was constructed. (B) Six patients were classified as genotype 1a (cases 1, 9, 31, 39, 44, and 45), six as genotype 1b (cases 4, 12, 19, 22, 40, and 43), and five as genotype 3a (cases 23, 29, 32, 38, and 42). (C) The proportion of HCV genotypes in 21 hemophilia patients co-infected HIV/HCV and positive for HCV RNA. HIV, human immunodeficiency virus; HCV, hepatitis C virus.
Characteristics of patients who were negative and positive for HCV RNA are shown in Table I. There was no significant difference in the ratio of median age between HCV RNA-positive and -negative patients. Furthermore, there was no significant difference in the ratio of patients with prior HBV infection and those who were HIV RNA-positive. White blood cell count, CD4 level, platelet count, and prothrombin activity were significantly lower in HCV RNA-positive than in-negative patients. Meanwhile, the level of alpha-fetoprotein, aspartate transaminase (AST)-platelet ratio index (APRI), and Fibrosis-4 score (Fib4 score) were significantly higher in positive than in negative patients. The cause of these differences is assumed to result from the higher rate of liver cirrhosis among HCV RNA-positive patients (Table II). There was no significant difference in the regimen of anti-HIV therapy on admission. Among anti-HIV therapy regimen, Boosted PI plus 2-NRTI regimen were the most used in HCV RNA-positive and negative patients. Interleukin (IL)28B was examined in 24 patients. There was no significant difference in the IL28B major allele (TT allele) ratio between HCV RNA-positive and-negative patients.
Table I.
Characteristics of study subjects negative and positive for HCV RNA.
| Characteristics | HCV RNA (−) (n=19, 43.2%) | HCV RNA (+) (n=25, 56.8%) | P-valuea |
|---|---|---|---|
| Age (years) | 45 (30–58) | 43 (30–68) | 0.7130 |
| Hemophilia A/B (%) | 16 (84.2)/3 (15.8) | 23 (92.0)/2 (8.0) | 0.7437 |
| non-LC/LC (%) | 14 (73.7)/5 (26.3) | 10 (40.0)/15 (60.0) | 0.0552 |
| Prior infection with HBVb (−/+) (%) | 4 (21.1)/15 (78.9) | 9 (36.0)/16 (64.0) | 0.4575 |
| HCV RNA (log IU/ml) | N.D.c | 6.3 (<1.2–7.5) | N.D. |
| HIV RNA (−/+) (%) | 16 (84.2)/3 (15.8) | 21 (84.0)/4 (16.0) | 0.6913 |
| WBC (/mm3) | 4,900 (3,200–8,800) | 3,900 (1,800–8600) | 0.0329 |
| CD4 number (/ml) | 502 (219–1081) | 359 (143–544) | 0.0041 |
| CD4% (%) | 29.7 (18.9–44.5) | 29.9 (13.3–46.5) | 0.1694 |
| Hb (g/dl) | 15.0 (11.3–17.1) | 14.5 (8.5–17.4) | 0.6351 |
| Plt (×104/mm3) | 17.0 (11.1–34.9) | 12.2 (4.4–33.1) | 0.0019 |
| PT (%) | 93 (63–116) | 81 (43–112) | 0.0215 |
| T. bil (mg/dl) | 0.9 (0.4–3.4) | 1.0 (0.4–4.3) | 0.9810 |
| AST (IU/l) | 30 (17–45) | 38 (18–173) | 0.0091 |
| ALT (IU/l) | 29 (11–74) | 41 (8–183) | 0.1150 |
| Cr (mg/dl) | 0.72 (0.46–1.19) | 0.79 (0.52–10.3) | 0.5530 |
| eGFR (ml/min/1.73 m2) | 88.9 (53.1–157.4) | 85.0 (4.6–138.6) | 0.3139 |
| AFP (ng/dl) | 2.7 (1.5–9.2) | 5.6 (1.5–654.4) | 0.0229 |
| HbA1c (%) | 5.0 (4.3–6.2) | 5.1 (4.4–7.6) | 0.2119 |
| APRI | 0.53 (0.18–0.91) | 1.08 (0.28–3.30) | 0.0001 |
| Fib4 score | 1.46 (0.53–3.44) | 3.27 (0.62–6.69) | 0.0040 |
| IL28Bd (major/minor) (%) | 10 (90.9)/1 (9.1) | 10 (76.9)/3 (23.1) | 0.7140 |
| Previous IFN therapy (−/+) (%) | 5 (26.3)/14 (73.7) | 10 (40.0)/15 (60.0) | 0.5303 |
| Anti-HIV therapy | 0.9321 | ||
| PI plus 2-NRTI | 2 (10.5) | 2 (8.0) | |
| Boosted PI plus 2-NRTI | 6 (31.6) | 9 (36.0) | |
| Boosted PI plus INSTI | 0 (0) | 2 (8.0) | |
| Boosted PI plus 2-NRTI and INSTI | 2 (10.5) | 2 (8.0) | |
| INSTI plus 2-NRTI | 5 (26.3) | 2 (8.0) | |
| NNRTI plus 2-NRTI | 0 (0) | 4 (16.0) | |
| Others | 4 (21.1) | 4 (16.0) | |
| TDF containing regimen (−/+) (%) | 11 (57.9)/8 42.1) | 14 (56.0)/11 (44.0) | 0.8559 |
LC, liver cirrhosis; HBV, hepatitis B virus; WBC, white blood cell; Hb, hemoglobin; Plt, platelet; PT, prothrombin time; T bil, total bilirubin; AST, Aspartate Aminotransferase; ALT, alanine transaminase; Cr, creatinine; eGFR, estimated glomerular filtration rate; AFP, alpha-fetoprotein; HbA1c, glycated hemoglobin A1c; APRI, aspartate transaminase (AST)-platelet ratio index; Fib4 score, Fibrosis-4 score; IL28B, interleukin 28 B; IFN, interferon; PI, protease inhibitors; NRTI, nucleoside/nucleotide reverse transcriptase inhibitor; INSTI, integrase strand transfer inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor; TDF, tenofovir disoproxil fumarate. Median and range presented, unless otherwise indicated.
P-value was calculated by statistical analysis between HCV non-LC and LC subjects.
Positive for ‘Prior infection with HBV’ indicates positive for any one among HBs antigen, HBs antibody, HBc antibody, and HBe antibody.
N.D. indicated non-detectable. dIL28B indicated IL28B single-nucleotide polymorphisms (rs8099917).
Table II.
Characteristics of non-LC and LC subjects.
| Characteristics | non-LC (n=24, 54.5%) | LC (n=20, 45.5%) | P-valuea |
|---|---|---|---|
| Age (years) | 42.5 (30–60) | 47 (30–68) | 0.4221 |
| Hemophilia A/B (%) | 21 (87.5)/3 (12.5) | 18 (90.0)/2 (10.0) | 0.8284 |
| Prior infection with HBVb (−/+) (%) | 6 (25.0)/18 (75.0) | 7 (35.0)/13 (65.0) | 0.6950 |
| HCV RNA (−/+) | 14 (58.3)/10 (41.7) | 5 (25.0)/15 (75.0) | 0.0552 |
| HIV RNA (−/+) (%) | 20 (83.3)/4 (16.7) | 17 (85.0)/3 (15.0) | 0.7923 |
| WBC (/mm3) | 4,850 (2,900–8,800) | 3,800 (1,800–6,300) | 0.0116 |
| CD4 number (/ml) | 473 (219–1081) | 414.5 (143–544) | 0.1439 |
| CD4% (%) | 30.2 (18.9–44.5) | 29.3 (13.3–46.5) | 0.6290 |
| Hb (g/dl) | 15.0 (10.2–17.4) | 14.4 (8.5–17.1) | 0.2158 |
| Plt (×104/mm3) | 16.9 (6.9–34.9) | 10.7 (4.4–33.1) | 0.0025 |
| PT (%) | 93.5 (69–116) | 74.5 (43–102) | 0.0002 |
| T. bil (mg/dl) | 0.9 (0.4–4.3) | 1.1 (0.4–3.4) | 0.3684 |
| AST (IU/l) | 30.5 (17–173) | 37 (18–82) | 0.2069 |
| ALT (IU/l) | 37.5 (12–183) | 33 (8–87) | 0.7682 |
| Cr (mg/dl) | 0.9 (0.46–1.19) | 0.84 (0.52–10.3) | 0.0504 |
| eGFR (ml/min/1.73 m2) | 71.9 (53.1–157.4) | 77.9 (4.33–138.6) | 0.0677 |
| AFP (ng/dl) | 2.5 (1.5–21.0) | 6.75 (1.6–654.4) | <0.0001 |
| HbA1c (%) | 5.1 (4.3–7.1) | 5.0 (4.4–7.6) | 0.6446 |
| APRI | 0.56 (0.18–3.30) | 0.90 (0.41–3.03) | 0.0196 |
| Fib4 score | 1.45 (0.53–3.43) | 3.35 (0.62–6.69) | 0.0016 |
| IL28Bc (major/minor) (%) | 10 (90.9)/1 (9.1) | 11 (78.6)/3 (21.4) | 0.7751 |
| Previous IFN therapy (−/+) (%) | 9 (37.5)/15 (62.5) | 6 (30.0)/14 (70.0) | 0.8390 |
| Anti-HIV therapy | 0.1753 | ||
| PI plus 2-NRTI | 2 (8.3) | 2 (10.0) | |
| Boosted PI plus 2-NRTI | 6 (25.0) | 9 (45.0) | |
| Boosted PI plus INSTI | 1 (4.2) | 1 (5.0) | |
| Boosted PI plus 2-NRTI and INSTI | 3 (12.5) | 1 (5.0) | |
| INSTI plus 2-NRTI | 4 (16.7) | 3 (15.0) | |
| NNRTI plus 2-NRTI | 2 (8.3) | 2 (10.0) | |
| Others/unknown | 6 (25.0) | 2 (10.0) | |
| TDF containing regimen (−/+) (%) | 16 (66.7)/8 (33.3) | 9 (45.0)/11 (55.0) | 0.1485 |
LC, liver cirrhosis; HBV, hepatitis B virus; WBC, white blood cell; Hb, hemoglobin; Plt, platelet; PT, prothrombin time; T bil, total bilirubin; AST, Aspartate Aminotransferase; ALT, alanine transaminase; Cr, creatinine; eGFR, estimated glomerular filtration rate; AFP, alpha-fetoprotein; HbA1c, glycated hemoglobin A1c; APRI, aspartate transaminase (AST)-platelet ratio index; Fib4 score, Fibrosis-4 score; IL28B, interleukin 28 B; IFN, interferon; PI, protease inhibitors; NRTI, nucleoside/nucleotide reverse transcriptase inhibitor; INSTI, integrase strand transfer inhibitor; NNRTI, non-nucleoside reverse transcriptase inhibitor; TDF, tenofovir disoproxil fumarate. Median and range presented, unless otherwise indicated.
P-value was calculated by statistical analysis between HCV non-LC and LC subjects.
Positive for ‘Prior infection with HBV’ indicates positive for any one among HBs antigen, HBs antibody, HBc antibody, and HBe antibody.
IL28B indicated IL28B single-nucleotide polymorphisms (rs8099917).
HCV RNA genotyping
HCV genotyping was performed for 21 patients. HCV genotypes of four patients were determined by commercial HCV core PCR on admission. Two patients were classified as genotype 1a, one as 2a, and one as 3a. HCV genotypes of 17 patients were determined by phylogenic analysis of the HCV core region. According to the phylogenic tree shown in Fig. 1B, six patients were classified as genotype 1a, six as 1b, and five as 3a. Altogether, there were four genotypes, 38.1% (8/21) were genotype 1a, 28.6% (6/21) were genotype 1b, 4.8% (1/21) were genotype 2a, and 28.6% (6/21) were genotype 3a (Fig. 1C).
Previous anti-HCV therapy in hemophilia patients co-infected with HIV/HCV
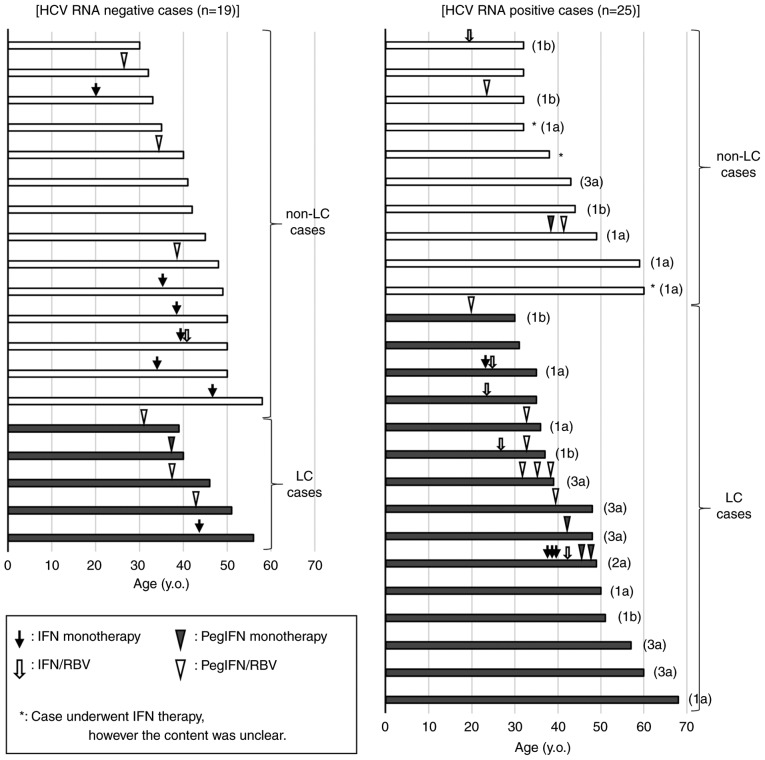
Of 44 patients, 29 (65.9%) previously underwent IFN-based anti-HCV therapy (Fig. 2). The contents of 37 IFN-based anti-HCV therapies administered to 26 patients were obtained, whereas the contents of anti-HCV therapy administered to three patients could not be obtained. Of the 37 anti-HCV therapies, 29.7% (11/37) were IFN mono-therapy, which included IFN α-2b and IFN β; 16.2% (6/37) were IFN plus ribavirin (RBV) combination therapy, which included IFN α-2b/RBV; 13.5% (5/37) were pegylated IFN (PegIFN) mono-therapy, which included PegIFN α-2a and PegIFN α-2b; and 40.5% (15/37) were PegIFN plus RBV combination therapy, which included PegIFN α-2a/RBV and PegIFN α-2b/RBV. Virus eradication was achieved in 48.3% (14/29) of patients. The sustained viral response rate of IFN mono-therapy, IFN plus RBV therapy, PegIFN mono-therapy, and PegIFN plus RBV therapy were 54.5% (6/11), 16.7% (1/6), 20% (1/5), and 40.0% (6/15), respectively. Five of the 44 (11.4%) patients who did not receive IFN-based anti-HCV therapy were negative for HCV RNA. In these cases, HCV was believed to have spontaneously resolved, and the IL28B major allele was detected in each.
Figure 2.
Previous interferon therapy history in hemophilia patients co-infected with HIV/HCV. Fig. 2 shows the previous interferon (IFN) therapy history among 19 hemophilia patients co-infected with HIV/HCV, but negative for HCV RNA (left column), and 25 patients positive for HCV RNA (right column). The vertical axis indicates all cases, which are divided into non-liver cirrhosis cases (white bar) and liver cirrhosis cases (black bar). The horizontal axis indicates age on admission. Previous IFN therapy episodes are shown on a bar graph with the therapy induction age. The black arrow, outlined arrow, black arrow head, and white arrow head indicates interferon monotherapy (IFN monotherapy), interferon plus ribavirin therapy (IFN/RBV), pegylated interferon monotherapy (PegIFN monotherapy), and pegylated interferon plus ribavirin therapy (PegIFN/RBV), respectively. The determined HCV genotypes of HCV RNA-positive cases are shown on the right side of the bar graph.
The ratio of liver cirrhosis in hemophilia patients co-infected with HIV/HCV
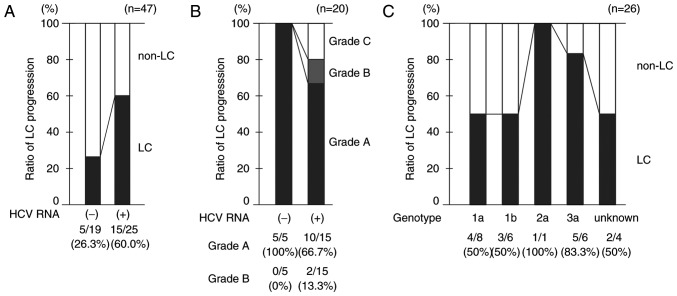
Liver cirrhosis was detected in 20 (45.5%) patients on admission. The median age of these patients was 43 years (age range, 30–68 years). Five of 19 (26.3%) HCV RNA-negative patients and 15 of 25 (60.0%) HCV RNA-positive patients were diagnosed with liver cirrhosis (Figs. 2 and 3A). The ratio of liver cirrhosis was higher in HCV RNA-positive patients than in HCV RNA-negative patients. All HCV RNA-negative patients with liver cirrhosis were Child-Pugh grade A, whereas 10 (66.7%), 2 (13.3%), and 3 (20.0%) HCV RNA-positive patients were Child-Pugh grades A, B and C, respectively (Fig. 3B). The ratio of liver cirrhosis for genotype 3a (83.3%) was relatively high compared with that for other genotypes, with the exception of genotype 2a (Fig. 3C). There was no significant difference in characteristics including the ratio of previous IFN therapy history among the patient positive for HCV RNA genotype 1a, 1b and 3a (Table III).
Figure 3.
Ratio of liver cirrhosis in hemophilia patients co-infected with HIV/HCV. (A) Comparison of liver cirrhosis (LC) ratios between hemophilia patients co-infected with HIV/HCV who are positive for HCV RNA and those who are negative for HCV RNA. (B) Comparison of Child-Pugh grade classification between LC-positive hemophilia patients co-infected with HIV/HCV who are positive for HCV RNA and those who are negative for HCV RNA. (C) Ratio of LC for each HCV genotype. HIV, human immunodeficiency virus; HCV, hepatitis C virus.
Table III.
Characteristics of study subjects positive for HCV RNA genotype 1a, 1b, and 3a.
| Characteristics | 1a (n=8) | 1b (n=6) | 3a (n=6) | P-valuea |
|---|---|---|---|---|
| Age (years) | 49 (35–66) | 36 (30–49) | 47 (39–66) | 0.1071 |
| Hemophilia A/B (%) | 7 (87.5)/1 (12.5) | 5 (83.3)/1 (16.7) | 6 (100)/0 (0) | 0.6009 |
| non-LC/LC (%) | 4 (50.0)/4 (50.0) | 3 (50.0)/3 (50.0) | 1 (16.7)/5 (83.3) | 0.3782 |
| Prior infection with HBVb (−/+) (%) | 3 (37.5)/5 (62.5) | 3 (50.0)/3 (50.0) | 1 (16.6)/5 (83.3) | 0.4719 |
| HCV RNA (log IU/ml) | 6.6 (5.3–7.1) | 6.3 (3.3–7.5) | 5.7 (4.6–6.6) | 0.3127 |
| HIV RNA (−/+) (%) | 7 (87.5)/1 (12.5) | 4 (66.6)/2 (33.3) | 5 (83.3)/1 (16.6) | 0.9346 |
| IL28Bc (major/minor) (%) | 3 (100)/0 (0) | 3 (75.0)/1 (25.0) | 4 (100)/0 (0) | 0.3819 |
| Previous IFN therapy (−/+) (%) | 3 (37.5)/5 (62.5) | 2 (33.3)/4 (66.6) | 3 (50.0)/3 (50.0) | 0.8262 |
LC, liver cirrhosis; HBV, hepatitis B virus; IL28B, interleukin 28 B; IFN, interferon. Median and range presented, unless otherwise indicated.
P-value was calculated by statistical analysis between HCV non-LC and LC subjects.
Positive for ‘Prior infection with HBV’ indicates positive for any one among HBs antigen, HBs antibody, HBc antibody, and HBe antibody.
IL28B indicated IL28B single-nucleotide polymorphisms (rs8099917).
Discussion
As of 1985, 1,431 hemophilia patients were infected with HIV in Japan, and 95% were co-infected with HCV due to contaminated or unheated blood products. A recent national survey of blood coagulation disorders identified 643 living hemophilia patients with HIV/HCV co-infection who had survived over 30 years after infection. Although this cross-sectional study was biased by the inclusion of subjects who were screened for liver dysfunction at their request, they were enrolled more recently (2009–2014). Therefore, the results of this study reflect the current HCV genoprevalence and characteristics among hemophilia patients co-infected with HIV/HCV in Japan.
It was reported that genotype 1b was dominant (60–70%) among HCV mono-infected patients in Japan (12,13), whereas the most common HCV genotypes in hemophilia patients co-infected with HIV/HCV were 1a, 1b, and 3a (2,14). Although the current proportions of HCV genotypes among hemophilia patients co-infected with HIV/HCV remains unknown, our data indicated that the major HCV RNA genotypes among hemophilia patients co-infected with HIV/HCV were 1a, 1b, and 3a, which was unchanged from earlier reports. Until recently, many hemophilia patients co-infected with HIV/HCV underwent IFN-based anti-HCV therapy and some achieved virus eradication. Considering that there have been few or no hemophilia patients newly co-infected with HIV/HCV, the current proportion of HCV genotype is probably affected by the effect of previous IFN-based anti-HCV therapy in each genotype. However, previous IFN-based anti-HCV therapy might not change the proportion of HCV genotypes. More detailed data are needed to clarify this issue.
Many studies have reported that co-infection with HIV is a risk factor for progression to decompensated liver cirrhosis and end-stage liver disease in hemophilia patients with HCV infection (2–5). Fransen van de Putte et al reported that 22% of patients co-infected with HIV/HCV developed end-stage liver disease at a median follow-up period of 31 years in a retrospective study that included 212 patients with inherited bleeding disorders and HIV/HCV co-infection (15). Furthermore, they reported that HCV eradication limited progression to decompensated liver cirrhosis and end-stage liver disease. It is difficult to confirm this findings in the present study because of the small number of cases and the timing of anti-HCV therapy induction differed in each case. However, the findings of this study showed that the development of liver cirrhosis and progression to decompensated liver cirrhosis were observed more often in hemophilia patients co-infected with HIV/HCV and were positive for HCV RNA than in those who were negative for HCV RNA in accordance with previous findings.
It is interesting to note that the frequency of liver cirrhosis was greater in hemophilia patients co-infected with HIV/HCV genotype 3. Two meta-analyses showed that HCV genotype 3 was associated with rapid fibrosis progression (16,17). Probst et al (16) reported that the odds ratio for the association of genotype 3 with accelerated fibrosis progression was 1.52 in single-biopsy studies and 1.37 in paired biopsy studies. Our observation is in agreement with these prior findings. On the other hand, the frequency of patients co-infected with HIV/HCV genotype 3a who underwent IFN therapy was 50.0% (3/6) in this study, which was lower than that with genotype 1a (62.5%, 5/8) and genotype 1b (66.6%, 4/6). The absence of prior IFN therapy might have contributed to the high frequency of liver cirrhosis in this study.
As shown in Fig. 2, hemophilia patients co-infected with HIV/HCV genotype 3 were relatively older. The median age of patients with genotype 1a/1b was 40.5 years, whereas the median age of those with genotype 3a was 48 years. The ratio of patients <40 years was 50.0% (7/14) and 16.7% (1/6) with genotype 1a/1b and 3a, respectively. It is possible that more hemophilia patients co-infected with HIV and HCV genotype 3 were ineligible for IFN-based anti-HCV therapy because of older age and fibrosis progression. Further, these characters might be correlated with the fact that the current ratio of HCV genotype 3a did not decrease, although sustained viral response rate in the patients infected with HCV genotype 3 was higher than that with HCV genotype 1a/1b (18,19). However, this study was biased by the inclusion of subjects admitted to a university hospital because of uncommon infections and refractory HCV genotype 3 with liver fibrosis progression. In addition, the genotypes of patients who achieved a sustained viral response have to be investigated in future studies.
In conclusion, the major HCV RNA genotypes among hemophilia patients with HIV/HCV co-infection are currently 1a, 1b, and 3a, which did not seem to change from previous reports. More than half of hemophilia patients with HIV/HCV co-infection and those positive for HCV RNA developed liver cirrhosis and some developed decompensated liver cirrhosis.
Acknowledgements
We would like to thank our colleagues in the Department of Gastroenterology and Hepatology, Graduate School of Biomedical Sciences, Nagasaki University, for their kind cooperation and support. We would also like to thank Enago (www.enago.jp) for the English language review.
References
- 1.Tatsunami S, Mimaya J, Shirahata A, Zelinka J, Horová I, Hanai J, Nishina Y, Ohira K, Taki M. Current status of Japanese HIV-infected patients with coagulation disorders: Coinfection with both HIV and HCV. Int J Hematol. 2008;88:304–310. doi: 10.1007/s12185-008-0144-x. [DOI] [PubMed] [Google Scholar]
- 2.Yotsuyanagi H, Kikuchi Y, Tsukada K, Nishida K, Kato M, Sakai H, Takamatsu J, Hige S, Chayama K, Moriya K, Koike K. Chronic hepatitis C in patients co-infected with human immunodeficiency virus in Japan: A retrospective multicenter analysis. Hepatol Res. 2009;39:657–663. doi: 10.1111/j.1872-034X.2009.00517.x. [DOI] [PubMed] [Google Scholar]
- 3.Benhamou Y, Bochet M, Di Martino V, Charlotte F, Azria F, Coutellier A, Vidaud M, Bricaire F, Opolon P, Katlama C, Poynard T. Liver fibrosis progression in human immunodeficiency virus and hepatitis C virus coinfected patients. Hepatology. 1999;30:1054–1058. doi: 10.1002/hep.510300409. [DOI] [PubMed] [Google Scholar]
- 4.Sagnelli C, Martini S, Pisaturo M, Pasquale G, Macera M, Zampino R, Coppola N, Sagnelli E. Liver fibrosis in human immunodeficiency virus/hepatitis C virus coinfection: Diagnostic methods and clinical impact. World J Hepatol. 2015;7:2510–2521. doi: 10.4254/wjh.v7.i24.2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ly KN, Xing J, Klevens RM, Jiles RB, Ward JW, Holmberg SD. The increasing burden of mortality from viral hepatitis in the United States between 1999 and 2017. Ann Intern Med. 2012;156:271–278. doi: 10.7326/0003-4819-156-4-201202210-00004. [DOI] [PubMed] [Google Scholar]
- 6.Antiretroviral Therapy Cohort Collaboration, corp-author. Life expectancy of individuals on combination antiretroviral therapy in high-income countries: A collaborative analysis of 14 cohort studies. Lancet. 2008;372:293–299. doi: 10.1016/S0140-6736(08)61113-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Palella FJ, Jr, Delaney KM, Moorman AC, Loveless MO, Fuhrer J, Satten GA, Aschman DJ, Holmberg SD. Declining morbidity and mortality among patients with advanced human immunodeficiency virus infection. HIV Outpatient Study Investigators. N Engl J Med. 1998;338:853–860. doi: 10.1056/NEJM199803263381301. [DOI] [PubMed] [Google Scholar]
- 8.Murphy EL, Collier AC, Kalish LA, Assmann SF, Para MF, Flanigan TP, Kumar PN, Mintz L, Wallach FR, Nemo GJ, Viral Activation Transfusion Study Investigators Highly active antiretroviral therapy decreases mortality and morbidity in patients with advanced HIV disease. Ann Intern Med. 2001;135:17–26. doi: 10.7326/0003-4819-135-1-200107030-00005. [DOI] [PubMed] [Google Scholar]
- 9.Puoti M, Moioli MC, Travi G, Rossotti R. The burden of liver disease in human immunodeficiency virus-infected patients. Semin Liver Dis. 2012;32:103–113. doi: 10.1055/s-0032-1316473. [DOI] [PubMed] [Google Scholar]
- 10.Data Collection on Adverse Events of Anti-HIV drugs (D:A:D) Study Group. Smith C, Sabin CA, Lundgren JD, Thiebaut R, Weber R, Law M, Monforte Ad, Kirk O, Friis-Moller N, et al. Factors associated with specific causes of death amongst HIV-positive individuals in the D:A:D Study. AIDS. 2010;24:1537–1548. doi: 10.1097/QAD.0b013e32833a0918. [DOI] [PubMed] [Google Scholar]
- 11.Kamo Y, Ichikawa T, Miyaaki H, Uchida S, Yamaguchi T, Shibata H, Honda T, Taura N, Isomoto H, Takeshima F, Nakao K. Significance of miRNA-122 in chronic hepatitis C patients with serotype 1 on interferon therapy. Hepatol Res. 2015;45:88–96. doi: 10.1111/hepr.12317. [DOI] [PubMed] [Google Scholar]
- 12.Tanaka H, Tsukuma H, Yamano H, Okubo Y, Inoue A, Kasahara A, Hayashi N. Hepatitis C virus 1b(II) infection and development of chronic hepatitis, liver cirrhosis and hepatocellular carcinoma: A case-control study in Japan. J Epidemiol. 1998;8:244–249. doi: 10.2188/jea.8.244. [DOI] [PubMed] [Google Scholar]
- 13.Toyoda H, Kumada T, Takaguchi K, Shimada N, Tanaka J. Changes in hepatitis C virus genotype distribution in Japan. Epidemiol Infect. 2014;142:2624–2628. doi: 10.1017/S0950268814000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fujimura Y, Ishimoto S, Shimoyama T, Narita N, Kuze Y, Yoshioka A, Fukui H, Tanaka T, Tsuda F, Okamoto H, et al. Genotypes and multiple infections with hepatitis C virus in patients with haemophilia A in Japan. J Viral Hepat. 1996;3:79–84. doi: 10.1111/j.1365-2893.1996.tb00085.x. [DOI] [PubMed] [Google Scholar]
- 15.van de Putte DE Fransen, Makris M, Fischer K, Yee TT, Kirk L, van Erpecum KJ, Patch D, Posthouwer D, Mauser-Bunschoten EP. Long-term follow-up of hepatitis C infection in a large cohort of patients with inherited bleeding disorders. J Hepatol. 2014;60:39–45. doi: 10.1016/j.jhep.2013.08.010. [DOI] [PubMed] [Google Scholar]
- 16.Probst A, Dang T, Bochud M, Egger M, Negro F, Bochud PY. Role of hepatitis C virus genotype 3 in liver fibrosis progression-a systematic review and meta-analysis. J Viral Hepat. 2011;18:745–759. doi: 10.1111/j.1365-2893.2011.01481.x. [DOI] [PubMed] [Google Scholar]
- 17.Bochud PY, Cai T, Overbeck K, Bochud M, Dufour JF, Müllhaupt B, Borovicka J, Heim M, Moradpour D, Cerny A, et al. Genotype 3 is associated with accelerated fibrosis progression in chronic hepatitis C. J Hepatol. 2009;51:655–666. doi: 10.1016/j.jhep.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 18.Shiffman ML, Suter F, Bacon BR, Nelson D, Harley H, Solá R, Shafran SD, Barange K, Lin A, Soman A, et al. Peginterferon alfa-2a and ribavirin for 16 or 24 weeks in HCV genotype 2 or 3. N Engl J Med. 2007;357:124–134. doi: 10.1056/NEJMoa066403. [DOI] [PubMed] [Google Scholar]
- 19.Di Marco V, Covolo L, Calvaruso V, Levrero M, Puoti M, Suter F, Gaeta GB, Ferrari C, Raimondo G, Fattovich G, et al. Who is more likely to respond to dual treatment with pegylated-interferon and ribavirin for chronic hepatitis C? A gender-oriented analysis. J Viral Hepat. 2013;20:790–800. doi: 10.1111/jvh.12106. [DOI] [PubMed] [Google Scholar]