Abstract
Allergic asthma is associated with airway inflammation and airway hyper-responsiveness. Macrophage polarization has been shown to have a profound impact on asthma pathogenesis. Upon exposure to local micro-environments, recruited macrophages can be polarized into either classically activated (or M1) or alternatively activated (or M2) phenotypes. Macrophage polarization has been heavily associated with development of asthma. The process of regulation of macrophage polarization involves an intricate interplay between various cytokines, chemokines, transcriptional factors, and immune-regulatory cells. Different signals from the microenvironment are controlled by different receptors on the macrophages to initiate various macrophage polarization pathways. Most importantly, there is an increased attention on the epigenetic changes (e.g., microRNAs, DNA methylation and histone modification) that impact macrophage functional responses and M1/M2 polarization through modulating cellular signaling and signature gene expression. Thus, modulation of macrophage phenotypes through molecular intervention by targeting some of those potential macrophage regulators may have therapeutic potential in the treatment of allergic asthma and other allergic diseases. In this review, we will discuss the origin of macrophages, characterization of macrophages, macrophage polarization in asthma, and the underlying mechanisms regarding allergen-induced macrophage polarization with emphasis on the regulation of epigenetics, which will provide new insights into the therapeutic strategy for asthma.
INTRODUCTION
Allergic disease prevalence has increased significantly in recent decades1. The majority of school age children and adults with asthma have concomitant allergic sensitization, which has been associated with asthma inception and severity2. In the United States, children living in inner-city households are exposed to higher levels of certain allergens (e.g., cockroach, house dust mite, and mouse) than those living in suburban homes3–5. The indoor environmental allergens are of particular interest in the study of determinants of allergic diseases because of constant exposure during early childhood and the potential for intervention6. However, the causal relationship between individual allergen exposure and the development of these conditions is currently not well-established. This may be due primarily to the complexity of interactions between various environmental factors, the timing and dose of exposure, and innate immune cells.
Macrophages are the most abundant immune cells in the lung (approximately 70% of the immune cells)7 and play an important role in environmental allergen-induced airway inflammation in asthma8,9. Although it is still unclear as to the origin of lung macrophages, recent studies suggest that these macrophages developed from either differentiation of blood monocytes or the proliferation of resident macrophages10. Furthermore, macrophages are an extremely heterogeneous population, displaying a combination of inflammatory and anti-inflammatory functions11. The two extremes in the spectrum of macrophage function are represented by the classically activated (or M1) and the alternatively activated (or M2) phenotypes12. M1 macrophages, induced by IFN-γ and lipopolysaccharide (LPS), upregulate the expression of genes involved with the clearance of pathogens and drive inflammation in response to intracellular pathogens. In contrast, M2 macrophages, induced by interleukin (IL)-4 and IL-13, up-regulate the expression of genes involved with wound healing, clearance of dead and dying cells and tissues, and are involved in anti-inflammatory responses13–15. These two macrophage states mirror the Th1-Th2 polarization of T cells10,14,16,17. Increased M2 macrophage polarization and activation was observed in asthma, which has been suggested to play an important role in allergic asthma18,19. Therefore, a better understanding of the molecular mechanisms regulating macrophage polarization is essential to understand the causal relationship between allergen exposure and development of allergic diseases like asthma.
Numerous studies have summarized the regulation of macrophage polarization by various cytokines, chemokines and transcriptional factors20–25. In this review, we will mainly discuss the advanced concepts of epigenetic changes involved with the macrophage polarization, including miRNAs, DNA methylation and histone modification, which may regulate macrophage polarization through modulating cellular signaling and signature gene expression. These studies on the modulation of macrophage polarization are critical to better understand the macrophage-mediated clearance of pathogens and innate immunity, and in developing treatment for asthma.
ORIGIN OF MACROPHAGES
Innate immunity comprises a number of components including the skin, enzymes, phagocytes and even the micro-biome26. Among the phagocytes, macrophages are the major effector cells of the innate immunity.
History of macrophages
Macrophage was first described by Elie Metchnikoff in 1893. In his experiments, ‘phagocytes’ attacking and engulfing the microbes were observed in starfish challenged by a rose thorn27. He classified these phagocytes into macrophages (“the big eaters”) and microphages (“the small eaters”, now better known as neutrophils). This led to development of theory of phagocytosis27. The next landmark came in 1924 when Aschoff defined macrophages as a part of the reticulo-endothelial system (RES)28. RES was defined as a network of phagocytic cells (reticulum) in various organs which was present in the vicinity of vascular endothelium (endothelial). It was postulated that macrophages originated from the RES and resided within the parent tissue. This finding was challenged in 1968 when Van Furth et al formulated the mononuclear phagocyte system, according to which all macrophages were derived from terminal differentiation of circulating monocytes29. The blood monocyte derivation of macrophages was confirmed by others in different parts of the world at that time30–32. This theory held true until recently when the dual origin of tissue macrophages was identified: macrophages developed from differentiation of circulating monocytes and are derivatives of primitive macrophages which were seen in embryonal yolk sac and fetal liver33. Seeding of tissues with these primitive macrophages occurred before birth in the embryonal period where they proliferated to form the resident tissue macrophage population.
Lung macrophages
Lung macrophages include a heterogeneous population of mononuclear phagocytes which are classified into two main categories: alveolar macrophages and interstitial macrophages34–36. Alveolar macrophages consist of airway macrophages and macrophages which truly reside in the alveoli; both these populations of cells are thought to arise from similar progenitor cells7. Alveolar macrophages reside on the epithelial surface of the lung, and in contrast to other resident macrophages, they are in direct contact with the environment (e.g., allergens, particulate matter, and commensal bacteria)10. Embryonically derived fetal monocytes appear to colonize the lung shortly after birth and differentiate into alveolar macrophages37. Alveolar macrophages normally live independently of blood monocyte input. However, if alveolar macrophages are damaged or depleted, monocytes recruited from the circulation become the dominant source of new macrophages38,39 and contribute to re-populate the alveolar macrophage niche, where both tissue-resident alveolar macrophages (TR-AMs) and monocyte-derived alveolar macrophages (Mo-AMs) contribute to repopulation (Figure 1)40. Interestingly, using a novel lineage tracing system in mice to identify TR-AMs and Mo-AMs during the development of fibrosis and over the subsequent life span of the animal, Misharin et al showed that Mo-AMs drive lung fibrosis and persist in the lung over the life span41. These results suggest heterogeneity in alveolar macrophage function during fibrosis with important implications for the design of targeted therapy. In contrast, interstitial macrophages are the macrophages which reside in the interstitium and are less well studied when compared to alveolar macrophages. Recent studies have challenged the current paradigm that interstitial macrophages (IMs) are one uniform cell population42. Three subpopulations were identified, including IM1 (MHCIIloCD206high), IM2 (MHCII+CD206high), and IM3 (MHCIIhighCD206loCCR2+). Limited studies suggest that the major function of interstitial macrophages is to maintain immune homeostasis in the respiratory tract and induce immune tolerance to benign antigens43.
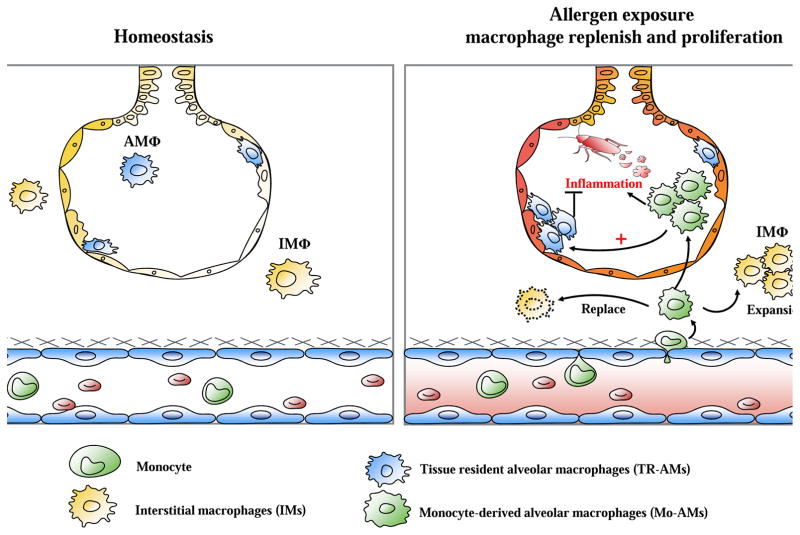
Figure 1.
Schematic diagram of repopulation strategies of macrophages. During homeostasis, majority of resident macrophages (i.e., alveolar macrophages) are derived from embryonic progenitors with small contribution from blood monocyte-derived macrophages. However, after allergen exposure inflammatory mediators produced by damaged epithelial cells or activated resident innate immune cells cause the influx of blood monocytes, which then differentiate and expand in the airway and re-populate the alveolar macrophage niche, where both tissue-resident alveolar macrophages (TR-AMs, suppress inflammation) and monocyte-derived alveolar macrophages (Mo-AMs, promote inflammation) contribute to repopulation.
Functionally, alveolar macrophages are the major effector cells of immune responses and take part in pro and anti-inflammatory functions. In allergic asthma, after allergen exposure, there is rapid recruitment of monocytes and increase in Mo-AMs that promote acute inflammatory responses, while TR-AMs proliferate locally and suppress allergic inflammation (Figure 1)9,44,45. It was suggested that these Mo-AMs fight the perceived danger signals from allergen challenge by promoting an inflammatory responses and subsequent expansion of suppressive TR-AMs to restore homeostasis. While it is still as yet unclear about the polarization of either TR-AMs or Mo-AMs and their influence on asthma phenotypes, the newly established lineage tracing system will provide a unique opportunity to trace macrophage polarization and determine their function in asthma in the future.
MACROPHAGE POLARIZATION IN ASTHMA
Macrophage polarization is defined as a dynamic process by which a macrophage expresses different functional phenotypes in response to different micro-environmental signals25. Upon exposure to pathogens, the microenvironment in the tissue is altered which leads to the polarization of macrophages. As summarized in Figure 2, we hypothesize that allergen exposure (i.e. cockroach allergen) activates lung epithelial cells and innate immune cells to produce a variety of cytokines, which not only recruit blood monocytes to lung tissues but also direct primary macrophage polarization into different subtypes with different roles in asthma. These macrophage subtypes differ in their stimulators, cell surface markers, and cytokine/chemokine secretions.
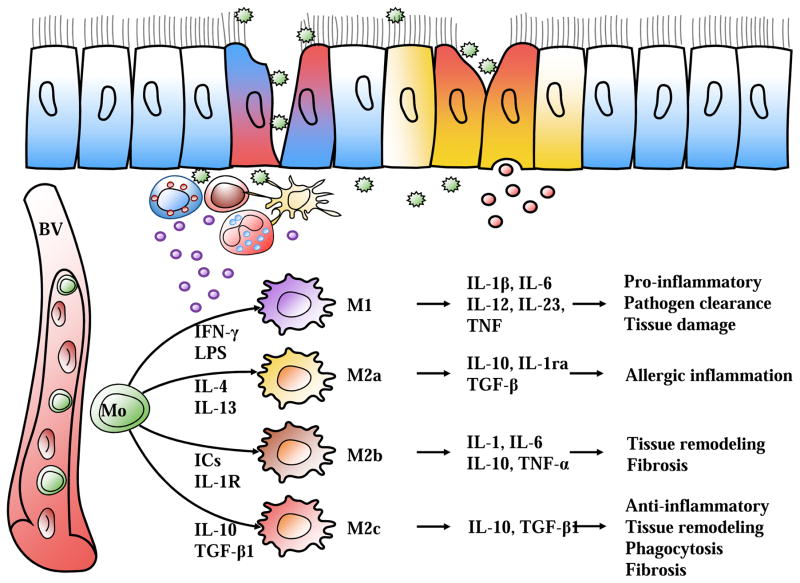
Figure 2.
Schematic diagram of macrophage subtypes. Allergen exposure (i.e. cockroach allergen) activates lung epithelial cells and innate immune cells to produce a variety of cytokines, which direct specific macrophage polarization subtype. M1 subtype is generally considered to be pro-inflammatory, M2a subtype is induced by IL-4 and IL-13, which are critical mediators of allergic inflammation. M2b and M2c subtypes predominately participate in tissue remodeling and fibrosis. BV (blood vessel), Mo (monocyte).
Classically (M1) activated macrophages and asthma
LPS, IFN-γ, and granulocyte-macrophage colony-stimulating factor (GM-CSF) are potent stimulators for the polarization of macrophages into the M1 phenotype25,46. Recent studies suggest that oxidized low density lipoprotein (ox-LDL)47, high-mobility group box 1 (HMGB1) protein48, and caveolin-1 (Cav-1) are also involved in M1 macrophage polarization49. Phenotypically, M1 macrophages highly express CD80, CD86, MHCII, TLR4, and iNOS, and produce high levels of pro-inflammatory Th1 cytokines (e.g., IL-6, IL-12, IL-1β, and TNF-α) and chemokines (e.g., CCL2, CCL5), which play a predominant role in the clearance of intracellular pathogens and recruitment and activation of T and B cells. Macrophages are increased in allergic and non-allergic asthma. In light of macrophage polarization, attempts were made to identify a predominant phenotype in either allergic or non-allergic asthma. A very interesting study was performed by Robbe et al. to identify the M1 or M2 cells in two distinct mouse models: house dust mite (HDM) induced allergic mouse model and farm dust extract (FDE) induced non-allergic mouse model50. Intriguingly, the FDE model showed M1 polarization with increased expression of Th1 and Th17 cells, whereas the HDM model showed M2 polarization with a predominant Th2 cell response. This raises the possibility that M1 cells are the major effector macrophages in non-allergic asthma whereas M2 cells predominate in allergic asthma. Furthermore, M1 macrophages have been linked with the pathophysiology of severe asthma, particularly for those with a poor response to systemic corticosteroids51.
Alternatively (M2) activated macrophages and asthma
In contrast to M1 macrophages that are activated by IFN-γ and LPS, M2 macrophages are induced by IL-4, IL-13 or IL-10. Further studies demonstrated that M2-polarized macrophages can be further divided into three subpopulations, M2a, M2b, and M2c, according to specific stimulators (Figure 2)17,52. The M2a subtype is induced by IL-4, IL-13, fungal and helminthic infections. M2b is elicited by IL-1 receptor ligands, immune complexes and LPS, whereas M2c is stimulated by IL- 10, TGFβ1 and glucocorticoids. Similarly, there is no unique surface marker for the identification of M2 macrophages. Phenotypically, they are characterized by high expression of MRC1, CD163, Arg-1 and low expression of iNOS, MHCII and CD8653,54. Recent studies found that both histamine receptor H1 (HRH1) and E-cadherin, highly expressed on M2 macrophages both in vitro and in bronchoalveolar lavage fluid (BALF) of asthmatic patients, may be additional markers for M2 macrophages19. M2 macrophages, in general, also express Ym1/2 (chitinase like protein) and FIZZ1 (resistin like molecule alpha)55,56. Characteristically, M2a cells secrete high levels of IL-13 and chemokines, including CCL-17, CCL-18, CCL-22 and CCL-24, which activate Th2 cells and promote eosinophil infiltration into the lungs57,58. M2c cells have greater expression of IL-10, an anti-inflammatory cytokine, but lower expression of NF-κβ and co-stimulator molecules including CD40, CD86, and HLA-DR59.
Given that both IL-4 and IL-13 are major inducers of M2 macrophage polarization, M2 cells are suggested to be the major macrophages in allergic asthma. Furthermore, IL-33 has been recently suggested to be a potent inducer of M2 macrophages60–62. In particular, IL-33 released from airway epithelial cells after antigen challenge can modulate M2 macrophage polarization through ST260. Intriguingly, ST2 also contains binding sites for the other cytokine IL-4, IL-5, and IL-13 and the chemokine CCL-17, CCL-18 and CCL-2461,62. In addition, eosinophils, innate lymphoid type 2 (ILC2), CD4+CD25+ regulatory (Treg) cells and mesenchymal stem cells (MSCs) were recently reported to be major regulatory cells in driving the polarization of M2 macrophages63–67. Recently, Girodet and colleagues provided evidence to support a direct contribution of M2 macrophages in allergic airway responses and the pathogenesis of asthma19. Particularly, they found that M2 macrophages were significantly increased with higher expression of both MRC1 and MHC-II in BALFs from patients with asthma when compared with healthy control subjects (>2.9 fold). These data point to a potentially important role for M2 macrophages in asthma, and pharmacologic interventions that target M2 development and function might be a promising approach in asthma therapy and synergistic with current asthma therapies.
Although the current M1/M2 paradigm with its polarized extremes is oversimplification, this in vitro construction provides a useful guide for studying microphage biology in vivo. Moreover, both M1 and M2 macrophages do exist in vivo. However, there are obvious limitations, e.g., macrophages placed into culture may no longer resemble these which exist in vivo, existence of an intermediate stage of macrophages that bridges the M1/M2 extremes. Recently, several other methods were suggested to delineate macrophage phenotypes. For example, naming macrophages according to the stimuli they encounter [e.g., M(IL-4), M(LPS)], according to what they do in their natural habitats (e.g., pruning macrophages, thermos-regulating macrophages), or according to their function which similar to Th1, Th2 or Th17. However, all of these descriptive systems have their limitations. In addition, we wish to emphasize that polarized M1/M2 macrophages act as antigen presenting cells that efficiently activate Th1, Th2, Th17, or Treg cells23. M1 macrophages cause activation of Th1 cells via production of TNFα and IL-12 mediated through CD86 and MHCII markers, leading to activation of non-allergic inflammation. Similarly, M2a cells activate Th2 cells via IL-4 and IL-13 production mediated by CCL17 and MRC1, leading to development of allergic asthma. M2b cells activate Treg cells via IL-10 and TGFβ production mediated by CCL24 and MRC1, leading to development of allergic tolerance and deceased inflammation.
REGULATION OF MACROPHAGE POLARIZATION
The process of regulation of macrophage polarization is a complex interplay between various cytokines, chemokines, and signaling molecules (Figure 3). Studies have revealed that different signals from the microenvironment are controlled by different receptors on the macrophages to initiate various macrophage polarization pathways.
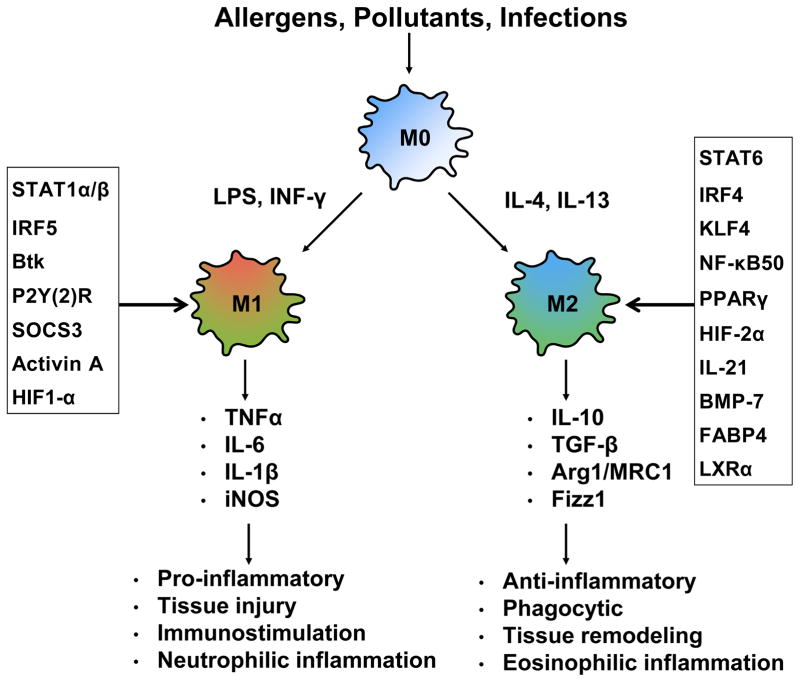
Figure 3.
Schematic diagram of macrophage activation with the most related signaling molecules involved in M1/M2 macrophage polarization. Lung primary macrophages are different into M1 and M2 phenotypes after exposed to allergens, pollutants, and infections, which are regulated by signaling molecules (box), respectively. M1 and M2 macrophages play distinct roles in inflammation and tissue remodeling by secreting different inflammatory mediators.
Polarization of macrophages to M1 phenotype
The LPS/TLR4 pathway has been considered as one of the major pathways in M1 macrophage polarization68. For example, The LPS/TLR4 pathway activates NF-κB, interferon regulatory factor 3 (IRF3) and promotes the secretion of pro-inflammatory cytokines (e.g., IL-6, TNFα, and iNOS), inducing M1 polarization69. This was supported by studies showing that the LPS/TLR4 pathway induced M1 polarization by activating STAT1 in a MyD88 independent fashion70. Additionally, IRF5 is also critical for M1 polarization and the induction of IL-12, IL-23 and TNF71. A role for Bruton’s tyrosine kinase (Btk) was implicated in macrophage polarization in response to LPS stimulation72. Specifically, absence of Btk was shown to skew macrophages towards the M2 phenotype. Several other molecules are also involved, including P2Y(2)R, G-protein coupled receptor, which plays a role in inducing the production of nitric oxide (NO) via iNOS73, SOCS3, which activates NF-κB/PI-3 kinase pathways to produce NO74, and Activin A, which down-regulates IL-1075.
Polarization of macrophages to M2 phenotype
IL-4 and IL-13 skew macrophages toward the M2 activation state via STAT676. Macrophage M2 phenotype is also promoted by several transcription factors, including IRF4, peroxisome proliferator activated receptor gamma (PPARγ)77–79, and Krueppel-like factor 4 (KLF-4)80. It has been suggested that STAT6, PPARγ, KLF-4, and IRF4 may coordinate the M2 polarization of macrophages. MRC1 has been suggested to mediate allergic sensitization and asthma to allergens, including cockroach allergens81,82. We found that MRC1 on macrophages tends to polarize macrophages towards M2 phenotype83.
EPIGENETIC REGULATION OF MACROPHAGE POLARIZATION
It is becoming increasingly clear that epigenetic mechanisms have a significant role in regulating macrophage activation and polarization in response to local environmental signal84,85. Studies have focused on the role of epigenetic changes in macrophage polarization that regulates immune responses and inflammatory gene expression in allergy and asthma. Epigenetic changes can occur in response to environmental stimuli, which may impact disease states86. Three main epigenetic mechanisms have been identified, including miRNAs, DNA methylation and histone modification, which may contribute to the altered cellular signaling and signature gene expression during M1 and M2 polarization.
miRNAs
miRNAs are a class of small and non-coding RNAs that regulate gene-expression programs by reducing the translation and stability of target mRNAs, thus regulating gene expression and cell function87–89. miRNAs have emerged as regulators of phagocyte activation90,91, Th2 polarization88,89, and pathogenic airway inflammation89,92–94. While miRNA-mediated macrophage polarization is a highly conserved process95, recent studies suggest that miRNAs may have the ability to control the balance of M1 and M2 macrophage polarization and skew immune responses96–98 through regulating different transcriptional factors99,100. Here, we will specifically review several of the most intensively studied immune system-related functional miRNAs that have been shown to play important roles in macrophage polarization and in the development of allergic diseases and asthma. (Table 1).
Table 1.
| miRNA | Targets | Function | References |
|---|---|---|---|
| M1 | |||
| mi-155 | SMAD2, C/EBP, IL13R 1, Pellino-1, SOCS1, BCL-6 | Inhibits SMAD2: TGF-α, | (131) |
| Akt dependent polarization via CEBP | (135) | ||
| Down-regulates IL-13 receptor | (36) | ||
| Increases expression of BCL6 | (134) | ||
| miR-127 | BCL6 | Activates JNK pathway | (136) |
| miR-125b | IRF4 | Enhances pro-inflammatory responses | (150) |
| miR-27b | PPAR-y | Increases LPS response | (151) |
| miR-223 | pknox1 | Pro-inflammatory activation of macrophages | (152) |
| miR-106a | IL-10 | Decreases IL-10 leading to inhibition of M2 polarization | (153) |
|
| |||
| M2 | |||
| miR-146a | Notch 1, PPARγ, NF-κB, IRAK, TRAF6 | Decreased inflammation | (38) |
| Suppresses pro-inflammatory cytokine production | (138–140) | ||
| miR-146b | IRF5 TLR4, MyD88, IRAK-1, TRAF6 | Suppresses M1 via IRF5 | (141) |
| Modulates TLR4 signaling pathway | (142) | ||
| Let 7c | p21-activated kinase 1(PAK1), CEBP | Inhibits PAK1 in M1 macrophages | (143) |
| Promotes M2 and inhibits M1 via CEBP | (144) | ||
| miR-21 | SIRPb1, Erk 1/2, NF-κB, PTEN, PDCD4, TLR4, STAT3 | Upstream of IL-10 | (146) |
| Downstream of CSF-1R | (145) | ||
| Prevents PGE2 mediated M2 polarization | |||
| miR-511-3p | PTGDS, ROCK2, LTBP1, CCL2, TLR4, C/EBP | M2 activation | (147), (39) (148) |
| miR-124 | C/EBP STAT3, TACE |
Upregulation of M2 associated markers like Arg1, FIZZ1, TGF and downregulation of M1 markers | (154) |
| miR-125a | KLF13 | Promotes M2 polarization | (155) |
| miR-125b | TNF-α; IRF4 | Decreases inflammation | (156) |
miR-155
Extensive studies have linked miR-155 to the development of allergic asthma101–106. Very recent studies suggested that miR-155 is a critical regulator of type 2 innate lymphoid cells (ILC2)104 and a novel target in allergic asthma107. Interestingly, studies have shown that miR-155 enhanced COX-2 expression in asthmatic human airway smooth muscle cells108. Most interestingly, miR-155 has been shown to stimulate M1 macrophage polarization and inhibit M2 macrophage polarization109. For example, miR-155 inhibits STAT6 by targeting IL-13Rα197 and suppresses TGFβ/Smad signaling pathway by inhibiting Smad2110. Studies also showed that miR-155 enhances M1 macrophage polarization by the repression of negative regulators of pro-inflammatory responses including SOCS1111, SHIP1112, and BCL6113. miR-155 promoted M1 macrophage polarization through the expression of BCL6, a negative transcription factor of the pro-inflammatory NF-kB pathway. Akt kinases differentially contribute to macrophage polarization, with Akt1 ablation giving rise to an M1 and Akt2 ablation resulting in an M2 phenotype. miR-155 was found to be essential in Akt isoform-dependent M1/M2 polarization of macrophages by targeting C/EBPβ114. However, it was also noted that miR-155 may act as either “pro-inflammatory” or “anti-inflammatory” miRNA by controlling the balance of M1 and M2 macrophages in some diseases84,115.
miR-146
miR-146a has also been linked to asthma and allergy116,117. miR-146 has two isoforms, miR-146a and miR-146b. miR-146a is highly expressed in M2 macrophages and plays a pivotal role in macrophage polarization. It was revealed that miR-146a modulated macrophage polarization by targeting Notch1 and PPARγ, which are highly expressed in M2 macrophages118. Furthermore, miR-146a acts as a negative regulator of classical NF-kB, IRAK, and TRAF6, which promotes M2 macrophage polarization and suppresses pro-inflammatory cytokine production119–121. In contrast to miR-146a, miR-146b is highly expressed in M1 macrophages and modulates the shaping of macrophage phenotypes. Mechanistically, miR-146b suppresses the M1 macrophage signature genes by targeting IRF5122. Furthermore, miR-146b modulates the TLR4 signaling pathway by direct targeting of multiple elements, including TLR4, MyD88, IRAK-1, and TRAF6123. Most importantly, these studies suggest that miR-146b may be a molecular effector of IL-10 anti-inflammatory activity.
Let-7c
Let-7c has been reported to promote M2 phenotype through targeting p21-activated kinase 1 (PAK1) that is up-regulated in M1 polarized macrophages124. Banerjee et al found that let – 7c was highly expressed in M2 macrophages, and over-expression of let-7c promoted M2 macrophage but suppressed M1 polarization via targeting C/EBPδ125. Let-7c levels are also greater in alveolar macrophages from fibrotic lungs as compared with those from normal lungs.
miR-21
miR-21 mediates homeostatic M2 macrophage polarization via CSF-1R signaling. Especially, the CSF-1R signaling suppresses the M1 inflammatory phenotype and promotes the M2 polarization by targeting pro-inflammatory molecules Erk1/2 and NF-κβ126. Furthermore, miR-21 has been shown to inhibit the pro-inflammatory tumor suppressor, PTEN and programmed cell death protein-4 (PDCD4) which favor pro-inflammatory response and enable M2 polarization of macrophages91. The regulation of LPS-TLR4 signaling by miR-21 increases IL-10 production which inhibits the transcriptional elongation of the TNFα gene127. The increased IL-10 promotes the activation of JAKs, STAT3 phosphorylation and subsequently favors M2 macrophage polarization. Interestingly, miR-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression96.
miR-511-3p
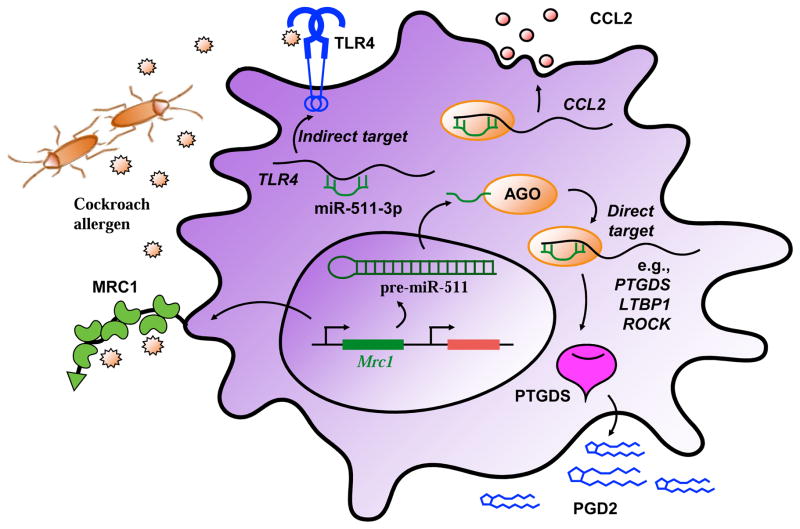
miR-511-3p, the active strand of miR-511, is an intronic miRNA encoded by both mouse and human MRC1 genes. miR-511-3p is transcriptionally co-regulated with Mrc1 in macrophages and regulates the activation of macrophages128,129. Recent studies suggest that miR-511-3p controls macrophage-mediated microbial responses and enhances intestinal inflammation130. Studies from our research group have demonstrated that MRC1 tends to polarize macrophages towards M2 phenotypes83. In agreement with these observations, miR-511-3p over-expression using miR-511-3p mimic suppressed M1 with reduced expression of IL-1β, IL-6, and iNOS and promoted M2 macrophage polarization with enhanced expression of Fizz1, Ym1, and Arg1. Mechanistically, our studies suggest that the miR-511-3p-modulated macrophage polarization may be through an interaction with PTGDS (prostaglandin D2 synthase). Other potential targets of miR-511-3p include ROCK2, LTBP1, CCL2 as direct targets, and TLR4 and C/EBPα as indirect targets (Figure 4). These findings imply that miR-511-3p has an effect on macrophage polarization that may control allergen-induced inflammatory responses.
Figure 4.
Schematic diagram of miR-511-3p in macrophage polarization. miR-511-3p is an intronic miRNA encoded by MRC1 gene. miR-511-3p is transcriptionally co-regulated with Mrc1 in macrophages. miR-511-3p can regulate macrophage polarization into M1 or M2 by binding its direct (e.g., PTGDS, LTBP1, ROCK, and CCL2) or indirect (e.g, TLR4) target genes. AGO (argonaute), PTGDS (prostaglandin D2 synthase), PGD2 (prostaglandin D2), LTBP1 (latent-transforming growth factor beta-binding protein 1), ROCK (Rho-associated protein kinase), CCL2 (C-C motif chemokine ligand 2), TLR4 (Toll-like receptor 4).
DNA methylation
DNA methylation is one of the major epigenetic regulatory systems and is generally linked with transcriptional silencing. DNA methylation occurs via DNA methyltransferases (DNMT) which adds methyl groups to 5′CpG dinycleotides leading to condensation of chromatin and prevention of transcription131. DNA methylation has been associated with asthma since altered DNA methylation status may cause differential gene expression of cytokines (e.g., IL-4, IFN-γ)132 and transcription factor (e.g., FOXP3)133. Early studies in human cohorts have demonstrated an association of DNA methylation in a few candidate genes with asthma phenotypes134,135. A study in African American inner-city children identified 81 differentially methylated regions (DMRs) in PBMCs associated with allergic asthma136. Several immune genes were hypomethylated in those asthmatic patients, including IL13, RUNX3, and TIGIT. However, these studies did not provide evidence for the role of DNA methylation in the control of gene expression and macrophage polarization137. Intriguingly, recent studies suggest a significant association for DNA methylation and differential expression of M1/M2 genes138. DNMT1, 3a and b are differentially expressed in M1 and M2 macrophages and play a critical role in gene silencing139. Moreover, DNMT3b knockdown promoted macrophage polarization to M2 phenotype and suppressed macrophage inflammation, whereas overexpressing DNMT3b did the opposite138. Mechanistic studies suggest that DNMT3b regulates macrophage polarization through binding to the methylation region of PPARγ1, a key transcriptional factor that regulates macrophage polarization. Furthermore, treatment with DNMT inhibitor 5-Aza 2 deoxycytidine (Aza) promotes an anti-inflammatory M2 macrophage phenotype and attenuation of acute lung injury140. These findings indicate that DNMT3b is crucial in regulating macrophage polarization through epigenetic mechanisms. So far, although there is no study on DNMT3b in asthma, we postulate that DNMT3b may be one of the major genes that are involved in regulating macrophage polarization and lung inflammation in asthma. In addition, several other asthma-associated genes were reported to be involved in controlling macrophage polarization by the regulation of promoter methylation (e.g., galectin-3141, BMP-2142). Thus, it would be of interest to identify these genes, which may provide a novel strategy for the prevention and therapy of allergic asthma.
Histone modification
Histone modification refers to the process in which histones may undergo divergent epigenetic changes, including methylation, acetylation, phosphorylation, ubiquitylation and SUMOylation, which could function as epigenetic markers of chromatin state linked with either transcriptional activation or repression84,85. Distinct histone-modifying enzymes have been identified that control macrophage activation and polarization to M1 or M2 phenotypes (e.g., methyltransferases, demethylases, acetyltransferases and deacetylases)139,117. Several M2 marker genes, e.g., Ym1 and arginase 1, are epigenetically regulated by reciprocal changes in H3K27 methylation. Under M2 condition, the expression of Jmjd3, a H3K27 demethylase, is increased, leading to a decreased methylation of H3K27 in the promoter of M2 marker genes143. Aryl hydrocarbon receptor (AhR)-mediated polarization has been associated with the development and resolution of several diseases such as bacterial and parasitic infections144. Most intriguingly, a recent study by Liao et al. suggests an existence of a new regulatory epigenetic synergism between AhR and IL-4 in macrophage polarization145. Jmjd3 expression was increased in M2 macrophages, which could be inhibited by IL-4 neutralizing antibody145. Importantly, GSKJ4, a selective and potent Jmjd3 inhibitor, blocked the binding of AhR to the promoter of CCL1, leading to a decreased expression of CCL1. Jmjd3 was also shown to be crucial for M2 macrophage polarization and host responses against helminth infection142. Jmjd3-deficient mice exhibit decreased expression of M2-associated genes143 and the inhibition of jmjd3 showed impaired pro-inflammatory macrophage response146, showing the importance of Jmjd3 in macrophage polarization and activation. Interestingly, histone methylations in several pro-inflammatory genes are also associated with macrophage polarization (e.g., TNF α147,148, IL-6147). Thus, targeting histone methylations of genes required for M1 or M2 polarization could be beneficial as a therapy for multiple inflammatory diseases. Indeed, emodin, a Chinese herb-derived compound with the potential to inhibit inflammation, is uniquely able to suppress the excessive response of macrophages to both M1 and M2 stimuli by inhibiting the removal of H3K27 trimethylation and the addition of H3K27 acetylation, respectively149. Furthermore, ornithine decarboxylase (ODC), the rate-limiting enzyme in polyamine metabolism, is a regulator of macrophage function. Odc-deficient macrophages had increased H3K4 monomethylation and H3K9 acetylation, accompanied by decreased H3K9 di/trimethylation in primary macrophages150.
In contrast, histone acetylation is the most studied histone modification where acetylation of lysine residues is achieved by histone acetyltransferases (HATs), but deacetylation occurs by histone deacetylases (HDACs). It appears that HATs are linked to transcriptional activity whereas HDACs are associated with transcriptional repression85. Thus, imbalance of HATs/HDACs may cause differential gene expression leading to different diseases like asthma. HDAC activity has been associated with steroid resistance and severity of asthma151,152. Moreover, HDAC3 is required for the activation of hundreds of, mainly STAT-1-dependent, inflammatory genes in M1 macrophages153. Macrophages lacking HDAC3 show an M2-line phenotype in the absence of external stimuli and are hyper-responsive to IL-4, suggesting that HDAC3 may promote M1 and inhibit M2 polarization154. Throughout the macrophage genome, HDAC3 deacetylates histone tails at regulatory regions, leading to repression of many IL-4-regulated genes, which are characteristics of M2 macrophages. Recent studies suggest that TGFβ transcriptionally downregulates TIMAP (TGFβ-inhibited membrane-associated protein) through HDAC3-associated Smad signaling, which is associated with TGFβ-induced macrophage polarization, suggesting that the strategies targeting the HDAC3/TIMAP axis might improve TGFβ-associated pathological processes in various diseases155. Thus, pharmacological blockade of HDAC3 functions could be of benefit in the treatment of inflammatory diseases. Additionally, the commensal microbe-derived butyrate, a novel activator of STAT6-mediated transcription, drives M2 macrophage polarization through H3K9 acetylation156.
CONCLUDING REMARKS
Significant progress has been made in identifying the origin, characteristics, and role of macrophages in physiology and pathophysiology over the last few decades. However, considering the fact that macrophages shows significant heterogeneity in function, a great deal of questions still remain unanswered. For example, although macrophages have been defined as two separate polarization states, M1 and M2, with inflammatory and anti-inflammatory functions, these states have been largely defined by in vitro studies, and tissue macrophages are possibly activated along a continuum between M1 and M2 states in vivo with mixed stimuli. Moreover, no specific M1/M2 markers have been identified for in vitro or in vivo studies and between human and mouse macrophage polarization. Furthermore, macrophages have been widely recognized as extremely attractive therapeutic targets for inflammatory diseases, and most of the therapies are targeted at macrophage markers and their related signaling pathways. Thus, better understanding of macrophage diversity and definition of these cells is essential. Most importantly, considerable progress has been made in understanding the underlying mechanisms associated with the regulation of macrophage polarization, which involves an intricate network of various regulatory cytokines, chemokines, transcriptional factors, and immune-regulatory cells. Due to this intricate network of cellular and marker interplay, it opens up an array of therapeutic targets for the future to modulate macrophage polarization and immune responses in asthma. Furthermore, there is an increased attention on the epigenetic changes that impact macrophage functional responses and M1/M2 polarization. Attempts have been made to study epigenetic mechanisms and identify epigenetic markers that regulate macrophage polarization and activation in asthma. Taken together, modulation of macrophage polarization through targeting those identified potential macrophage regulators may have therapeutic potential in the treatment of allergic asthma and other allergic diseases.
Acknowledgments
This work was supported by grants from the US National Institute of Health (NIH) RO1ES021739, R21 AI109062, R21 AI121768, and National Science Foundation of China (NSFC) No. 81628001 (to P Gao). All authors have read the journal’s policy on disclosure of potential conflicts of interest, and there is no conflict of interest. All authors have read the journal’s authorship agreement and that the manuscript has been reviewed by and approved by all named authors.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Burbank AJ, Sood AK, Kesic MJ, Peden DB, Hernandez ML. Environmental determinants of allergy and asthma in early life. The Journal of allergy and clinical immunology. 2017;140(1):1–12. doi: 10.1016/j.jaci.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Togias A, Fenton MJ, Gergen PJ, Rotrosen D, Fauci AS. Asthma in the inner city: the perspective of the National Institute of Allergy and Infectious Diseases. The Journal of allergy and clinical immunology. 2010;125(3):540–4. doi: 10.1016/j.jaci.2010.01.040. [DOI] [PubMed] [Google Scholar]
- 3.Gergen PJ, Togias A. Inner city asthma. Immunology and allergy clinics of North America. 2015;35(1):101–14. doi: 10.1016/j.iac.2014.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sohn MH, Kim KE. The cockroach and allergic diseases. Allergy Asthma Immunol Res. 2012;4(5):264–9. doi: 10.4168/aair.2012.4.5.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olmedo O, Goldstein IF, Acosta L, et al. Neighborhood differences in exposure and sensitization to cockroach, mouse, dust mite, cat, and dog allergens in New York City. The Journal of allergy and clinical immunology. 2011;128(2):284–92. e7. doi: 10.1016/j.jaci.2011.02.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Do DC, Zhao Y, Gao P. Cockroach allergen exposure and risk of asthma. Allergy. 2016;71(4):463–74. doi: 10.1111/all.12827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cai Y, Sugimoto C, Arainga M, Alvarez X, Didier ES, Kuroda MJ. In vivo characterization of alveolar and interstitial lung macrophages in rhesus macaques: implications for understanding lung disease in humans. J Immunol. 2014;192(6):2821–9. doi: 10.4049/jimmunol.1302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fehervari Z. Alveolar macrophages in asthma. Nature immunology. 2014;16(1):64. [Google Scholar]
- 9.Lee YG, Jeong JJ, Nyenhuis S, et al. Recruited alveolar macrophages, in response to airway epithelial-derived monocyte chemoattractant protein 1/CCl2, regulate airway inflammation and remodeling in allergic asthma. American journal of respiratory cell and molecular biology. 2015;52(6):772–84. doi: 10.1165/rcmb.2014-0255OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epelman S, Lavine KJ, Randolph GJ. Origin and functions of tissue macrophages. Immunity. 2014;41(1):21–35. doi: 10.1016/j.immuni.2014.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Li H, Ciric B, Yang J, et al. Intravenous tolerance modulates macrophage classical activation and antigen presentation in experimental autoimmune encephalomyelitis. Journal of neuroimmunology. 2009;208(1–2):54–60. doi: 10.1016/j.jneuroim.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gordon S. Alternative activation of macrophages. Nature reviews Immunology. 2003;3(1):23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 13.Liu C, Li Y, Yu J, et al. Targeting the shift from M1 to M2 macrophages in experimental autoimmune encephalomyelitis mice treated with fasudil. PloS one. 2013;8(2):e54841. doi: 10.1371/journal.pone.0054841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sica A, Mantovani A. Macrophage plasticity and polarization: in vivo veritas. The Journal of clinical investigation. 2012;122(3):787–95. doi: 10.1172/JCI59643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Spence S, Fitzsimons A, Boyd CR, et al. Suppressors of cytokine signaling 2 and 3 diametrically control macrophage polarization. Immunity. 2013;38(1):66–78. doi: 10.1016/j.immuni.2012.09.013. [DOI] [PubMed] [Google Scholar]
- 16.Biswas SK, Mantovani A. Macrophage plasticity and interaction with lymphocyte subsets: cancer as a paradigm. Nature immunology. 2010;11(10):889–96. doi: 10.1038/ni.1937. [DOI] [PubMed] [Google Scholar]
- 17.Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M. Macrophage plasticity and polarization in tissue repair and remodelling. The Journal of pathology. 2013;229(2):176–85. doi: 10.1002/path.4133. [DOI] [PubMed] [Google Scholar]
- 18.Melgert BN, ten Hacken NH, Rutgers B, Timens W, Postma DS, Hylkema MN. More alternative activation of macrophages in lungs of asthmatic patients. The Journal of allergy and clinical immunology. 2011;127(3):831–3. doi: 10.1016/j.jaci.2010.10.045. [DOI] [PubMed] [Google Scholar]
- 19.Girodet P-OO, Nguyen D, Mancini JD, et al. Alternative Macrophage Activation Is Increased in Asthma. American journal of respiratory cell and molecular biology. 2016;55(4):467–75. doi: 10.1165/rcmb.2015-0295OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hazlett LD, McClellan SA, Barrett RP, et al. IL-33 shifts macrophage polarization, promoting resistance against Pseudomonas aeruginosa keratitis. Investigative ophthalmology & visual science. 2010;51(3):1524–32. doi: 10.1167/iovs.09-3983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wynn TA, Chawla A, Pollard JW. Macrophage biology in development, homeostasis and disease. Nature. 2013;496(7446):445–55. doi: 10.1038/nature12034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sierra-Filardi E, Nieto C, Dominguez-Soto A, et al. CCL2 shapes macrophage polarization by GM-CSF and M-CSF: identification of CCL2/CCR2-dependent gene expression profile. J Immunol. 2014;192(8):3858–67. doi: 10.4049/jimmunol.1302821. [DOI] [PubMed] [Google Scholar]
- 23.Jiang Z, Zhu L. Update on the role of alternatively activated macrophages in asthma. Journal of asthma and allergy. 2016;9:101–7. doi: 10.2147/JAA.S104508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carson WF, Salter-Green SE, Scola MM, Joshi A, Gallagher KA, Kunkel SL. Enhancement of macrophage inflammatory responses by CCL2 is correlated with increased miR-9 expression and downregulation of the ERK1/2 phosphatase Dusp6. Cellular immunology. 2017;314:63–72. doi: 10.1016/j.cellimm.2017.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Murray PJ. Macrophage Polarization. Annu Rev Physiol. 2017;79:541–66. doi: 10.1146/annurev-physiol-022516-034339. [DOI] [PubMed] [Google Scholar]
- 26.Hinde K, Lewis ZT. MICROBIOTA. Mother’s littlest helpers. Science. 2015;348(6242):1427–8. doi: 10.1126/science.aac7436. [DOI] [PubMed] [Google Scholar]
- 27.Gordon S. Elie Metchnikoff: father of natural immunity. European journal of immunology. 2008;38(12):3257–64. doi: 10.1002/eji.200838855. [DOI] [PubMed] [Google Scholar]
- 28.Normann SJ. Function of the Reticuloendothelial System IV. Evidence for Two Types of Particle-Induced Reticuloendothelial Paralysis. Infection and immunity. 1970;1(4):327–33. doi: 10.1128/iai.1.4.327-333.1970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Furth R, Cohn ZA, Hirsch JG, Humphrey JH, Spector WG, Langevoort HL. The mononuclear phagocyte system: a new classification of macrophages, monocytes, and their precursor cells. Bull World Health Organ. 1972;46(6):845–52. [PMC free article] [PubMed] [Google Scholar]
- 30.Carrel A, Ebeling AH. The Fundamental Properties of the Fibroblast and the Macrophage: I. The Fibroblast. The Journal of experimental medicine. 1926;44(2):261–84. doi: 10.1084/jem.44.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Marchesi VT, Florey HW. Electron micrographic observations on the emigration of leucocytes. Q J Exp Physiol Cogn Med Sci. 1960;45:343–8. doi: 10.1113/expphysiol.1960.sp001489. [DOI] [PubMed] [Google Scholar]
- 32.Volkman A, Gowans JL. The Origin of Macrophages from Bone Marrow in the Rat. Br J Exp Pathol. 1965;46:62–70. [PMC free article] [PubMed] [Google Scholar]
- 33.Yona S, Kim KW, Wolf Y, et al. Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis. Immunity. 2013;38(1):79–91. doi: 10.1016/j.immuni.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Van den Bossche J, Malissen B, Mantovani A, De Baetselier P, Van Ginderachter JA. Regulation and function of the E-cadherin/catenin complex in cells of the monocyte-macrophage lineage and DCs. Blood. 2012;119(7):1623–33. doi: 10.1182/blood-2011-10-384289. [DOI] [PubMed] [Google Scholar]
- 35.Satoh T, Kidoya H, Naito H, et al. Critical role of Trib1 in differentiation of tissue-resident M2-like macrophages. Nature. 2013;495(7442):524–8. doi: 10.1038/nature11930. [DOI] [PubMed] [Google Scholar]
- 36.McCubbrey AL, Barthel L, Mohning MP, et al. Deletion of c-FLIP from CD11bhi Macrophages Prevents Development of Bleomycin-induced Lung Fibrosis. American journal of respiratory cell and molecular biology. 2017 doi: 10.1165/rcmb.2017-0154OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guilliams M, De Kleer I, Henri S, et al. Alveolar macrophages develop from fetal monocytes that differentiate into long-lived cells in the first week of life via GM-CSF. The Journal of experimental medicine. 2013;210(10):1977–92. doi: 10.1084/jem.20131199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Okuma T, Terasaki Y, Kaikita K, et al. C-C chemokine receptor 2 (CCR2) deficiency improves bleomycin-induced pulmonary fibrosis by attenuation of both macrophage infiltration and production of macrophage-derived matrix metalloproteinases. The Journal of pathology. 2004;204(5):594–604. doi: 10.1002/path.1667. [DOI] [PubMed] [Google Scholar]
- 39.Tsou CL, Peters W, Si Y, et al. Critical roles for CCR2 and MCP-3 in monocyte mobilization from bone marrow and recruitment to inflammatory sites. The Journal of clinical investigation. 2007;117(4):902–9. doi: 10.1172/JCI29919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hashimoto D, Chow A, Noizat C, et al. Tissue-resident macrophages self-maintain locally throughout adult life with minimal contribution from circulating monocytes. Immunity. 2013;38(4):792–804. doi: 10.1016/j.immuni.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Misharin AV, Morales-Nebreda L, Reyfman PA, et al. Monocyte-derived alveolar macrophages drive lung fibrosis and persist in the lung over the life span. The Journal of experimental medicine. 2017;214(8):2387–404. doi: 10.1084/jem.20162152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gibbings SL, Thomas SM, Atif SM, et al. Three Unique Interstitial Macrophages in the Murine Lung at Steady State. American journal of respiratory cell and molecular biology. 2017;57(1):66–76. doi: 10.1165/rcmb.2016-0361OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ji WJ, Ma YQ, Zhou X, et al. Temporal and spatial characterization of mononuclear phagocytes in circulating, lung alveolar and interstitial compartments in a mouse model of bleomycin-induced pulmonary injury. Journal of immunological methods. 2014;403(1–2):7–16. doi: 10.1016/j.jim.2013.11.012. [DOI] [PubMed] [Google Scholar]
- 44.Zaslona Z, Przybranowski S, Wilke C, et al. Resident alveolar macrophages suppress, whereas recruited monocytes promote, allergic lung inflammation in murine models of asthma. J Immunol. 2014;193(8):4245–53. doi: 10.4049/jimmunol.1400580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Draijer C, Peters-Golden M. Alveolar Macrophages in Allergic Asthma: the Forgotten Cell Awakes. Current allergy and asthma reports. 2017;17(2):12. doi: 10.1007/s11882-017-0681-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sierra-Filardi E, Vega MA, Sanchez-Mateos P, Corbi AL, Puig-Kroger A. Heme Oxygenase-1 expression in M-CSF-polarized M2 macrophages contributes to LPS-induced IL-10 release. Immunobiology. 2010;215(9–10):788–95. doi: 10.1016/j.imbio.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 47.van Tits LJ, Stienstra R, van Lent PL, Netea MG, Joosten LA, Stalenhoef AF. Oxidized LDL enhances pro-inflammatory responses of alternatively activated M2 macrophages: a crucial role for Kruppel-like factor 2. Atherosclerosis. 2011;214(2):345–9. doi: 10.1016/j.atherosclerosis.2010.11.018. [DOI] [PubMed] [Google Scholar]
- 48.Tian S, Zhang L, Tang J, Guo X, Dong K, Chen SY. HMGB1 exacerbates renal tubulointerstitial fibrosis through facilitating M1 macrophage phenotype at the early stage of obstructive injury. American journal of physiology Renal physiology. 2015;308(1):F69–75. doi: 10.1152/ajprenal.00484.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shivshankar P, Halade GV, Calhoun C, et al. Caveolin-1 deletion exacerbates cardiac interstitial fibrosis by promoting M2 macrophage activation in mice after myocardial infarction. Journal of molecular and cellular cardiology. 2014;76:84–93. doi: 10.1016/j.yjmcc.2014.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Robbe P, Draijer C, Borg TR, et al. Distinct macrophage phenotypes in allergic and nonallergic lung inflammation. American journal of physiology Lung cellular and molecular physiology. 2015;308(4):L358–67. doi: 10.1152/ajplung.00341.2014. [DOI] [PubMed] [Google Scholar]
- 51.Oriss TB, Raundhal M, Morse C, et al. IRF5 distinguishes severe asthma in humans and drives Th1 phenotype and airway hyperreactivity in mice. JCI Insight. 2017;2(10) doi: 10.1172/jci.insight.91019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mantovani A, Sica A, Sozzani S, Allavena P, Vecchi A, Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends in immunology. 2004;25(12):677–86. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- 53.Zhong B, Yang X, Sun Q, et al. Pdcd4 modulates markers of macrophage alternative activation and airway remodeling in antigen-induced pulmonary inflammation. Journal of leukocyte biology. 2014;96(6):1065–75. doi: 10.1189/jlb.3A0313-136RRR. [DOI] [PubMed] [Google Scholar]
- 54.Bhatia S, Fei M, Yarlagadda M, et al. Rapid host defense against Aspergillus fumigatus involves alveolar macrophages with a predominance of alternatively activated phenotype. PloS one. 2011;6(1):e15943. doi: 10.1371/journal.pone.0015943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Raes G, De Baetselier P, Noel W, Beschin A, Brombacher F, Hassanzadeh Gh G. Differential expression of FIZZ1 and Ym1 in alternatively versus classically activated macrophages. Journal of leukocyte biology. 2002;71(4):597–602. [PubMed] [Google Scholar]
- 56.Raes G, Noel W, Beschin A, Brys L, de Baetselier P, Hassanzadeh GH. FIZZ1 and Ym as tools to discriminate between differentially activated macrophages. Dev Immunol. 2002;9(3):151–9. doi: 10.1080/1044667031000137629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Byers DE, Holtzman MJ. Alternatively activated macrophages and airway disease. Chest. 2011;140(3):768–74. doi: 10.1378/chest.10-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Siddiqui S, Secor ER, Jr, Silbart LK. Broncho-alveolar macrophages express chemokines associated with leukocyte migration in a mouse model of asthma. Cellular immunology. 2013;281(2):159–69. doi: 10.1016/j.cellimm.2013.03.001. [DOI] [PubMed] [Google Scholar]
- 59.Lu J, Cao Q, Zheng D, et al. Discrete functions of M2a and M2c macrophage subsets determine their relative efficacy in treating chronic kidney disease. Kidney Int. 2013;84(4):745–55. doi: 10.1038/ki.2013.135. [DOI] [PubMed] [Google Scholar]
- 60.Nabe T, Wakamori H, Yano C, et al. Production of interleukin (IL)-33 in the lungs during multiple antigen challenge-induced airway inflammation in mice, and its modulation by a glucocorticoid. European journal of pharmacology. 2015;757:34–41. doi: 10.1016/j.ejphar.2015.03.015. [DOI] [PubMed] [Google Scholar]
- 61.Joshi AD, Oak SR, Hartigan AJ, et al. Interleukin-33 contributes to both M1 and M2 chemokine marker expression in human macrophages. BMC Immunol. 2010;11:52. doi: 10.1186/1471-2172-11-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kurowska-Stolarska M, Stolarski B, Kewin P, et al. IL-33 amplifies the polarization of alternatively activated macrophages that contribute to airway inflammation. J Immunol. 2009;183(10):6469–77. doi: 10.4049/jimmunol.0901575. [DOI] [PubMed] [Google Scholar]
- 63.Tiemessen MM, Jagger AL, Evans HG, van Herwijnen MJ, John S, Taams LS. CD4+CD25+Foxp3+ regulatory T cells induce alternative activation of human monocytes/macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(49):19446–51. doi: 10.1073/pnas.0706832104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wu D, Molofsky AB, Liang HE, et al. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science. 2011;332(6026):243–7. doi: 10.1126/science.1201475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Molofsky AB, Nussbaum JC, Liang HE, et al. Innate lymphoid type 2 cells sustain visceral adipose tissue eosinophils and alternatively activated macrophages. The Journal of experimental medicine. 2013;210(3):535–49. doi: 10.1084/jem.20121964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Song X, Xie S, Lu K, Wang C. Mesenchymal Stem Cells Alleviate Experimental Asthma by Inducing Polarization of Alveolar Macrophages. Inflammation. 2014 doi: 10.1007/s10753-014-9954-6. [DOI] [PubMed] [Google Scholar]
- 67.Braza F, Dirou S, Forest V, et al. Mesenchymal Stem Cells Induce Suppressive Macrophages Through Phagocytosis in a Mouse Model of Asthma. Stem cells. 2016;34(7):1836–45. doi: 10.1002/stem.2344. [DOI] [PubMed] [Google Scholar]
- 68.Sica A, Erreni M, Allavena P, Porta C. Macrophage polarization in pathology. Cellular and Molecular Life Sciences. 2015;72(21):4111–26. doi: 10.1007/s00018-015-1995-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mills CD, Ley K. M1 and M2 macrophages: the chicken and the egg of immunity. Journal of innate immunity. 2014;6(6):716–26. doi: 10.1159/000364945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Toshchakov V, Jones BW, Perera PY, et al. TLR4, but not TLR2, mediates IFN-beta-induced STAT1alpha/beta-dependent gene expression in macrophages. Nature immunology. 2002;3(4):392–8. doi: 10.1038/ni774. [DOI] [PubMed] [Google Scholar]
- 71.Krausgruber T, Blazek K, Smallie T, et al. IRF5 promotes inflammatory macrophage polarization and TH1-TH17 responses. Nature immunology. 2011;12(3):231–8. doi: 10.1038/ni.1990. [DOI] [PubMed] [Google Scholar]
- 72.Ni Gabhann J, Hams E, Smith S, et al. Btk regulates macrophage polarization in response to lipopolysaccharide. PloS one. 2014;9(1):e85834. doi: 10.1371/journal.pone.0085834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Eun SY, Seo J, Park SW, Lee JH, Chang KC, Kim HJ. LPS potentiates nucleotide-induced inflammatory gene expression in macrophages via the upregulation of P2Y2 receptor. International immunopharmacology. 2014;18(2):270–6. doi: 10.1016/j.intimp.2013.11.026. [DOI] [PubMed] [Google Scholar]
- 74.Arnold CE, Whyte CS, Gordon P, Barker RN, Rees AJ, Wilson HM. A critical role for suppressor of cytokine signalling 3 in promoting M1 macrophage activation and function in vitro and in vivo. Immunology. 2014;141(1):96–110. doi: 10.1111/imm.12173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sierra-Filardi E, Puig-Kroger A, Blanco FJ, et al. Activin A skews macrophage polarization by promoting a proinflammatory phenotype and inhibiting the acquisition of anti-inflammatory macrophage markers. Blood. 2011;117(19):5092–101. doi: 10.1182/blood-2010-09-306993. [DOI] [PubMed] [Google Scholar]
- 76.Sica A, Bronte V. Altered macrophage differentiation and immune dysfunction in tumor development. The Journal of clinical investigation. 2007;117(5):1155–66. doi: 10.1172/JCI31422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chawla A. Control of macrophage activation and function by PPARs. Circulation research. 2010;106(10):1559–69. doi: 10.1161/CIRCRESAHA.110.216523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Bouhlel MA, Derudas B, Rigamonti E, et al. PPARgamma activation primes human monocytes into alternative M2 macrophages with anti-inflammatory properties. Cell Metab. 2007;6(2):137–43. doi: 10.1016/j.cmet.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 79.Luzina IG, Keegan AD, Heller NM, Rook GA, Shea-Donohue T, Atamas SP. Regulation of inflammation by interleukin-4: a review of “alternatives”. Journal of leukocyte biology. 2012;92(4):753–64. doi: 10.1189/jlb.0412214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liao X, Sharma N, Kapadia F, et al. Kruppel-like factor 4 regulates macrophage polarization. The Journal of clinical investigation. 2011;121(7):2736–49. doi: 10.1172/JCI45444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Emara M, Royer PJ, Abbas Z, et al. Recognition of the major cat allergen Fel d 1 through the cysteine-rich domain of the mannose receptor determines its allergenicity. The Journal of biological chemistry. 2011;286(15):13033–40. doi: 10.1074/jbc.M111.220657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Royer PJ, Emara M, Yang C, et al. The mannose receptor mediates the uptake of diverse native allergens by dendritic cells and determines allergen-induced T cell polarization through modulation of IDO activity. J Immunol. 2010;185(3):1522–31. doi: 10.4049/jimmunol.1000774. [DOI] [PubMed] [Google Scholar]
- 83.Zhou Y, Do DC, Ishmael FT, et al. Mannose Receptor Regulates Macrophage Polarization and Allergic Inflammation through MiR-511-3p. Journal of Allergy and Clinical Immunology. 2017 doi: 10.1016/j.jaci.2017.04.049. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhou D, Yang K, Chen L, et al. Promising landscape for regulating macrophage polarization: epigenetic viewpoint. Oncotarget. 2017 doi: 10.18632/oncotarget.17027. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Van den Bossche J, Neele AE, Hoeksema MA, de Winther MP. Macrophage polarization: the epigenetic point of view. Curr Opin Lipidol. 2014;25(5):367–73. doi: 10.1097/MOL.0000000000000109. [DOI] [PubMed] [Google Scholar]
- 86.Seibold MA, Schwartz DA. The lung: the natural boundary between nature and nurture. Annu Rev Physiol. 2011;73:457–78. doi: 10.1146/annurev-physiol-012110-142212. [DOI] [PubMed] [Google Scholar]
- 87.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Lu TX, Hartner J, Lim EJ, et al. MicroRNA-21 limits in vivo immune response-mediated activation of the IL-12/IFN-gamma pathway, Th1 polarization, and the severity of delayed-type hypersensitivity. J Immunol. 2011;187(6):3362–73. doi: 10.4049/jimmunol.1101235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Simpson LJ, Patel S, Bhakta NR, et al. A microRNA upregulated in asthma airway T cells promotes TH2 cytokine production. Nature immunology. 2014;15(12):1162–70. doi: 10.1038/ni.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Bazzoni F, Rossato M, Fabbri M, et al. Induction and regulatory function of miR-9 in human monocytes and neutrophils exposed to proinflammatory signals. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(13):5282–7. doi: 10.1073/pnas.0810909106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Das A, Ganesh K, Khanna S, Sen CK, Roy S. Engulfment of apoptotic cells by macrophages: a role of microRNA-21 in the resolution of wound inflammation. J Immunol. 2014;192(3):1120–9. doi: 10.4049/jimmunol.1300613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mattes J, Collison A, Plank M, Phipps S, Foster PS. Antagonism of microRNA-126 suppresses the effector function of TH2 cells and the development of allergic airways disease. Proceedings of the National Academy of Sciences of the United States of America. 2009;106(44):18704–9. doi: 10.1073/pnas.0905063106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Solberg OD, Ostrin EJ, Love MI, et al. Airway epithelial miRNA expression is altered in asthma. American journal of respiratory and critical care medicine. 2012;186(10):965–74. doi: 10.1164/rccm.201201-0027OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Comer BS, Camoretti-Mercado B, Kogut PC, Halayko AJ, Solway J, Gerthoffer WT. MicroRNA-146a and microRNA-146b expression and anti-inflammatory function in human airway smooth muscle. American journal of physiology Lung cellular and molecular physiology. 2014;307(9):L727–34. doi: 10.1152/ajplung.00174.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Melton DW, Lei X, Gelfond JA, Shireman PK. Dynamic macrophage polarization-specific miRNA patterns reveal increased soluble VEGF receptor 1 by miR-125a-5p inhibition. Physiol Genomics. 2016;48(5):345–60. doi: 10.1152/physiolgenomics.00098.2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lu TX, Munitz A, Rothenberg ME. MicroRNA-21 is up-regulated in allergic airway inflammation and regulates IL-12p35 expression. J Immunol. 2009;182(8):4994–5002. doi: 10.4049/jimmunol.0803560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Martinez-Nunez RT, Louafi F, Sanchez-Elsner T. The interleukin 13 (IL-13) pathway in human macrophages is modulated by microRNA-155 via direct targeting of interleukin 13 receptor alpha1 (IL13Ralpha1) The Journal of biological chemistry. 2011;286(3):1786–94. doi: 10.1074/jbc.M110.169367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Graff JW, Dickson AM, Clay G, McCaffrey AP, Wilson ME. Identifying functional microRNAs in macrophages with polarized phenotypes. The Journal of biological chemistry. 2012;287(26):21816–25. doi: 10.1074/jbc.M111.327031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Lawrence T, Natoli G. Transcriptional regulation of macrophage polarization: enabling diversity with identity. Nature reviews Immunology. 2011;11(11):750–61. doi: 10.1038/nri3088. [DOI] [PubMed] [Google Scholar]
- 100.Yoon WH, Meinhardt H, Montell DJ. miRNA-mediated feedback inhibition of JAK/STAT morphogen signalling establishes a cell fate threshold. Nature cell biology. 2011;13(9):1062–9. doi: 10.1038/ncb2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Plank MW, Maltby S, Tay HL, et al. MicroRNA Expression Is Altered in an Ovalbumin-Induced Asthma Model and Targeting miR-155 with Antagomirs Reveals Cellular Specificity. PLoS One. 2015;10(12):e0144810. doi: 10.1371/journal.pone.0144810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Sonkoly E, Janson P, Majuri ML, et al. MiR-155 is overexpressed in patients with atopic dermatitis and modulates T-cell proliferative responses by targeting cytotoxic T lymphocyte-associated antigen 4. J Allergy Clin Immunol. 2010;126(3):581–9. e1–20. doi: 10.1016/j.jaci.2010.05.045. [DOI] [PubMed] [Google Scholar]
- 103.Malmhall C, Alawieh S, Lu Y, et al. MicroRNA-155 is essential for T(H)2-mediated allergen-induced eosinophilic inflammation in the lung. The Journal of allergy and clinical immunology. 2014;133(5):1429–38. 38e1–7. doi: 10.1016/j.jaci.2013.11.008. [DOI] [PubMed] [Google Scholar]
- 104.Johansson K, Malmhäll C, Ramos-Ramírez P, Rådinger M. MicroRNA-155 is a critical regulator of type 2 innate lymphoid cells and IL-33 signaling in experimental models of allergic airway inflammation. The Journal of allergy and clinical immunology. 2017;139(3):1007. doi: 10.1016/j.jaci.2016.06.035. [DOI] [PubMed] [Google Scholar]
- 105.Okoye IS, Czieso S, Ktistaki E, et al. Transcriptomics identified a critical role for Th2 cell-intrinsic miR-155 in mediating allergy and antihelminth immunity. Proceedings of the National Academy of Sciences of the United States of America. 2014;111(30):E3081–90. doi: 10.1073/pnas.1406322111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zech A, Ayata CK, Pankratz F, et al. MicroRNA-155 modulates P2R signaling and Th2 priming of dendritic cells during allergic airway inflammation in mice. Allergy. 2015;70(9):1121–9. doi: 10.1111/all.12643. [DOI] [PubMed] [Google Scholar]
- 107.Zhou H, Li J, Gao P, Wang Q, Zhang J. miR-155: A Novel Target in Allergic Asthma. Int J Mol Sci. 2016;17(10) doi: 10.3390/ijms17101773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Comer BS, Camoretti-Mercado B, Kogut PC, Halayko AJ, Solway J, Gerthoffer WT. Cyclooxygenase-2 and microRNA-155 expression are elevated in asthmatic airway smooth muscle cells. Am J Respir Cell Mol Biol. 2015;52(4):438–47. doi: 10.1165/rcmb.2014-0129OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Ruggiero T, Trabucchi M, De Santa F, et al. LPS induces KH-type splicing regulatory protein-dependent processing of microRNA-155 precursors in macrophages. FASEB journal: official publication of the Federation of American Societies for Experimental Biology. 2009;23(9):2898–908. doi: 10.1096/fj.09-131342. [DOI] [PubMed] [Google Scholar]
- 110.Louafi F, Martinez-Nunez RT, Sanchez-Elsner T. MicroRNA-155 targets SMAD2 and modulates the response of macrophages to transforming growth factor-{beta} The Journal of biological chemistry. 2010;285(53):41328–36. doi: 10.1074/jbc.M110.146852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ye J, Guo R, Shi Y, Qi F, Guo C, Yang L. miR-155 Regulated Inflammation Response by the SOCS1-STAT3-PDCD4 Axis in Atherogenesis. Mediators of inflammation. 2016;2016:8060182. doi: 10.1155/2016/8060182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lu ZJ, Wu JJ, Jiang WL, et al. MicroRNA-155 promotes the pathogenesis of experimental colitis by repressing SHIP-1 expression. World J Gastroenterol. 2017;23(6):976–85. doi: 10.3748/wjg.v23.i6.976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nazari-Jahantigh M, Wei Y, Noels H, et al. MicroRNA-155 promotes atherosclerosis by repressing Bcl6 in macrophages. The Journal of clinical investigation. 2012;122(11):4190–202. doi: 10.1172/JCI61716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Arranz A, Doxaki C, Vergadi E, et al. Akt1 and Akt2 protein kinases differentially contribute to macrophage polarization. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(24):9517–22. doi: 10.1073/pnas.1119038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Li X, Kong D, Chen H, et al. miR-155 acts as an anti-inflammatory factor in atherosclerosis-associated foam cell formation by repressing calcium-regulated heat stable protein 1. Scientific reports. 2016;6:21789. doi: 10.1038/srep21789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Panganiban RP, Wang Y, Howrylak J, et al. Circulating microRNAs as biomarkers in patients with allergic rhinitis and asthma. The Journal of allergy and clinical immunology. 2016;137(5):1423–32. doi: 10.1016/j.jaci.2016.01.029. [DOI] [PubMed] [Google Scholar]
- 117.Malmhäll C, Johansson K, Winkler C, Alawieh S, Ekerljung L, Rådinger M. Altered miR-155 Expression in Allergic Asthmatic Airways. Scandinavian journal of immunology. 2017;85(4):300–7. doi: 10.1111/sji.12535. [DOI] [PubMed] [Google Scholar]
- 118.Huang C, Liu XJ, Zhou Qun, et al. MiR-146a modulates macrophage polarization by inhibiting Notch1 pathway in RAW264. 7 macrophages. International immunopharmacology. 2016;32:46–54. doi: 10.1016/j.intimp.2016.01.009. [DOI] [PubMed] [Google Scholar]
- 119.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proceedings of the National Academy of Sciences of the United States of America. 2006;103(33):12481–6. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Vergadi E, Vaporidi K, Theodorakis EE, et al. Akt2 deficiency protects from acute lung injury via alternative macrophage activation and miR-146a induction in mice. J Immunol. 2014;192(1):394–406. doi: 10.4049/jimmunol.1300959. [DOI] [PubMed] [Google Scholar]
- 121.Zeng Z, Gong H, Li Y, et al. Upregulation of miR-146a contributes to the suppression of inflammatory responses in LPS-induced acute lung injury. Exp Lung Res. 2013;39(7):275–82. doi: 10.3109/01902148.2013.808285. [DOI] [PubMed] [Google Scholar]
- 122.Peng L, Zhang H, Hao Y, et al. Reprogramming macrophage orientation by microRNA 146b targeting transcription factor IRF5. EBioMedicine. 2016;14:83–96. doi: 10.1016/j.ebiom.2016.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Curtale G, Mirolo M, Renzi TA, Rossato M, Bazzoni F, Locati M. Negative regulation of Toll-like receptor 4 signaling by IL-10-dependent microRNA-146b. Proceedings of the National Academy of Sciences of the United States of America. 2013;110(28):11499–504. doi: 10.1073/pnas.1219852110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang W, Liu H, Liu W, Liu Y, Xu J. Polycomb-mediated loss of microRNA let-7c determines inflammatory macrophage polarization via PAK1-dependent NF-kappaB pathway. Cell death and differentiation. 2015;22(2):287–97. doi: 10.1038/cdd.2014.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Banerjee S, Xie N, Cui H, et al. MicroRNA let-7c regulates macrophage polarization. J Immunol. 2013;190(12):6542–9. doi: 10.4049/jimmunol.1202496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Caescu CI, Guo X, Tesfa L, et al. Colony stimulating factor-1 receptor signaling networks inhibit mouse macrophage inflammatory responses by induction of microRNA-21. Blood. 2015;125(8):e1–13. doi: 10.1182/blood-2014-10-608000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Smallie T, Ricchetti G, Horwood NJ, Feldmann M, Clark AR, Williams LM. IL-10 inhibits transcription elongation of the human TNF gene in primary macrophages. The Journal of experimental medicine. 2010;207(10):2081–8. doi: 10.1084/jem.20100414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Squadrito ML, Pucci F, Magri L, et al. miR-511-3p modulates genetic programs of tumor-associated macrophages. Cell reports. 2012;1(2):141–54. doi: 10.1016/j.celrep.2011.12.005. [DOI] [PubMed] [Google Scholar]
- 129.Squadrito ML, Etzrodt M, De Palma M, Pittet MJ. MicroRNA-mediated control of macrophages and its implications for cancer. Trends in immunology. 2013;34(7):350–9. doi: 10.1016/j.it.2013.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Heinsbroek SE, Squadrito ML, Schilderink R, et al. miR-511-3p, embedded in the macrophage mannose receptor gene, contributes to intestinal inflammation. Mucosal immunology. 2016;9(4):960–73. doi: 10.1038/mi.2015.113. [DOI] [PubMed] [Google Scholar]
- 131.Wu SC, Zhang Y. Active DNA demethylation: many roads lead to Rome. Nature reviews Molecular cell biology. 2010;11(9):607–20. doi: 10.1038/nrm2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kwon NH, Kim JS, Lee JY, Oh MJ, Choi DC. DNA methylation and the expression of IL-4 and IFN-gamma promoter genes in patients with bronchial asthma. J Clin Immunol. 2008;28(2):139–46. doi: 10.1007/s10875-007-9148-1. [DOI] [PubMed] [Google Scholar]
- 133.Runyon RS, Cachola LM, Rajeshuni N, et al. Asthma discordance in twins is linked to epigenetic modifications of T cells. PloS one. 2012;7(11):e48796. doi: 10.1371/journal.pone.0048796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Morales E, Bustamante M, Vilahur N, et al. DNA hypomethylation at ALOX12 is associated with persistent wheezing in childhood. American journal of respiratory and critical care medicine. 2012;185(9):937–43. doi: 10.1164/rccm.201105-0870OC. [DOI] [PubMed] [Google Scholar]
- 135.Breton CV, Byun HM, Wang X, Salam MT, Siegmund K, Gilliland FD. DNA methylation in the arginase-nitric oxide synthase pathway is associated with exhaled nitric oxide in children with asthma. American journal of respiratory and critical care medicine. 2011;184(2):191–7. doi: 10.1164/rccm.201012-2029OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Yang IV, Pedersen BS, Liu A, et al. DNA methylation and childhood asthma in the inner city. The Journal of allergy and clinical immunology. 2015;136(1):69–80. doi: 10.1016/j.jaci.2015.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yang IV, Lozupone CA, Schwartz DA. The environment, epigenome, and asthma. Journal of Allergy and Clinical Immunology. 2017;140(1):14–23. doi: 10.1016/j.jaci.2017.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Yang X, Wang X, Liu D, Yu L, Xue B, Shi H. Epigenetic regulation of macrophage polarization by DNA methyltransferase 3b. Molecular endocrinology. 2014;28(4):565–74. doi: 10.1210/me.2013-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kittan NA, Allen RM, Dhaliwal A, et al. Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes. PloS one. 2013;8(10):e78045. doi: 10.1371/journal.pone.0078045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Thangavel J, Samanta S, Rajasingh S, et al. Epigenetic modifiers reduce inflammation and modulate macrophage phenotype during endotoxemia-induced acute lung injury. J Cell Sci. 2015;128(16):3094–105. doi: 10.1242/jcs.170258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Cokus SJ, Feng S, Zhang X, et al. Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature. 2008;452(7184):215–9. doi: 10.1038/nature06745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Satoh T, Takeuchi O, Vandenbon A, et al. The Jmjd3-Irf4 axis regulates M2 macrophage polarization and host responses against helminth infection. Nature immunology. 2010;11(10):936–44. doi: 10.1038/ni.1920. [DOI] [PubMed] [Google Scholar]
- 143.Ishii M, Wen H, Corsa CA, et al. Epigenetic regulation of the alternatively activated macrophage phenotype. Blood. 2009;114(15):3244–54. doi: 10.1182/blood-2009-04-217620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Climaco-Arvizu S, Domínguez-Acosta O, Cabañas-Cortés MAA, et al. Aryl hydrocarbon receptor influences nitric oxide and arginine production and alters M1/M2 macrophage polarization. Life sciences. 2016;155:76–84. doi: 10.1016/j.lfs.2016.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Liao W-TT, Lu J-HH, Wang W-TT, Hung C-HH, Sheu C-CC, Huang S-KK. Epigenetic synergism between interleukin-4 and aryl-hydrocarbon receptor in human macrophages. Journal of molecular medicine (Berlin, Germany) 2017;95(4):395–404. doi: 10.1007/s00109-016-1493-1. [DOI] [PubMed] [Google Scholar]
- 146.Kruidenier L, Chung CW, Cheng Z, et al. A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response. Nature. 2012;488(7411):404–8. doi: 10.1038/nature11262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Xu G, Liu G, Xiong S, Liu H, Chen X, Zheng B. The histone methyltransferase Smyd2 is a negative regulator of macrophage activation by suppressing interleukin 6 (IL-6) and tumor necrosis factor alpha (TNF-alpha) production. The Journal of biological chemistry. 2015;290(9):5414–23. doi: 10.1074/jbc.M114.610345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Chen CH, Wang CZ, Wang YH, et al. Effects of low-level laser therapy on M1-related cytokine expression in monocytes via histone modification. Mediators of inflammation. 2014;2014:625048. doi: 10.1155/2014/625048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Iwanowycz S, Wang J, Altomare D, Hui Y, Fan D. Emodin Bidirectionally Modulates Macrophage Polarization and Epigenetically Regulates Macrophage Memory. The Journal of biological chemistry. 2016;291(22):11491–503. doi: 10.1074/jbc.M115.702092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Hardbower DM, Asim M, Luis PB, et al. Ornithine decarboxylase regulates M1 macrophage activation and mucosal inflammation via histone modifications. Proceedings of the National Academy of Sciences of the United States of America. 2017;114(5):E751–E60. doi: 10.1073/pnas.1614958114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Kabesch M, Adcock IM. Epigenetics in asthma and COPD. Biochimie. 2012;94(11):2231–41. doi: 10.1016/j.biochi.2012.07.017. [DOI] [PubMed] [Google Scholar]
- 152.Barnes PJ. Corticosteroid resistance in patients with asthma and chronic obstructive pulmonary disease. The Journal of allergy and clinical immunology. 2013;131(3):636–45. doi: 10.1016/j.jaci.2012.12.1564. [DOI] [PubMed] [Google Scholar]
- 153.Chen X, Barozzi I, Termanini A, et al. Requirement for the histone deacetylase Hdac3 for the inflammatory gene expression program in macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2012;109(42):E2865–74. doi: 10.1073/pnas.1121131109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Mullican SE, Gaddis CA, Alenghat T, et al. Histone deacetylase 3 is an epigenomic brake in macrophage alternative activation. Genes & development. 2011;25(23):2480–8. doi: 10.1101/gad.175950.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Yang J, Yin S, Bi F, et al. TIMAP repression by TGFβ and HDAC3-associated Smad signaling regulates macrophage M2 phenotypic phagocytosis. Journal of molecular medicine (Berlin, Germany) 2017;95(3):273–85. doi: 10.1007/s00109-016-1479-z. [DOI] [PubMed] [Google Scholar]
- 156.Ji J, Shu D, Zheng M, et al. Microbial metabolite butyrate facilitates M2 macrophage polarization and function. Scientific reports. 2016;6:24838. doi: 10.1038/srep24838. [DOI] [PMC free article] [PubMed] [Google Scholar]