Abstract
Clinical and genetic heterogeneity associated with retinal diseases makes stem cell-based therapies an attractive strategy for personalized medicine. However, we have limited understanding of the timing of key events in the developing human retina, and in particular the factors critical for generating the unique architecture of the fovea and surrounding macula. Here we define three key epochs in the transcriptome dynamics of human retina from fetal day (D) 52 to 150. Coincident histological analyses confirmed the cellular basis of transcriptional changes and highlighted the dramatic acceleration of development in the fovea compared to peripheral retina. Human and mouse retinal transcriptomes show remarkable similarity in developmental stages, though morphogenesis was greatly expanded in humans. Integration of DNA accessibility data allowed us to reconstruct transcriptional networks controlling photoreceptor differentiation. Our studies provide insights into human retinal development and serve as resource for molecular staging of human stem cell-derived retinal organoids.
eTOC blurb
Hoshino et al. describe transcriptome dynamics and spatiotemporal analyses of developing human retina, demonstrating distinct periods of neurogenesis in the fovea versus nasal retina. Comparison to mouse retinal transcriptomes and integration with open chromatin datasets reveal evolutionary conservation of developmental regulatory networks and unique characteristics of human retinal differentiation.
INTRODUCTION
Retinal and macular degeneration are a major cause of vision impairment, with immense social and economic burden globally. Extensive genetic heterogeneity and a plethora of clinical manifestations observed for retinal diseases (https://sph.uth.edu/Retnet/) present considerable challenge for diagnosis, counseling, and disease management. Gene replacement and gene editing have emerged as promising treatments for retinopathy patients carrying specific genetic loss-of-function mutations (Dalkara et al., 2016; Scholl et al., 2016; Tabebordbar et al., 2016; Yu et al., 2017), and novel therapeutic designs are being attempted for broader disease and population spectrum. Pioneering discoveries of induced pluripotent stem cells (iPSCs) (Yamanaka, 2012) and organoid cultures (Nakano et al., 2012; Sasai, 2013) have brought stem cell-based approaches to the forefront of personalized medicine, and patient-specific treatment paradigms appear feasible for retinal regeneration, photoreceptor replacement, and/or drug design (Kaewkhaw et al., 2016; Nakamura et al., 2016). Rapid advancement of stem cell therapies is hampered, at least in part, by limited understanding of networks and pathways underlying retinogenesis and natural history of disease progression in humans.
The sense of vision occupies a major part of the central nervous system in humans, providing exceptional evolutionary advantage for complex tasks, such as learning, memory, and behavior. The peripheral retina of humans shares the laminated cellular organization and basic developmental events with other vertebrates (La Vail et al., 1991; Prada et al., 1991; Wong and Rapaport, 2009; Young, 1985); however, the retina of humans and other simian primates possesses a unique central architecture with an unusual spatial distribution of neurons and a highly ordered synaptic configuration designed for high acuity vision (Provis and Hendrickson, 2008). This central part of the retina, called the fovea centralis, spans 1.5 mm, which includes an avascular pit comprised of long and medium wavelength (L/M)-cone photoreceptors and a few short wavelength (S)-cones, but no rods (Curcio et al., 1990). The unique structure of the fovea likely develops by mechanisms not present in the peripheral retina, and remarkable histology and immunohistochemical (IHC) studies have suggested a more rapid differentiation of the fovea compared to the rest of the retina in humans (Hendrickson, 2016; Hendrickson et al., 2012; Hendrickson and Yuodelis, 1984) and in non-human primates (Hendrickson et al., 2016; Hendrickson et al., 2009; La Vail et al., 1991; Sears et al., 2000). However, at present, we know very little about the mechanisms that might orchestrate the singular architecture of this region. The elucidation of intrinsic factors, as well as signaling pathways, that guide human retinal development would establish a critical foundation for understanding retinal features specific to primates.
The advent of next generation sequencing has led to global insights into gene regulatory networks and provided opportunities to integrate transcriptome and epigenome with morphogenesis and function (Trapnell et al., 2013; Yang et al., 2015), even at the level of a single cell (Darmanis et al., 2015; Liu and Trapnell, 2016; Macosko et al., 2015). In the retina, transcriptome analyses of developing mouse photoreceptors have uncovered possible evolutionary relationships (Kim et al., 2016) and the role of epigenome in gene regulation (Hughes et al., 2017; Kim, 2016; Mo et al., 2016). The human transcriptome studies have generally focused on adult retina (Farkas et al., 2013; Hornan et al., 2007; Li et al., 2014; Mustafi et al., 2016; Pinelli et al., 2016; Whitmore et al., 2014) but few are available for fetal retina (Aldiri et al., 2017; Kozulin et al., 2009).
As a prelude to delineate genetic networks underlying cellular morphogenesis, we have generated transcriptome profiles of human fetal retina at twelve different time points, from day (D) 52 (corresponding to Carnegie stage 22) to D136 of development, temporally spanning the major stages of early differentiation. Coordinated and somewhat concomitant expression of several key cell-type specific transcription factors, together with cell proliferation-associated genes, suggested a regionally-restricted differentiation blueprint within the developing retina. By integrating gene profiling data with in situ and immunofluorescence analyses, we have correlated transcriptome dynamics to retinal morphogenesis and demonstrated transcriptome signatures associated with rapid developmental progression of central fovea/macula relative to the nasal retina of the same distance from the optic nerve, with complete cell organization and lamination in the fovea by D96. Comparative transcriptome dynamics of developing human and mouse retina has revealed a high degree of conservation in key regulatory genes; however, observed distinctions in several signaling pathways indicate human retina-specific specializations. Furthermore, integration of DNase-seq data at two distinct stages of human retinal development with the corresponding transcriptomes has identified novel promoter/enhancer regions associated with gene expression and allowed us to reconstruct potential regulatory networks. Finally, this is the first study to provide comprehensive molecular analysis on retina younger than 10 fetal weeks. Thus, in addition to expanding the molecular understanding of human retinal development, our studies provide a much-needed reference for staging of retinal organoids from human stem cells to design treatments of retinal and macular diseases.
RESULTS
Transcriptome dynamics of developing human retina
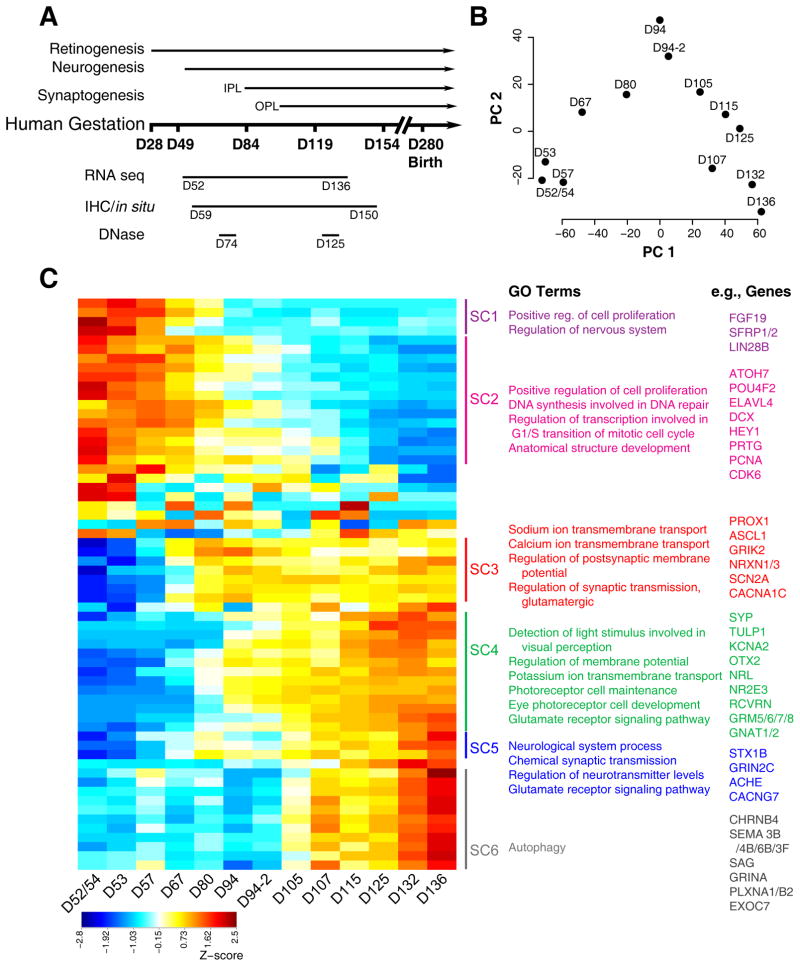
Total RNA from 17 fetal retina samples (D52 to D136) spanning distinct stages of development (Figure 1A, Table S1) were used for transcriptome profiling by RNA-seq. After quality control, normalization and principal components analysis (PCA) (Figures S1A–C), four samples—one each at D57, D67, D70, and D80—were deemed outliers and removed from subsequent analysis. PCA of the remaining 13 samples indicated age as the first principal component explaining almost 60% of the variance (Figure 1B). Differential expression (DE) analysis between the first two (D52/54 and D53) and the last two (D132 and D136) time points identified alterations in 3072 genes. Affinity propagation clustering of DE genes yielded 62 clusters of genes with specific patterns of developmental expression. The dendrogram of 62 clusters was trimmed at a height of 0.125 yielding 6 super clusters (SCs), showing distinct gene expression signatures (Figure 1C, Table S2).
Figure 1. Global analysis of RNA-seq data of human fetal retina ages D52–D136.
A. Schematic of the experimental paradigm. The timing of developmental events was inferred using published studies of human fetal retina and birthdating studies of monkey retinae by shifting proportionately by length of gestation. Whole retinae were collected for RNA-seq (D52–D136; 17 samples) and DNase-seq (D74 and D125) studies. Whole eyes (D59–D150) were processed for immunohistochemistry and in situ hybridization. B. Principle component (PC) analysis indicated that the largest source of variation among the RNA-seq samples was age accounting for 59.3% of the variance in the data. Biological replicates for D94 were included in this study and were differentiated from each other with suffixes (e.g., D94-2). C. Z-score of the exemplar genes from the 62 clusters found by Affinity Propagation (AP) clustering analysis. Blue to red indicates a gradient from low to high gene expression. Six superclusters were obtained by cutting the dendrograms at a height 0.125 of 62 clusters. Pathway enrichment analysis was carried out for the genes in each supercluster. The top GO categories and examples of genes found in the cluster are given. See also Figures S1 and S4.
The overall pattern of gene expression from the heatmap revealed three main “epochs”. The first epoch included approximately 30% of the clustered genes, which fell into superclusters (SCs) 1 and 2. These genes were highly expressed from D52 to D67, but were rapidly downregulated thereafter. Gene Ontology analysis (GO) identified enrichment of genes associated with early developmental processes, such as mitosis and cell proliferation (Figure 1C); these included early retinal progenitor cell (RPC) genes (e.g., FGF19, LIN28B, PRTG, and SFRP2), and those associated with early-generated cell types, such as retinal ganglion cells (RGCs) (e.g., ATOH7, DLX2, and POU4F2). A second major epoch was evident in SC3, which included genes whose expression began between D67 and D80 and maintained their expression to at least D136. SC3 contained transcription factors important for producing horizontal cells and amacrine cells, such as PROX1 and ASCL1, and genes implicated in synapse formation, such as NRXN1 and NRXN3, as well as neurotransmitter receptors and ion channels. The GO analysis of the genes in this epoch highlighted the switch from proliferation to differentiation of the early generated neurons, with 4 of the top 5 most significant Biological Process terms being related to synaptic transmission (e.g., nervous system development; anterograde trans-synaptic signaling; chemical synaptic transmission; synaptic signaling; and trans-synaptic signaling). Several of these synapse-related genes have not been previously described in the retina (e.g., UNC13C, UNC13D, DLGAP1, and LRRTM2) and warrant additional study. The third major epoch, with gene expression beginning at D90–D100, was highlighted by the genes related to photoreceptor and bipolar cells that were clustered in SC4, and those involved in synaptic connectivity and neurotransmission that were present in SC5 and SC6. The GO analysis for these SCs identified a group of 137 genes in the Biological Process category, “neuron projection development” (p = 1.18E–07). These included EPHB2 and SEMA5B—both with known roles in ganglion cell guidance and inner plexiform layer (IPL) lamination—as well as additional members of these and other gene families implicated in human retinal synaptogenesis (e.g., SEMA7A).
Thus, broad RNA-seq profile of the developing human retina nicely mirrors the predicted sequence of neurogenesis identified in model organisms (Cepko, 2014; He et al., 2012; Sanes and Zipursky, 2010), and the SCs allow us to clearly define the timing of key developmental events in human retinal development.
Correlation of transcriptome to early retinal development
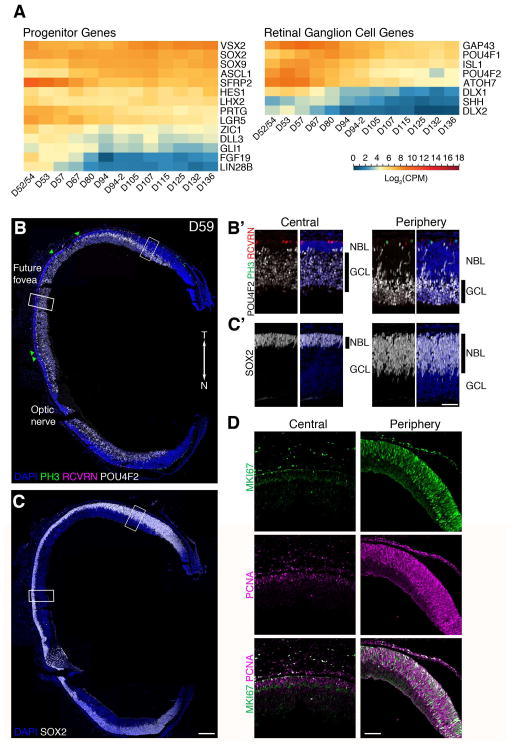
The RNA-seq results from the early stages of development show that the early retina is dominated by two main cell types: progenitor cells and RGCs (Figure 2A). Genes required for cell proliferation, such as PCNA, are expressed in the early SCs, as are several progenitor genes (such as LIN28B, FGF19, PRTG, and SFRP2), consistent with the expansive growth of the early retina. Not all progenitor genes peak at the early stages; some well-characterized progenitor genes (such as ASCL1, a gene expressed in progenitors after RGC differentiation is complete in mouse) have a later peak in expression, at D90–D100. Nevertheless, the major GO terms for this stage include G1/S transition of mitotic cycle and “Positive regulation of cell proliferation.” The RGC genes also exhibit peak expression prior to D94, and many genes known to be important for ganglion cell development, like POU4F2 and ATOH7, peak in the samples from D52–D57.
Figure 2. D59 human fetal retina is mainly composed of retinal ganglion cells and progenitors.
A. Expression profile of select marker genes of retinal progenitors and ganglion cells. Genes were organized from high (red) to low (blue) based on its expression level at D136. B. PH3 (green, mitotic cells), RCVRN (Magenta, photoreceptors), and POU4F2 (white, ganglion cells), and C. SOX2 (white, progenitor cells). Green triangles point to PH3+ cells. B′ and C′. Higher magnification views of regions boxed in B and C. D. Immunostaining with MKI67 (green) and PCNA (magenta) show reduced proliferation in D59 central retina, however most cells in the NBL are positive for these makers in the periphery. Scale bars for B&C: 200 μm; B′, C′: 25 μm. T: Temporal; N: Nasal; NBL: Neuroblastic layer; GCL: Ganglion Cell Layer. See also Figure S2.
We performed IHC using specific markers of progenitors [phosphohistone 3 (PH3) and SOX2] and RGCs (POU4F2) to correlate cellular composition with the transcriptome (Figure 2B and 2C). The overall portrait provided by the IHC is similar to that of the RNA-seq, but the regional differences in the cellular composition are already evident in our earliest samples. At D59, the central retina is comprised of 10–15 rows of RGCs with only a few layers of SOX2+ progenitor cells (Figure 2B′). The most central retina (the “future” foveal region) at D59 was identified structurally; this area in the temporal retina had a visible IPL and only a single layer of cone nuclei was present (Hendrickson, 2016). This central area was free of PH3+ mitotic cells (flanked by green triangles), had fewer Ki67+ and PCNA+ cells, and only a few RCVRN+ photoreceptor cells (Figures 2B′ and 2D). At this stage, the distance from the optic nerve to the presumptive foveal region was 1.3 mm, consistent with previous estimation of 1.4 mm distance from the optic nerve head to the incipient foveal region in D77 fetal retina (Xiao and Hendrickson, 2000). Thus, the “future” foveal region does not seem to expand nearly as much as the peripheral retina after D59.
In contrast to the central retina, at this early stage, progenitor cells constitute a majority of cells in the peripheral retina (Figure 2C′) with only a few layers of RGCs. The peripheral retina at D59 had a thicker neuroblastic layer (NBL) compared to the RGC layer, and a majority of the cells in the NBL within 1 mm of the presumptive ora serrata were positive for MKI67 and PCNA (Figure 2D). POU4F2+ RGCs were present and migrating through the NBL. RCVRN+ photoreceptors had not yet appeared in the most peripheral regions of the retina at this age (Figure 2B′).
From the total RNA-seq data, we were not able to assign previously uncharacterized genes to progenitors or ganglion cells; nonetheless, we compared our data with a single cell RNA-seq dataset from a published study (Pollen et al., 2015), in which cells of the developing fetal human brain and 84 cells from a D63 retina were sequenced. We clustered the single cell data using either SOX2 (for progenitors) or POU4F2 (for ganglion cells), and the cells fell into one or another of these two classes (Figure S2A).
Studies in mice have shown that the retinal progenitors can be further subdivided in their competence to generate different types of retinal neurons. In one model, so-called “Early Progenitors” or ERPs produce ganglion cells, cone photoreceptors, and horizontal cells, whereas “Late Progenitors” or LRPs generate amacrine cells, rod photoreceptors, bipolar cells, and Müller glia. At D63, we would predict that both ERPs and LRPs are present. Hierarchical clustering of the SOX2+ cells generated three main clusters, two of which expressed cell cycle genes (Figure S2B, red, blue) and were likely progenitors, while the other cluster may represent a subtype of amacrine cells. We used a previously reported marker of ERPs, PRTG, to distinguish between the two SOX2+ progenitor clusters; cells in the red cluster expressed this gene, while the cells in the blue cluster did not. We therefore classified these two clusters as ERPs and LRPs, respectively.
We further characterized the ERPs and LRPs by taking their respective 1000 most highly differentially-expressed genes and assigning them to one of the SCs from Figure 1. We show that the genes differentially expressed in the ERPs were more highly represented in SCs 1 and 2 than the LRP genes; on the contrary, the genes expressed in LRPs were more likely to be in SCs 3 and 4. Combining the single cell RNA-seq with our comprehensive dataset thus provides a powerful approach to assigning unknown genes to specific cell types, and will become increasingly important as additional single cell data becomes available.
Finally, the study of Pollen et al. focused on identifying progenitor cell subtypes in the cerebral cortex. Cortical progenitors can be classified as ventricular radial glia (vRG), which are the most stem-like progenitor in the brain, and two more limited progenitor types: intermediate progenitors (tRG) and outer radial glia (oRG). To assess the similarities between the two types of retinal progenitors and those from the cerebral cortex, we compared the genes from each cortical progenitor subtype to a supercluster. Notably, the genes expressed in vRG were more highly expressed in SCs 1 and 2, whereas those specific to the oRGs were more highly represented in the later SCs 3 and 4. Thus, ERPs and vRGs seem to share a common gene expression profile, and the oRGs (and tRGs) may exhibit more commonality with the LRPs.
Transcriptome and the development of inner retinal neurons
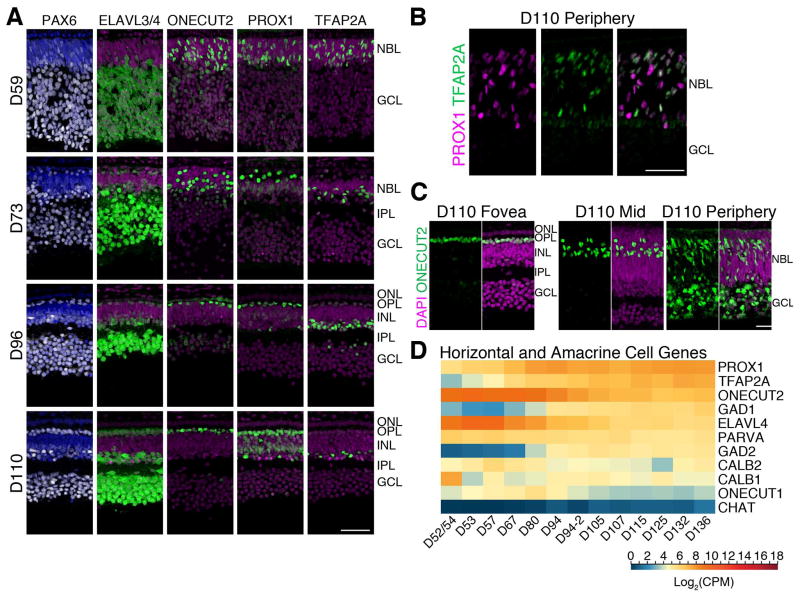
The second major epoch apparent from the RNA-seq data was reflected in the genes of SC3. Many of the genes in this cluster were characteristic of inner retinal neurons. PROX1, a key gene associated with both horizontal and amacrine cell differentiation, was present in SC3, showing increased expression from D67 to D80. IHC analysis confirmed the presence of both horizontal and amacrine cells as early as D59 in the presumptive fovea though a majority of PROX1+ and TFAP2A+ (a second marker of amacrine) cells were still migrating and scattered throughout the NBL (Figure 3A). By D73, immature horizontal and amacrine cell layers were observed, with the brightest PROX1+ cells lined near the outer plexiform layer (OPL) (horizontal cells) and those with lower PROX1 expression in the deeper layers of the INL (possible Müller glia and amacrine cells). The horizontal and amacrine cell layers were apparent by D96 in the foveal region, but had not yet developed even by D110 in the peripheral retina; a majority of PROX1+ and TFAP2A+ cells were still migrating through the NBL similar to the stage of maturity in the D59 fovea (Figure 3B).
Figure 3. Horizontal and amacrine cell development in the human fetal fovea.
A. D59–D110 foveas were immunolabeled with PAX6 (white), ELAVL3/4 (green, horizontal cells, amacrine cells, ganglion cells), ONECUT2 (green, cones, horizontal cells, ganglion cells), PROX1 (green, horizontal cells, amacrine cells, and Müller glia), and TFAP2A (amacrine cells). At early stages the horizontal and amacrine cells are intermixed, but by D96, the cells had migrated to the appropriate layer. B. Peripheral retina at D110 showing the PROX1+ horizontal and TFAP2A+ amacrine cells have still not formed a distinct layer. C. ONECUT2 is expressed by different cells during development; ONECUT2 is initially expressed in ganglion cells, cones, and horizontal cells but is later restricted to horizontal and a subset of amacrine cells. This range of expression was observed in a single section of a D110 retina, from the fovea to the peripheral edge. D. Log2 transformed Counts Per Million (CPM) values of selected horizontal and amacrine-related gene expression from RNA-seq were plotted as a heatmap. Blue to red represents low to high gene expression. Scale bars: 50 μm. INL: Inner Nuclear Layer; IPL; Inner Plexiform Layer; ONL: Outer Nuclear Layer; OPL: Outer Plexiform Layer.
To further evaluate cellular compositions, we performed IHC with ONECUT2, a marker of horizontal cells (Sapkota et al., 2014). At D59, weak ONECUT2 immunostaining was detected in RGC layer of the fovea with somewhat increased labeling throughout the NBL. By D96, ONECUT2+ horizontal cells were present in a single row. Albeit at a lower level of expression, ONECUT2 was detected in a subset of amacrine cells, particularly in those that were CALB1+ (CALB1 staining not shown). In a single section of D110 retina (Figure 3C), from the fovea to the peripheral edge, the expression of ONECUT2 was visibly limited to the HC layer and few amacrine cells in the fovea, but detected throughout the NBL and RGC layer in the periphery, reminiscent of a D59 fovea. A similar profile was observed for ONECUT1 expression (data not shown).
Although the second major period of development is largely characterized by genes associated with amacrine and horizontal cell generation, genes involved in their maturation were expressed somewhat later and detected in SC4 (Figures 1C and 3D); e.g., GAD1/2 increased in expression from D70 onward followed by CHAT. Nevertheless, the large relative increase of genes related to amacrine and horizontal cell production appears to drive this SC3 formation, and the retina undergoes a major shift away from RGC production to the generation of amacrine and horizontal cells during this period.
Temporal sequence of outer retina differentiation
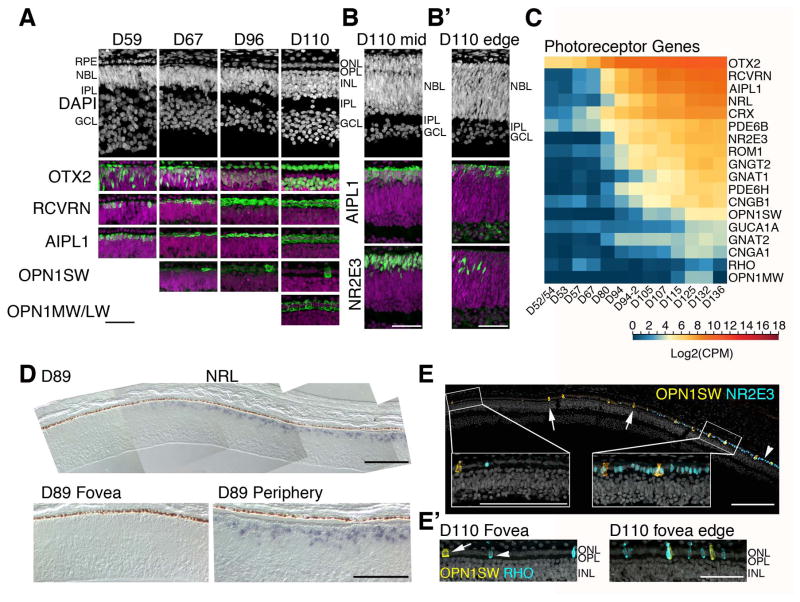
The last major epoch revealed by the RNA-seq clustering was characterized by a large increase in expression of genes related to photoreceptor development, in concordance with the appearance of these cells in histological sections. Several photoreceptor genes present in SC4, such as OTX2, AIPL1 (Kirschman et al., 2010; Ramamurthy et al., 2004), and RCVRN, were among the earliest markers detected by immunofluorescence. A small number of OTX2+ and RCVRN+ photoreceptors were detected as early as D59 in the central retina (Figure 4A). The OTX2+ cells in the photoreceptor layer were confined to a single layer in the presumptive fovea throughout the ages examined. A majority of OTX2+ cells at D59 and all OTX2+ cells in the photoreceptor layer by D67 also expressed AIPL1 and RCVRN. The first photoreceptors are presumably cones, though S-Opsin+ cells were not detected until D67 (Figure 4A). We consistently observed a low-level S-Opsin staining in all of the remaining cone photoreceptors, and this faint expression persisted until D96 (Figure 4A). By D110, this faint S-Opsin staining was no longer observed in the fovea and the cones expressed M/L-Opsin, consistent with the OPN1MW expression profile in the RNA-seq data.
Figure 4. Photoreceptor development in the human fetal retina.
A. Human fetal retinae from ages D59–D110 were stained with photoreceptor markers. Each of the photoreceptor markers are shown in green and nuclei were counterstained with DAPI (magenta). A single layer of photoreceptors was already present at D59, some of which were AIPL1+ and RCVRN+. The first OPN1SW+ cell was detected at D67 and OPN1MW/LW was detected at D110. Foveas older than D67 were generally defined as a zone free of S-Opsin and NR2E3 immunoreactivity. B. Retinal structure and AIPL1 and NR2E3 expression were compared between mid-temporal (B) and temporal peripheral edge (B′) of a D110 retina. Nuclear layers were not separated yet and NR2E3 expression was just beginning at the retinal edge. C. Heatmap for RNA-seq analysis of pan photoreceptor, cone, and rod genes show a steady increase over time. Blue indicates low and red indicates high gene expression. D. In situ hybridization of NRL mRNA at D89. The composite image shows the NRL-free fovea; NRL expression rapidly increases from the foveal edge and reaches the periphery (bottom right panel). E. A composite image of D110 fovea and 450 μm perifovea towards temporal retina. Immunostaining with OPN1SW (yellow) and a rod-specific marker NR2E3 (blue) showed a sharp increase in rod generation adjacent to the fovea. E′. A subset of these rods expressed RHO (blue). Scale bars for A–C: 50 μm.
In contrast to the fovea where all nuclear layers were present at D110, NBL in the mid-retina had just started to separate into the future ONL and INL (Figure 4B). At this stage, the immature ONL was only 3–4 layers and the presumptive INL was about 10 cell layers; however, the deepest few layers near the RGCs had the more rounded nuclear morphology characteristic of amacrine cells and the IPL was evident. Multiple cells in the presumptive ONL were AIPL1+ indicating the expansion of the photoreceptor layer outside of the fovea. The retinal edge was even more developmentally delayed, with ¾ of the retinal thickness comprising of a large NBL (Figure 4B). Despite this relative immaturity, AIPL1 expression had reached the retinal edge albeit with fewer cells.
SC4 also included NRL and NR2E3, two genes important for rod specification; their expression was detected at D80, whereas RHO (Rhodopsin) expression was observed at D115 (Figure 4C). Since rods are not present in the fovea, we examined NRL mRNA expression in the central retinal region by in situ hybridization. NRL-free zone was evident at D89, and NRL was expressed from the foveal edge and to the periphery (Figure 4D). We performed immunostaining of NR2E3, a downstream target of NRL (Oh et al., 2008), and generated a composite image of D110 fovea and the fovea/macula edge on the temporal side to examine the boundary between S-Opsin− and NR2E3− fovea from the rest of the rod-rich retina (Figure 4E). A few S-cones and rods were observed in the fovea/macula, but NR2E3+ photoreceptors increased dramatically at the fovea/macula edge on the temporal side where the ONL was still a single layer of cells. A few of these rods were already labeled for RHO (Figure 4E′) and had smaller, oval-shaped nuclei compared to cones with larger, cuboid nuclei. Several rows of NR2E3+ cells were detected in D110 mid-retina (Figure 4B) and the NR2E3 immunoreactivity decreased towards the retinal edge (Figure 4B′).
Accelerated development of the fovea/macula
The third major epoch defined by clustering also included genes characteristic of bipolar cells, Müller glia, and synapse formation. We analyzed these three classes of genes with IHC to gain a better appreciation of regional differences in development. We observed that like photoreceptors, the bipolar cells and Müller glia develop much earlier in the fovea compared to the rest of the retina. Although the RNA-seq results show the genes associated with these cells have their greatest increases between D80 and D105, cells with immunoreactivity for these markers appear much earlier in the fovea.
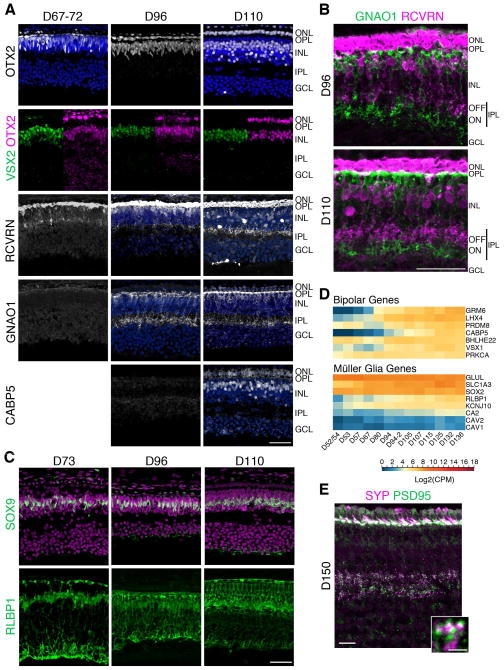
To identify the first bipolar cells, we used co-labeling of OTX2 and a variety of bipolar markers, such as VSX2, RCVRN, GNAO1, and CABP5. By this metric, as early as D67, most of the bipolar cells in the fovea have already been generated (Figure 5A, top two rows). These cells did not make their appearance in peripheral retina until after D110. The maturation of the bipolar cells is also accelerated in the fovea relative to other retinal regions. RCVRN+ flat midget Cone OFF bipolar cells (Milam et al., 1993) were first observed in the fovea at D67 and GNAO1+ (ON bipolar cells) appeared by D96; thus, the main components of the midget pathway were present in the fovea by D96. Furthermore, by this stage, foveal bipolar cells had axonal arborizations in the IPL, and the axons were stratified into ON and OFF sublaminae (Figure 5B). The processes of these cells were also observed in the OPL. Notably, CABP5 (Figure 5A) immunoreactivity was not detected until D110.
Figure 5. Later cell types are generated by D73 in the fovea and bipolar cell process stratification is observed by D96.
A. Fovea from D67–110 retinae were immunostained with OTX2 (first row: white, photoreceptors, bipolars); VSX2 and OTX2 (green and magenta, respectively; bipolar cells); RCVRN (white, OFF bipolar cells); GNAO1 (white, ON bipolar cells); and CABP5 (white, cone bipolar cells). Arrow heads point to VSX2+OTX2+ double-labeled bipolar cells. GNAO1 was first observed at D96 and CABP5 at D110. B. Higher magnification of D96 and D110 fovea double-labeled with GNAO1 and RCVRN show axonal arborizations in the IPL that were stratified into ON and OFF sublaminae by D96. C. Sections of D73–110 foveas were immunostained with SOX9 and RLBP1 (both green) revealed that a row of Müller glia was present in the fovea at D73 and RLBP1 expression got more elaborated over time. D. Log2 transformed Counts Per Million (CPM) values of select bipolar and Müller glia genes show a steady increase in gene expression beginning at D67–D80. Expression values plotted from high to low based on expression at D136. Blue to red indicates low to high gene expression. E. Immunostaining of the fovea of a D150 retina. In the IPL, SYP and PSD95 were expressed opposed to each other indicative of putative synapses. Scale bars: 50 μm. See also Figure S3.
In agreement with early foveal maturation, Müller glial-specific genes, such as SLC1A3, RLBP1, and CA2 present in SC4, also exhibited progressive increases in expression (Figure 5D). IHC analysis identified the earliest generated Müller glia in the human fovea at D73, with a row of fovea-restricted SOX9+ and RLBP1+ cells (Figure 5C). RLBP1 expression increased with time and the glial structure became more elaborate. However, Müller glia genesis had not yet reached the periphery even by D110 (data not shown). We noted that RLBP1 expression was detected very early in retinal pigment epithelium (RPE) and then in Müller cells, but was still lacking in the Müller cells in the periphery of D122 retina (data not shown).
The third epoch was also characterized by a large increase in genes related to synapse formation (e.g., SYN1, NRXN1/3) (Table S2). IHC confirmed the timing of expression of synaptic genes, and highlighted the accelerated development of the fovea. Pre-synaptic markers, SYP, CTBP2, and SLC17A7, were expressed first in the fovea in the IPL as early as D67 and in both IPL and OPL by D96 (Figure S3). As shown in previous reports of synaptogenesis (Hendrickson and Zhang, 2017), we observed the glutamatergic post-synaptic marker, PSD95, from D67 in the IPL. In addition, SYP and PSD95 were expressed opposed to one another, indicative of putative synapses at D150 (Figure 5E).
RNA-seq analysis of the developing fovea/macula
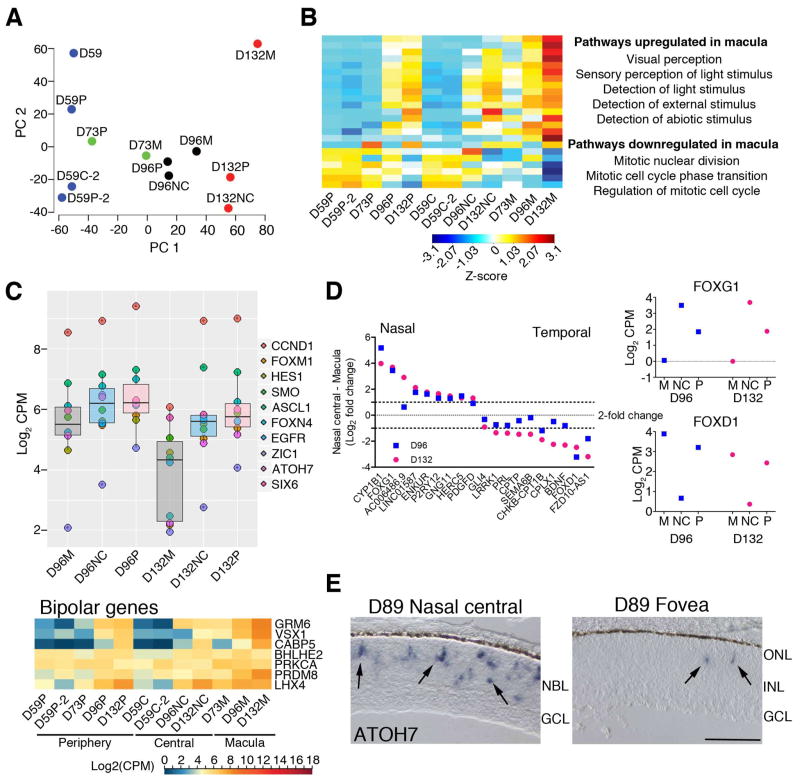
To further substantiate developmental divergence between the fovea/macula and the rest of the retina, we performed transcriptome analyses of specific regions dissected from the fetal retina (Figure 6, S5). Given that the macular region was not yet visibly distinct for accurate dissection, we isolated the central third from the peripheral two thirds in D59 retina. At later stages (D73, D96, and D132), we dissected the fovea/macula and the central nasal region from the peripheral retina. PCA of RNA-seq data revealed the most variance in PC1 (Figure 6A), which separated the samples by age. At each age, the peripheral retinal samples were “less mature” (i.e., closer to the earlier age) than the macular samples, thus confirming the IHC results. The nasal-central and peripheral retina regions at each stage were typically closer together on the PC1 axis, suggesting that the maturity of the fovea/macula is not simply because of the overall central to peripheral gradient of retinal development.
Figure 6. RNA-seq of the macula, nasal central, and peripheral retina dissected at different time points of fetal development.
A. Principle component (PC) analysis of these samples showed that the macula was developmentally ahead (further right on the PC1 ‘age’ axis) of the nasal central and peripheral regions of the same age. B. Z-score of the Affinity Propagation (AP) clustering exemplar genes from differential expression analysis of D59 central and D132 macula samples showed that genes that were upregulated were enriched for GO terms related to visual perception, while cell cycle-related genes were downregulated. C. A subset of progenitor and cell cycle genes were plotted to show that their downregulation over time occurs first in the macula. Heatmap of bipolar cell genes show that their gene expression of a late born cell type increases in the macula before the rest of the retina. D. There were some genes that were expressed highly in the macula and periphery, but not in the nasal retina (e.g., FOXG1), and others that were expressed higher in the nasal central and periphery but not in the macula (e.g., FOXD1). The differences in their Log2 Counts Per Million (CPM) values were calculated as the fold change and is plotted such that genes that are expressed more highly in nasal retina are on the left. E. In situ hybridization of ATOH7 in a D89 retina. ATOH7 mRNA expression can be seen in the NBL of the nasal central retina, whereas only a few ATOH7+ cells were observed in the fovea. Scale bar = 50 μm. See also Figures S5–8.
We performed DE analysis of RNA-seq data from the central retina of the earliest (D59C) and the oldest fovea/macula (D132M) samples to identify pathways that may be important for macula development. The resulting 360 genes were further subjected to AP clustering and gene ontology analysis (Figures 6B, S5B and C, Table S3). Almost 80% of the DE genes showed higher expression in the fovea/macula samples, and the onset of expression changes were initiated in the fovea. Gene Ontology analysis revealed the enrichment of visual perception and photoreceptor cell development in the upregulated genes in the fovea/macula, which included NRL, NR2E3, OPN1SW, RCVRN, USH2A, CRX, and AIPL1, consistent with the precocious development of this region. The downregulated genes generally belonged to mitosis-related terms (e.g., MKI67, CCND1, and TOP2A) but also included early cell fate specification genes, such as ATOH7, LEF1, and GLI1, and progenitor genes, such as HES1 and ASCL1, that showed relatively less expression in the fovea/macula compared to other retinal regions (Figure 6C, top panel). Consistent with the DE analysis, nearly all genes expressed in the mature fovea (including those specific for bipolar and Müller glia) were expressed by D73 (Figure 6C, bottom panel)—the first time point where we could differentiate macula from rest of the retina. Heat maps of various other cell types are presented in Figure S6.
DE analysis of macula vs. periphery at each time point yielded very few to no DE genes; this is probably due to the very small differences and lack of replicates for the time points. To further examine regional differences, we analyzed the top 50% of the expressed genes using K-median clustering, which yielded 10 clusters (Figure S7). Two of these clusters showed a nasal- or temporal-specific expression pattern (Figure 6D and S7); these included FOXG1 and FOXD1, two transcription factors demonstrated in other species to set up the nasal-temporal axis in the retina. Genes having a similar pattern as FOXG1 revealed high expression in the nasal central retina but were lower in the fovea/macula and periphery (Figure S7, top right panel), which was a mixture of nasal and temporal periphery. In contrast, temporal-specific genes, such as FOXD1, were highly expressed in the fovea/macula, but not in the nasal central retina (Figure S7, bottom right panel). ATOH7, a gene important for RGC specification, also showed differential expression between nasal and temporal central retina; by D89, only a few cells in the fovea expressed ATOH7, while ATOH7+ cells were observed throughout the NBL on the nasal side and towards the periphery (Figures 6E, S8). We also examined regional differences in FGF signaling, previously implicated in cone maturation and elongation during foveal pit formation in macaque retina (Cornish et al., 2005). Interestingly, this morphological change occurs in a central-fovea gradient; consistent with this, at D132, we detected increased expression of FGF2, FGFR1, and FGFR3 (but not FGFR2) in the nasal central and periphery relative to the fovea. These differences appeared to be age-dependent (Figure S6).
Comparative analyses of human and mouse transcriptome during retinal development
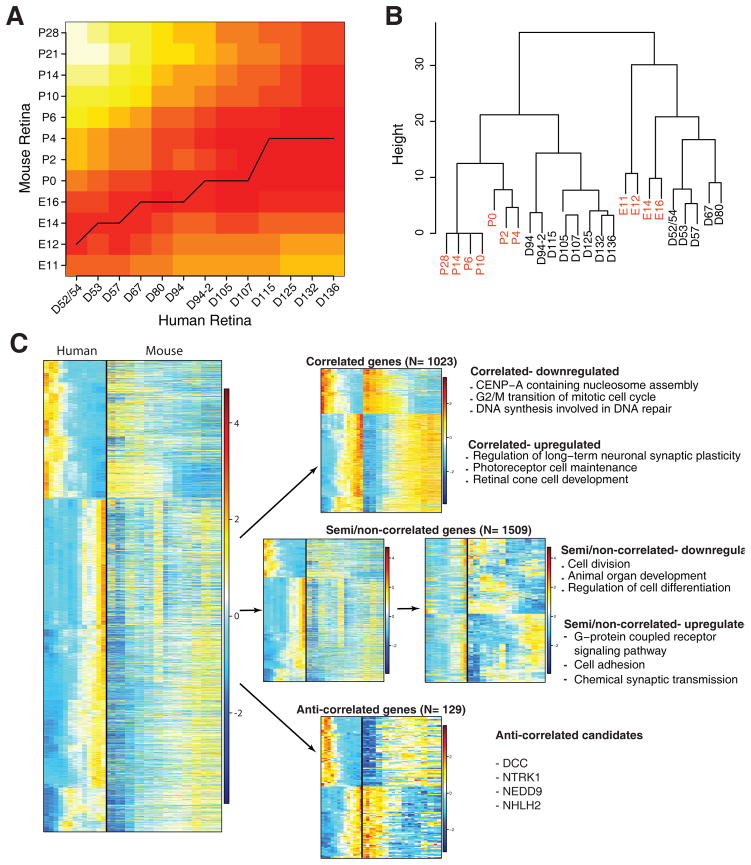
To decipher possible human-specific developmental regulation, we performed Open-Ended Dynamic Time Warping (OE-DTW) analysis of 2661 DE human genes for which we could identify mouse homologs in our mouse retinal development transcriptome dataset, collected from C57BI/6J mice at embryonic days, E11, E12, E14, E16 and postnatal days P0, P2, P4, P6, P10, P14, P21 and P28 (Brooks, Chaitankar and Swaroop, unpublished data) (Figure 7A). At early stages of development, we observed a tighter temporal correlation between mouse (E12–E14) and human genes (D52–57); however, an expansion of correlated temporal window was evident as development proceeded in human retina. For example, the E16–P0 stage in developing mouse retina was concordant with human fetal retina from D67 to D107; the period when human peripheral retina exhibited both cell proliferation and differentiation (as in the mouse) but foveal neurogenesis and lamination was complete. Expression of genes in D115–D136 human fetal retina was correlated to mouse P2–P4 retina. An independent Spearman correlation analysis of a set of 109 genes (Table S4) expressed in distinct retinal cell types further corroborated the similarity of human D52–D80 retina profiles with the mouse embryonic retina (E11–E16), and of D94–D136 human fetal retina with the postnatal P0–P4 mouse retina (Figure 7B). None of the human samples clustered with maturing mouse retina from P6–P28, probably because the fetal peripheral retina had not yet reached the developmental maturity of late postnatal mouse retina.
Figure 7. Comparison between developing mouse and human retina.
A. Open-Ended Dynamic time warping analysis for comparison of embryonic (E) 11–Postnatal (P) 28 retinae with human D52–D136 retinae. There were 2661 differentially expressed genes in humans that had 1:1 mapping with mouse. Expression data of these genes were subjected to Open-Ended Dynamic Time Warping analysis and the results are plotted as a heatmap. B. Spearman correlation of 109 retinal genes reveal that early human retinae are similar to embryonic mouse retinae, but at later developmental times, the mouse and human retinae segregate. C. Pearson correlation of human and mouse genes. Approximately 38% of the genes were highly correlated between mouse and human (Pearson correlation coefficient ≥ 0.7), approximately 57% of the genes were semi/non-correlated (Pearson correlation coefficient > −0.7 and < 0.7), and approximately 5% of the genes were anti-correlated (Pearson correlation coefficient ≤ −0.7). Enriched GO terms for the correlated and semi/non-correlated are presented as well as several significantly expressed non-correlated genes. Heatmaps were generated using Z-scores of expressed genes and AP clustering.
We then performed Pearson correlation to more precisely correlate different time periods and rates of differentiation in these two-mammalian species. Hierarchical clustering of human DE genes with corresponding expression in developing mouse retina (shown as heatmaps in Figure 7C) provided several distinct patterns: highly “correlated” genes (Pearson correlation coefficient ≥ 0.7, N = 1023, 38.4%) that can be further sub-divided in correlated-upregulated and correlated-downregulated; “semi/non-correlated” genes (Pearson correlation coefficient > −0.7 and < 0.7, N = 1509, 56.7%), which can be sub-divided as up- and downregulated genes; and “anti-correlated” genes showing opposite patterns in developing human and mouse retina (Pearson correlation coefficient ≤ −0.7, N = 129, 4.8%). GO analysis of genes in these clusters revealed that the genes with the highest degree of correlation between the species were G2/M transition cell cycle genes (correlated-downregulated) and photoreceptor development genes (correlated-upregulated). Approximately half of the genes fall into the semi/non-correlated category and can be better evaluated by clustering of expression patterns in the mouse retina (right middle panel in Figure 7C) rather than expression in human (left middle panel). While GO terms associated with semi/non-correlated-downregulated genes largely reflected cell proliferation, those corresponding to upregulated genes were enriched for cell signaling and neurotransmission. The anti-correlated genes, corresponding to the smallest group, did not show enrichment for any specific GO category. Nevertheless, a few genes in this cluster exhibited high levels of expression and showed very distinct patterns in the two species; especially, two genes in this category—DCC, encoding the netrin receptor, and NTRK1, encoding the NGF receptor—are known to be involved in mouse retinal development (Llamosas et al., 1997; Shi et al., 2010), and it is not clear why they would be anti-correlated in their pattern of expression in the two species. Further investigations are needed to determine whether these molecules have fundamentally different roles in developing human retina than they do in mouse.
Integration of chromatin accessibility at D74 and D125 to RNA-seq data
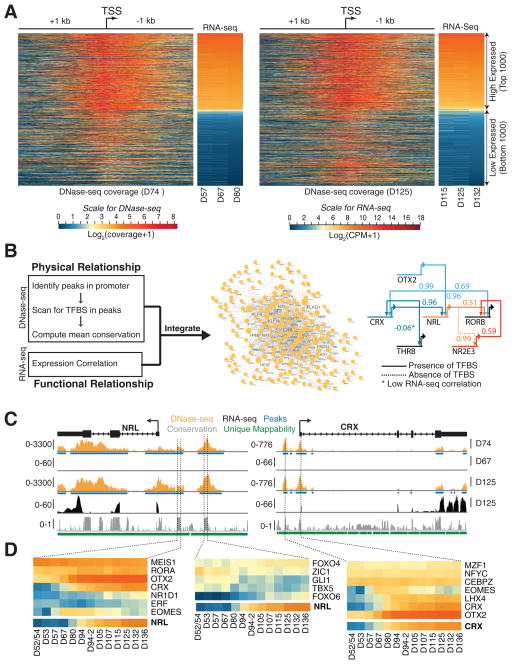
To develop correlations between cis-elements and gene expression and to construct gene regulatory networks, we integrated DNase-seq data from D74 and D125 retina produced by the ENCODE project (https://www.encodeproject.org/) with the corresponding temporal transcriptome data. The ‘findPeaks’ utility in Homer (v4.8.2) (Heinz et al., 2010) identified 319,828 and 318,459 variable length peaks in D74 and D125 samples, respectively. The distribution of the DNase-seq signal was computed 1 kb upstream and downstream of the transcription start site (TSS) for every gene and visualized as a heatmap (Figure 8A) along with the corresponding gene expression profile. The genome-wide comparison of DNase-seq and transcriptome data revealed a substantial association between gene expression and chromatin accessibility. Further integrative analysis with transcription factor binding site (TFBS) models yielded a predicted regulatory network specific to development of the human retina (Figure 8B). This network consisted of 219 genes and 1237 edges. Focusing on photoreceptor-specific genes in the network, we extracted possible upstream regulators of NRL and CRX (Figure 8C). We observed two conserved domains (CD) within the promoter of NRL with open chromatin signatures; of these, the first CD had TFBS for OTX2 and CRX, consistent with previous studies (Kautzmann et al., 2011; Montana et al., 2011; Roger et al., 2014), along with MEIS1, RORA, and EOMES. The second CD suggested novel binding proteins at the NRL promoter, including FOXO4, ZIC1, GLI1, TBX5, and FOXO6. Similarly, the CRX promoter regions included potential binding sites for several known and novel TFs. Expression of candidate regulatory proteins correlated well with NRL and CRX (Figure 8D).
Figure 8. DNase-seq shows changes in chromatin landscape between D74 and D125 retinae.
Integrative analysis of RNA-seq and DNase-seq data yields putative regulators of NRL and CRX. A. The 1000 highest and lowest expressed genes show a strong correlation between DNase-seq signal at Transcription Start Site (TSS) of a gene and its expression at D74 (left panel) and D125 (right panel). B. To build the gene regulatory network, the promoter region of every gene (3 kb upstream and 100 bp downstream from the TSS) was scanned for Transcription Factor Binding site (TFBS) in conserved regions to obtain putative binding evidence, and the presence of potential binding was augmented with expression correlation to infer high confidence regulatory interactions (left panel). Scanning for putative regulators of 135 retina-expressed genes yielded a network with 219 nodes and 1237 edges (middle panel). The network inferred several known regulatory interactions between photoreceptor-specific transcription factors (right panel). C. Genome snapshot for NRL (left panel) and CRX (right panel) promoter regions that includes DNase-seq and RNA-seq tracks are shown, along with peak regions and conservation. We observed two distinct conserved domains (CDs) in NRL and one in CRX promoter region. D. Expression heatmaps of putative regulators of NRL (left and middle panel) and CRX (right panel). Only TFBS with >99% mean conservation confidence were plotted (middle panel).
DISCUSSION
The basic cellular architecture and functional circuitry in the vertebrate retina is remarkably conserved (Hoon et al., 2014), as is the overall developmental sequence that produces this complex neural structure (Cayouette et al., 2006; Cepko, 2014; Sanes and Zipursky, 2010). However, the retinae of several birds and simian primates (including humans) possess a specialized cone-only fovea that is structurally and functionally distinct from the generally rod-rich periphery, and much less is known about how such retinal specializations arise during development. Degeneration of photoreceptors (and the underlying RPE) in the fovea and surrounding macula can lead to macular degeneration, a major cause of untreatable blindness worldwide, and a better understanding of the mechanisms controlling foveal development will permit the design of novel stem cell-based treatment paradigms for this devastating disease.
Here, we report a comprehensive profile of developing human retina by integrating next generation sequencing technology with a detailed anatomical characterization to illustrate several unique features, including the genesis of the fovea. We find that the human retina emerges in three fundamental “epochs”. The early retina is dominated by progenitor proliferation and ganglion cell production. Nearly all the cells in the D50–D60 retina are one of these two types. The second epoch is characterized by the emergence of horizontal cells and amacrine cells. Synapse-related genes also show strong increases during this period, likely due to the onset of synaptogenesis between ganglion cells and the newly differentiating amacrine cells. The third major epoch that is apparent from the RNA-seq reflects the production and differentiation of photoreceptors, bipolar cells, and Müller glia. The basic sequence of gene expression follows that of birthdating studies in model organisms (particularly that of Macaque), but in human retina it is surprising how the different cell types and their characteristic genes come to dominate the phases of retinal development. These basic epochs now provide a way to stage development in retina derived from human pluripotent stem cells.
Recently, RNA-seq data was published (Aldiri et al., 2017) for human fetal retina overlapping with the time course presented in this report. We thus analyzed the data using our pipeline and performed an initial comparison to evaluate whether addition of the samples would enhance our analysis (Figure S4A and B). Notwithstanding the two apparent outlier samples (FW10 and FW18), the data between the two reports are concordant with some reservations. Our data is very robust, showing the age of the samples to be perfectly sequential using PCA (Figure 1B, Figure S4B). The data in the Aldiri et al. report shows a similar trend though less of the variance is represented by this component. We observed similar expression pattern in a majority of key developmental genes from each of the SCs highlighted in Figure 1C and known photoreceptor genes shown in Figure 4C between the current report and the Aldiri, et al. data (Figure S4C–D).
A second important finding, which stemmed from the characterization of spatiotemporal genesis of the major cell types, was that the morphological differentiation of the fovea is complete much earlier than other regions for all cell types. In fact, molecular markers of distinct neurons could distinguish boundaries of “future” fovea by accelerated developmental timing and establish the temporal sequence of differentiation, lamination, and synaptogenesis in the developing human retina. By contrast, nasal retina at a similar distance from the optic disc and the peripheral retina have a much more protracted development.
The sequential appearance of genes associated with specific cell types that we observed in the human retina also occurs in the development of the mouse retina, and the overall pattern of gene expression is quite similar between the species. Although at early stages of development the similarity in timing of gene expression between the species is quite high, the temporal correlation between mouse and human becomes less distinct over time. We initially ascribed this difference to the greater central-to-peripheral developmental gradient in human retina, due in part to the more protracted gestational period; however, we were surprised by the very early onset of genes associated with late born types in the human samples and this led us to explore divergence in the differentiation of the fovea and the retinal periphery, as suggested previously by anatomical and IHC studies (Hendrickson and Yuodelis, 1984; Provis et al., 1985). Detailed in situ hybridization and IHC analyses of fetal retina confirmed the accelerated development of the fovea/macula and the early appearance of even late-generated cell types, such as bipolar cells. The foveal region was also precocious in the loss of mitotic retinal progenitors and the cessation of neurogenesis. RNA-seq of microdissected retinal regions validated these findings. Strikingly, fovea/macula differentiation precedes that of even more central regions of the nasal retina, despite the substantial central-to-peripheral gradient of development in the human retina.
The accelerated developmental timing of the fovea may partly explain its unique composition, particularly the lack of rod photoreceptors, since these cells are among the later generated cell types. Given evidence of heterogeneity in retinal progenitor cells, as well as progenitors elsewhere in the CNS, it is tempting to speculate that progenitors in the fovea/macula may have repressor(s) of the rod determination genes NRL and NR2E3 or be devoid of specific signals that induce these genes. Single cell RNA-seq studies may shed light on this issue. The rapid development of the fovea/macula is also of significance when considering the factors that control developmental timing in the nervous system. One important regulator of developmental time is the heterochronic pathway (La Torre et al., 2013), which seems to be important in the transition from production of early cell fates (e.g., RGCs) to the genesis of late cell types in mouse retina (Georgi and Reh, 2010; La Torre et al., 2013). Components of this pathway (e.g., LIN28B, PRTG) appear to be similarly expressed at the appropriate time and may carry out a similar function in human retinal development.
In addition to the accelerated foveal development, we were also able to better characterize other developmental gradients, like the central-to-peripheral gradient in developmental timing, and the nasal-temporal developmental gradient of positional determinants. IHC analysis further showed that the peripheral human retina is delayed by at least 50 days from central regions in the appearance of most cell types defined by specific cell markers, e.g., ONECUT2, TFAP2A, PROX1, and synaptic markers, e.g., SLC17A7. In many cases, the expression pattern of these markers in the periphery at D110 was similar to D59 fovea. The large central-to-peripheral gradient in human retina was also clear from the RNA-seq of microdissected retinal regions, and consistent with previous studies comparing gene expression in adult human tissue between the fovea and peripheral retina (Li et al., 2014). The differences in the timing of gene expression between central and peripheral retina and nasal vs. temporal retina can also be factored into assessment of the stage and position of retinal organoids. For example, the onset of rod genes, like NRL and NR2E3, could be used to define the approximate region of retina produced in vitro, while the relative expression of FOXG1 and FOXD1 can define its position along the nasal-temporal axis.
Further integration of transcriptome data, being presented here, should help in constructing developmental gene networks that are associated with functional maturation of specific cell types in human retina. The combination of RNA-seq, IHC, and DNase-seq provides the first detailed analysis of human fetal retinal development. Previous RNA-seq studies of human retina have focused on adult tissue or retinal organoids, highlighting novel transcripts, splice variants, and potential gene networks (Farkas et al., 2013; Kaewkhaw et al., 2015; Kaewkhaw et al., 2016; Karali et al., 2016; Li et al., 2014; Pinelli et al., 2016; Whitmore et al., 2014). The generation of mature retinal circuits from pluripotent cells is still a work-in-progress; however, some types of retinal diseases can already be modeled in iPSC-derived retinal tissue. It is not yet known what developmental stage these cultured cells can reach, partly because of a lack of reliable data to stage them. Up until now, these studies have relied on a few markers to stage the ESC-derived retinae, and define their regional specification, but the information in this report will now provide a benchmark to more accurately compare retinal tissue derived from pluripotent stem cells with that developed in vivo.
STAR Methods
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resource and reagents should be directed to and will be fulfilled by the Lead Contact, Dr. Thomas Reh (tomreh@uw.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Human fetal retina
Human conceptal eyes with no identifiers between post-conception D52–150 were recovered under an approved protocol through the University of Washington (UW5R24HD000836). The age for human specimens was estimated by a combination of clinic intakes, gestational ultrasound, crown-rump, and fetal foot length measurements where possible (FitzSimmons et al., 1994; Shepard, 1975). The gender was determined by either anatomic identification or PCR detection of Y-specific sequences using standard methods.
Mice
All procedures involving the use of mice were approved by the Animal Care and Use Committee of the National Eye Institute of the NIH. Mice of C57BL/6 genetic background were purchased from Jackson Laboratory (Bar Harbor, ME).
METHOD DETAILS
RNA-seq
Whole human fetal retina samples [spanning 14 time points: D52/54, D53, D57 (2 samples), D67 (2 samples), D70, D80, D87, D94 (2 samples), D105, D107, D115, D125, D132 and D136] or dissected retinal regions [at four time points: D59 (periphery and central, 2 samples each), D73 (periphery and fovea/macula), D96 (periphery, fovea/macula, and nasal central) and D132 (periphery, fovea/macula, and nasal central)] were dissected and homogenized in Trizol (Invitrogen, Carlsbad, CA) and stored at −80°C. Total RNA was extracted following manufacturer’s instructions. RNA integrity number (RIN) and quantity of RNA was assessed using Bioanalyzer RNA 6000 Pico Assay (Agilent Technologies, Santa Clara, CA). Average RIN for all samples was 8.6 and ranged from 6.8–10. Strand-specific libraries were constructed with 100 ng of total RNA using TruSeq Stranded mRNA Library Preparation Kit (Illumina, San Diego, CA). Paired-end sequencing was performed at a length of 125 bases on HiSeq2500 (Illumina).
Retinas of C57Bl/6 mice for embryonic days E11, E12, E14, E16 and postnatal days P0, P2, P4, P6, P10, P14, P21, and P28 were excised rapidly, frozen on dry ice, and stored at −80°C until use. Fresh frozen mouse retinas were lysed with a mortar and pestle in Trizol, and total RNA was isolated per manufacture’s protocol (Invitrogen). RNA quality and quantity were assessed with RNA 6000 Nano Kit (Agilent). Quantitative data from RNA-seq analysis was performed as previously stated (Veleri et al., 2017).
Analysis of RNA-seq data
An overview of the RNA-seq data analysis workflow is provided in Figure S1A. FASTQ files were evaluated for quality control using FastQC (v0.11.5) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/), and raw reads were trimmed for Illumina adapter (SLIDINGWINDOW:4:5; LEADING:5; TRAILING:5; MINLEN:25) in Trimmomatic (v0.36) (Bolger et al., 2014). Sequence alignment was performed using STAR (v2.5.2a) with reference assembly GRCh38.p3 and Ensembl annotation 82 with ENCODE options. Per sample, 2-pass transcriptome guided mapping (Dobin et al., 2013) was used. RSEM (v1.2.31) was used to estimate gene and transcript expression levels (Li and Dewey, 2011).
All secondary analyses were performed in the R statistical environment (www.r-project.org). Genes with more than 1 count per million (CPM) in at least four samples were retained for further analysis. PCA was performed using normalized log2 CPM values with pca3d (v0.8) package. Expected counts from RSEM output were TMM normalized using edgeR (v3.16.5) (McCarthy et al., 2012; Robinson et al., 2010). Statistical analysis for differentially expressed genes was performed with limma (v3.30.12) (Law et al., 2014). A generalized linear model was set up with time as cofactor, dispersion estimation was performed using the voom function, and statistical analysis was performed using eBayes. A gene was considered differentially expressed (DE) if absolute value of the log2 fold change was > 1 and Benjamini-Hochberg false discovery rate was ≤ 5%.
Clustering and pathway analysis
Affinity propagation (AP) clustering was performed on Z-score of the expression from DE genes using R package apcluster (v1.4.3) with “corSimMat” function. The resulting dendrogram was cut at a height of 0.125 yielding 16 aggregated SCs; six largest SCs were non-trivial and analyzed further. GO and KEGG pathway analysis was performed using goana and kegga function in limma. To reduce redundancy of the significant GO terms (< 1% FDR), the end-of-branch terms were chosen as representatives.
IHC and microscopy
Whole eyes were fixed in fresh 4% paraformaldehyde for 30 minutes to 1 hour at room temperature. The eyes were cryoprotected, embedded in OCT compound for freezing, then serially sectioned at 14–20 μm thickness. Only sections containing the fovea were used for this study. Sections were rinsed and incubated in blocking solution containing 10% normal horse serum and 0.5% Triton X-100 in phosphate buffered solution (PBS) for 1 hour at room temperature. Slides were labeled with primary antibodies diluted in blocking buffer overnight at 4°C. The primary antibodies used in this study are listed in the Key Resources Table 1. The following day, sections were washed with PBS and incubated for 1 hour at room temperature with appropriate secondary antibodies conjugated to 488/568/647 fluorophores and 4′,6-Diamidino-2-Phenylindole (DAPI, Sigma, St. Louis, MO) to counterstain nuclei. Subsequently, sections were rinsed in PBS, coverslipped, mounted in Fluoromount-G (SouthernBiotech, Birmingham, AL), and imaged using an Olympus FluoView FV1000 confocal microscope (Olympus, Tokyo, Japan). Images were processed using ImageJ (NIH, Bethesda, MD) and Adobe Photoshop. We defined the fovea of younger retinae (D59–D73) as an area where the subset of gene expression was higher than that of the periphery, the nuclear layers were clearly more developed, and only a single layer of cone photoreceptors were present (Hendrickson, 2016; O’Brien et al., 2003; Xiao and Hendrickson, 2000). At later stages, the fovea was identified as the area containing a single layer of cone photoreceptors that were mostly negative for S-Opsin and NR2E3 immunoreactivity (Bumsted O’Brien et al., 2004; Xiao and Hendrickson, 2000).
KEY RESOURCES TABLE.
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| AIPL1 | Gift from Dr. V. Ramamurthy | N/A |
| CABP5 | Gift from Dr. F. Haeseleer | N/A |
| CTBP2 | BD Biosciences | 612044 |
| DLG4 (6G6-1C9) | Abcam | B2723 |
| ELAVL3/4 | Invitrogen | A-21271 |
| GNAO1 | Chemicon | Mab3073 |
| MKI67 | Abcam | ab15580 |
| NR2E3 | Perseus Proteomics | PP-H7223-00 |
| ONECUT2 | R&D | AF6294 |
| OPN1MW/LW | Gift from J. Saari | N/A |
| OPN1SW (N-20) | Santa Cruz | sc-14363 |
| OTX2 | R&D Systems | BAF1979 |
| OTX2 | Abcam | 21990 |
| PAX6 | Developmental Systems Hybridoma Bank | N/A |
| PAX6 | Covance | PRB-278P |
| PCNA(PC10) | Santa Cruz | sc56 |
| PH3 | Novus | NB600-1168 |
| POU4F2 | Santa Cruz Biotechnology | SC-6026 |
| PROX1 | R&D Systems | AF2727 |
| RCVRN | Chemicon | AB5585 |
| RHO Clone 4D2 | Gift from Dr. R. Molday | N/A |
| RLBP1 | Abcam | ab15051 |
| SLC17A7 | Synaptic Systems | 135304 |
| SOX2 | Santa Cruz | sc-17320 |
| SOX9 | Temecula/Millipore | AB5535 |
| SYP | Sigma | S5768 |
| TFAP2A | Developmental Systems Hybridoma Bank | 5E4 |
| VSX2 | Exalpha Biologicals | X1179P |
| Donkey anti-goat 488 | Life Technologies | A11055 |
| Donkey anti-goat 568 | Life Technologies | A11057 |
| Donkey anti-goat 647 | Jackson Immuno | 705-605-147 |
| Donkey anti-mouse 488 | Jackson Immuno | 715-546-151 |
| Donkey anti-mouse 568 | Life Technologies | A10037 |
| Donkey anti-mouse 647 | Jackson Immuno | 715-605-150 |
| Donkey anti-rabbit 488 | Life Technologies | A21206 |
| Donkey anti-rabbit 568 | Life Technologies | A10042 |
| Donkey anti-rabbit 647 | Thermo Fisher Scientific | A-31573 |
| Donkey anti-sheep 568 | Life Technologies | A21099 |
| Donkey anti-sheep FITC | ICN | 55811 |
| DAPI | Sigma | D9542 |
| Biological Samples | ||
| Fetal eyes | Birth Defects Research Laboratory at the University of Washington | N/A |
| C57BL/6 | Jackson Laboratory | 000664 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Trizol | Invitrogen | 15596026 |
| Paraplast X-tra | McCormick Scientific | 39503002 |
| Paraformaldehyde | Electron Microscopy Sciences | RT-15710 |
| Digoxigenin-11-UTP | Sigma | 11209256910 |
| Roche Blocking Reagent | Sigma | 11096176001 |
| Normal goat serum | Millipore | S26-100ML |
| anti-Digoxigenin-AP, Fab fragments | Sigma | 11093274910 |
| BCIP/NBT Liquid Substrate | Sigma | B1911-100ML |
| Formamide, deionized | Fisher | AM9344 |
| 50% Dextran Sulfate | Millipore | S4031 |
| 50x Denhardt’s | Fisher | 750018 |
| Yeast tRNA | Invitrogen | 15401-029 |
| 20x SSC | Sigma | 11666681001 |
| Critical Commercial Assays | ||
| miRNeasy Micro kit | Qiagen | 217084 |
| TruSeq RNA Sample Prep Kit-v2 | Illumina | RS-122-2001 |
| MAXIscript kit | Ambion | AM1324 or AM1320 |
| Deposited Data | ||
| RNA sequencing of human fetal whole retina | This study | |
| RNA sequencing of human fetal dissected retina | This study | |
| RNA sequencing of mouse whole retina | GSE101986 | |
| Software and Algorithms | ||
| R version 3.4.0 | https://www.r-project.org | |
| Adobe Creative Suite 4 | Adobe | |
| Microsoft Office 2013 | Microsoft | |
| Image J | NIH | N/A |
| Other | ||
| FastQC version 0.11.5 | ||
| Trimmomatic, version 0.36 | Bolger et al., 2014 | |
| STAR, version 2.5.2a | Dobin et al., 2013 | |
| RSEM, version 1.2.31 | Li and Dewey,2011 | |
In situ hybridization
Whole eyes were fixed in Modified Carnoy’s Fix (60% ethanol, 30% formaldehyde, 10% glacial acetic acid) overnight at 4°C, then dehydrated in 70% ethanol (2 × 15 minute washes), 90% ethanol (2 × 15 minute washes), and 100% ethanol (2 × 30 minute washes followed by an overnight wash at 4°C). The e yes were then washed in xylene (3 × 15 minutes) and paraffin at 65°C (3 × 30 minutes) before being embedded in paraffin. Blocks were stored at 4°C until use.
Clones for human NRL and ATOH7 were obtained from Dharmacon, Clone ID numbers: 3866588 and 5580461, respectively. The plasmids were linearized with Sal1 and the anti-sense probe made using digoxigenin-11-UTP with MAXIscript kit (Invitrogen) and T7 RNA polymerase.
Eyes were sectioned at 8 μm, dewaxed, and hybridized at 68°C as previously described (Hayashi et al., 2007). In situ product was visualized using anti-DIG alkaline phosphatase conjugated secondary antibody (Roche, Basel, Switzerland) with color development using NBT (nitro blue tetrazolium)/BCIP (5-bromo-4-chloro-3-indolyl phosphate) solution (Sigma). After color development, the slides were washed and fixed with 4% PFA for 15 minutes and mounted with Flouromount-G (Southern Biotech).
Comparison of human and mouse retinal transcriptomes
Open-Ended Dynamic time warping (OE-DTW) analysis (Giorgino, 2009) was performed using the R package dtw (v1.18-1) on expression profiles of human development spanning from D52 through D136 days and mouse retina development between E11 through P28 (Brooks, Chaitankar and Swaroop, unpublished data). Utilizing the 3072 genes identified as differentially expressed in the human fetal development data, we selected the mouse orthologs using one-to-one mapping from HOM_MouseHumanSequence.rpt file obtained from Mouse Genome Informatics (http://www.informatics.jax.org). OE-DTW analysis was performed on the resulting 2661 genes to find an optimal alignment between the human and mouse retina development. To independently validate this observation, we performed Spearman correlation with 109 genes involved in retinal development (Table S4) that exhibited mouse-human homology.
A gene-wise Pearson’s correlation coefficient (r) was calculated for the orthologous DE genes between the human fetal development and mouse development samples. The spline function in R was used to interpolate the mouse samples so that they would be of the same sample number as the human data. Genes were assigned to correlated (r ≥ 0.7), semi/non-correlated (−0.7 < r < 0.7), or anti-correlated (r ≤ −0.7) groups. The AP clustered heatmap presented was as previously mentioned with the exception of the semi/non-correlated genes, which were then clustered with reference to the mouse samples.
DNase hypersensitivity and network analysis
Details of protocols and standards for DNase-seq are described by ENCODE (https://www.encodeproject.org/). All replicates of DNase-seq samples were combined prior to analysis for D74 and D125 time points. BWA-backtrack algorithm was implemented to align 36-bp paired-end reads to human (GRCh38.p3) reference genome. One mismatch was allowed during alignment and reads that were uniquely and pair-mapped (read mapped n proper pair) were used for further downstream analysis. An additional filter for mapping quality (q ≥ 30) was applied to eliminate low quality alignments. The ‘MarkDuplicates’ utility in Picard (v2.9.2) (http://broadinstitute.github.io/picard) was used to eliminate PCR or optical duplicates.
Gene regulatory network was inferred based on the presence of potentially functional (expression correlation) and physical binding (presence of TFBS in the promoter region) evidence. For inferring physical binding, TFBS using models from HOCOMOCO database (Kulakovskiy et al., 2016) were scanned in the open chromatin within the promoter region (defined as 3 kb upstream and 100 bp downstream) of 135 retinal genes (Table S5) (Kim, 2016; Xiang, 2013) using the ‘FIMO’ utility in MEME (v4.11.2) suite (Bailey et al., 2009; Grant et al., 2011). For every TFBS, mean conservation score was generated for alignments between six vertebrate genomes (Chimp, Rhesus, Dog, Mouse, Rat, and Opossum) and the human genome. Only those TFBS with mean conservation score ≥ 0.9 were selected for further downstream analysis. For every pair of genes that had evidence for potential physical binding, we computed the Pearson’s correlation across the time-series expression data. An absolute expression correlation score ≥ 0.75 was applied to infer the final network.
DATA AND SOFTWARE AVAILABILITY
Data was deposited in the Gene Expression Omnibus (GEO) 101986 (mouse RNA-seq) and Sequence Read Archive (SRA) with BioProject accession number ####. Additionally, the FASTQ and BAM files, as well as interactive data accession, are available at NEI Commons (https://neicommons.nei.nih.gov).
Supplementary Material
Highlights.
Transcriptomes of developing human retina reveal compartmentalized neurogenesis
Fovea differentiation, including lamination, is complete much before nasal retina
Combined RNA-seq and DNase analysis reveals developmental regulatory networks
Reference for molecular staging of retinal organoids derived from human stem cells
Acknowledgments
The authors thank the members of the Bermingham-McDonogh, Birth Defects Research, Reh, and Swaroop laboratories for helpful discussions. This work was supported by Intramural Research Program of the National Eye Institute (EY000450, EY000474 to A.S.); NIH grants 5F32 EY025117-02 (A.H.), 1PO1 GM081619-01 (T.A.R.), and 5R24 HD000836 (I.A.G); and Allen Distinguished Investigator Award (T.A.R, R.O.L.W, and F.R). We thank Dr. Tom Nowakowski for assistance with the analysis of the cerebral cortical data. This research utilized the high-performance computational capabilities of the Biowulf Linux cluster at NIH (http://biowulf.nih.gov). The authors would also like to acknowledge the pioneering contributions of Professor Anita Hendrickson to our understanding of human retinal development.
Footnotes
Author Contributions
Overall Conceptualization, A.H., R.R., A.L.T., T.A.R. and A.S.; General Methodology and Investigation, A.H., R.R., M.N.; Tissue acquisition and processing, A.H., O.B., M.N., M.S.W., I.A.G.; Immunohistochemistry and Microscopy, A.H., C.Z.; In situ hybridization and microscopy, O.B-McD., T.A.R.; Transcriptome Data Generation, R.R., L.G.; Transcriptome and DNase Data Analysis, M.J.B., V.C., R.R., M.R.S., M.S.W., A.W.; Data Curation, M.J.B., M.R.S., R.R.; Network Analysis, M.S.W., V.C.; Writing – Original Draft, A.H., R.R., T.A.R., A.S.; Writing – Review & Editing, A.H., R.R., R.O.L.W., T.A.R., A.S; Funding Acquisition, A.H., I.A.G., R.O.L.W., T.A.R., A.S.; Supervision and Project Administration, T.A.R., A.S.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aldiri I, Xu B, Wang L, Chen X, Hiler D, Griffiths L, Valentine M, Shirinifard A, Thiagarajan S, Sablauer A, et al. The Dynamic Epigenetic Landscape of the Retina During Development, Reprogramming, and Tumorigenesis. Neuron. 2017;94:550–568. e510. doi: 10.1016/j.neuron.2017.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bumsted O’Brien KM, Cheng H, Jiang Y, Schulte D, Swaroop A, Hendrickson AE. Expression of photoreceptor-specific nuclear receptor NR2E3 in rod photoreceptors of fetal human retina. Investigative ophthalmology & visual science. 2004;45:2807–2812. doi: 10.1167/iovs.03-1317. [DOI] [PubMed] [Google Scholar]
- Cayouette M, Poggi L, Harris WA. Lineage in the vertebrate retina. Trends Neurosci. 2006;29:563–570. doi: 10.1016/j.tins.2006.08.003. [DOI] [PubMed] [Google Scholar]
- Cepko C. Intrinsically different retinal progenitor cells produce specific types of progeny. Nat Rev Neurosci. 2014;15:615–627. doi: 10.1038/nrn3767. [DOI] [PubMed] [Google Scholar]
- Cornish EE, Madigan MC, Natoli R, Hales A, Hendrickson AE, Provis JM. Gradients of cone differentiation and FGF expression during development of the foveal depression in macaque retina. Vis Neurosci. 2005;22:447–459. doi: 10.1017/S0952523805224069. [DOI] [PubMed] [Google Scholar]
- Curcio CA, Sloan KR, Kalina RE, Hendrickson AE. Human photoreceptor topography. J Comp Neurol. 1990;292:497–523. doi: 10.1002/cne.902920402. [DOI] [PubMed] [Google Scholar]
- Dalkara D, Goureau O, Marazova K, Sahel JA. Let There Be Light: Gene and Cell Therapy for Blindness. Human gene therapy. 2016;27:134–147. doi: 10.1089/hum.2015.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darmanis S, Sloan SA, Zhang Y, Enge M, Caneda C, Shuer LM, Hayden Gephart MG, Barres BA, Quake SR. A survey of human brain transcriptome diversity at the single cell level. Proceedings of the National Academy of Sciences of the United States of America. 2015;112:7285–7290. doi: 10.1073/pnas.1507125112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas MH, Grant GR, White JA, Sousa ME, Consugar MB, Pierce EA. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC genomics. 2013;14:486. doi: 10.1186/1471-2164-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- FitzSimmons J, Fantel A, Shepard TH. Growth parameters in mid-trimester fetal Turner syndrome. Early human development. 1994;38:121–129. doi: 10.1016/0378-3782(94)90223-2. [DOI] [PubMed] [Google Scholar]
- Georgi SA, Reh TA. Dicer is required for the transition from early to late progenitor state in the developing mouse retina. J Neurosci. 2010;30:4048–4061. doi: 10.1523/JNEUROSCI.4982-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giorgino T. Computing and Visualizing Dynamic Time Warping Alignments in R: The dtw Package. Journal of Statistical Software. 2009;31:1–24. [Google Scholar]
- Grant CE, Bailey TL, Noble WS. FIMO: scanning for occurrences of a given motif. Bioinformatics. 2011;27:1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Cunningham D, Bermingham-McDonogh O. Loss of Fgfr3 leads to excess hair cell development in the mouse organ of Corti. Developmental dynamics: an official publication of the American Association of Anatomists. 2007;236:525–533. doi: 10.1002/dvdy.21026. [DOI] [PubMed] [Google Scholar]
- He J, Zhang G, Almeida AD, Cayouette M, Simons BD, Harris WA. How variable clones build an invariant retina. Neuron. 2012;75:786–798. doi: 10.1016/j.neuron.2012.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, Cheng JX, Murre C, Singh H, Glass CK. Simple combinations of lineage-determining transcription factors prime cisregulatory elements required for macrophage and B cell identities. Molecular cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson A. Development of Retinal Layers in Prenatal Human Retina. American journal of ophthalmology. 2016;161:29–35. e21. doi: 10.1016/j.ajo.2015.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson A, Possin D, Kwan WC, Huang J, Bourne JA. The temporal profile of retinal cell genesis in the marmoset monkey. J Comp Neurol. 2016;524:1193–1207. doi: 10.1002/cne.23897. [DOI] [PubMed] [Google Scholar]
- Hendrickson A, Possin D, Vajzovic L, Toth CA. Histologic development of the human fovea from midgestation to maturity. American journal of ophthalmology. 2012;154:767–778. e762. doi: 10.1016/j.ajo.2012.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson A, Troilo D, Djajadi H, Possin D, Springer A. Expression of synaptic and phototransduction markers during photoreceptor development in the marmoset monkey Callithrix jacchus. J Comp Neurol. 2009;512:218–231. doi: 10.1002/cne.21893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson A, Zhang C. Development of cone photoreceptors and their synapses in the human and monkey fovea. J Comp Neurol. 2017 doi: 10.1002/cne.24170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendrickson AE, Yuodelis C. The morphological development of the human fovea. Ophthalmology. 1984;91:603–612. doi: 10.1016/s0161-6420(84)34247-6. [DOI] [PubMed] [Google Scholar]
- Hoon M, Okawa H, Della Santina L, Wong RO. Functional architecture of the retina: development and disease. Progress in retinal and eye research. 2014;42:44–84. doi: 10.1016/j.preteyeres.2014.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hornan DM, Peirson SN, Hardcastle AJ, Molday RS, Cheetham ME, Webster AR. Novel retinal and cone photoreceptor transcripts revealed by human macular expression profiling. Investigative ophthalmology & visual science. 2007;48:5388–5396. doi: 10.1167/iovs.07-0355. [DOI] [PubMed] [Google Scholar]
- Hughes AE, Enright JM, Myers CA, Shen SQ, Corbo JC. Cell Type-Specific Epigenomic Analysis Reveals a Uniquely Closed Chromatin Architecture in Mouse Rod Photoreceptors. Scientific reports. 2017;7:43184. doi: 10.1038/srep43184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewkhaw R, Kaya KD, Brooks M, Homma K, Zou J, Chaitankar V, Rao M, Swaroop A. Transcriptome Dynamics of Developing Photoreceptors in Three-Dimensional Retina Cultures Recapitulates Temporal Sequence of Human Cone and Rod Differentiation Revealing Cell Surface Markers and Gene Networks. Stem cells. 2015;33:3504–3518. doi: 10.1002/stem.2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaewkhaw R, Swaroop M, Homma K, Nakamura J, Brooks M, Kaya KD, Chaitankar V, Michael S, Tawa G, Zou J, et al. Treatment Paradigms for Retinal and Macular Diseases Using 3-D Retina Cultures Derived From Human Reporter Pluripotent Stem Cell Lines. Investigative ophthalmology & visual science. 2016;57:ORSFl1–ORSFl11. doi: 10.1167/iovs.15-17639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karali M, Persico M, Mutarelli M, Carissimo A, Pizzo M, Singh Marwah V, Ambrosio C, Pinelli M, Carrella D, Ferrari S, et al. High-resolution analysis of the human retina miRNome reveals isomiR variations and novel microRNAs. Nucleic Acids Res. 2016;44:1525–1540. doi: 10.1093/nar/gkw039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kautzmann MA, Kim DS, Felder-Schmittbuhl MP, Swaroop A. Combinatorial regulation of photoreceptor differentiation factor, neural retina leucine zipper gene NRL, revealed by in vivo promoter analysis. The Journal of biological chemistry. 2011;286:28247–28255. doi: 10.1074/jbc.M111.257246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Yang HJ, Oel AP, Brooks MJ, Jia L, Plachetzki DC, Li W, Allison WT, Swaroop A. Recruitment of Rod Photoreceptors from Short-Wavelength-Sensitive Cones during the Evolution of Nocturnal Vision in Mammals. Developmental cell. 2016;37:520–532. doi: 10.1016/j.devcel.2016.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JW, Yang HJ, Brooks MJ, Zelinger L, Karakulah G, Gotoh N, Boleda A, Gieser L, Giuste F, Whitaker DT, Walton A, Vilasmil R, Barb JJ, Munson PJ, Kaya KD, Chaitankar V, Cogliati T, Swaroop A. NRL-regulated transcriptome dynamics of developing rod photoreceptors. Cell Rep. 2016;17:2460–2473. doi: 10.1016/j.celrep.2016.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschman LT, Kolandaivelu S, Frederick JM, Dang L, Goldberg AF, Baehr W, Ramamurthy V. The Leber congenital amaurosis protein, AIPL1, is needed for the viability and functioning of cone photoreceptor cells. Human molecular genetics. 2010;19:1076–1087. doi: 10.1093/hmg/ddp571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozulin P, Natoli R, O’Brien KM, Madigan MC, Provis JM. Differential expression of anti-angiogenic factors and guidance genes in the developing macula. Mol Vis. 2009;15:45–59. [PMC free article] [PubMed] [Google Scholar]
- Kulakovskiy IV, Vorontsov IE, Yevshin IS, Soboleva AV, Kasianov AS, Ashoor H, Ba-Alawi W, Bajic VB, Medvedeva YA, Kolpakov FA, et al. HOCOMOCO: expansion and enhancement of the collection of transcription factor binding sites models. Nucleic Acids Res. 2016;44:D116–125. doi: 10.1093/nar/gkv1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Torre A, Georgi S, Reh TA. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2362–2370. doi: 10.1073/pnas.1301837110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Vail MM, Rapaport DH, Rakic P. Cytogenesis in the monkey retina. J Comp Neurol. 1991;309:86–114. doi: 10.1002/cne.903090107. [DOI] [PubMed] [Google Scholar]
- Law CW, Chen Y, Shi W, Smyth GK. voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome biology. 2014;15:R29. doi: 10.1186/gb-2014-15-2-r29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12:323. doi: 10.1186/1471-2105-12-323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Jia C, Kazmierkiewicz KL, Bowman AS, Tian L, Liu Y, Gupta NA, Gudiseva HV, Yee SS, Kim M, et al. Comprehensive analysis of gene expression in human retina and supporting tissues. Human molecular genetics. 2014;23:4001–4014. doi: 10.1093/hmg/ddu114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Trapnell C. Single-cell transcriptome sequencing: recent advances and remaining challenges. F1000Research. 2016;5 doi: 10.12688/f1000research.7223.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llamosas MM, Cernuda-Cernuda R, Huerta JJ, Vega JA, Garcia-Fernandez JM. Neurotrophin receptors expression in the developing mouse retina: an immunohistochemical study. Anatomy and embryology. 1997;195:337–344. doi: 10.1007/s004290050053. [DOI] [PubMed] [Google Scholar]
- Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, Tirosh I, Bialas AR, Kamitaki N, Martersteck EM, et al. Highly Parallel Genome-wide Expression Profiling of Individual Cells Using Nanoliter Droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNASeq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milam AH, Dacey DM, Dizhoor AM. Recoverin immunoreactivity in mammalian cone bipolar cells. Vis Neurosci. 1993;10:1–12. doi: 10.1017/s0952523800003175. [DOI] [PubMed] [Google Scholar]
- Mo A, Luo C, Davis FP, Mukamel EA, Henry GL, Nery JR, Urich MA, Picard S, Lister R, Eddy SR, et al. Epigenomic landscapes of retinal rods and cones. eLife. 2016;5:e11613. doi: 10.7554/eLife.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montana CL, Lawrence KA, Williams NL, Tran NM, Peng GH, Chen S, Corbo JC. Transcriptional regulation of neural retina leucine zipper (Nrl), a photoreceptor cell fate determinant. The Journal of biological chemistry. 2011;286:36921–36931. doi: 10.1074/jbc.M111.279026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustafi D, Kevany BM, Bai X, Golczak M, Adams MD, Wynshaw-Boris A, Palczewski K. Transcriptome analysis reveals rod/cone photoreceptor specific signatures across mammalian retinas. Human molecular genetics. 2016 doi: 10.1093/hmg/ddw268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura PA, Tang S, Shimchuk AA, Ding S, Reh TA. Potential of Small Molecule- Mediated Reprogramming of Rod Photoreceptors to Treat Retinitis Pigmentosa. Investigative ophthalmology & visual science. 2016;57:6407–6415. doi: 10.1167/iovs.16-20177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano T, Ando S, Takata N, Kawada M, Muguruma K, Sekiguchi K, Saito K, Yonemura S, Eiraku M, Sasai Y. Self-formation of optic cups and storable stratified neural retina from human ESCs. Cell stem cell. 2012;10:771–785. doi: 10.1016/j.stem.2012.05.009. [DOI] [PubMed] [Google Scholar]
- O’Brien KM, Schulte D, Hendrickson AE. Expression of photoreceptor-associated molecules during human fetal eye development. Mol Vis. 2003;9:401–409. [PubMed] [Google Scholar]
- Oh EC, Cheng H, Hao H, Jia L, Khan NW, Swaroop A. Rod differentiation factor NRL activates the expression of nuclear receptor NR2E3 to suppress the development of cone photoreceptors. Brain Res. 2008;1236:16–29. doi: 10.1016/j.brainres.2008.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinelli M, Carissimo A, Cutillo L, Lai CH, Mutarelli M, Moretti MN, Singh MV, Karali M, Carrella D, Pizzo M, et al. An atlas of gene expression and gene co-regulation in the human retina. Nucleic Acids Res. 2016;44:5773–5784. doi: 10.1093/nar/gkw486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollen AA, Nowakowski TJ, Chen J, Retallack H, Sandoval-Espinosa C, Nicholas CR, Shuga J, Liu SJ, Oldham MC, Diaz A, et al. Molecular identity of human outer radial glia during cortical development. Cell. 2015;163:55–67. doi: 10.1016/j.cell.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prada C, Puga J, Perez-Mendez L, Lopez, Ramirez G. Spatial and Temporal Patterns of Neurogenesis in the Chick Retina. Eur J Neurosci. 1991;3:1187. doi: 10.1111/j.1460-9568.1991.tb00053.x. [DOI] [PubMed] [Google Scholar]
- Provis JM, Hendrickson AE. The foveal avascular region of developing human retina. Archives of ophthalmology. 2008;126:507–511. doi: 10.1001/archopht.126.4.507. [DOI] [PubMed] [Google Scholar]
- Provis JM, van Driel D, Billson FA, Russell P. Development of the human retina: patterns of cell distribution and redistribution in the ganglion cell layer. J Comp Neurol. 1985;233:429–451. doi: 10.1002/cne.902330403. [DOI] [PubMed] [Google Scholar]
- Ramamurthy V, Niemi GA, Reh TA, Hurley JB. Leber congenital amaurosis linked to AIPL1: a mouse model reveals destabilization of cGMP phosphodiesterase. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:13897–13902. doi: 10.1073/pnas.0404197101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roger JE, Hiriyanna A, Gotoh N, Hao H, Cheng DF, Ratnapriya R, Kautzmann MA, Chang B, Swaroop A. OTX2 loss causes rod differentiation defect in CRX-associated congenital blindness. The Journal of clinical investigation. 2014;124:631–643. doi: 10.1172/JCI72722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanes JR, Zipursky SL. Design principles of insect and vertebrate visual systems. Neuron. 2010;66:15–36. doi: 10.1016/j.neuron.2010.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota D, Chintala H, Wu F, Fliesler SJ, Hu Z, Mu X. Onecut1 and Onecut2 redundantly regulate early retinal cell fates during development. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:E4086–4095. doi: 10.1073/pnas.1405354111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasai Y. Next-generation regenerative medicine: organogenesis from stem cells in 3D culture. Cell stem cell. 2013;12:520–530. doi: 10.1016/j.stem.2013.04.009. [DOI] [PubMed] [Google Scholar]
- Scholl HP, Strauss RW, Singh MS, Dalkara D, Roska B, Picaud S, Sahel JA. Emerging therapies for inherited retinal degeneration. Science translational medicine. 2016;8:368rv366. doi: 10.1126/scitranslmed.aaf2838. [DOI] [PubMed] [Google Scholar]
- Sears S, Erickson A, Hendrickson A. The spatial and temporal expression of outer segment proteins during development of Macaca monkey cones. Investigative ophthalmology & visual science. 2000;41:971–979. [PubMed] [Google Scholar]
- Shepard TH. Growth and Development of the Human Embryo and Fetus. Philadelphia, PA: W. B. Saunders Company; 1975. [Google Scholar]
- Shi M, Zheng MH, Liu ZR, Hu ZL, Huang Y, Chen JY, Zhao G, Han H, Ding YQ. DCC is specifically required for the survival of retinal ganglion and displaced amacrine cells in the developing mouse retina. Developmental biology. 2010;348:87–96. doi: 10.1016/j.ydbio.2010.09.013. [DOI] [PubMed] [Google Scholar]
- Tabebordbar M, Zhu K, Cheng JK, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veleri S, Nellissery J, Mishra B, Manjunath SH, Brooks MJ, Dong L, Nagashima K, Qian H, Gao C, Sergeev YV, et al. REEP6 mediates trafficking of a subset of Clathrin-coated vesicles and is critical for rod photoreceptor function and survival. Human molecular genetics. 2017;26:2218–2230. doi: 10.1093/hmg/ddx111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitmore SS, Wagner AH, DeLuca AP, Drack AV, Stone EM, Tucker BA, Zeng S, Braun TA, Mullins RF, Scheetz TE. Transcriptomic analysis across nasal, temporal, and macular regions of human neural retina and RPE/choroid by RNA-Seq. Exp Eye Res. 2014;129:93–106. doi: 10.1016/j.exer.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LL, Rapaport DH. Defining retinal progenitor cell competence in Xenopus laevis by clonal analysis. Development. 2009;136:1707–1715. doi: 10.1242/dev.027607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang M. Intrinsic control of mammalian retinogenesis. Cellular and molecular life sciences: CMLS. 2013;70:2519–2532. doi: 10.1007/s00018-012-1183-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao M, Hendrickson A. Spatial and temporal expression of short, long/medium, or both opsins in human fetal cones. J Comp Neurol. 2000;425:545–559. [PubMed] [Google Scholar]
- Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell stem cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- Yang HJ, Ratnapriya R, Cogliati T, Kim JW, Swaroop A. Vision from next generation sequencing: multi-dimensional genome-wide analysis for producing gene regulatory networks underlying retinal development, aging and disease. Progress in retinal and eye research. 2015;46:1–30. doi: 10.1016/j.preteyeres.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young RW. Cell differentiation in the retina of the mouse. Anat Rec. 1985;212:199–205. doi: 10.1002/ar.1092120215. [DOI] [PubMed] [Google Scholar]
- Yu W, Mookherjee S, Chaitankar V, Hiriyanna S, Kim JW, Brooks M, Ataeijannati Y, Sun X, Dong L, Li T, et al. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nature communications. 2017;8:14716. doi: 10.1038/ncomms14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.