Abstract
Background
Pseudomonas chlororaphis HT66 isolated from the rice rhizosphere is an important plant growth-promoting rhizobacteria that produce phenazine-1-carboxamide (PCN) in high yield. Phenazine production is regulated by a quorum sensing (QS) system that involves the N-acylated homoserine lactones (AHLs)—a prevalent type of QS molecule.
Results
Three QS signals were detected by thin layer chromatography (TLC) and high-performance liquid chromatography–mass spectrometry (HPLC–MS/MS), which identified to be N-(3-hydroxy hexanoyl)-l-homoserine lactone (3-OH-C6-HSL), N-(3-hydroxy octanoyl)-l-homoserine lactone (3-OH-C8-HSL) and N-(3-hydroxy decanoyl)-l-homoserine lactone (3-OH-C10-HSL). The signal types and methods of synthesis were different from that in other phenazine-producing Pseudomonas strains. By non-scar deletion and heterologous expression techniques, the biosynthesis of the AHL-signals was confirmed to be only catalyzed by PhzI, while other AHLs synthases i.e., CsaI and HdtS were not involved in strain HT66. In comparison to wild-type HT66, PCN production was 2.3-folds improved by over-expression of phzI, however, phzI or phzR mutant did not produce PCN. The cell growth of HT66∆phzI mutant was significantly decreased, and the biofilm formation in phzI or phzR inactivated strains of HT66 decreased to various extents.
Conclusion
In conclusion, the results demonstrate that PhzI–PhzR system plays a critical role in numerous biological processes including phenazine production.
Electronic supplementary material
The online version of this article (10.1186/s12934-017-0854-y) contains supplementary material, which is available to authorized users.
Keywords: Pseudomonas chlororaphis, Phenazine-1-carboxamide, Quorum sensing, N-Acylated homoserine lactones, phzI, Biofilm formation
Background
Quorum sensing (QS) is a well-studied form of communication process used by a large variety of bacteria to regulate diverse cellular functions such as antibiotic production, biofilm development, gene expression, surface attachment and virulence in a cell-population density-dependent manner [1, 2]. The Gram-negative bacteria use N-acylated homoserine lactones (AHLs) to sense cell density, which is composed of a homoserine lactone ring (HSL) with varying acyl chain [3]. These diffusible small signaling molecules are synthesized by a member of the LuxI protein family. Bacteria can monitor cell-population density by measuring the concentration of small secreted signaling molecules, so-called AHLs. When AHL concentration reaches a certain threshold value, the cells can switch on the expression of a set of genes responsible for the production of bioluminescence [4], antibiotics [5], plasmid transfer [6], and symbiosis [7] etc. Therefore, the AHL-mediated QS play an essential role in numerous biological processes.
Phenazines are a class of pigmented heterocyclic metabolites, which are produced by the genera Pseudomonas, Burkholderia, and Streptomyces [8]. Phenazine and its derivatives, such as a phenazine-1-carboxylic acid (PCA), phenazine-1-carboxamide (PCN), and 2-hydroxy-phenazine (2-OH-PHZ), exhibit potent antifungal activities against a wide range of eukaryotic microbes and therefore could be employed as a fungicide in agriculture production [9]. For example, PCA greatly reduces the risk of a severe wheat root disease caused by Gaeumannomyces graminis var. tritici [10, 11]. PCN possesses notable antifungal activity against Fusarium oxysporum f. sp. radices-lycopersici [12, 13]. Intriguingly, almost all of the phenazine compounds secreted by bacteria display antimicrobial, anti-tumor, antimalarial, and antiparasitic effects compared with some chemically synthesized phenazine derivatives [14–19]. In addition, natural phenazine products show great promise for use as electron acceptors and donors, components of microbial fuel cells (MFC), and environmental sensors and biosensors [20–23]. Most phenazine-producing microorganisms have been isolated from diverse terrestrial, freshwater, and marine environments [24, 25]. Fluorescent pseudomonads, which are members of the gamma subclass of the proteobacteria, are the best-studied phenazine producers, with strains of Pseudomonas fluorescens, P. chlororaphis, and P. aeruginosa known to produce these compounds [26]. PhzI, a LuxI homolog, is an AHL synthase mainly responsible for the synthesis of AHL signals in phenazine-producing Pseudomonas. Subsequently, the AHL-receptor protein, PhzR, binds to AHL-signals and activates the expression of downstream phenazine biosynthetic gene cluster [27].
In this study, an isolated microbe from rice rhizosphere namely Pseudomonas chlororaphis HT66 has been selected. Based on whole genome sequencing, three possible AHL synthesis genes phzI, csaI and hdtS were investigated. Among them, the phzI gene is located upstream of the phz cluster like other Pseudomonas. However, the types and biosynthesis of AHL signals are unclear and the regulation of phenazine biosynthesis needs to be studied. The AHLs were identified by Thin Layer Chromatography (TLC) and high-performance liquid chromatography-mass spectrometry (HPLC–MS/MS) analyses. Additionally, we elucidated the relationship between phenazine production, biofilm formation and AHLs biosynthesis, including which gene is responsible for the AHLs synthesis. The biocontrol activity of P. chlororaphis HT66 and its derivatives were also evaluated.
Methods
Bacterial strains and growth conditions
Selected bacterial strains and plasmids used in this study are summarized in Table 1. Bacterial strains were grown in Luria–Bertani (LB) medium at 37 °C (for Escherichia coli) or King’s B (KB) medium at 28 °C (for P. chlororaphis HT66). The bioreporter strains, Agrobacterium tumefaciens NTL4 (pZLR4) [28] and Chromobacterium violaceum CV026 [29], were grown at 28 °C in AB minimal medium (ABM) [30] and LB medium, respectively. When required, antibiotics were used at the following concentrations: ampicillin (100 μg/ml), kanamycin (50 μg/ml) or tetracycline (20 μg/ml) for E. coli and ampicillin (100 μg/ml), kanamycin (50 μg/ml) and tetracycline (200 μg/ml) for P. chlororaphis; whereas the gentamicin (30 μg/ml) and kanamycin (20 μg/ml) were used for NTL4 (pZLR4) and CV026, respectively.
Table 1.
Bacterial strains, and plasmids used in this study
| Strain, plasmid | Characteristics | Reference or source |
|---|---|---|
| Escherichia coli strains | ||
| E.coli DH5α | supE44∆lacU169(ɸ80lacZ∆M15)recAhsdR17 recA1 endA1 gyrA96 thi-1 relA-1 | Hanahan [45] |
| E. coli S17 | res- pro mod+ integrated copy of RP4, mob+, used for incorporating constructs in P. chlororaphis | Hoffmann et al. [46] |
| A. tumefaciens NTL4 (pZLR4) | Biosensor strain for AHLs | Cha et al. [28] |
| C. violaceum CV026 | Biosensor strain for AHLs | McClean et al. [29] |
| Pythium ultimum | The pathogen causes ripe fruit rot of tomato | Pearson et al. [33] |
| Pseudomonas chlororaphis | ||
| HT66 | PCN, wild-type, AmprSpr | This study |
| ∆phzI | HT66 derivative, phzI deleted | This study |
| ∆phzR | HT66 derivative, phzR deleted | This study |
| ∆csaI | HT66 derivative, csaI deleted | This study |
| ∆hdtS | HT66 derivative, hdtS deleted | This study |
| Plasmids | ||
| pK18mobsacB | Broad-host-range gene replacement vector; sacB, Kmr | Schafer et al. [47] |
| pME6032 | IPTG-inducible expression vector, Tcr | Heeb et al. [48] |
| pME-phzI | phzI from HT66 in pME6032,Tcr | This study |
| pME-csaI | csaI from HT66 in pME6032,Tcr | This study |
| pME-hdtS | hdtS from HT66 in pME6032,Tcr | This study |
r resistance
DNA manipulation and mutant construction
The non-scar gene deletion was carried out as previously described [31]. First, two pairs of primers designed to clone 100–600 bp fragments located upstream or downstream of the target gene. These two 100–600 bp fragments were amplified by polymerase chain reaction (PCR) from the genomic DNA of HT66. Next, an overlap PCR was used to combine the two fragments, which created a new sequence excluding the target gene. After digesting the overlap PCR product by restriction enzymes EcoRI and HindIII, the new sequence was ligated into pK18mobsacB by T4 DNA ligase. The resulting plasmid was first transferred into E. coli S17 and then mobilized into HT66 by conjugation. Afterwards, the colony carrying pK18mobsacB was inoculated into 15% sucrose counter-selection plate, only the marked mutant population and spontaneous SucR colonies grew. Finally, the colonies were tested and verified by PCR analysis and sequencing, to make sure a double crossover had occurred and the target part had been replaced by the new sequence. All the other gene deletions were carried out following the same strategy.
Taking the over-expression and complementation of phzI as an example, we used primers to amplify the 591-bp phzI gene from HT66. The resulting PCR-amplified fragment was digested with restriction enzymes EcoRI and XhoI and then cloned into vector pME6032, yielding pME-phzI. Through electroporation, plasmid pME-phzI was separately transformed into wild-type HT66 and ∆phzI for over-expression and trans-complementation. In a similar way, pME-csaI and pME-hdtS were also constructed.
Extraction of AHLs from culture supernatants
Strain HT66 was grown in KB medium (enrichment media for Pseudomonas) at 28 °C for 24 h with shaking at 180 rpm. For the expression of PhzI, CsaI and HdtS in E. coli, strains were grown in LB medium to an OD600 of 0.1 and induced with Isopropyl β-d-1-Thiogalactopyranoside (IPTG) (final concentration, 1 mM) for 4–8 h. After growth, the bacterial cells were eliminated by centrifugation (8000×g for 10 min) and culture supernatants were extracted twice with equal volumes of ethyl acetate. The organic phase was pooled, dried over anhydrous magnesium sulfate. The ethyl acetate was removed by reduced pressure distillation at 30 °C and the residue contained the AHL extracts. The residue was stored at − 20 °C or dissolved with an appropriate volume of HPLC-grade acetonitrile for bioassay or HPLC–MS/MS analysis.
TLC separation and visualization of AHLs
The AHL plate assay using A. tumefaciens NTL4 was performed as reported earlier [30]. Briefly, AHL extracts (1–5 μl), were applied to C18 reversed-phase (RP)-TLC plates (Merck, Germany) and the chromatograms were developed with methanol/water (60:40, v/v). The plates were removed from the chromatography tank when the solvent front reaches the top of the TLC plate. The air-dried plates were overlaid with ABM medium soft gel (0.8% agar), followed by the addition of X-Gal (5-Bromo-4-chloro-3-indolyl-β-d-galactopyranoside; 60 μg/ml) and overnight culture of A. tumefaciens NTL4. After the agar solidification, the chromatography plates were incubated at 28 °C for 12–18 h and observed the blue spots indicating the location of AHLs. TLC analysis was repeated at least three times.
Analysis of AHLs by HPLC–MS/MS
AHL extracts from 100 ml cultures were partially purified by preparative TLC. For this, preparative TLC plate was cut into small strips and the compounds were carefully removed by scraping off the silica gel at the appropriate Rf and extracted three times with ethyl acetate. The dissolved compounds were centrifuged (at 12,000g for 10 min), and clear supernatant thus obtained was filtrated by syringe filters (0.22 µm). The supernatant was concentrated and the residue was dried and re-dissolved in acetonitrile for HPLC–MS/MS analysis at Instrumental Analysis Center of Shanghai Jiao Tong University, Shanghai China. A reverse phase column (5 μm; 4.6 × 250 mm, Shimadzu, Japan) with a detection wavelength of 254 nm was used. The AHL separation was performed with water containing 0.1% formic acid (analytical grade) and acetonitrile (AcN) (HPLC-grade). A flow rate of 0.5 mL/min was used, with increasing concentrations of 5 to 95% AcN in 15 min. Then, the flow was held for 3 min, and equilibration was performed for 7 min. All the mass spectra were recorded in the positive-ion mode. MS parameters were a spray voltage of 5 kV, a capillary temperature of 230 °C, and a sheath gas rate of 12 units N2.
Determination of the PCN production
HT66 and its mutants were grown overnight and inoculated into the fresh KB medium (1% inoculum ratio) followed by incubation for 24 h at an agitation speed of 180 rpm. Fermentation broth (300 μl) was acidified to pH 2.0 with 6 M HCl, and then 2.7 ml of ethyl acetate was added. The sample was vigorously agitated and centrifuged at 13,000g for 5 min. A 300-μl portion of the upper layer was collected and evaporated to complete dryness in a rotary evaporator. The resulting residue was dissolved in 1 ml acetonitrile and PCN concentration was determined by HPLC (Agilent Technologies 1200 series, Santa Clara, USA) with mobile phase acetonitrile (component A) and 5 mM ammonium acetate solution (component B), starting with 8% A for 2 min, then changing A from 8 to 90% in 18 min, at last returning to 8% A in 1 min. The column temperature was maintained 30 °C, the flow rate was 1.0 ml/min and the detection wavelength was 254 nm.
Biofilm assay
The ability to form biofilms was analyzed by a highly reproducible 96-well plate assay as described earlier [32]. Overnight cultures of P. chlororaphis HT66 and its mutants were diluted with 0.01 M phosphate-buffered saline (PBS) to 106 CFU/ml, and 100 μl diluted cultures were seeded into a 24-well plate followed by incubation at 28 °C for 24 h under static conditions. After the designated time, the cultures were gently removed by pipetting and each well was washed twice with 150 μl sterile PBS. Afterwards, 200 μl of 1% (w/v) crystal violet (CV) was added to each well to stain bacterial biofilm, and the plate was incubated at 28 °C for 20 min. The CV was then rinsed with 500 μl 95% ethyl alcohol and the amount of biofilm was quantified by measuring the optical density at 595 nm using Microtitre plate reader (Victor™ X series multi-label plate reader).
In-vitro antifungal activity
Pythium ultimum [33] was used to detect the antifungal activity of HT66 and its mutant derivatives. A mycelium disc of fungus was scrapped from the colonies grown on an agar plate, and spotted at the left of the agar plate, whereas the HT66 and its mutants were inoculated at the right of the agar plate. Fungal growth was monitored following incubation in darkness for 2 weeks at 28 °C. Three replicate plates were tested for each treatment.
Results
Detection and identification of AHLs produced by HT66
Chromobacterium violaceum CV026 and A. tumefaciens NTL4 plate assay were used to analyze the AHL-containing extracts of strain HT66. The CV026 indicator strain responds only to medium-chain alkanoyl-homoserine lactones (alkanoyl-HSLs) by producing purple spots, while NTL4 (pZLR4) responds strongly to 3-OH and 3-oxo-substituted HSLs, which react with X-gal resulting in blue pigment production. After the addition of AHL extracts to CV026 and NTL4 plates, the NTL4 assay showed blue pigment, while CV026 did not produce purple pigment, suggesting that the AHLs produced by HT66 might be 3-OH and 3-oxo-acyl-HSLs.
Further, ethyl acetate extracts from HT66 culture supernatants were separated by C18-reverse TLC and then overlaid with the biosensor NTL4 strain. Notably, three blue spots were detected from the extracts of HT66 (Fig. 1). The detected compounds were compared with reported known Rf values [34] and speculated that AHLs might be 3-oxo or 3-OH-acyl-HSLs with side chains ranging from 6 to 10 carbons. To identify the signals more precisely, extracts were separated by preparative TLC, and then the corresponding parts were scrapped and dissolved in the sample for HPLC–MS/MS analysis. It is known that every member of the AHL family gives a particular lactone ring at a nominal mass of m/z 102.05 [35], which results from the cleavage of homoserine lactone ring at the N-acyclic side chain. This characteristic ion was the key to analyze AHLs by HPLC–MS/MS. After MS/MS analysis, we found that three ions generated a peak for the characteristic ion at m/z 102.05 and some other peaks of fragmentations (Fig. 2a–c). For example, the 3-OH-C8-HSL has a molecular weight of 243, and the detected mass fragmentation [M + H]+ was 244.1546, almost same as the calculated mass 244.1543. Moreover, the ion [M + H–H2O]+ (226.1456) and [M + Na]+ (266.1363) were also observed and confirmed that the AHL was 3-OH-C8-HSL. Combined with the results of TLC and bio-reporter experiments, we concluded that the AHLs produced by HT66 were 3-OH-C6-HSL, 3-OH-C8-HSL, and 3-OH-C10-HSL.
Fig. 1.
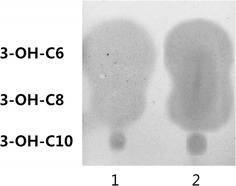
Detection of AHLs produced by Pseudomonas chlororaphis HT66 and E. coli with the plasmid pME-phzI using Agrobacterium tumefaciens NTL4. Lane 1, AHLs extract from HT66 and lane 2, AHLs from E. coli (pME-phzI)
Fig. 2.
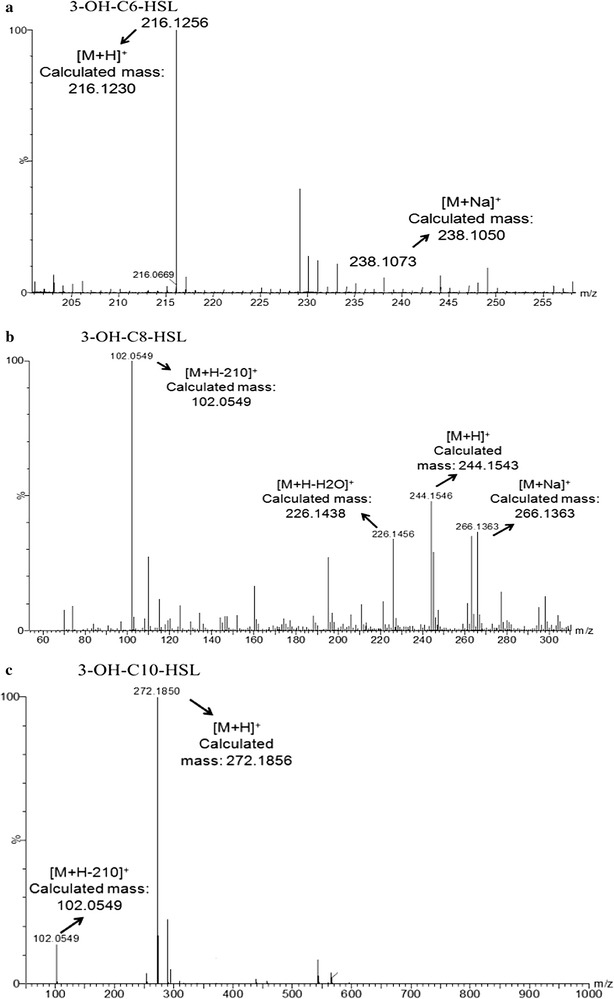
Identification of AHLs produced by Pseudomonas chlororaphis HT66 using LC–MS/MS analysis, a 3-OH-C6-HSL b 3-OH-C8-HSL and c 3-OH-C10-HSL
PhzI of strain HT66 directs the synthesis of AHLs
The whole-genome shotgun sequence of the HT66 has been deposited in National Center for Biotechnology Information (NCBI) databases under accession number ATBG00000000. In order to study which gene participated in quorum sensing and phenazine biosynthesis, luxI and luxR homologous genes were searched in the genome of HT66. According to the comparison results, one gene displayed extremely high identity (99%) to the AHL synthesis gene of PCL1391. This gene encodes 196 amino acids, of which only one amino acid is different from phzI of PCL1391. At the same time, an HdtS homolog was discovered, which has a similarity of 83% with the HdtS in P. fluorescens F113. The hdtS encodes 257 amino acids that direct the synthesis of C6-HSL, C10-HSL, and 3OH-C14:1-HSL in F113. Additionally, we also found another possible AHL synthesis gene in HT66 genome, CsaI, which had been reported to produce C4-, C5- and C6-HSL in P. chlororaphis 30-84 [36]. Although CsaI gene only contains 204 bp but shows high identity (over 93%) to the csaI (1416 bp) in strain 30-84. The CsaI–CsaR is a second quorum sensing system in 30-84, which is supposed to regulate the cell surface properties in response to the signals produced by CsaI.
To determine which genes produce the AHLs in HT66, the genes phzI, casI and hdtS were cloned and heterologous expressed in E. coli DH5α. A broad-host-range expression vector pME 6032 was used and the target gene was inserted between the EcoRI and XhoI sites, which was located in the downstream of the Ptac promoter. After induction with IPTG, the DH5α with plasmid pME-phzI, pME-csaI or pME-hdtS were cultivated overnight; the culture broths were then extracted with ethyl acetate and detected by A. tumefaciens NTL4 plate assay. Only the pME-phzI culture sample contained a detectable amount of AHLs (Fig. 3) and the AHLs produced by DH5α/pME-phzI was same as wild-type (Fig. 1). Whereas, the expression of csaI or hdtS in E. coli did not produce any signals judged by NTL4, which was totally different from the AHL synthase in strain 30-84 and F113 as discussed above.
Fig. 3.
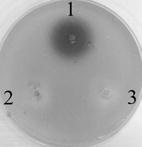
Detection of AHL extracts using Agrobacterium tumefaciens NTL4 plate bioassay. Samples contain extracts from cultures as follows (1) DH5α (pME-phzI) (2) DH5α (pME-csaI) and (3) DH5α (pME-hdtS)
To confirm our conclusion, we also constructed ∆phzI, ∆csaI and ∆hdtS mutants of HT66 through a non-scar gene knock-out method. Results showed that the phzI-deleted mutant completely loses the production of AHLs. On the other hand, as we assumed, the ∆csaI and ∆hdtS mutants exhibited no obvious difference with the wild-type and NTL4 assay showed blue pigment. Our results confirmed that PhzI was responsible for the synthesis of three AHLs (3-OH-C6, C8, C10-HSL) in HT66, which are irrelevant with CsaI or HdtS.
PhzI and PhzR are essential for phenazine biosynthesis and over-expression of phzI improves the PCN production
These two genes, phzI, and phzR are located just upstream of the phenazine biosynthesis gene cluster. In this work, we knocked out phzI from the HT66 genome to construct HT66∆phzI mutant through a non-scar deletion method. The results showed that deletion of phzI caused the complete loss of PCN biosynthesis as well as the AHLs synthesis (Fig. 4); whereas the constitutive expression of phzI restored the production of PCN and AHLs in ∆phzI mutant by transforming into the pME6032-phzI plasmid, which could produce up to 83% of the PCN concentration in wild-type. These facts suggested that PCN production of HT66 was regulated by phzI and is also responsible for the synthesis of AHLs. Similar to HT66∆phzI strain, the HT66∆phzR derivative was also unable to produce PCN, demonstrating that the phenazine biosynthetic cluster was regulated by PhzR receptor protein.
Fig. 4.
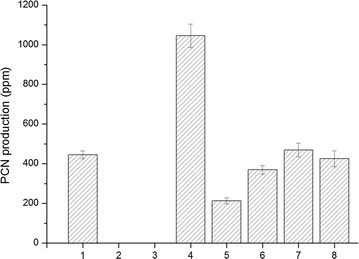
PCN production of HT66 and its mutants; (1) wild-type HT66, (2) HT66 ∆phzI, (3) HT66 ∆phzR, (4) HT66 (pME-phzI), (5) HT66 (pME6032), (6) HT66∆phzI (pME-phzI), (7) HT66 ∆hdtS and (8) HT66 ∆csaI
To investigate the effects of exogenous AHLs on PCN production in HT66, we transformed the phzI over-expressed plasmid into the wild-type strain, and the empty plasmid was also transformed as a control. Results evidenced that the HT66/pME-phzI had a higher level of PCN up to 1045 mg/l, which was almost 2.3- and 4.9-folds higher than wild-type and control strain (HT66 carrying the empty pME6032 plasmid). Thus, the results implied that exogenous AHLs production had improved the PCN production.
The absence of PCN production in ∆phzI or ∆phzR mutants indicated that a functional expressed quorum-sensing system was essential for the activation of the phz operon. The continuous over-expression of phzI resulted in an elevated level of PCN in wild-type HT66 strain. In addition, the PCN titers of ∆csaI and ∆hdtS mutants were detected to be 425 and 469 mg/l, respectively, and had statistically no difference in contrast with the wild-type HT66 strain.
Effect of phzI and phzR on growth and biofilm formation
A comparative growth profile results portrayed that the HT66 strain reached stationary phase at almost 24 h, and started to decline after 48 h (Fig. 5a). No noticeable difference was observed between the growth of wild-type strain and its ∆phzR mutant. While ∆phzI mutant had a lower cell density in stationary phase compared to wild-type, which might be correlated with the absence of signals and negative impact on communication between microorganisms.
Fig. 5.
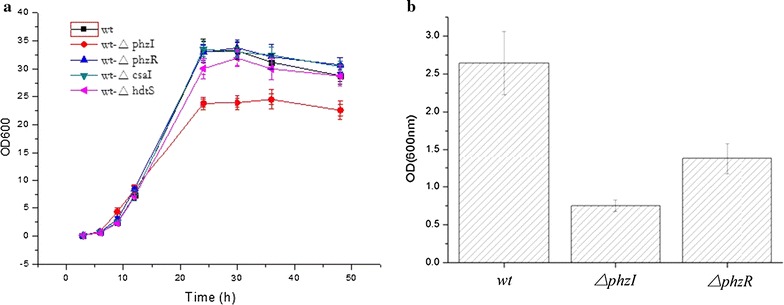
a The growth curves of HT66 and its derivatives and b biofilm formation in HT66 and its derivatives
Biofilm structures, which protect bacteria from various physical and chemical stresses, are the major reason for bacterial persistence during chronic infections [37]. Quorum sensing regulation of swarming and DNA release has been reported, which play important roles in P. aeruginosa biofilm development. On the other hand, QS signaling and transcription of genes are connected with the biofilm matrix biosynthesis [38]. Reports have shown that in some bacteria phenazines were not limited to secondary metabolites but also could function as cell signals [39]. We used 24-well plate assay to assess the biofilm formation ability of HT66 strains and its mutant derivatives. Compared with the wild-type, a significant decrease in biofilm production was observed in ∆phzI and ∆phzR mutants (Fig. 5b). The biofilm formation of ∆phzI was markedly damaged and recorded only 34% amount of the wild-type. It also appeared delicate and loose in a structure following stained with crystal violet. The findings indicated that PhzI–PhzR system plays a significant role in biofilm formation in strain HT66.
Effect on colonial morphology and antifungal activity
On specialized colonial morphology medium (KB medium), the mutants had almost the identical growth rate as the wild-type to form colonies (Additional file 1: Figure S1). The colonies of HT66 were observed to be protruded, neat edge, and round with a semi-diameter of about 0.6 cm after 36 h growth. The surface was smooth with little viscosity. After inoculated on a plate for 48 h or a longer period, the wild-type colonies turned yellow and produced green pigment on the surface. The ∆phzI and ∆phzR mutants had no discernible changes in the morphology of colonies; however, their colonies appeared milky white and had no green pigment due to the lack of PCN production.
Pythium ultimum was chosen for antifungal activity experiment. HT66 and its mutants were inoculated on PDA agar plate, simultaneously with the pathogenic fungi on the other side. After cultivation together for 2 weeks, the development of pathogenic fungi was considerably inhibited by the wild-type presumably due to PCN production (Additional file 1: Figure S1). In contrary, the PCN-deficient mutant’s ∆phzI and ∆phzR could not inhibit the growth of mycelia. It is illustrated that the absence of phzI–phzR system reduced the antifungal activity of strain HT66 indicating the significance of PCN production in the biological control of soil-borne crop diseases.
Discussion
In this study, thin-layer chromatography and liquid chromatography-mass/mass spectrometry techniques were applied to identify three AHLs (3-OH-C6, C8, C10-HSL) in P. chlororaphis HT66. Strain HT66 was a newly discovered phenazine-producing P. chlororaphis from rice rhizosphere, and the characteristic AHLs synthesis potential renders it distinguishing from other Pseudomonads. At first, we found three possible genes from the genome sequence that might produce AHLs. By heterologous expression in E. coli and non-scar deletion, phzI was evidenced to be the only responsible gene for the synthesis of these signals in HT66, while negating the contribution of csaI or hdtS. The knocking out of csaI or hdtS in HT66 has no obvious effect on PCN production and growth profile of HT66. In P. aureofaciens 30-84, CsaI–CsaR was not required for the expression of the gene involved in phenazine biosynthesis [36]. In P. fluorescens F113, hdtS was able to direct the synthesis of AHLs in strain F113. Nevertheless, the protein HdtS not belongs to either of the known AHL synthase families (LuxI or LuxM) and has a relationship with the lysophosphatidic acid acyltransferase family [40].
In Table 2, phzI in several Pseudomonas were compared with that in HT66, including their signals products. PhzI proteins of HT66 and PCL1391 were found to be similar as high as 99% (Fig. 6). It was interesting to note that C4 and C6-HSL were the products of the phzI gene in PCL1391, while HT66 produces 3-OH-C6-HSL, 3-OH-C8-HSL, and 3-OH-C10-HSL. The AHLs discrepancy was due to a different amino acid in PhzI proteins of HT66 and PCL1391 and noted to be threonine and alanine respectively, at position 125. Additionally, PhzI of strain 30-84 was also compared with that of HT66. The results show they shared almost 94% identical sequences (Fig. 6). Among the 197 amino acids sequence, there are five differences between PhzI HT66 and PhzI 30-84, and these five changes were: Glycine to Glutamine at position 10, Proline to Alanine at position 42, Threonine to Alanine at position 125, Glycine to Serine at position 179 and Alanine to Valine at position 181. As a consequence, AHLs produced by PhzI 30-84 were not only 3-OH-C6, C7, C8, C10-HSL but also C6-HSL and small amounts of C5- and C8-HSL. Regarding strain StFRB 508, there were 7 amino acids changes between PhzI 508 and PhzI HT66. PhzI 508 catalyzes the biosynthesis of C6-HSL as well as 3-OH-C6-HSL; whereas the second AHL synthase, AurI produces C4 and C6-HSL. In addition, we compared the phzI similarity of several phenazine-producing strains with HT66. The results revealed that these phzI genes were functionally same and were strongly conserved with the high sequence identity (Table 3).
Table 2.
Primers used in this study
| Primer name | Primer sequence (5′–3′) |
|---|---|
| phzI-F1 | TCGGAATTCATGCACATGGAAGAGCACA |
| phzI-R1 | TCGCTCGAGTCAAGCTATCTCTTTCAATAATGT |
| csaI-F1 | TCGGAATTCATGGCGCGGAGCCGGCT |
| csaI-R1 | TCGCTCGAGCTACTCCCTGAGCGCCTGA |
| hdtS-F1 | TCGGAATTCATGTCGATCCTGCAGGCAATCAGAA |
| hdtS-R1 | TCGCTCGAGTCAGATGGCCATTTTGTCCG |
| ΔphzI-F1 | CCGGAATTCCGGACTGAAGGTTGCTGAGAG |
| ΔphzI-R1 | TTACTATCTCCGAGTCGACCATCGAAGGCGACAGTTT |
| ΔphzI-F2 | GGTCGACTCGGAGATAGTAAATGCCCCTC |
| ΔphzI-R2 | CCCAAGCTTCGGTTTGATTTCTTTGCCTACGG |
| ΔphzR-F1 | CCGGAATTCATGGAAGAGCACACACTGAG |
| ΔphzR-R1 | TGTCACATTGAGGGTCTTGCATTTACTATCTCCGAGT |
| ΔphzR-F2 | GCAAGACCCTCAATGTGACAGCCGTAAA |
| ΔphzR-R2 | CCCAAGCTTTTGGCGAAGTTCAAGATGATCATT |
| ΔcsaI-F1 | CCGGAATTCCAGTTGACCGAGGAAGGC |
| ΔcsaI-R1 | TCTACTCCCTGAGCGCCTGAGTAGGTAAAGACACTTG |
| ΔcsaI-F2 | TCAGGCGCTCAGGGAGTAGACCAGCG |
| ΔcsaI-R2 | CCCAAGCTTCGATCCTGTCGTACCTGGC |
| ΔhdtS-F1 | CCGGAATTCACGACTCCGACGCTTACATC |
| ΔhdtS-R1 | CGGTACACAGGTTATCCACAAGGATGTCAGAAGAACT |
| ΔhdtS-F2 | TGTGGATAACCTGTGTACCGTTTTAGCGGAAATCGC |
| ΔhdtS-R2 | CCCAAGCTTCCTCGACGACGATGCC |
Fig. 6.

PhzI protein sequences of Pseudomonas chlororaphis HT66, PCL1391, P. chlororaphis 30-84 and P. chlororaphis subsp. aurantiaca StFRB508. The PhzI proteins were highly similar and closely related. Amino acids that are different in these four proteins are shown in black blocks
Table 3.
AHLs produced by different Pseudomonas strains
| Pseudomonas | Similarity of PhzI compared with HT66 (%) | AHLs synthesized by PhzI | References |
|---|---|---|---|
|
Pseudomonas chlororaphis
HT66 |
N/A | 3-OH-C6, C8, C10-HSL | This study |
|
Pseudomonas chlororaphis
PCL1391 |
99 | C6-HSL | Gigard et al. [49] |
|
Pseudomonas chlororaphis
30-84 |
95 | C6-HSL, 3-OH-C6, C7, C8, C10-HSL | Khan et al. [50] |
|
Pseudomonas chlororaphis subsp. aurantiaca StFRB508 |
94 | C6-HSL, 3-OH-C6-HSL | Morohoshi et al. [41] |
| Pseudomonas sp. G5(2008b) | 94 | C4, C6, C8-HSL | Li et al. [51] |
|
Pseudomonas chlororaphis
GP72 |
94 | C4, C6-HSL | Huang et al. [52] |
Apart from the identification and production of AHLs in HT66, this study found that the over-expression of phzI had improved the level of PCN by 2.3-folds compared with the wild-type strain. More importantly, PCN production was abolished following the inactivation of phzI or phzR genes in HT66 indicating their roles in regulating the expression of the phenazine biosynthesis gene cluster. Previously, Morohoshi et al. [41] reported that the mutation in phzI caused a considerable reduction in phenazine biosynthesis by P. chlororaphis subsp. aurantiaca. On the other hand, no phenazine production was recorded in the triple mutant of phzI, aurI, and csaI (508ΔPACI). Noticeably, phenazine production was supposed to be strongly stimulated by PhzI-mediated AHLs than produced by AurI and CsaI that only induced a slight stimulation. Though, Pseudomonas sp. CMR12a and P. chlororaphis subsp. aureofaciens 30-84 display a second QS system apart from phzI/phzR system, phenazine biosynthesis was merely regulated by the AHLs produced by PhzI [36, 42, 43].
In order to appraise the involvement of AHL production to the biocontrol capacity of StFRB508, P. ultimum was used in this study. After cultivation together for 2 weeks, the development of pathogenic fungi was considerably inhibited by the wild-type presumably due to PCN production confirming the strain as an effective biocontrol agent. In P. chlororaphis PA23, it was examined that the biofilm development was reduced by approximately fivefold in the QS-deficient mutants and the motility was also altered [44]. Similarly, the biofilm formation in phzI and phzR deletion strains of HT66 had a decrease in different extents. With the absence of PCN, the antimicrobial ability was influenced when grew against the P. ultimum in vitro.
Conclusions
In conclusion, we identified three kinds of AHLs (3-OH-C6-HSL, 3-OH-C8-HSL, 3-OH-C10-HSL) in P. chlororaphis HT66 by TLC and HPLC–MS/MS analyses. The production of AHLs exhibiting the activating role in phenazine biosynthesis gene cluster was only catalyzed by PhzI. In comparison to wild-type HT66, PCN production was 2.3-folds improved by over-expression of phzI, however, phzI or phzR mutant did not produce PCN. In addition, the growth, morphology and biofilm formation were all under the control of the PhzI–PhzR regulatory system. These results revealed that PhzI–PhzR system plays a pivotal role in PCN production and the biocontrol activity of HT66.
Additional files
Additional file 1: Figure S1. a Colonial morphology changes in wild-type HT66, HT66∆phzI and HT66∆phzR strains during 7 days and b Influence of antifungal activity of HT66 and its mutants on the growth of Pythium ultimum. P. ultimum was spotted at the left of the PDA plate, whereas HT66 and its mutants were inoculated on the right side.
Authors’ contributions
PZ carried out the experimental-based work and drafted the manuscript. HBH, WW, and XHZ analyzed and interpreted the data. MB analyzed the data and assisted in drafting the manuscript. All the research work was carried out under the dynamic guidance and supervision of PZ who designed, conceived and coordinated the experiments. All authors read and approved the final manuscript.
Acknowledgements
We would like to thank “Instrumental Analysis Centre of SJTU for providing technical and experimental facilities.
Competing interests
The authors declare that they have no competing interests.
Availability of supporting data
The dataset(s) supporting the conclusions of this article is (are) included within the article (and its additional file(s)).
Consent for publication
All authors read and approved the final manuscript.
Ethics approval and consent to participate
We state this is not applicable. The manuscript does not report on or involve the use of any animal or human data or tissue.
Funding
This work was supported by the National Key Basic Research Program of China (No. 2012CB721005), the National Natural Science Foundation of China (No. 31270084), and the National High Technology Research and Development Program of China (No. 2012AA022107).
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Footnotes
Electronic supplementary material
The online version of this article (10.1186/s12934-017-0854-y) contains supplementary material, which is available to authorized users.
Contributor Information
Huasong Peng, Phone: +86 021 3420 7047, Email: hspeng@sjtu.edu.cn.
Yi Ouyang, Email: Ouyang132@sjtu.edu.cn.
Muhammad Bilal, Email: bilaluaf@sjtu.edu.cn.
Wei Wang, Email: weiwang100@sjtu.edu.cn.
Hongbo Hu, Email: hbhu@sjtu.edu.cn.
Xuehong Zhang, Email: xuehzhang@sjtu.edu.cn.
References
- 1.Ng WL, Bassler BL. Bacterial quorum-sensing network architectures. Annu Rev Genet. 2009;43:197–222. doi: 10.1146/annurev-genet-102108-134304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Li Z, Nair SK. Quorum sensing: how bacteria can coordinate activity and synchronize their response to external signals? Protein Sci. 2012;21:1403–1417. doi: 10.1002/pro.2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fuqua C, Greenberg EP. Listening in on bacteria: acyl-homoserine lactone signalling. Nat Rev Mol Cell Biol. 2002;3:685–695. doi: 10.1038/nrm907. [DOI] [PubMed] [Google Scholar]
- 4.Waiters M, Sircili MP, Sperandio V. AI-3 syntheses is not dependent on luxS in Escherichia coli. J Bacteriol. 2006;188:5668–5681. doi: 10.1128/JB.00648-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jones S, Yu B, Bainton NJ, Birdsall M, Bycroft BW, Chhabra SR, Cox AJ, Golby P, Reeves PJ, Stephens S. The lux autoinducer regulates the production of exoenzyme virulence determinants in Erwinia carotovora and Pseudomonas aeruginosa. EMBO J. 1993;12:2477–2482. doi: 10.1002/j.1460-2075.1993.tb05902.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Houdt R, Aertsen A, Moons P, Vanoirbeek K, Michiels CW. N-Acyl-l-homoserine lactone signal interception by Escherichia coli. FEMS Microbiol Lett. 2006;256:83–89. doi: 10.1111/j.1574-6968.2006.00103.x. [DOI] [PubMed] [Google Scholar]
- 7.Riedel K, Hentzer M, Geisenberger O, Huber B, Steidle A, Wu H, Hoiby N, Givskov M, Molin S, Eberl LN. Acylhomoserine-lactone-mediated communication between Pseudomonas aeruginosa and Burkholderia cepacia in mixed biofilms. Microbiology. 2001;147:3249–3262. doi: 10.1099/00221287-147-12-3249. [DOI] [PubMed] [Google Scholar]
- 8.Turner JM, Messenger AJ. Occurrence, biochemistry and physiology of phenazine pigment production. Adv Microb Physiol. 1986;27:211–275. doi: 10.1016/S0065-2911(08)60306-9. [DOI] [PubMed] [Google Scholar]
- 9.Mazzola M, Cook RJ, Thomashow LS, Weller DM, Pierson LS. Contribution of phenazine antibiotic biosynthesis to the ecological competence of fluorescent pseudomonads in soil habitats. Appl Environ Microbiol. 1992;58:2616–2624. doi: 10.1128/aem.58.8.2616-2624.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thomashow LS, Weller DM. Role of a phenazine antibiotic from Pseudomonas fluorescens in biological control of Gaeumannomyces graminis var. tritici. J Bacteriol. 1988;170:3499–3508. doi: 10.1128/jb.170.8.3499-3508.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Daval S, Lebreton L, Gazengel K, Boutin M, Guillerm-Erckelboudt AY, Sarniguet A. The biocontrol bacterium Pseudomonas fluorescens Pf29Arp strain affects the pathogenesis-related gene expression of the take-all fungus Gaeumannomyces graminis var. tritici on wheat roots. Mol Plant Pathol. 2011;12:839–854. doi: 10.1111/j.1364-3703.2011.00715.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.ChinAWoeng TFC, Bloemberg GV, van der Bij AJ, van der Drift K, Schripsema J, Kroon B, Scheffer RJ, Keel C, Bakker P, Tichy HV, de Bruijn FJ, Thomas-Oates J, Lugtenberg B. Biocontrol by phenazine-1-carboxamide-producing Pseudomonas chlororaphis PCL1391 of tomato root rot caused by Fusarium oxysporum f. sp. radicis-lycopersici. Mol Plant-Microbe Interact. 1998;11:1069–1077. doi: 10.1094/MPMI.1998.11.11.1069. [DOI] [Google Scholar]
- 13.Shanmugaiah V, Mathivanan N, Varghese B. Purification, crystal structure and antimicrobial activity of phenazine-1-carboxamide produced by a growth-promoting biocontrol bacterium, Pseudomonas aeruginosa MML2212. J Appl Microbiol. 2010;108:703–711. doi: 10.1111/j.1365-2672.2009.04466.x. [DOI] [PubMed] [Google Scholar]
- 14.Laursen JB, Nielsen J. Phenazine natural products: biosynthesis, synthetic analogues, and biological activity. Chem Rev. 2004;104:1663–1686. doi: 10.1021/cr020473j. [DOI] [PubMed] [Google Scholar]
- 15.Liu H, He Y, Jiang H, Peng H, Huang X, Zhang X, Thomashow LS, Xu Y. Characterization of a phenazine-producing strain Pseudomonas chlororaphis GP72 with broad-spectrum antifungal activity from green pepper rhizosphere. Curr Microbiol. 2007;54:302–306. doi: 10.1007/s00284-006-0444-4. [DOI] [PubMed] [Google Scholar]
- 16.Huang X, Zhang X, Xu Y. PltR expression modulated by the global regulators GacA, RsmA, LasI and RhlI in Pseudomonas sp. M18. Res Microbiol. 2008;159:128–136. doi: 10.1016/j.resmic.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 17.Dasgupta D, Kumar A, Mukhopadhyay B, Sengupta TK. Isolation of phenazine 1,6-di-carboxylic acid from Pseudomonas aeruginosa strain HRW.1-S3 and its role in biofilm-mediated crude oil degradation and cytotoxicity against bacterial and cancer cells. Appl Microbiol Biotechnol. 2015;99:8653–8665. doi: 10.1007/s00253-015-6707-x. [DOI] [PubMed] [Google Scholar]
- 18.Hu H, Li Y, Liu K, Zhao J, Wang W, Zhang X. Production of trans-2,3-dihydro-3-hydroxyanthranilic acid by engineered Pseudomonas chlororaphis GP72. Appl Microbiol Biotechnol. 2017;17:6607–6613. doi: 10.1007/s00253-017-8408-0. [DOI] [PubMed] [Google Scholar]
- 19.Jin X, Peng H, Hu H, Huang X, Wang W, Zhang X. iTRAQ-based quantitative proteomic analysis reveals potential factors associated with the enhancement of phenazine-1-carboxamide production in Pseudomonas chlororaphis P3. Sci. Rep. 2016;6:27393. doi: 10.1038/srep27393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeykumari DRS, Narayanan SS. Covalent modification of multiwalled carbon nanotubes with neutral red for the fabrication of an amperometric hydrogen peroxide sensor. Nanotechnology. 2007;18:125501–125510. doi: 10.1088/0957-4484/18/12/125501. [DOI] [Google Scholar]
- 21.Pierson LS, Pierson EA. Metabolism and function of phenazines in bacteria: impacts on the behavior of bacteria in the environment and biotechnological processes. Appl Microbiol Biotechnol. 2010;86:1659–1670. doi: 10.1007/s00253-010-2509-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Torres CI, Marcus AK, Lee HS, Parameswaran P, Krajmalnik-Brown R, Rittmann BE. A kinetic perspective on extracellular electron transfer by anode-respiring bacteria. FEMS Microbiol Rev. 2010;34:3–17. doi: 10.1111/j.1574-6976.2009.00191.x. [DOI] [PubMed] [Google Scholar]
- 23.Rabaey K, Boon N, Hofte M, Verstraete W. Microbial phenazine production enhances electron transfer in biofuel cells. Environ Sci Technol. 2005;39:3401–3408. doi: 10.1021/es048563o. [DOI] [PubMed] [Google Scholar]
- 24.Shen X, Chen M, Hu H, Wang W, Peng H, Xu P, Zhang X. Genome sequence of Pseudomonas chlororaphis GP72, a root-colonizing biocontrol strain. J Bacteriol. 2012;194:1269–1270. doi: 10.1128/JB.06713-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang X, Wang Z, Liu Y, Zhang X. Complete genome sequence of Pseudomonas protegens H78, a plant growth-promoting rhizobacterium. Genome Announc. 2017;5:e00233–e00317. doi: 10.1128/genomeA.00233-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mavrodi DV, Blankenfeldt W, Thomashow LS. Phenazine compounds in fluorescent Pseudomonas spp. biosynthesis and regulation. Annu Rev Phytopathol. 2006;44:417–445. doi: 10.1146/annurev.phyto.44.013106.145710. [DOI] [PubMed] [Google Scholar]
- 27.Steindler L, Venturi V. Detection of quorum-sensing N-acyl homoserine lactone signal molecules by bacterial biosensors. FEMS Microbiol Lett. 2007;266:1–9. doi: 10.1111/j.1574-6968.2006.00501.x. [DOI] [PubMed] [Google Scholar]
- 28.Cha C, Gao P, Chen YC, Shaw PD, Farrand SK. Production of acyl-homoserine lactone quorum-sensing signals by Gram-negative plant-associated bacteria. Mol Plant-Microbe Interact. 1998;11:1119–1129. doi: 10.1094/MPMI.1998.11.11.1119. [DOI] [PubMed] [Google Scholar]
- 29.McClean KH, Winson MK, Fish L, Taylor A, Chhabra SR, Camara M, Daykin M, Lamb JH, Swift S, Bycroft BW, Stewart GS, Williams P. Quorum sensing and Chromobacterium violaceum: exploitation of violacein production and inhibition for the detection of N-acylhomoserine lactones. Microbiology. 1997;143:3703–3711. doi: 10.1099/00221287-143-12-3703. [DOI] [PubMed] [Google Scholar]
- 30.Chilton MD, Currier TC, Farrand SK, Bendich AJ, Gordon MP, Nester EW. Agrobacterium tumefaciens DNA and PS8 bacteriophage DNA not detected in crown gall tumors. PNAS. 1974;71:3672–3676. doi: 10.1073/pnas.71.9.3672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Du X, Li Y, Zhou W. Phenazine-1-carboxylic acid production in a chromosomally non-scar triple-deleted mutant Pseudomonas aeruginosa using statistical experimental designs to optimize yield. Appl Microbiol Biotechnol. 2013;97:7767–7778. doi: 10.1007/s00253-013-4921-y. [DOI] [PubMed] [Google Scholar]
- 32.O’Toole GA, Kolter R. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signaling pathways: a genetic analysis. Mol Microbiol. 1998;28:449–461. doi: 10.1046/j.1365-2958.1998.00797.x. [DOI] [PubMed] [Google Scholar]
- 33.Pearson RC, Hall DH. Ripe fruit rot of tomato caused by Pythium ultimum and Pythium aphanidermatum. Plant Dis. 1973;57:1066–1069. [Google Scholar]
- 34.Shaw PD, Ping G, Daly SL, Cha C, Cronan JE, Rinehart KL, Farrand SK. Detecting and characterizing N-acyl-homoserine lactone signal molecules by thin-layer chromatography. PNAS. 1997;94:6036–6041. doi: 10.1073/pnas.94.12.6036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cataldi TRI, Bianco G, Fonseca J, Schmitt-Kopplin P. Perceiving the chemical language of Gram-negative bacteria: listening by high-resolution mass spectrometry. Anal Bioanal Chem. 2013;405:493–507. doi: 10.1007/s00216-012-6371-2. [DOI] [PubMed] [Google Scholar]
- 36.Zhang Z, Pierson LS., III A second quorum-sensing system regulates cell surface properties but not phenazine antibiotic production in Pseudomonas aureofaciens. Appl Environ Microbiol. 2001;67:4305–4315. doi: 10.1128/AEM.67.9.4305-4315.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maddula VSRK, Zhang Z, Pierson EA. Quorum sensing and phenazines are involved in biofilm formation by Pseudomonas chlororaphis (aureofaciens) strain 30-84. Microb Ecol. 2006;52:289–301. doi: 10.1007/s00248-006-9064-6. [DOI] [PubMed] [Google Scholar]
- 38.Sakuragi Y, Kolter R. Quorum-sensing regulation of the biofilm matrix genes (pel) of Pseudomonas aeruginosa. J Bacteriol. 2007;189:5383–5386. doi: 10.1128/JB.00137-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dietrich LEP, Teal TK, Price-Whelan A, Newman DK. Redox-active antibiotics control gene expression and community behavior in divergent bacteria. Science. 2008;321:1203–1206. doi: 10.1126/science.1160619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Laue BE, Jiang Y, Chhabra SR, Jacob S, Stewart GSAB, Hardman A, Downie JA, O’Gara F, Williams P. The biocontrol strain Pseudomonas fluorescens F113 produces the rhizobium small bacteriocin, N-(3-hydroxy-7-cis-tetradecenoyl) homoserine lactone, via HdtS, a putative novel N-acylhomoserine lactone synthase. Microbiology. 2000;146:2469–2480. doi: 10.1099/00221287-146-10-2469. [DOI] [PubMed] [Google Scholar]
- 41.Morohoshi T, Wang WZ, Suto T. Phenazine antibiotic production and antifungal activity are regulated by multiple quorum-sensing systems in Pseudomonas chlororaphis subsp. aurantiaca StFRB508. J Biosci Bioeng. 2013;116:580–584. doi: 10.1016/j.jbiosc.2013.04.022. [DOI] [PubMed] [Google Scholar]
- 42.Morohoshi T, Yamaguchi T, Xie X, Wang W, Takeuchi K, Someya N. Complete genome sequence of Pseudomonas chlororaphis subsp. Aurantiaca reveals a triplicate quorum-sensing mechanism for regulation of phenazine production. Microbes Environ. 2017;32:47–53. doi: 10.1264/jsme2.ME16162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Maeyer K, D’Aes J, Hua GK, Perneel M, Vanhaecke L, Noppe H, Hofte M. N-Acyl homoserine lactone quorum sensing signalling in antagonistic phenazine-producing Pseudomonas isolates from the red cocoyam rhizosphere. Microbiology. 2011;157:459–472. doi: 10.1099/mic.0.043125-0. [DOI] [PubMed] [Google Scholar]
- 44.Selin C, Fernando WGD, de Kievit T. The PhzI/PhzR quorum-sensing system is required for pyrrolnitrin and phenazine production, and exhibits cross-regulation with RpoS in Pseudomonas chlororaphis PA23. Microbiology. 2012;158:896–907. doi: 10.1099/mic.0.054254-0. [DOI] [PubMed] [Google Scholar]
- 45.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/S0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 46.Hoffmann A, Thimm T, Droge M, Moore ER, Munch JC, Tebbe CC. Intergeneric transfer of conjugative and mobilizable plasmids harbored by Escherichia coli in the gut of the soil microarthropod Folsomia candida (Collembola) Appl Environ Microbiol. 1998;64(7):2652–2659. doi: 10.1128/aem.64.7.2652-2659.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schafer A, Tauch A, Jager W, Kalinowski J, Thierbach G, Puhler A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 1994;145(1):69–73. doi: 10.1016/0378-1119(94)90324-7. [DOI] [PubMed] [Google Scholar]
- 48.Heeb S, Blumer C, Haas D. Regulatory RNA as mediator in GacA/RsmA-dependent global control of exoproduct formation in Pseudomonas fluorescens CHA0. J Bacteriol. 2002;184:1046–1056. doi: 10.1128/jb.184.4.1046-1056.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Girard G, Barends S, Rigali S, van Rij ET, Lugtenberg BJ, Bloemberg GV. Pip, a novel activator of phenazine biosynthesis in Pseudomonas chlororaphis PCL1391. J Bacteriol. 2006;188(23):8283–8293. doi: 10.1128/JB.00893-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Khan SR, Herman J, Krank J, Serkova NJ, Churchill MEA, Suga H, Farrand SK. N-(3-Hydroxyhexanoyl)-l-homoserine lactone is the biologically relevant quormone that regulates the phz operon of Pseudomonas chlororaphis strain 30-84. Appl Environ Microbiol. 2007;73:7443–7455. doi: 10.1128/AEM.01354-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H, Liu X, Gao K, Jia J. Cloning of phzIR from the endophytic Pseudomonas sp. G5 and its expression in Escherichia coli. Chin. J Biotech. 2009;6:832–839. [PubMed] [Google Scholar]
- 52.Huang L, Chen MM, Wang W, Hu HB, Peng HS, Xu YQ, Zhang XH. Enhanced production of 2-hydroxyphenazine in Pseudomonas chlororaphis GP72. Appl. Microbiol. Biotechnol. 2011;89:169–177. doi: 10.1007/s00253-010-2863-1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. a Colonial morphology changes in wild-type HT66, HT66∆phzI and HT66∆phzR strains during 7 days and b Influence of antifungal activity of HT66 and its mutants on the growth of Pythium ultimum. P. ultimum was spotted at the left of the PDA plate, whereas HT66 and its mutants were inoculated on the right side.


