Significance
Autoimmune disease pathogenesis is driven by inflammation, induced partly by IgG autoantibody-containing immune complexes binding to Fc gamma receptors (FcγRs). These receptors are valid therapeutic targets in the treatment of autoimmunity. FcγRIIIa is one of a family of highly homologous receptors for IgG antibodies; previous attempts at therapeutic blockade have resulted in off-target effects involving cells that express the almost identical protein FcγRIIIb. Here we report the identification of functionally specific protein-based inhibitors (Affimer proteins) of FcγRIIIa and the structural/functional basis of their selectivity. As molecular research tools FcγRIIIa-specific Affimer proteins provide the ability to block IgG interaction with a single receptor. Our findings suggest that highly selective protein-based blocking agents that may have therapeutic applications can be readily produced.
Keywords: Fc gamma receptor IIIa, specific inhibitor, Affimer, allosteric, competitive
Abstract
Protein–protein interactions are essential for the control of cellular functions and are critical for regulation of the immune system. One example is the binding of Fc regions of IgG to the Fc gamma receptors (FcγRs). High sequence identity (98%) between the genes encoding FcγRIIIa (expressed on macrophages and natural killer cells) and FcγRIIIb (expressed on neutrophils) has prevented the development of monospecific agents against these therapeutic targets. We now report the identification of FcγRIIIa-specific artificial binding proteins called “Affimer” that block IgG binding and abrogate FcγRIIIa-mediated downstream effector functions in macrophages, namely TNF release and phagocytosis. Cocrystal structures and molecular dynamics simulations have revealed the structural basis of this specificity for two Affimer proteins: One binds directly to the Fc binding site, whereas the other acts allosterically.
Improved understanding of genetic, genomic, and cellular processes underpinning human disease has led to the identification of a multitude of protein–protein interactions that represent potentially important therapeutic targets, frequently for multiple diseases. Drug discovery has traditionally focused on classical enzyme pockets, and chemical libraries are screened to identify inhibitors using biochemical and biophysical assays. Protein–protein interactions are notoriously difficult to target by this approach, since the interfaces frequently comprise large contact surfaces, which generally lack the deep pockets required for traditional medicinal chemistry approaches. In recent years, alternative strategies have emerged including fragment-based approaches to explore the chemical space or the use of peptide-based recognition molecules, such as hydrocarbon-stapled peptides, alpha mimetics, nonantibody protein scaffolds, and antibody-aided technologies (reviewed in ref. 1). Proteomimetic molecules have inherently greater potential to bind to critical interaction interfaces and sterically block protein–protein interactions. Traditional computational-based design tools have tended to focus on orthosteric inhibitors that directly target the interaction site, such as the receptor ligand-binding domain or active site of an enzyme. Approaches for therapeutic development include stabilization of protein complexes and identification of allosteric modulators that bind at sites distant to the interacting proteins (2, 3).
Currently, antibodies are the best-studied group of protein-based inhibitors with a wide range of therapeutic humanized monoclonal antibodies already in clinical use (4). However, antibodies are not always ideal as molecular tools due to their multiple domains and chains, poor stability, high production costs, and batch-to-batch variation, some of which may be due to glycosylation heterogeneity (5). Artificial binding reagents (protein, RNA, and DNA aptamers) are relatively small and make attractive alternatives to antibodies. We have recently established a scaffold consensus protein based on plant cystatins, called “Affimer,” also known as “Adhiron” (12 kDa), which provides a highly stable scaffold (melting temperature = 101 °C) for presenting one to three variable amino acid sequence regions for molecular recognition (6). These variable regions (VRs) form a binding interface analogous to that presented by the complementarity-determining regions of an antibody. Affimer proteins are selected from phage display libraries (>3 × 1010), allowing rapid identification of highly specific reagents that selectively bind to a target and often act as competitive or allosteric inhibitors (7–9). Non-antibody–binding proteins tend to recognize binding hot spots, which are small groups of amino acids on the target protein that contribute the majority of the interaction free energy (10). We propose that Affimer proteins can be used to study protein function and to disrupt protein–ligand interactions. This unbiased approach may also increase the potential for introducing selectivity where multiple receptors bind to a single ligand or, conversely, where multiple ligands bind to a single receptor. We have explored the potential utility of this approach using human Fc gamma receptors (FcγRs) as a model system.
Human FcγR–ligand interactions constitute a biological system in which multiple layers of complexity facilitate the fine-tuning of immune responses to infections. IgG is the major ligand and mediates both pro- and antiinflammatory effects following immune complex formation and engagement with different FcγRs. These activating and inhibitory receptors play a central role in the initiation and regulation of many immunological processes, including setting thresholds for B cell activation, recruitment of leukocytes, proinflammatory mediator release, phagocytosis, and antibody-dependent cellular cytotoxicity (ADCC) (11, 12). Our genetic studies have demonstrated a number of independent associations with genes in the FCGR locus in different autoimmune and inflammatory diseases (13–15). We have also described higher expression levels of FcγRIIIa on circulating CD14++ monocytes in rheumatoid arthritis patients compared with healthy controls; these higher levels correlated with increased TNF release on exposure to immune complexes and inferior treatment outcomes (16). Animal models also provide a strong rationale for targeting FcγRs in autoantibody-mediated inflammatory diseases, including autoantibody/immune complex-induced arthritis (17, 18).
There are six functional human FcγRs subdivided into three classes (FcγRI, FcγRIIa, FcγRIIb, FcγRIIc, FcγRIIIa, and FcγRIIIb). Multiple segmental duplications and deletions during hominid evolution have resulted in a family of highly homologous receptors with significant divergence of biological functions from those observed in rodents (19, 20). The level of homology has been a major obstacle for the development of FcγR-specific therapeutics.
A number of FcγR class-specific monoclonal antibodies have been tested in humans, predominantly to block ADCC in immune thrombocytopenia purpura (21). An antibody against FcγRIII (CD16-3G8) led to transient increases in platelet count, demonstrating early efficacy. However, in addition to immunogenicity, a number of infusion and atypical hypersensitivity reactions were observed in conjunction with neutrophil and monocyte cytopenias that led to early termination of this program. Although these were believed to be secondary to unwanted engagement of the therapeutic Fc region with FcγRs, these were not abrogated when a humanized anti-FcγR with an aglycosylated Fc was used, suggesting that alternative approaches may be required (reviewed in ref. 22). Blockade of the critical proximal signaling molecule spleen tyrosine kinase (SYK) downstream of several FcγRs initially showed promising efficacy in rheumatoid arthritis (23), chronic lymphocytic leukemia, and non-Hodgkin’s lymphoma (24), providing clinical support for therapeutic FcγR blockade in human disease. However, further development in rheumatoid arthritis has been suspended, principally due to adverse off-target events (25).
In this proof-of-principle study, we have screened artificial binding protein libraries against a recombinant, glycosylase-treated FcγRIIIa ectodomain and identified several FcγRIIIa-specific Affimer proteins. We present two Affimer proteins and their structures derived from X-ray crystallography of their complexes with FcγRIIIa, allowing structures to be solved at atomic resolution. Molecular dynamics (MD) simulations based on the X-ray crystallographic models supported the molecular basis of Affimer protein's mode of action and selectivity for FcγRIIIa. One Affimer protein [Protein Data Bank (PDB) ID code 5ML9] bound close to the Fc-binding domain, acting as a steric inhibitor, whereas the other (PDB ID code 5MN2) recognized an allosteric site and bound in the interdomain hinge region.
Our results demonstrate the feasibility of generating highly specific inhibitors of protein–ligand interactions that bind unexplored sites and illustrate the utility of Affimer proteins in the study of protein function at both a molecular and cellular level.
Results
Identification and Characterization of FcγRIIIa-Specific Affimer Proteins.
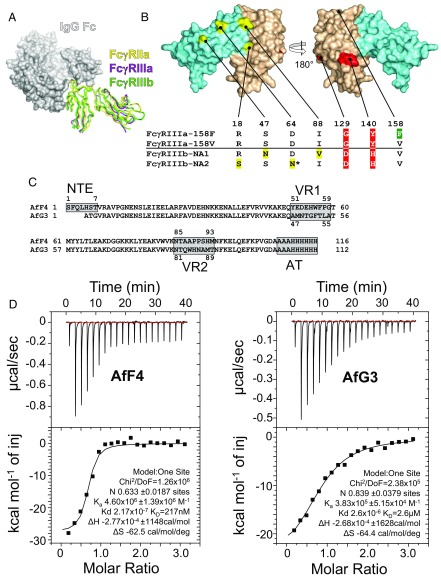
The extent of the challenge faced when developing specific agents against FcγRIIIa is illustrated by the structural alignment of FcγR crystal structures, demonstrating the high degree of target homology (Fig. 1A). This is particularly true for FcγRIIIa and FcγRIIIb; only two amino acids are consistently different between FcγRIIIa and both common human neutrophil alloantigen types of FcγRIIIb (NA1 and NA2, highlighted in red in Fig. 1B), but FcγRIIIb has four further polymorphic amino acids (highlighted in yellow in Fig. 1B): NA2 has one more site for N-linked glycosylation than FcγRIIIa (Asn64) and differs in having serine at residue 18, whereas FcγRIIIa and FcγRIIIb-NA1 have arginine at this locus.
Fig. 1.
The challenge of structural homology and the selection of specific protein-based inhibitors of FcγRIIIa. (A) Superimposed crystal structures of three FcγR ectodomains are shown as ribbon diagrams in complex with a space-filling model of the Fc domain of IgG1. FcγRIIa (PDB ID code 3RY6) is shown in yellow, FcγRIIIa (PDB ID code 3AY4) is shown in purple, and FcγRIIIb (PDB ID code 1T83) is shown in green. (B) Structural homology between FcγRIIIa and FcγRIIIb. The four amino acids in yellow differ in the FcγRIIIb NA1 and NA2 allotypes; the two amino acids in red discriminate FcγRIIIa from FcγRIIIb; and the location of the FcγRIIIa-158F/V allotype is green. The FcγRIIIb-NA2 allotype has an extra N-linked glycosylation site at Asn64. Extracellular domains 1 and 2 are depicted in aquamarine (D1 residues 1–89) and wheat (D2 residues 90–174), respectively. (C) The aligned amino acid sequences of AfF4 and AfG3 highlighting the positions of VR1, VR2, and the affinity tag (AT). Note that AfF4 has an additional NTE. Residue numbering within the VRs is indicated. (D) ITC of the FcγRIIIa–AfF4 and –AfG3 interactions with isotherms and data fits. FcγRIIIa was at 10 µM in the sample cell, and Affimer proteins were injected in stepwise additions of 2 μL to a final concentration of 100 µM.
For phage display we used FcγRIIIa ectodomain bait that had been produced in HEK293T cells in the presence of kifunensine and treated with endoglycosidase F1 to facilitate crystallization and to allow valid comparisons between structural and biophysical data. A total of 72 randomly chosen Affimer proteins were tested for binding to FcγRIIIa using phage ELISA after three rounds of selection. Of these, 52 gave positive results, and DNA sequencing revealed six unique clones. The most frequently recovered Affimer proteins were expressed as soluble proteins. AfG3 differed from AfF4 in terms of primary sequence, being derived from different libraries; AfF4 has an extra VR on an N-terminal extension (NTE) (Fig. 1C). We measured the FcγRIIIa–AfF4 and –AfG3 interactions by isothermal titration calorimetry (ITC), fitting a 1:1 binding model, which gave estimated Kds of 217 nM and 2.6 µM, respectively (Fig. 1D). These ITC measurements may represent underestimates of the actual Kd, as the N-values of 0.6 and 0.8 for AfF4 and AfG3, respectively, may indicate the presence of an inactive proportion of the analyte.
When amine-coupled to carboxymethylated dextran sensor chips, the soluble ectodomain of FcγRIIIa interacted with both AfF4 and AfG3 with rapid association and dissociation rates (Fig. S1A) and 1:1 stoichiometry, allowing fitting to a Langmuir kinetic model, with calculated Kds of 963 nM for AfF4 and 253 nM for AfG3. Since kinetic measurements at high analyte concentrations were around the detection limits of the instrument used, we calculated steady-state affinity from the same interactions, estimating the Kds to be 1.03 µM for AfF4 and 2.77 µM for AfG3. In addition, we performed surface plasmon resonance (SPR) assays on fully glycosylated and endoglycosidase F1-treated FcγRIIIa immobilized via a biotinylated C-terminal Avitag on streptavidin-coated chips. The orientated receptor displayed steady-state affinity for AfF4 and AfG3 at ∼860 and ∼680 nM, respectively, with negligible difference conferred by glycosylation (Fig. S1B).
Affimer Proteins Block IgG Binding with a High Degree of FcγR Specificity.
Since primary cells expressing FcγRIIIa also express a number of other FcγRs, HEK293 cells stably expressing individual FcγRs were constructed to test the specificity of each Affimer protein. Each gene was fused to a C-terminal SNAP tag (26), except for FcγRIIIb, which was GPI-linked to the membrane. For FcγRIIIa allotypes each C-terminal SNAP domain fusion was coexpressed with the common γ-chain of FcεR to facilitate cell-surface expression.
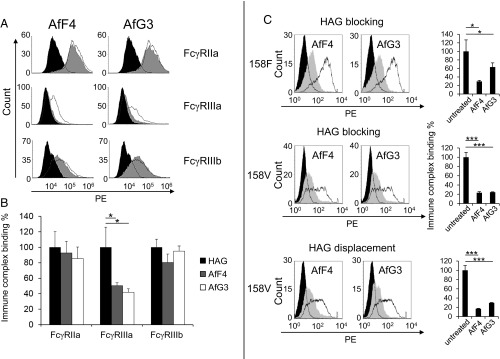
We then assessed blockade of heat-aggregated IgG1 (HAG) binding on stably transfected HEK293 cells. Both AfF4 and AfG3 significantly reduced (P = 0.01) HAG binding to FcγRIIIa (158V) (Fig. 2 A and B). Our cellular assays on FcγR specificity demonstrated that AfF4 and AfG3 did not have a significant effect on the binding of HAG to ectopically expressed FcγRIIa and FcγRIIIb, confirming considerably weaker interactions between the Affimer proteins and these homologous FcγRs (Fig. 2A).
Fig. 2.
Affimer proteins AfF4 and AfG3 specifically reduce immune complex binding to FcγRIIIa. Heat-aggregated HAG-binding assays utilized HEK293 cells stably expressing the full-length Fcγ receptors. Cells were treated with each Affimer protein before the addition of HAG followed by anti-human F(ab′)2 fragments labeled with phycoerythrin (PE); binding was measured by flow cytometry. (A, Top Row) Representative examples of the effect of AfF4 and AfG3 on immune complex (HAG) binding on cells expressing FcγRIIa; the unfilled distribution represents the untreated cells, and the gray distribution represents binding to cells pretreated with Affimer protein. The filled black region represents background binding of F(ab′)2 fragments to the cells. (Middle Row) Both Affimer proteins reduce HAG binding to cells expressing FcγRIIIa alone. (Bottom Row) Affimer proteins had little effect on HAG binding to cells expressing FcγRIIIb. (B) Histograms showing the reproducibility of AfF4 and AfG3 inhibition of HAG binding to FcγRs expressed stably on HEK293 cells. Values are normalized to Affimer proteins-untreated (HAG only) measurements. Error bars indicate the SD within three biological replicates. (C, Top and Middle) Representative experiments that demonstrate both Affimer proteins (AfF4 and AfG3) reduce the binding of heat-aggregated IgG1 to both common allotypes of FcγRIIIa, -158F and -158V. The open black histograms show the binding of HAG to cells expressing FcγRIIIa. The effect of preloading the receptors with Affimer proteins for 1 h before adding HAG is shown as a solid gray histogram. The filled black histograms are controls without HAG. (Bottom) Both Affimer proteins displaced HAG from the 158V allotype when added after HAG. *P < 0.05, ***P < 0.001, two-tailed Student’s t test.
Importantly for therapeutic applications, AfF4 and AfG3 inhibited HAG binding to both the FcγRIIIa-158F and -158V allotypes. These Affimer proteins also displaced bound HAG from FcγRIIIa-158V, the allotype with a greater affinity for IgG1 complexes (Fig. 2C).
To understand how the Affimer proteins blocked FcγRIIIa function and achieved such high specificity, we determined the crystal structures of AfF4 and AfG3 in complex with the FcγRIIIa ectodomain. The crystals belonged to space groups P212121 (AfF4) and P21 (AfG3), both diffracting to a resolution of 2.35 Å, and were refined to convergence (Rwork/Rfree of 21.9/27.2% and 20.6/24.6%, respectively) (Table 1). All structures demonstrated that the core Affimer protein scaffold maintained its compact structure while the VRs formed contacts with FcγRIIIa (rmsd <0.5 Å in all cases). For clarity of referencing amino acid positions in FcγRs and Affimer proteins, FcγRIIIa and FcγRIIIb will be referred to henceforth as “γa-” and “γb-,” whereas Affimer proteins will be referred to as “AfG3-” and “AfF4-,” respectively. The two selected Affimer proteins bound to opposite faces of the FcγRIIIa ectodomain, with AfF4 interfacing with the two FcγRIIIa-discriminating residues γa-Gly129 and γa-Tyr140 and AfG3 making contacts with FcγRIIIa extracellular domains 1 and 2.
Table 1.
Scaling and refinement statistics for crystallographic Affimer protein–FcγRIIIa complexes
| Statistic | AfF4–FcγRIIIa (5ML9) | AfG3–FcγRIIIa (5MN2) | ||||
| Average unit cell | 56.48 Å, 72.49 Å, 96.45 Å | 64.99 Å, 59.92 Å, 100.0 Å; β = 102.1° | ||||
| Space group | P212121 | P21 | ||||
| Overall | Inner shell | Outer shell | Overall | Inner shell | Outer shell | |
| Low-resolution limit, Å | 56.48 | 56.48 | 2.43 | 51.09 | 51.09 | 2.45 |
| High-resolution limit, Å | 2.34 | 9.08 | 2.34 | 2.35 | 8.47 | 2.35 |
| Rmerge (all I+ and I−) | 0.148 | 0.059 | 1.020 | 0.040 | 0.014 | 0.807 |
| Rmeas (all I+ and I−) | 0.179 | 0.071 | 1.235 | 0.049 | 0.018 | 0.997 |
| Rpim (all I+ and I−) | 0.097 | 0.038 | 0.678 | 0.028 | 0.010 | 0.578 |
| Rmerge in top intensity bin | 0.054 | 0.015 | ||||
| Total no. of observations | 47,492 | 806 | 4,301 | 82,177 | 1,702 | 9,183 |
| Total no. unique observations | 16,390 | 296 | 1,548 | 30,227 | 653 | 3,431 |
| Mean, (I)/sd(I) | 7.1 | 24.2 | 1.5 | 15.7 | 55.9 | 1.2 |
| Completeness, % | 95.4 | 84.3 | 92.5 | 95.8 | 90.6 | 97.2 |
| Multiplicity | 2.9 | 2.7 | 2.8 | 2.7 | 2.6 | 2.7 |
| Average mosaicity, ° | 0.18 | 0.14 | ||||
| Wilson B factor, Å2 | 37.3 | 58.5 | ||||
| Refinement | ||||||
| Reflections used in refinement | 16,154 | 30,207 | ||||
| Reflections used for R-free | 832 | 1,510 | ||||
| R-work/R-free | 0.219/0.272 | 0.206/0.246 | ||||
| Protein residues | 275 | 529 | ||||
| Ligands/ions | 11 | 11 | ||||
| Solvent molecules | 84 | 146 | ||||
| Rms bonds, Å | 0.002 | 0.002 | ||||
| Rms angles, ° | 0.49 | 0.49 | ||||
| Ramachandran favored, % | 97.05 | 97.43 | ||||
| Ramachandran allowed, % | 2.95 | 2.57 | ||||
| Ramachandran outliers, % | 0.00 | 0.00 | ||||
| Rotamer outliers, % | 0.00 | 0.00 | ||||
| Clash score | 0.46 | 0.37 | ||||
| Average B-factor, Å2 | 45.51 | 73.64 | ||||
| Protein | 45.05 | 73.46 | ||||
| Ligands | 63.17 | 91.92 | ||||
| Solvent | 41.67 | 62.75 | ||||
The FcγRIIIa residues contributing to IgG binding, described in Ferrara et al. (27), are depicted in green on the receptor surface in Figs. 3A and 4A. Analysis of PDB ID code 3SGJ coordinates using PISA [European Molecular Biology Laboratory-European Bioinformatics Institute (EMBL-EBI)] estimated the total buried surface area of this interaction interface to be ∼950 Å2 and estimated a solvation-free energy gain of −8.4 kcal/mol, with the formation of 10 hydrogen bonds.
Fig. 3.
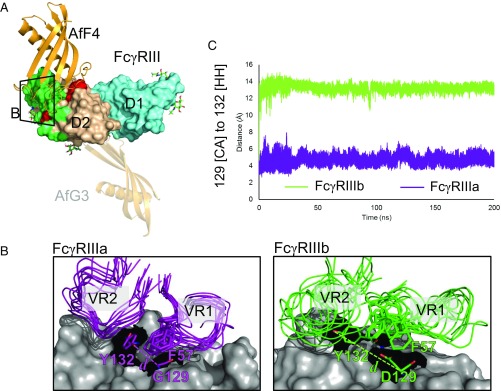
Molecular basis of interaction specificity of AfF4 for FcγRIIIa over FcγRIIIb. (A) Overview of AfF4 (orange) interaction with FcγRIIIa/b (domain 1 in aquamarine and domain 2 in wheat) showing FcγRIIIa/b-discriminating residues in red and IgG contacts in green. N-linked glycans with distinguishable electron density in the crystal structures are depicted as sticks and are listed in Table S6. The zoomed area for B is also shown. The AfG3-binding position is depicted as a transparent cartoon to aid comparison. (B) The AfF4 VR interface in FcγRIIIa and FcγRIIIb from MD simulations. AfF4 VR1 and VR2 are depicted as ensembles of snapshots taken at 20-ns intervals in representative simulations of the FcγRIIIa– and FcγRIIIb–AfF4 interactions. Interatomic distances between γa/b-Gly/Asp129 [CA] and γa/b-Tyr132 [HH] are illustrated as dashed lines and are represented in dynamic measurements in C. In FcγRIIIa the γa-Tyr132 sidechain orientates toward γa-Gly129, enabling AfF4 VR2 to form stable hydrophobic interactions involving γa-Tyr132 and AfF4-Phe57. In FcγRIIIb the γb-Asp129 sidechain clashes with γb-Tyr132, causing the ring to orientate away from γb-Asp129. γb-Asp129 also clashes with AfF4-Phe57, leading to disruption of the AfF4 VR2 hydrophobic interaction and higher mobility of VR2 and AfF4-Phe57 in FcγRIIIb. (C) Interatomic distance d averaged over triplicate MD simulations of FcγRIIIa– and FcγRIIIb–AfF4 interactions.
Fig. 4.
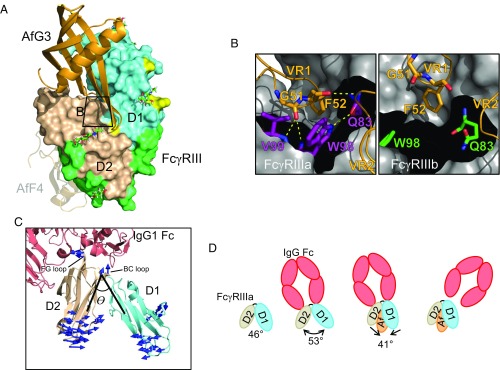
Molecular basis of AfG3 selectivity for FcγRIIIa. (A) Overview of the binding position of AfG3 (orange cartoon) to FcγRIII (D1 in aquamarine, D2 in wheat, IgG contacts in light green, polymorphic residues in yellow, FcγRIIIa discriminatory residues in red). N-linked glycans with distinguishable electron density in the crystal structures are depicted as sticks and listed in Table S6. The zoomed window for B is indicated by the black box. The AfF4-binding position is depicted as a transparent cartoon to aid comparison. (B) MD simulation snapshot of the interaction of AfG3 with FcγRIIIa (purple sticks) and FcγRIIIb (green sticks). γa-Trp98 intercalates between AfG3 VR1 and VR2, resulting in several stable intermolecular H-bonds, whereas these contacts did not form in the MD simulations in FcγRIIIb. (C) Cartoon representation of FcγRIIIa (aquamarine and wheat) interacting with IgG Fc (salmon). The interdomain angle θ is described by lines connecting the [CA] of γa-Trp90 at the top of the hinge and the [CA] of Asn169 in D2 and the [CA] of Gln83 in D1. Mode vectors describing the allosteric change from the IgG-bound state to the AfG3-bound state are shown as blue arrows. Mode vectors shorter than 3 Å are not shown. (D) Schematic representation of the allosteric change induced by AfG3. Unbound FcγRIII (PDB ID code 1FNL) describes a D1–D2 interdomain angle θ of 46°, which opens to 53° on interaction with IgG Fc. FcγRIIIa interaction with AfG3 narrows the D1–D2 angle by 12° to 41°, and we hypothesize that this allosteric shift causes sufficient deformation of the IgG Fc binding site to induce IgG Fc displacement.
AfF4–FcγRIIIa Cocrystal Structure Reveals a Steric Mode of Inhibition.
AfF4 residues in the two VRs, VR1 and VR2, interacted with γa-Ile106-His107 and γa-His119-Asp148. Several residues in the AfF4 NTE (His5–Ala10) also interfaced with FcγRIIIa. Analysis with PISA (EMBL-EBI) estimated the total buried surface area of the FcγRIIIa–AfF4 interaction to be ∼940 Å2 and estimated a solvation-free energy gain of −9.0 kcal/mol with the formation of 12 direct hydrogen bonds. All hydrogen-bond pairings are listed in Table S1.
The overlapping buried surface area between IgG Fc and AfF4 totaled about half of the individual interfaces, suggesting that AfF4 probably acts as a competitive inhibitor of IgG (Fig. S2).
The AfF4–FcγRIIIa crystal structure shows that the AfF4-binding region includes two amino acids that discriminate between FcγRIIIa and FcγRIIIb γa-Gly129/γb-Asp129 and γa-Tyr140/γb-His140 (Fig. 3A). To provide atomistic insight into the preference of AfF4 for FcγRIIIa, we used MD simulations to compare the interactions of AfF4 with both FcγRIIIa and FcγRIIIb by mutating the AfF4–FcγRIIIa complex in silico to resemble AfF4–FcγRIIIb-NA2. Simulations were performed in triplicate for 200 ns for each complex. Calculations of the rmsd (Fig. S3) showed that triplicates remained stable during the timescale of the simulations and that 200 ns was sufficient for the rmsd to converge to a stable value, which indicates that no significant global conformational changes took place.
Simulations were first subjected to atomic fluctuation analysis (Fig. S4), a measure of the average per-residue mobility throughout the simulations, which identified that VR2 of AfF4 was more mobile. Visual inspection of simulations around the AfF4 VR2–FcγRIIIa interface confirmed the mobility of VR2 and that the aromatic ring of γa-Y132 orientates toward γa-Gly129 in FcγRIIIa simulations and away from γb-Asp129 in FcγRIIIb NA2 simulations (Fig. 3 B and C). This is likely due to steric clash of the tyrosine ring with the γb-Asp129 side chain, causing the ring to move position. As observed in the X-ray structure, simulations confirmed that the absence of a sidechain in γa-Gly129 allows the AfF4-Phe57 (VR1) sidechain to sit on top of γa-Gly129. AfF4-Phe57 in this position may also contribute to a hydrophobic pocket centered on AfF4 VR2 and γa-Tyr132. Conversely, the presence of a sidechain in γb-Asp129 leads to a steric clash with the AfF4-Phe57 sidechain. AfF4-Phe57 is therefore more mobile in FcγRIIIb-containing simulations (Fig. 3B), which may further weaken the AfF4 VR2-γa-Tyr132 binding pocket. In summary, AfF4 inhibits IgG binding to FcγRIIIa by steric blocking of the IgG binding site, and the specificity mechanism of AfF4 is likely due to the variation at position 129 in FcγRIIIa/b, which leads to steric clash with a number of important binding residues.
AfG3–FcγRIIIa Cocrystal Structures Revealed Allosteric Mode of Inhibition.
As described above, crystals of FcγRIIIa–AfG3 belonged to space group P21 with four chains in the asymmetric unit (chains A and B: FcγRIIIa; chains C and D: AfG3), with chain A and D forming one FcγRIIIa–AfG3 complex and chain B and C the other. Because there are fewer crystallization contacts than in chain A, chain B, and in particular D2 of chain B, is highly flexible, resulting in poor local quality of the electron density map and a high average B factor (Table S2). The overall rmsd per alpha carbon (Cα) between the AD and BC complexes is nonetheless only 0.62 Å (217 aligned atoms), with the differences entirely distal to the binding interface. Thus, the FcγRIIIa–AfG3 complex formed by chain A and D was used for all following analyses and as the template for MD simulations.
Analysis of the FcγRIIIa–AfG3 cocrystal with PISA (EMBL-EBI) gave a total buried surface area in the interface of ∼710 Å2 (estimated solvation-free energy gain of −7.5 kcal/mol). All hydrogen-bond pairings are listed in Table S3. AfG3 bound to the interdomain hinge region of FcγRIIIa, and there was no overlap with the IgG-binding site (Fig. 4A). AfG3 residue Phe52 (VR1) sits in a hydrophobic pocket formed by the main chain of γa-Arg97 and the sidechains of γa-Gln83, Trp98, and in particular forms CH-π interactions with γa-Tyr17. AfG3 Gly51 [O] forms an H-bond with γa-Val99 [N]. γa-Trp98 intercalates between VR1 and VR2, forming water-mediated interactions to AfG3-Phe52 [O] and to AfG3-Gln83 [O]. The residues in VR2 that form sidechain interactions with FcγRIIIa are Gln83–Asn86. AfG3-Trp84 stacks on top of AfG3-His85, which stacks on top of γa-His87. In addition, they interact with the sidechains of γa-Arg18, γa-Gln83, γa-Glu85, and γa-Thr167. Importantly, γa-Arg18 is a discriminating residue between FcγRIIIa/FcγRIIIb-NA1 and FcγRIIIb-NA2 (γa-Arg18/γb-Ser18) and is key in the binding of AfG3, even though it does not interact directly with AfG3. Most of the hydrophilic interactions are via bridging water molecules. For instance, Wat520 is coordinated by γa-Tyr17, γa-Glu85, and the [O] of AfG3-His85 such that it is forced into an uncommon, but not disallowed, torsion angle conformation of φ = 59.2° and ψ = −100.1°. This positions the sidechain so that it interacts again via a water molecule (Wat615) with the backbone of γa-Val86 and is able to form the π–π–stacking interaction mentioned above. The discriminatory γa-Arg18 is held in place by an ion pair with γa-Glu85 and interacts with Wat520 and Wat506, which in turn interact with the VR2 loop.
Triplicate MD simulations (200 ns) of FcγRIIIa and FcγRIIIa, mutated in silico to resemble FcγRIIIb, in complex with AfG3 were performed. H-bond analysis of the simulations identified a number of intramolecular H-bonds that formed between γa-Arg18 and D2 residues of FcγRIIIa, which facilitated the narrowing of the D1–D2 interdomain angle. Specifically, γa-Arg18 [O] participated in an intramolecular H-bond with γa-Gln94 [Nε2], γa-Arg18 [H] with γa-Ala95 [O], and γa-Arg18 [NH1/NH2] with γa-Glu166 [OE1/OE2] (Table S4). In the MD simulations, γa-Arg18 was observed interacting with AfG3-VR2 through an H-bond between the γa-Arg18 [NH1/NH2] atoms and AfG3-Asn86 [OD1]. Conversely, in FcγRIIIb, γa-Ser18 preferred participating in intramolecular H-bonds with neighboring residues from D1 of FcγRIIIb (γb-Glu21 and γb-Leu20). H-bonds between FcγRIIIa and Wat520 and Wat615, as seen in the crystal, and between AfG3 and Wat517 were also observed in the MD simulations.
Narrowing of the interdomain angle likely allowed γa-Trp99 to move closer to AfG3, leading to intercalation of γa-Trp99 between VR1 and VR2 and the formation of several intermolecular H-bonds and an additional intramolecular H-bond (Fig. 4B and Fig. S5). Conversely, as γb-Ser18 in FcγRIIIb formed only a single weak interdomain contact, the interdomain angle did not narrow, and γb-Trp99 was unable to form these contacts.
Measurement of the D1–D2 interdomain angles (described by Cα atoms in γa-Gln83, -Trp90, and -Asn169) in unbound FcγRIIIb (PDB ID code 1FNL), IgG-FcγRIIIa (3SGJ), and AfG3-FcγRIIIa (5MN2) identified hinge angles of 46°, 53°, and 41°, respectively (Fig. 4 C and D).
To analyze how this change in the interdomain angle may affect IgG binding, we superimposed the D2 domains (γa-Trp90–Gln174) derived from our X-ray structure 5MN2 on the FcγRIIIa structure bound to IgG (3SGJ). This shows that the overall shape of the IgG-binding site is not disturbed and that only D2 is involved in binding. However, very subtle small conformational changes upon AfG3 interaction prevent IgG binding. For example, the stacking interaction of AfG3-Trp84, AfG3-His85, and γa-His87 stabilizes the BC loop (Ile88–Trp90) in a conformation that prevents AfG3-Trp90 from moving, leading to a steric clash with Pro329 in chain B of IgG. In summary, simulations demonstrated that the presence of γa-Arg18 in FcγRIIIa-AfG3, but not of γb-Ser18 in FcγRIIIb-AfG3, allows direct interaction of γa-Arg18 with AfG3 and leads to narrowing of the D1–D2 interdomain angle through multiple γa-Arg18–mediated interdomain contacts. This narrowing effectively forms the AfG3-binding interface by bringing VR1, VR2, and γa-Trp99 into close proximity.
Our proposed mechanism of IgG blocking is thus allosteric restraint of the interdomain angle that typically opens to accommodate IgG binding (28).
Affimer Proteins Block Downstream Effector Functions in FcγRIIIa-Expressing Monocytic Cells.
We sought to demonstrate that AfF4 and AfG3 could block clinically relevant FcγRIIIa effector functions using the THP-1 monocytic cell line. We characterized the cell line and determined that THP-1 cells were of the FCGR3A-158FF, FCGR2A-131HH, and FCGR2C-STP/STP genotype, rendering them incapable of functional FcγRIIc expression. This allowed us to select suitable monoclonal antibodies for evaluation of FcγR expression under different experimental conditions using flow cytometry. Staining with CD32-3D3 (which recognizes FcγRIIa-131R, FcγRIIb, and FcγRIIc but not FcγRIIa-131H) represents FcγRIIb expression in this cell line. Transcriptional analysis also confirmed that FCGR2B transcript variant 3 (RefSeq NM_001002274) and FCGR3A, but not FCGR3B, were transcribed, thus confirming that the anti-CD16 (3G8) staining was a true reflection of FcγRIIIa expression.
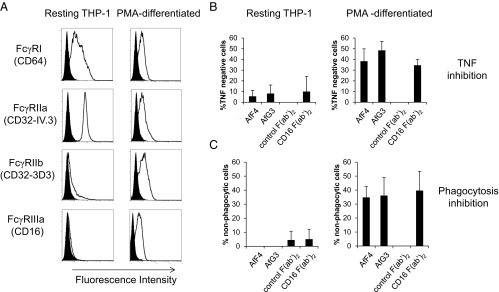
Following phorbol myristate acetate (PMA) differentiation, THP-1 cells demonstrated marked up-regulation of FcγRIIIa (CD16) and increased expression of FcγRIIb (CD32-3D3) along with decreased expression of FcγRIIa (CD32-IV.3) and, to a lesser extent, FcγRI (CD64), compared with resting cells (Fig. 5A). The marked increase in FcγRIIIa expression following culture with PMA allowed us to test the ability of the Affimer proteins to inhibit effector functions in the presence or absence of FcγRIIIa expression.
Fig. 5.
Affimer proteins are effective inhibitors of FcγRIIIa-dependent functions. (A) FACS profiles for resting monocytic THP-1 cells (Left) and PMA-differentiated, macrophage-like THP-1 cells (Right). In each case, the specific antibody staining is shown as an unfilled distribution, and the isotype control staining is shown as a filled distribution. The high-affinity FcγRIa (CD64) is reduced in differentiated THP-1 cells, along with the activatory FcγRIIa (CD32-IV.3). The inhibitory FcγRIIb is up-regulated in differentiated cells along with FcγRIIIa. (B) Affimer proteins were more effective in blocking HAG-induced TNF release in differentiated cells (Right) than in undifferentiated cells (Left), confirming their specificity for FcγRIIIa. This was represented as an increase in the percentage of TNF− cells. Blocking F(ab′)2 fragments against FcγRIIIa inhibited TNF production only in differentiated THP-1 cells. F(ab′)2 fragments against FcγRIII (CD16) were far more effective in differentiated cells, reflecting the differences in FcγRIIIa expression shown in A. F(ab′)2 fragments from preimmune serum had no effect on TNF production. (C) Both Affimer proteins were as effective as F(ab′)2 fragments in reducing phagocytosis of IgG opsonized E. coli in differentiated THP-1 cells.
The contribution of FcγRIIIa to HAG-induced TNF production was determined in both resting and PMA-differentiated THP-1 by assessing the level of inhibition obtained with FcγRIII-specific F(ab′)2 fragments (Fig. 5B). Our results showed that FcγRIIIa blockade with F(ab′)2 fragments resulted in a 34.5% increase in cells showing no TNF production following differentiation with PMA and no demonstrable inhibition in TNF production in resting THP-1 cells that do not express appreciable amounts of FcγRIIIa (Fig. 5B). We then assessed the ability of AfF4 and AfG3 to inhibit HAG-mediated TNF production. Resting THP-1 and PMA-differentiated THP-1 cells were pretreated with the Affimer proteins and assessed for their ability to produce TNF in response to HAG. Each Affimer protein demonstrated inhibition of TNF production in PMA-differentiated THP-1 at a level comparable to that observed with the FcγRIIIa-specific F(ab′)2 fragment. Resting THP-1 cells that do not express FcγRIIIa display less than 10% inhibition of HAG-induced TNF production, consistent with the levels seen following blockade with the FcγRIIIa-specific F(ab′)2 fragment.
We then compared the ability of Affimer proteins to inhibit phagocytosis of IgG-opsonized Escherichia coli in both resting and PMA-differentiated THP-1 cells and compared this with the level of inhibition observed following pretreatment with FcγRIII-specific F(ab′)2. Inhibition of phagocytosis by each of the Affimer proteins was observed only in PMA-differentiated THP-1, where FcγRIIIa was expressed, and at a level comparable to that in cells treated with FcγRIII F(ab′)2, consistent with data on inhibition of TNF production (Fig. 5C).
Discussion
We describe the isolation of highly specific steric and allosteric inhibitors of FcγRIIIa using Affimer protein technology. These Affimer proteins specifically block IgG immune complex (HAG) binding to FcγRIIIa but not the closely related FcγRIIIb and FcγRIIa and also inhibit downstream effector functions such as TNF release and phagocytosis. While some FcγR class-specific monoclonal antibodies recognize epitopes in the IgG binding site, no commercially available antibody is specific for FcγRIIIa. This lack of specificity has been demonstrated in vivo when both monocyte and neutrophil cytopenias were observed in clinical trials of the CD16-3G8 monoclonal antibody that recognizes both FcγRIIIa (expressed on natural killer cells and some peripheral blood monocytes) and FcγRIIIb (expressed on neutrophils) (29). Preservation of neutrophil function offers the potential to dampen inflammatory processes orchestrated by macrophages while leaving host immunity to infections afforded by neutrophils intact.
We have identified an Affimer protein (AfF4) that binds within the IgG-binding site and acts as a highly specific steric inhibitor of IgG binding to FcγRIIIa but not to FcγRIIa or FcγRIIIb, as shown by HAG-binding assays using HEK293 cells expressing a single FcγR allotype. Elucidation of the structural basis for this specificity may facilitate engineering of CD16 therapeutic antibodies to achieve increased selectivity for FcγRIIIa over FcγRIIIb. Through X-ray crystallography and MD simulations, we have shown that AfF4 specificity for FcγRIIIa is likely focused around the region containing the FcγRIIIa/b-discriminating residue (γa-Gly129/γb-Asp129), showing that subtle differences in primary sequence can lead to local changes in topology that can have knock-on effects on molecular recognition.
An allosteric site in the hinge region of FcγRIIIa was recognized by AfG3, which holds the receptor ectodomain in a restricted conformation, preventing the opening of the structure associated with IgG Fc binding and in particular γa-Trp90. A major attraction of targeting allosteric sites is that they may be less evolutionarily conserved, and therefore allosteric inhibitors can potentially be more selective. Interestingly, AfG3, although binding to FcγRIIIa at a seemingly conserved region to FcγRIIIb, showed high cellular specificity to FcγRIIIa. Our proposed mechanism of AfG3 specificity is that the presence of γaArg18 can organize a large number of intramolecular H-bonds that, when AfG3 binds, create a tight FcγRIIIa–AfG3 interface that cannot occur with FcγRIIIb.
This highlights that allosteric regulation could provide a valid method for modulating biological function even in highly conserved proteins. Reduced binding of HAG to both FcγRIIIa-158F and -158V allotypes, but not to FcγRIIa or FcγRIIIb, was observed. Pertinently, both Affimer proteins disrupted complexes of prebound HAG to FcγRIIIa-158V as well as blocking HAG binding to receptor pretreated with Affimer protein.
The molecular design of Affimer protein, which employs a stable scaffold for the constraint of flexible regions of variable amino acid sequences, uses the same successful strategy for generating specific protein–protein interactions as antibodies. The high plasticity of the VRs combined with the chemical heterogeneity achievable through the wide variety of sequences generated by phage display ensure that sufficient biochemical space is explored and conformational space is sufficient to discover Affimer proteins capable of discriminating between highly homologous receptors. However, the delicate balance of interactions involved implies that rational design of future inhibitors based on structural information alone may not be adequate and that each Affimer protein identified as a binder should be considered unique. Indeed, this may be why loop grafting can result in affinity differences between different scaffolds, for example as observed in ref. 30.
The effect of each Affimer protein on clinically relevant FcγRIIIa effector functions was confirmed by the TNF production and phagocytosis assays. Although FcγRI, FcγRIIa, and FcγRIIIa are expressed on the monocyte-like THP-1 cells, the inhibition of these downstream functions was correlated with the greatly increased FcγRIIIa expression in THP-1 cells differentiated with PMA. TNF release is a relevant in vitro model of receptor signaling, since immune complex-activated macrophages have been shown to release large amounts of TNF in rheumatoid arthritis (31).
Affimer protein technology therefore represents a promising methodological approach for the generation of highly stable, easily expressed antibody mimetic reagents with capabilities to modulate protein function and protein–protein interactions. X-ray structures and molecular dynamics simulations of Affimer protein/FcγRIIIa complexes provide a structural basis for understanding the potential mechanism of inhibition. Written informed consent was provided for the use of healthy human donor cDNA as cloning template and was approved by the Leeds (East) National Health Service Research Ethics Committee (Ref: 04/Q1206/107).
Supplementary Material
Acknowledgments
We thank Mr. Thomas Taylor of the Leeds Biomedical Health Research Centre BioScreening Technology Group (BSTG) for excellent technical support. ITC was performed in the Astbury Biomolecular Interactions Facility supported by Wellcome Trust Grant 094232/Z/10/Z. The work on beamline I24 at Diamond Light Source was undertaken under proposal NT5969. The Oxford Protein Production Facility was supported by Medical Research Council Grant MR/K018779/1. The research was also supported by Arthritis Research UK Grant 19764, the National Institute for Health Research (NIHR) Leeds Biomedical Research Centre, the Ann Wilks Memorial Fund, and Biotechnology and Biological Sciences Research Council Grant BB/M021610/1. This work was further supported by a grant by Marie Skłodowska-Curie Actions in Horizon 2020 (to M.T.). The Leeds Biomedical Health Research Centre BSTG received funding from the University of Leeds and Leeds Teaching Hospitals NHS Trust. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, or the Department of Health.
Footnotes
Conflict of interest statement: The University of Leeds has filed a patent application on Adhiron (referred to here as Affimer protein) that is licensed to Avacta Life Sciences Ltd.
This article is a PNAS Direct Submission.
Data deposition: Coordinates and structure factors have been deposited in the Protein Data Bank under the accession codes 5ML9 and 5MN2. Molecular dynamics simulation set-up files and compressed trajectories are available at https://doi.org/10.5518/258.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1707856115/-/DCSupplemental.
References
- 1.Milroy LG, Grossmann TN, Hennig S, Brunsveld L, Ottmann C. Modulators of protein-protein interactions. Chem Rev. 2014;114:4695–4748. doi: 10.1021/cr400698c. [DOI] [PubMed] [Google Scholar]
- 2.Higueruelo AP, Jubb H, Blundell TL. Protein-protein interactions as druggable targets: Recent technological advances. Curr Opin Pharmacol. 2013;13:791–796. doi: 10.1016/j.coph.2013.05.009. [DOI] [PubMed] [Google Scholar]
- 3.Jubb H, Higueruelo AP, Winter A, Blundell TL. Structural biology and drug discovery for protein-protein interactions. Trends Pharmacol Sci. 2012;33:241–248. doi: 10.1016/j.tips.2012.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Reichert JM, Rosensweig CJ, Faden LB, Dewitz MC. Monoclonal antibody successes in the clinic. Nat Biotechnol. 2005;23:1073–1078. doi: 10.1038/nbt0905-1073. [DOI] [PubMed] [Google Scholar]
- 5.Vlasak J, Ionescu R. Heterogeneity of monoclonal antibodies revealed by charge-sensitive methods. Curr Pharm Biotechnol. 2008;9:468–481. doi: 10.2174/138920108786786402. [DOI] [PubMed] [Google Scholar]
- 6.Tiede C, et al. Adhiron: A stable and versatile peptide display scaffold for molecular recognition applications. Protein Eng Des Sel. 2014;27:145–155. doi: 10.1093/protein/gzu007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharma R, et al. Label-free electrochemical impedance biosensor to detect human interleukin-8 in serum with sub-pg/ml sensitivity. Biosens Bioelectron. 2016;80:607–613. doi: 10.1016/j.bios.2016.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rawlings AE, et al. Phage display selected magnetite interacting Adhirons for shape controlled nanoparticle synthesis. Chem Sci. 2015;6:5586–5594. doi: 10.1039/c5sc01472g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kyle HF, et al. Exploration of the HIF-1α/p300 interface using peptide and Adhiron phage display technologies. Mol Biosyst. 2015;11:2738–2749. doi: 10.1039/c5mb00284b. [DOI] [PubMed] [Google Scholar]
- 10.Modell AE, Blosser SL, Arora PS. Systematic targeting of protein-protein interactions. Trends Pharmacol Sci. 2016;37:702–713. doi: 10.1016/j.tips.2016.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nimmerjahn F, Ravetch JV. Fcgamma receptors as regulators of immune responses. Nat Rev Immunol. 2008;8:34–47. doi: 10.1038/nri2206. [DOI] [PubMed] [Google Scholar]
- 12.Hogarth PM, Pietersz GA. Fc receptor-targeted therapies for the treatment of inflammation, cancer and beyond. Nat Rev Drug Discov. 2012;11:311–331. doi: 10.1038/nrd2909. [DOI] [PubMed] [Google Scholar]
- 13.Morgan AW, et al. Association of FCGR2A and FCGR2A-FCGR3A haplotypes with susceptibility to giant cell arteritis. Arthritis Res Ther. 2006;8:R109. doi: 10.1186/ar1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robinson JI, et al. BRAGGSS Confirmation of association of FCGR3B but not FCGR3A copy number with susceptibility to autoantibody positive rheumatoid arthritis. Hum Mutat. 2012;33:741–749. doi: 10.1002/humu.22031. [DOI] [PubMed] [Google Scholar]
- 15.Willcocks LC, et al. Copy number of FCGR3B, which is associated with systemic lupus erythematosus, correlates with protein expression and immune complex uptake. J Exp Med. 2008;205:1573–1582. doi: 10.1084/jem.20072413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cooper DL, et al. YEAR Consortium FcγRIIIa expression on monocytes in rheumatoid arthritis: Role in immune-complex stimulated TNF production and non-response to methotrexate therapy. PLoS One. 2012;7:e28918. doi: 10.1371/journal.pone.0028918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ji H, et al. Arthritis critically dependent on innate immune system players. Immunity. 2002;16:157–168. doi: 10.1016/s1074-7613(02)00275-3. [DOI] [PubMed] [Google Scholar]
- 18.Kleinau S, Martinsson P, Heyman B. Induction and suppression of collagen-induced arthritis is dependent on distinct fcgamma receptors. J Exp Med. 2000;191:1611–1616. doi: 10.1084/jem.191.9.1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qiu WQ, de Bruin D, Brownstein BH, Pearse R, Ravetch JV. Organization of the human and mouse low-affinity Fc gamma R genes: Duplication and recombination. Science. 1990;248:732–735. doi: 10.1126/science.2139735. [DOI] [PubMed] [Google Scholar]
- 20.Machado LR, et al. Evolutionary history of copy-number-variable locus for the low-affinity Fcγ receptor: Mutation rate, autoimmune disease, and the legacy of helminth infection. Am J Hum Genet. 2012;90:973–985. doi: 10.1016/j.ajhg.2012.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clarkson SB, et al. Treatment of refractory immune thrombocytopenic purpura with an anti-Fc gamma-receptor antibody. N Engl J Med. 1986;314:1236–1239. doi: 10.1056/NEJM198605083141907. [DOI] [PubMed] [Google Scholar]
- 22.Bosques CJ, Manning AM. Fc-gamma receptors: Attractive targets for autoimmune drug discovery searching for intelligent therapeutic designs. Autoimmun Rev. 2016;15:1081–1088. doi: 10.1016/j.autrev.2016.07.035. [DOI] [PubMed] [Google Scholar]
- 23.Weinblatt ME, et al. Effects of fostamatinib (R788), an oral spleen tyrosine kinase inhibitor, on health-related quality of life in patients with active rheumatoid arthritis: Analyses of patient-reported outcomes from a randomized, double-blind, placebo-controlled trial. J Rheumatol. 2013;40:369–378. doi: 10.3899/jrheum.120923. [DOI] [PubMed] [Google Scholar]
- 24.Friedberg JW, et al. Inhibition of Syk with fostamatinib disodium has significant clinical activity in non-Hodgkin lymphoma and chronic lymphocytic leukemia. Blood. 2010;115:2578–2585. doi: 10.1182/blood-2009-08-236471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacFarlane LA, Todd DJ. Kinase inhibitors: The next generation of therapies in the treatment of rheumatoid arthritis. Int J Rheum Dis. 2014;17:359–368. doi: 10.1111/1756-185X.12293. [DOI] [PubMed] [Google Scholar]
- 26.Keppler A, et al. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- 27.Ferrara C, et al. Unique carbohydrate-carbohydrate interactions are required for high affinity binding between FcgammaRIII and antibodies lacking core fucose. Proc Natl Acad Sci USA. 2011;108:12669–12674. doi: 10.1073/pnas.1108455108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Woof JM, Burton DR. Human antibody-Fc receptor interactions illuminated by crystal structures. Nat Rev Immunol. 2004;4:89–99. doi: 10.1038/nri1266. [DOI] [PubMed] [Google Scholar]
- 29.Nakar CT, Bussel JB. 3G8 and GMA161, anti FcγRIII inhibitory monoclonal antibodies in the treatment of chronic refractory ITP. (Summary of 2 pilot studies) Blood. 2009;114:2404. [Google Scholar]
- 30.Vita C, et al. Novel miniproteins engineered by the transfer of active sites to small natural scaffolds. Biopolymers. 1998;47:93–100. doi: 10.1002/(SICI)1097-0282(1998)47:1<93::AID-BIP10>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 31.Cassatella MA, et al. Soluble TNF-like cytokine (TL1A) production by immune complexes stimulated monocytes in rheumatoid arthritis. J Immunol. 2007;178:7325–7333, and erratum (2007) 179:1390. doi: 10.4049/jimmunol.178.11.7325. [DOI] [PubMed] [Google Scholar]
- 32.Berrow NS, Alderton D, Owens RJ. The precise engineering of expression vectors using high-throughput in-fusion PCR cloning. Methods Mol Biol. 2009;498:75–90. doi: 10.1007/978-1-59745-196-3_5. [DOI] [PubMed] [Google Scholar]
- 33.Bird LE. High throughput construction and small scale expression screening of multi-tag vectors in Escherichia coli. Methods. 2011;55:29–37. doi: 10.1016/j.ymeth.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 34.Nettleship JE, Rahman-Huq N, Owens RJ. The production of glycoproteins by transient expression in mammalian cells. Methods Mol Biol. 2009;498:245–263. doi: 10.1007/978-1-59745-196-3_16. [DOI] [PubMed] [Google Scholar]
- 35.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 36.Tiede C, et al. Affimer proteins are versatile and renewable affinity reagents. Elife. 2017;6:e24903. doi: 10.7554/eLife.24903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Warmerdam PAM, et al. Interaction of a human Fc gamma RIIb1 (CD32) isoform with murine and human IgG subclasses. Int Immunol. 1993;5:239–247. doi: 10.1093/intimm/5.3.239. [DOI] [PubMed] [Google Scholar]
- 38.Daëron M, et al. The same tyrosine-based inhibition motif, in the intracytoplasmic domain of Fc gamma RIIB, regulates negatively BCR-, TCR-, and FcR-dependent cell activation. Immunity. 1995;3:635–646. doi: 10.1016/1074-7613(95)90134-5. [DOI] [PubMed] [Google Scholar]
- 39.Morgan AW, et al. Analysis of Fcgamma receptor haplotypes in rheumatoid arthritis: FCGR3A remains a major susceptibility gene at this locus, with an additional contribution from FCGR3B. Arthritis Res Ther. 2006;8:R5. doi: 10.1186/ar1847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Metes D, et al. Expression of functional CD32 molecules on human NK cells is determined by an allelic polymorphism of the FcgammaRIIC gene. Blood. 1998;91:2369–2380. [PubMed] [Google Scholar]
- 41.Walter TS, et al. A procedure for setting up high-throughput nanolitre crystallization experiments. Crystallization workflow for initial screening, automated storage, imaging and optimization. Acta Crystallogr D Biol Crystallogr. 2005;61:651–657. doi: 10.1107/S0907444905007808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Evans PR, Murshudov GN. How good are my data and what is the resolution? Acta Crystallogr D Biol Crystallogr. 2013;69:1204–1214. doi: 10.1107/S0907444913000061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Afonine PV, et al. Towards automated crystallographic structure refinement with phenix.refine. Acta Crystallogr D Biol Crystallogr. 2012;68:352–367. doi: 10.1107/S0907444912001308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen VB, et al. MolProbity: All-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr. 2010;66:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tina KG, Bhadra R, Srinivasan N. PIC: Protein interactions calculator. Nucleic Acids Res. 2007;35:W473–W476. doi: 10.1093/nar/gkm423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Case DA, et al. 2014. AMBER 14 (University of California, San Francisco)
- 50.Maier JA, et al. ff14SB: Improving the accuracy of protein side chain and backbone parameters from ff99SB. J Chem Theory Comput. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kirschner KN, et al. GLYCAM06: A generalizable biomolecular force field. Carbohydrates. J Comput Chem. 2008;29:622–655. doi: 10.1002/jcc.20820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prokop M, Adam J, Kríz Z, Wimmerová M, Koča J. TRITON: A graphical tool for ligand-binding protein engineering. Bioinformatics. 2008;24:1955–1956. doi: 10.1093/bioinformatics/btn344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graph. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 54. Schrödinger LLC (2010) The PyMOL Molecular Graphics System (Schrödinger, LLC, New York), Version 1.2r3pre.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.