Significance
Mimicking protein-like specific interactions and functions has been a long-pursued goal in nanotechnology. The key challenge is to precisely organize nonfunctional surface groups on nanoparticles into specific 3D conformations to function in a concerted and orchestrated manner. Here, we develop a method to graft the complementary-determining regions of natural antibodies onto nanoparticles and reconstruct their “active” conformation to create nanoparticle-based artificial antibodies that recognize the corresponding antigens. Our work demonstrates that it is possible to create functions on nanoparticles by conformational engineering, namely tuning flexible surface groups into specific conformations. Our straightforward strategy could be used further to create other artificial antibodies for various applications and provides a new tool to understand the structure and folding of natural proteins.
Keywords: artificial antibody, conformation engineering, nanoparticle, Goldbody, EGFR
Abstract
To impart biomedical functions to nanoparticles (NPs), the common approach is to conjugate functional groups onto NPs by dint of the functions of those groups per se. It is still beyond current reach to create protein-like specific interactions and functions on NPs by conformational engineering of nonfunctional groups on NPs. Here, we develop a conformational engineering method to create an NP-based artificial antibody, denoted “Goldbody,” through conformational reconstruction of the complementary-determining regions (CDRs) of natural antibodies on gold NPs (AuNPs). The seemingly insurmountable task of controlling the conformation of the CDR loops, which are flexible and nonfunctional in the free form, was accomplished unexpectedly in a simple way. Upon anchoring both terminals of the free CDR loops on AuNPs, we managed to reconstruct the “active” conformation of the CDR loops by tuning the span between the two terminals and, as a result, the original specificity was successfully reconstructed on the AuNPs. Two Goldbodies have been created by this strategy to specifically bind with hen egg white lysozyme and epidermal growth factor receptor, with apparent affinities several orders of magnitude stronger than that of the original natural antibodies. Our work demonstrates that it is possible to create protein-like functions on NPs in a protein-like way, namely by tuning flexible surface groups to the correct conformation. Given the apparent merits, including good stability, of Goldbodies, we anticipate that a category of Goldbodies could be created to target different antigens and thus used as substitutes for natural antibodies in various applications.
Nanotechnology is one of the most active fields, with great expectations such as revolutionizing cancer diagnosis and therapy (1, 2). After decades of extensive studies, nowadays scientists can easily control the size and shape of many nanoparticles (NPs), yet there seems still a long way to go to fulfill their expectations (3). Apparently, nanotechnology must go beyond size and shape for its further booming development. As shown by natural proteins, the most miraculous nanomachines in nature, it is not their size or shape, not even their covalent bonds, but the unique conformation of their peptide chains that makes proteins special and marvelous. Although unfolded proteins are generally inactive, billions of years of evolution have endowed proteins with the ability to fold their peptide chains into unique active conformations, putting individual “nonfunctional” groups into the right place to work in a concerted and orchestrated manner. In addition, contrary to the usually rigid amorphous surface or symmetrical crystal facets of NPs, natural proteins have relatively flexible and “irregular” surfaces, the major characteristics that make proteins recognize/interact with their partners with high specificity.
At present, to impart biomedical functions (such as tumor targeting for drug delivery) to NPs, the common approach is to conjugate NPs directly with functional groups, such as folic acid (4), aptamers (5), and antibodies (6), relying on the multivalency effect to enhance the function of the conjugated groups per se (7, 8). A key challenge in nanoscience is to engineer NP surfaces to achieve protein-like highly specific and reversible binding properties (9, 10), especially with nonfunctional groups. Though chemists have synthesized millions of molecules through chemical bond engineering, it is still beyond current reach to precisely control the conformation of big molecules (such as polymers, peptides, and NPs), with the exception of some success in protein design (11–14). Potentially, conformational engineering of the surface groups on NPs will bring nanotechnology to an additional dimension. To explore this new dimension, we demonstrate here that it is possible to precisely tune the conformation of the surface groups on NPs to create NP-based artificial antibodies with protein-like highly specific and reversible binding properties.
An interesting fact of natural proteins is that a protein may have hundreds of amino acid residues but only a few key residues play a major role in its functions and specific interactions with other molecules (15–18). These key residues alone do not have any function; the function emerges only when all key residues are at precisely the right positions. One major role of the rest of residues of the protein is to keep those key residues at the right places. Interestingly, the function of a protein may be reproduced by grafting its key residues or fragment (short peptide) onto another heterologous protein skeleton that is suitable to anchor them in the right positions (19–23). Inspired by this fact, we hypothesized that NPs could serve as such skeletons, and the key residues of a protein could be grafted onto NPs to reconstruct the function of the original protein.
We chose gold NPs (AuNPs) as the “skeleton” for the easy control of its size (24) and potential biomedical applications (6), and chose natural antibodies as the original function to be reconstructed on AuNPs. Our goal was to create an AuNP-based artificial antibody (denoted “Goldbody”) that can specifically bind to the same antigen as the natural antibody.
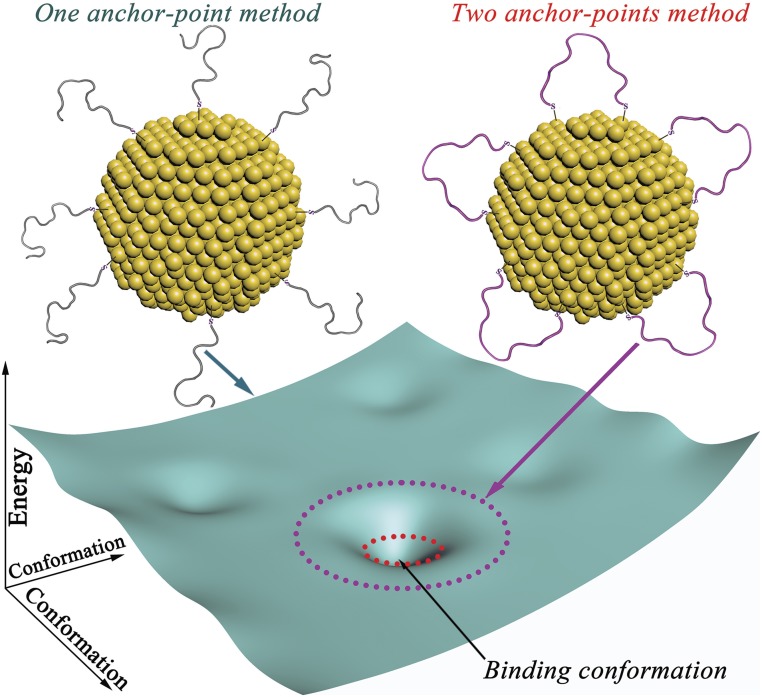
To create a Goldbody, our idea was to graft the complementary-determining region (CDR) loops of natural antibodies onto AuNPs and reconstruct their “active” conformation. The CDR loops are the recognition regions for antigens, and also the most flexible parts of natural antibodies. The common method that functionalizes an AuNP with a peptide is conjugating the peptide by forming one Au–S bond with a Cys residue. This traditional “one-anchor-point” method has no control over the conformation of the conjugated peptide. As shown in Fig. 1, the conformational space (the cyan sheet) of this one Au–S bond-linked peptide on AuNP is enormous, and its energy landscape is relatively flat. Therefore, even though the “binding” conformation might be enthalpically favored, keeping the CDR peptide on AuNPs in the specific binding conformation (for the antigen) is a mission that is entropically impossible. We further hypothesized that if we conjugated both terminals of the CDR peptide on AuNPs, then its conformational space would be greatly reduced (magenta dotted circle in Fig. 1) and, if the span of the CDR peptide could be tuned close to that in natural antibodies, the conformational space might be further restricted in a small area (red dotted circle in Fig. 1) that is ready for induced complementation to the unique binding conformation after binding with its partners. So, the impossible mission of precise conformational control is simplified to tuning the span of CDR loops in the suitable range, which is achieved here by controlling the loop density on the surface of AuNPs (SI Appendix, Fig. S1). To demonstrate the general applicability of this strategy, here we designed and synthesized two Goldbodies that bind specifically with hen egg white lysozyme (HEWL) or epidermal growth factor receptor (EGFR).
Fig. 1.
Comparison of the energy landscapes of the surface peptides on AuNPs conjugated by the traditional one-anchor-point method and our two-anchor-point method. The conformational space of the peptide by the two-anchor-point method is only a small subset (magenta circle) of that by the one-anchor point method (whole cyan sheet), and can be further reduced to the approximation of the binding conformation (red circle) by adjusting the peptide density on the surface of AuNPs.
Results
Design and Synthesis of an Anti-Lysozyme Goldbody.
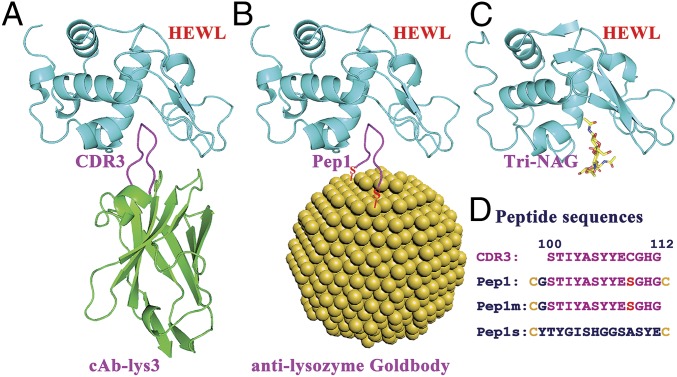
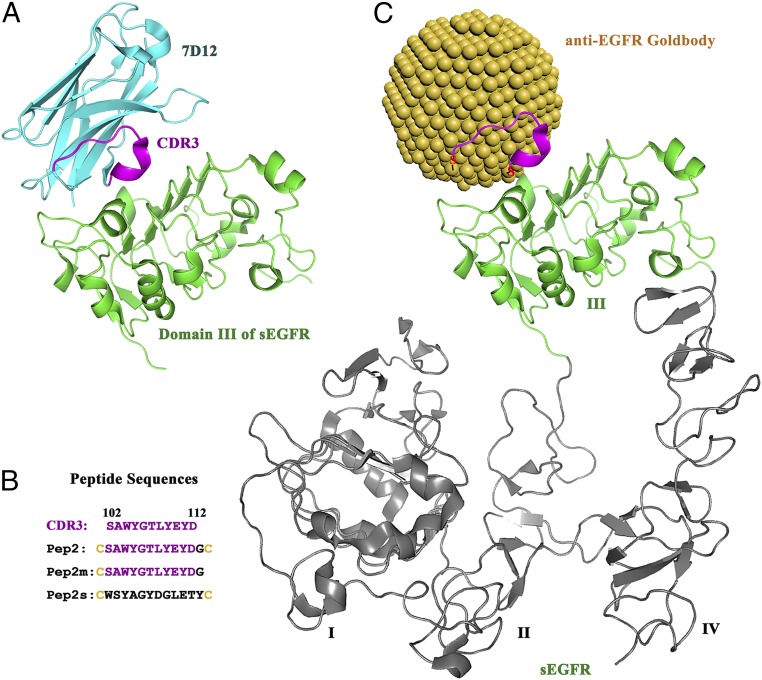
HEWL was selected as the first target antigen for the Goldbody design. Among a large number of lysozyme antibodies, camelid anti-lysozyme antibody cAb-Lys3 was chosen as the natural antibody to be reproduced as an anti-lysozyme Goldbody. cAb-Lys3 binds HEWL strongly, with a long loop (CDR3) penetrating deeply into the active-site cleft of HEWL (25, 26) (Fig. 2A). The penetrating fragment 100 to 112 of CDR3 was chosen for grafting. A Cys residue (C) was added at both terminals of this peptide. In addition, a Gly residue (G) was inserted after the N-terminal Cys to prevent potential tension caused by the Cys–AuNP interaction, and Cys109 in the middle of the fragment was changed to Ser (S) to avoid forming an unwanted S–Au bond. This peptide is referred to as Pep1. Pep1 was anchored on the AuNP surface simply by mixing AuNPs with Pep1 in solution [see SI Appendix, Fig. S2 for transmission electron microscopy (TEM) images of AuNPs and AuNP–Pep, and SI Appendix, Fig. S3 for the conjugating efficiency]. The Pep1-conjugated AuNP is referred to as AuNP–Pep1, which is the designed anti-lysozyme Goldbody when the right conformation is reconstructed (Fig. 2B).
Fig. 2.
Scheme of the design of the anti-lysozyme Goldbody. (A) Structure of cAb-Lys3 (green, with the CDR3 loop shown in magenta) in complex with HEWL (cyan) [Protein Data Bank (PDB) ID code 1MEL]. (B) Model of the anti-lysozyme Goldbody (with the grafted peptide loop shown in magenta) in complex with HEWL (cyan); for clarity, only one peptide is shown on the AuNP. (C) Structure of HEWL (cyan) in complex with its inhibitor Tri-NAG (shown as sticks) (PDB ID code 1HEW). (D) Sequences of the CDR3 fragment (100 to 112) of HEWL and the three designed peptides.
For comparison, Pep1m, a peptide having only one terminal Cys (corresponding to the one-anchor-point method), and Pep1s, a peptide having two terminal Cys residues and the same amino acid composition as Pep1 but with a scrambled sequence, were conjugated onto AuNPs as controls to show the effect of conformational engineering and the specificity of binding, respectively. The sequences of these peptides are shown in Fig. 2D.
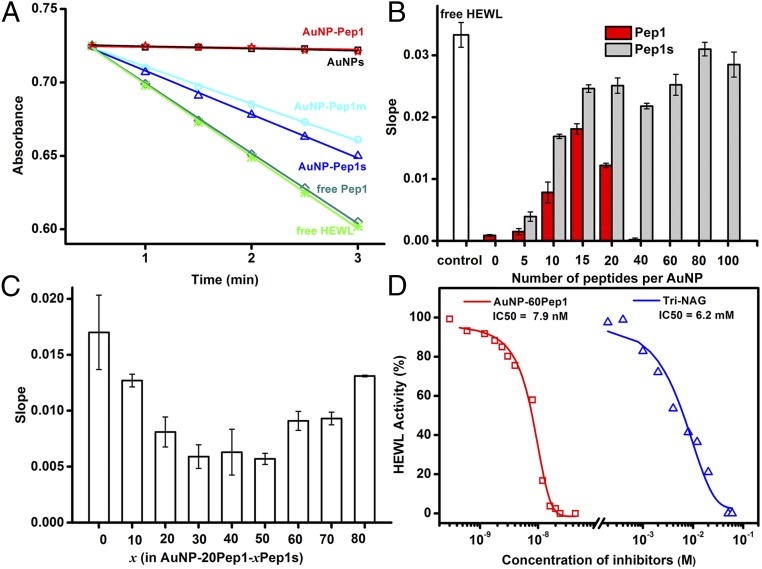
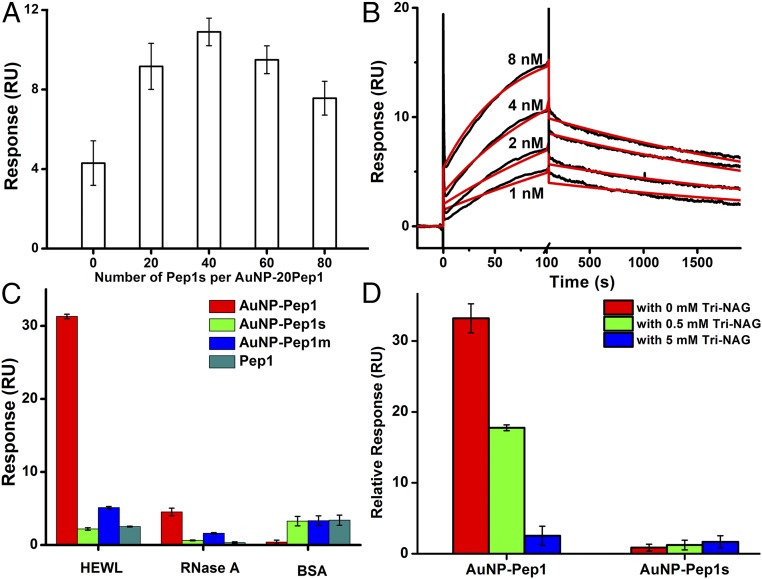
AuNP–Pep1 was designed to bind specifically into the active cleft of HEWL (Fig. 2B) as cAb-Lys3 (Fig. 2A), so the inhibition of HEWL’s activity can be used to quantify the binding between AuNP–Pep1 and HEWL. Fig. 3A shows the activity profiles of HEWL after incubation with free Pep1 or AuNPs (3.6 nm) conjugated with different peptides. The decreased slopes represent the inhibition of HEWL activity due to the binding with different species. It can be seen that free Pep1 does not affect the activity of HEWL, indicating that free Pep1 does not bind to HEWL. It is unsurprising that the nonfunctionalized AuNPs could inhibit the activity of HEWL completely, because it is well-known that there is strong nonspecific binding between the nonfunctionalized AuNPs and proteins, forming so-called protein corona on the surface of AuNPs (27–30). When the AuNP surface is conjugated with peptides, the strong nonspecific binding between the AuNP surface and HEWL could be suppressed. Therefore, the inhibition of HEWL activity by AuNPs decreases while increasing the coverage of Pep1s until the coverage reaches around 15 peptides per AuNP (3.6 nm) (Fig. 3B). This point represents the minimal coverage of peptides to passivate the strong nonspecific binding surface of AuNPs. Similar to Pep1s, when Pep1 is grafted on AuNPs, the activity inhibition first decreases (corresponding to reducing strong nonspecific interaction) until reaching a minimal coverage around 15 peptides per AuNP (3.6 nm). However, then the activity inhibition increases with the further increase of Pep1 (Fig. 3B), indicating that the increased inhibition may come from the specific interaction between AuNP–Pep1 and HEWL.
Fig. 3.
Interaction between the anti-lysozyme Goldbody and HEWL. (A) Enzymatic activity assays of 30 nM HEWL alone, in the presence of 24 nM nonfunctionalized AuNPs (3.6 nm), or in the presence of 24 nM AuNPs (3.6 nm) functionalized with 60 Pep1, 60 Pep1m, or 60 Pep 1s per AuNP, respectively. Free Pep1 at the equivalent peptide concentration (24 × 60 = 1,440 nM) was used as the control. (B) Slopes of 30 nM HEWL assay curves in the presence of 24 nM AuNPs (3.6 nm) functionalized with different numbers of Pep1 (red) or Pep1s (gray). The white column denotes the free HEWL control. (C) Slopes of 30 nM HEWL assay curves in the presence of 24 nM different AuNP–20Pep1–xPep1s, showing the effect of peptide density [changed by the number (x) of the nonactive Pep1s]. (D) IC50 of AuNP–Pep1 and Tri-NAG for 30 nM HEWL. Error bars indicate SDs.
Reconstructing the Conformation of the CDR3 Loop on AuNPs.
To finely tune the peptide conformation on AuNPs, we changed the peptide density on the AuNP surface to adjust the span of the two anchor points of peptides. For a convincing comparison, AuNPs (3.6 nm) were functionalized with different mixtures of Pep1 and Pep1s, while the number of the potential active Pep1 was fixed at 20 per AuNP and the peptide density on the AuNP surface was adjusted by adding different numbers (x) of the “inactive” Pep1s, forming different samples of AuNP–20Pep1–xPep1s. Fig. 3C shows the influence of peptide density on the activity of HEWL. It is clear that 60 peptides (20 Pep1 + 40 Pep1s) per AuNP (3.6 nm), or about one peptide per 0.68-nm2 AuNP surface {equal to the surface area of an AuNP [4 × 3.14 × (3.6/2)2 nm2] divided by 60 peptides}, is the optimal peptide density to keep the grafted CDR loop in the active conformation for the specific binding with HEWL. Three different-sized AuNPs (3.6, 6.9, and 15.0 nm) were tested for grafting Pep1. By keeping similar peptide density (one peptide per 0.68 nm2), all three different-sized AuNP–Pep1 can inhibit the activity of lysozyme (SI Appendix, Figs. S4 and S5). Since 3.6-nm AuNP–Pep1 has a size similar to cAb-Lys3, and possesses much higher specific surface area than the larger ones, in all the following experiments 3.6-nm AuNPs were used. And, in the following, AuNP–60Pep1 was taken to represent the anti-lysozyme Goldbody.
Interestingly, the conformation of the peptide on AuNPs can be tuned sequentially. As shown in SI Appendix, Fig. S6, instead of functionalizing AuNPs with the mixture of Pep1 and Pep1s, the conformation of Pep1 on AuNPs can also be tuned by functionalizing 20 Pep1 first and then adding 40 Pep1s later. This result implies that the peptide loops or the Au–S bonds are movable and adjustable on the AuNP surface. The underlying mechanisms for the mobility of Au–S are the same as the reported mobility of gold adatoms on the surface (31–34) and the formation of transient Au···S···Au bonds (35). This movable feature of Au–S bonds is critical for the success of the Goldbody, because it decreases the energy barrier between different peptide conformations on AuNPs and makes the enthalpically favored binding conformation accessible from any initial conjugated state. In addition, it also makes possible further binding-induced mutually complementary conformation adjustment after binding with the corresponding antigen.
Strong Binding Affinity and Specificity of the Anti-Lysozyme Goldbody Toward HEWL.
As shown in Fig. 3D, AuNP–60Pep1 has an extremely low IC50 of 7.9 nM (when the concentration of HEWL is 30 nM), which is six orders of magnitude lower than that of the small molecular inhibitor tri-N-acetyl-d-glucosamine (Tri-NAG) (6.2 mM), indicating that the anti-lysozyme Goldbody is a strong inhibitor of HEWL. In comparison, free Pep1 shows almost no noticeable inhibitory effect on HEWL even at much higher concentrations up to 1 mM, which is evidence that the free Pep1 is flexible and inactive.
AuNP–60Pep1s has the same amino acid composition, and the same peptide density and span, as AuNP–60Pep1; however, the scrambled sequence of Pep1s eliminates the specificity toward HEWL. As expected, only weak nonspecific interaction between AuNP–60Pep1s and HEWL is observed (Fig. 3A). As for Pep1m, it has the same sequence as Pep1 except only one anchor point on AuNP, and thus can move more freely on the AuNP surface than Pep1 (SI Appendix, Fig. S1). Since fixing the more flexible Pep1m to the binding conformation will cause a large conformational entropy penalty, the activity inhibition of HEWL by AuNP–60Pep1m is only comparable to that by AuNP–60Pep1s, much lower than that by AuNP–60Pep1 (Fig. 3A).
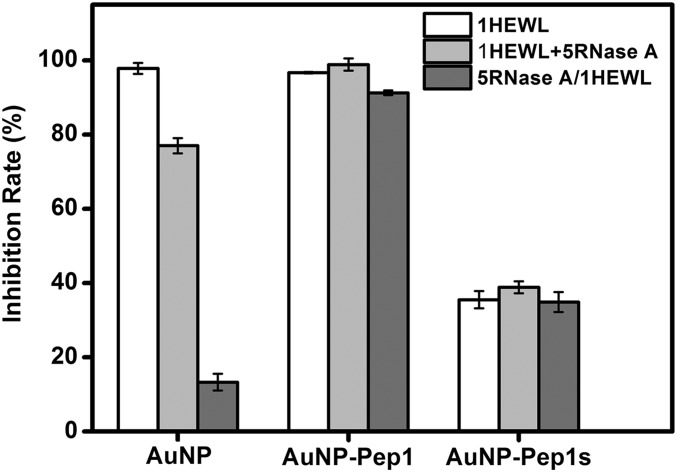
To further demonstrate that the strong binding between AuNP–60Pep1 and HEWL is a specific interaction, ribonuclease A (RNase A) from bovine pancreas was used to compete with HEWL. RNase A and HEWL have similar molecular mass and size, and both are positively charged at neutral and acidic conditions. Therefore, RNase A serves as an ideal decoy for HEWL to discriminate specific binding from nonspecific. If the binding between HEWL and AuNPs is nonspecific, incubation with the mixture of RNase A and HEWL will “dilute” or reduce the binding of HEWL on AuNPs, and the preincubation of AuNPs with RNase A may even cause covering the NP surface first by RNase A, preventing the binding of AuNPs with HEWL. This is exactly what happened for the nonfunctionalized AuNPs (Fig. 4). On the contrary, if the binding between HEWL and AuNPs was specific, RNase A would have little effect. As shown in Fig. 4, even in the presence of fivefold more RNase A than HEWL, AuNP–60Pep1 can still inhibit HEWL’s activity strongly, indicating a highly specific interaction between AuNP–60Pep1 and HEWL [see SI Appendix, Fig. S7 for dynamic light scattering (DLS) data].
Fig. 4.
Effects of RNase A on the inhibition rate of HEWL activity by nonfunctionalized AuNPs and AuNP–Pep1. Gray columns: NPs were incubated with the mixture of HEWL and RNase A (in a 1:5 ratio); dark columns: NPs were preincubated with RNase A, and then HEWL was added into the mixture. Error bars indicate SDs.
The highly specific interaction between AuNP–60Pep1 and HEWL was further characterized by surface plasmon resonance (SPR) analysis on a Biacore T200 with HEWL immobilized on a CM5 chip. RNase A and BSA were immobilized as dummy ligands on CM5 chips in different channels. To demonstrate the effect of conformational control of the grafted peptide on the binding affinity, the binding of different AuNP–20Pep1–xPep1s species with conformation tuned by adding different numbers of Pep1s is shown in Fig. 5A. The results confirm those obtained from the enzymatic activity inhibition assays (Fig. 3C), namely that the surface peptide density on AuNPs affects the binding affinity of AuNP–Pep1 with HEWL, with an optimal density corresponding to 60 peptides per AuNP (3.6 nm).
Fig. 5.
SPR characterization of the specific interaction between AuNP–Pep1 and HEWL. (A) Binding of AuNPs functionalized with 20 Pep1 and different numbers of Pep1s per AuNP onto the immobilized HEWL. (B) SPR kinetics of the interaction between AuNP–60Pep1 and immobilized HEWL. Red curves are the fitting lines (see SI Appendix, Fig. S8A for the residue plot). (C) SPR binding of AuNP–60Pep1 (red), AuNP–60Pep1s (green), AuNP–60Pep1m (blue), and free Pep1 (cyan) with the same “peptide concentration” onto the immobilized HEWL, RNase A, and BSA. (D) SPR binding of AuNP–60Pep1 and AuNP–60Pep1s onto the immobilized HEWL in the absence (red) and presence of 0.5 mM (green) or 5 mM (blue) Tri-NAG. Error bars indicate SDs. RU, resonance unit.
The kinetics of the binding between the anti-lysozyme Goldbody and HEWL was also measured by SPR, with HEWL immobilized on a CM5 chip at a very low level so that the SPR specific binding data could be fitted with the simple 1:1 model. As shown in Fig. 5B, the fitting gives an apparent affinity (KD) of 1.5 × 10−10 M, with a kon of 2.0 × 106 M−1⋅s−1 and a koff of 2.9 × 10−4 s−1. This apparent affinity is two orders of magnitude stronger than that of the original cAb-Lys3–HEWL interaction (KD 2∼5 × 10−8 M) (26, 36). Fitting SPR kinetic data is complicated, and the accuracy of the fitting is dependent on the system and the fitting models (37). In a strict sense, systems involving AuNPs are generally heterogeneous, due to the size distribution of AuNPs. Nevertheless, the kinetic fitting is quite good (see SI Appendix, Fig. S8A for residue plots), and the two-order-of-magnitude stronger affinity than that of the original antibody is definitely far more than the possible fitting errors. Therefore, the strong binding unambiguously indicates that our reconstruction of the conformation and activity of the CDR on AuNPs is successful. It should be pointed out that ideally the binding affinity for a single binding site (one CDR3 peptide on an AuNP) should be comparable to that of the original antibody, and therefore the much stronger apparent affinity of Goldbody is likely due to the avidity effects or the multivalency effects, which accounts for the slow dissociation processes (Fig. 5B).
It is worthwhile to mention that grafting protein loops onto different protein scaffolds usually results in weaker binding affinities, due to local conformational change affected by long-range interactions (38). For example, the same CDR3 loop of cAb-Lys3 has been grafted onto different proteins, and grafted protein–HEWL interactions are several orders of magnitude weaker than the original cAb-Lys3–HEWL interaction (21, 22), making the high affinity between the anti-lysozyme Goldbody and HEWL even more remarkable.
Except for AuNP–60Pep1, other species (free Pep1, AuNP–60Pep1m, and AuNP–60Pep1s) only display weak nonspecific interactions with HEWL. Thus, only qualitative data were obtained from these systems in the SPR experiments (Fig. 5C). As expected, in the RNase A and BSA channels, all species including AuNP–60Pep1 only show weak nonspecific binding.
The anti-lysozyme Goldbody was designed to specifically bind into the active cleft of HEWL, as shown in Fig. 2B. To determine whether or not the binding site of the anti-lysozyme Goldbody is the active cleft of HEWL, we used Tri-NAG, an inhibitor of HEWL that binds into the same cleft of HEWL like cAb-Lys3 (Fig. 2C) (39), as a competing reagent in the SPR experiment. As shown in Fig. 5D and SI Appendix, Fig. S9, Tri-NAG does not influence the nonspecific binding between HEWL and AuNP–60Pep1s; however, Tri-NAG does suppress the binding of AuNP–60Pep1 to HEWL, indicating that AuNP–60Pep1 and Tri-NAG compete for the same binding site on HEWL, that is, the anti-lysozyme Goldbody does bind to the active cleft of HEWL as designed.
Design and Synthesis of an Anti-EGFR Goldbody.
To prove that our Goldbody design strategy is generally applicable to create artificial antibodies against different antigens, we chose EGFR as our next target. EGFR is overexpressed in many epithelial tumors, and thus has been an extensively studied target of antitumor treatment (40). Several anti-EGFR monoclonal antibodies, including cetuximab, have been approved for the treatment of various malignant tumors by the Food and Drug Administration.
We chose 7D12, a heavy chain-only antibody that binds to domain III of the extracellular soluble part of EGFR (sEGFR) (Fig. 6A) (41), as the natural antibody to be reproduced as the anti-EGFR Goldbody. Different from the cAb-Lys3–lysozyme interaction mentioned above, which has a CDR loop penetrating into the active cleft of lysozyme, the interaction between 7D12 and sEGFR is a typical antigen–antibody interaction with a relatively flat interface. Following the same strategy, we grafted the CDR3 of 7D12, the major binding loop, onto AuNPs (3.6 nm). Similarly, Pep2 was designed for the anti-EGFR Goldbody, and Pep2m and Pep2s were two control peptides for displaying conformational control and binding specificity, respectively (see Fig. 6B for their sequences). Fig. 6C shows the binding model for the designed anti-EGFR Goldbody with sEGFR.
Fig. 6.
Scheme of the design of the anti-EGFR Goldbody. (A) Structure of the antibody 7D12 (cyan, with CDR3 shown in magenta) in complex with domain III of sEGFR (green) (PDB ID code 4KRL). (B) Sequences of CDR3 of 7D12 and synthesized peptides. (C) Model of the anti-EGFR Goldbody (with the grafted peptide loop shown in magenta) in complex with sEGFR (domain III shown in green, with domains I, II, and IV in gray).
Strong Binding Affinity and Specificity of the Anti-EGFR Goldbody Toward sEGFR.
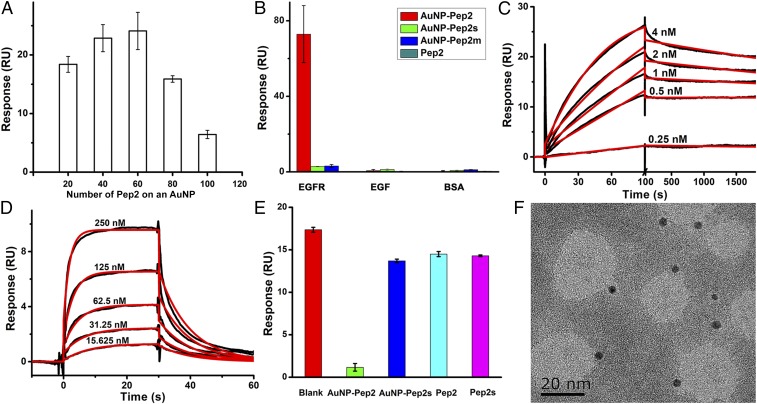
The interaction between the anti-EGFR Goldbody (AuNP–Pep2) and sEGFR was quantitatively investigated by SPR. Fig. 7A shows the binding between AuNPs functionalized with different numbers of Pep2 and the immobilized sEGFR, suggesting that 40∼60 Pep2 per AuNP (3.6 nm) is the optimal peptide density for reconstruction of the binding conformation (considering that more peptides on AuNPs means more multivalency effects, the optimal density would be close to 40). Since the original span of Pep1 in cAb-Lys3 is about 1.1 nm (26) and the original span of Pep2 in 7D12 is about 1.3 nm (42), the difference in optimal density for AuNP–Pep1 and AuNP–Pep2 is thus in reasonably good agreement with the peptide spans in the original antibodies, suggesting that changing peptide density on AuNPs does change the span of peptides on the AuNP surface. For the convenience of comparison with the previous results, 60 Pep2 per AuNP (3.6 nm) were used for the following experiments.
Fig. 7.
Interaction between the anti-EGFR Goldbody and sEGFR at the molecular level. (A) Binding of AuNPs functionalized with different numbers of Pep2 onto immobilized sEGFR. (B) Binding of AuNP–Pep2 (red), AuNP–Pep2s (green), AuNP–Pep2m (blue), and free Pep2 (cyan) onto immobilized sEGFR, EGF, and BSA. (C) SPR kinetics of the interaction between AuNP–Pep2 and immobilized sEGFR. Red curves are the fitting lines (see SI Appendix, Fig. S8B for the residue plot). (D) SPR kinetics of the interaction between EGF and immobilized sEGFR. Red curves are the fitting lines (see SI Appendix, Fig. S8C for the residue plot). (E) Binding of sEGFR onto immobilized EGF in the absence (red) or the presence of AuNP–Pep2 (green), AuNP–Pep2s (blue), free Pep2 (cyan), or free Pep2s (magenta). (F) TEM image of the 1:1 complex of anti-EGFR Goldbody (dark balls) and sEGFR. Error bars indicate SDs. RU, resonance unit.
As demonstrated in Fig. 7B, the binding of AuNP–60Pep2 with sEGFR is specific (see SI Appendix, Fig. S7 for DLS data). AuNP–60Pep2s, AuNP–60Pep2m, and free Pep2 only show weak nonspecific binding with sEGFR. In the two control channels immobilized with EGF or BSA, respectively, all species including AuNP–60Pep2 show only weak nonspecific binding.
Similar to the previous example, the kinetics of the binding between the anti-EFGR Goldbody and sEGFR was determined with sEGFR immobilized on a CM5 chip at a very low level, and the binding data (Fig. 7C) could be fitted with a 1:1 model, with an apparent affinity of 1.2 × 10−11 M (kon 9.0 × 106 M−1⋅s−1 and koff 1.1 × 10−4 s−1). This affinity is four orders of magnitude stronger than that of the original antibody 7D12–sEGFR interaction (KD 2.19 × 10−7 M) (42), three orders of magnitude stronger than that of the EGF–sEGFR interaction (KD 6.0 × 10−8 M) (Fig. 7D), and two orders of magnitude stronger than that of the cetuximab–sEGFR interaction [KD ranges from 2.3 × 10−9 M (43) to 8.4 × 10−9 M] (SI Appendix, Fig. S10).
AuNP–60Pep2 was designed to mimic 7D12. Due to binding sites overlapping, 7D12 can sterically block ligand (EGF) binding to EGFR (42) (see SI Appendix, Fig. S11 for their binding models). A successful reconstruction of the CDR3 of 7D12 on AuNPs would make AuNP–60Pep2 function like 7D12 to compete with EGF for the binding with sEGFR. The competitive SPR experiments (Fig. 7E) demonstrate that AuNP–60Pep2 does inhibit the EGF–sEGFR interaction, while other species do not.
The large size of sEGFR (with a molecular mass of 110 kDa plus glycosylation) makes it possible to distinguish the binding between the anti-EGFR Goldbody and sEGFR by TEM. As shown in Fig. 7F, the TEM image clearly shows 1:1 anti-EGFR Goldbody–sEGFR complexes.
Interaction of the Anti-EGFR Goldbody with Cells and Its Inhibition of EGF-Induced Cell Proliferation.
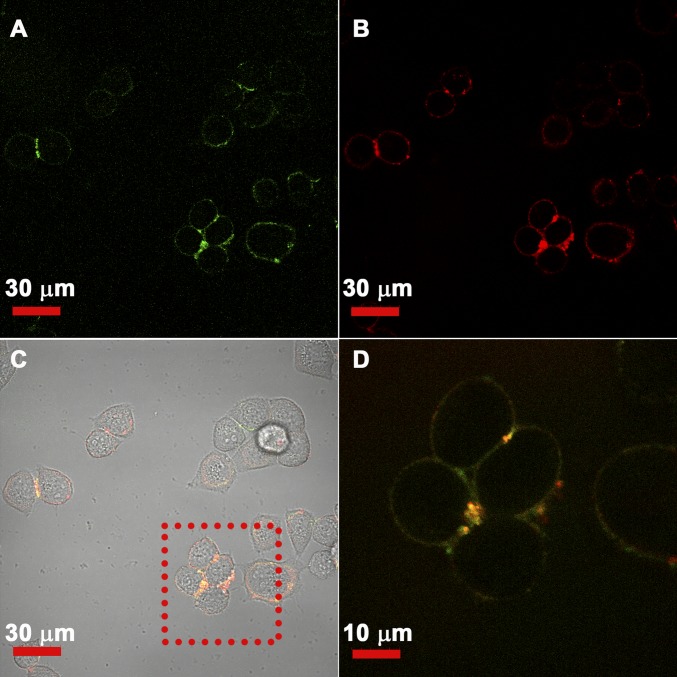
To verify the interaction between the anti-EGFR Goldbody and EGFR at the cellular level, FITC-labeled anti-EGFR Goldbody and Cy5-labeled natural anti-EGFR antibody were cocultured with HeLa cells, a human cervix adenocarcinoma cell line with EGFR expression on the membrane, and then investigated by confocal fluorescence microscopy. As shown in Fig. 8, the green and red fluorescences overlap very well on the membranes of HeLa cells, especially those bright spots where high copies of EGFR might be expressed. This result unambiguously demonstrates that our anti-EGFR Goldbody indeed binds to the EGFR on the cell membrane.
Fig. 8.
Colocalization of the anti-EGFR Goldbody and natural anti-EGFR antibody demonstrated by confocal fluorescence images. (A) Green fluorescence (FITC) image showing the location of the anti-EGFR Goldbody on the membrane of HeLa cells. (B) Red fluorescence (Cy5) image showing the location of the natural anti-EGFR antibody. (C) Colocalization of the anti-EGFR Goldbody and natural anti-EGFR antibody (yellow) with cells in a bright-field image. (D) Enlarged image of the square area in C showing the overlap of green and red fluorescence.
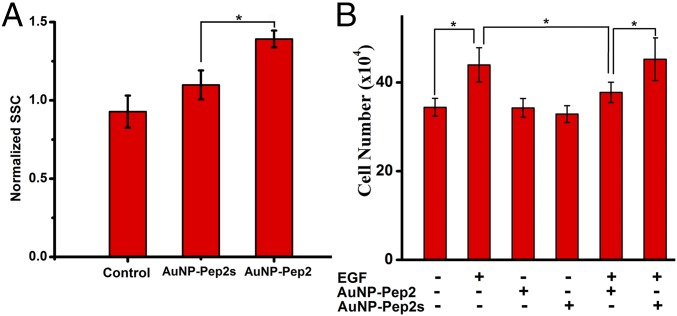
To provide statistically significant evidence, flow cytometry was used to investigate the different binding with HeLa cells between AuNP–60Pep2 and the nonspecific control AuNP–60Pep2s. The incorporation of AuNPs into cells may induce the increase of the granularity of the cells, which could be reflected by the increased intensity of the side scatter parameter (SSC). As shown in Fig. 9A (see also SI Appendix, Fig. S12), the granularity of AuNP–60Pep2–treated cells is significantly larger than that of AuNP–60Pep2s–treated cells, which do not differ significantly from the control cells without AuNP treatment. In addition, the anti-EGFR Goldbody on the HeLa cell membrane could also be identified by TEM (SI Appendix, Figs. S13 and S14).
Fig. 9.
Interaction between the anti-EGFR Goldbody and EGFR at the cellular level. (A) Normalized flow cytometry SSC of control cells, cells incubated with AuNP–Pep2s, and cells incubated with the anti-EGFR Goldbody, respectively. (B) Inhibition of EGF-induced cell proliferation by the anti-EGFR Goldbody. Error bars indicate SDs. *P < 0.05.
To show the potential biological functions and applications, the inhibition of EGF-induced cell proliferation by the anti-EGFR Goldbody was tested by counting cell numbers. Neither the anti-EGFR Goldbody nor AuNP–60Pep2 influenced the morphology and proliferation of HeLa Cells. But, as shown in Fig. 9B, the anti-EGFR Goldbody does significantly inhibit EGF-induced cell proliferation, while the AuNP–60Pep2s control does not. This result is consistent with the previous SPR results that the anti-EGFR Goldbody can inhibit the binding of EGF to EGFR (Fig. 7E). Combination of all these results demonstrates the successful reconstruction of the conformation and function of the CDR3 loops of 7D12 on AuNPs.
Thermal Stability of Goldbodies.
Conventional antibodies have been widely used as clinical therapeutics, including the recent breakthrough of the “cancer immunogram” (44, 45), as well as indispensable workhorses of biological experiments (46). However, there are serious problems with conventional antibodies, in that some of them may unfold and lose their activity during manufacture and storage processes, causing problems including safety, problematic reproducibility (46), and loss of activity, particularly at high temperatures. Many conventional antibodies are produced by animals, thus also raising the animal welfare problem (47). Besides, clinical antibodies usually have to be humanized to reduce the risk of immunogenicity. For Goldbodies, the peptide sequences only involve the essential short fragments needed for binding, thus minimizing the humanization problem. AuNPs are solid particles, so that the scaffolds do not collapse under protein-denaturing conditions. This is why Goldbodies can be stored at room temperature. As demonstrated in SI Appendix, Fig. S15, Goldbodies can maintain their binding capabilities even after being boiled at 100 °C for 1 h. The excellent thermal stability of AuNPs not only benefits storage but also reliably maintains their binding specificity, avoiding the problematic reproducibility of conventional antibody products.
Discussion
Here, we demonstrate that the active conformation of the CDR loops of natural antibodies can be reconstructed on AuNPs to create Goldbodies, which bind the corresponding antigens specifically with apparent affinities several orders of magnitude stronger than those of the original natural antibodies. Up to now, both NP functionalization (4–8) and protein mimicking (48) have relied on the incorporation of functional groups. Herein we show that new functions of NPs can be created by controlling the conformation of the surface groups on NPs. Self-assembled monolayers on gold surfaces have shown certain molecular density effects on the packing of the immobilized groups (49–51), yet those self-assembling methods aim to create certain “macro” patterns consisting of numerous chains. Precisely manipulating conformations of individual groups for specific functions is still beyond reach. The seemingly insurmountable task of tuning the conformation of individual peptides for specific interactions is accomplished here by anchoring both ends of the peptide on AuNPs and tuning the span of the two ends by optimizing peptide density on the surface.
There are two critical factors for the success of our approach. One is the mobility of Au–S bonds on the surface of AuNPs, which makes it possible to tune the peptide span on AuNPs close to the native span in the original proteins by changing the peptide density on the AuNP surface. The other, which makes it possible for the peptide to adopt the right conformation when the peptide span on the AuNP surface is tuned right, is that the native conformation of proteins is likely the most stable one among its enormous conformational space, which might be called the “localized” Anfinsen’s dogma (52). It was reported that some isolated peptide fragments of natural proteins were more likely to adopt their native conformation (53). We also reported previously that by covalently anchoring multiple surface groups of proteins to rigid silica shells, the stability of silica-encapsulated proteins could be significantly enhanced (54–56). The enhanced stability is likely attributed to those anchor positions keeping the fragments of proteins in the right spans, that is, by keeping the right spans for individual fragments during denaturation, all those fragments (and hence the whole protein) would automatically adopt their native conformation after removing the denaturing conditions. This preference of certain conformations for peptide fragments has also been successfully used for protein structure prediction (57) and design (11). Most isolated peptide fragments are flexible and do not adopt their “native” conformations, because without the constraints in the original proteins, the enthalpy favor for the native conformation of the free peptide cannot compensate the entropy disfavor over the enormous number of other conformations. Therefore, the fundamental mechanism for our successful tuning of peptide conformation is mainly entropic (58). Tuning the peptide span on AuNPs to its native one can dramatically reduce the conformational space of the anchored peptides by excluding the huge conformational space corresponding to all other peptide spans, thus achieving an entropic effect and making the enthalpically favored native/active conformation more stable. In other words, our results imply that the so-called peptide fragments’ structural preference is actually based on the right peptide span. Thus, our approach might be used as a tool to tailor the structure and understand the folding of natural proteins.
It has to be pointed out that, in the absence of binding partners, the peptides on AuNP surfaces are not fixed in the exact binding conformation but in the proximity of the binding conformation with certain flexibility (corresponding to the red circle in Fig. 1) that is ready for mutually induced complementation with its binding partners. In fact, the association processes (Figs. 5B and 7C) of Goldbodies seem a little bit slower than expected for such strong binding, implying the possible existence of induced conformational fitting for binding. This restricted flexibility and induced fitting of Goldbodies resemble those of natural antibodies (59–61), and are unique features that distinguish natural proteins from most synthesized materials. The spatial positions of the active residues of proteins are defined, but not as confined as those in crystalline and amorphous inorganic materials, which are unable to adjust their shape to maximize interface for binding. Also, the active residues of proteins are certainly not too flexible as those in long organic/polymer chains, causing a big entropic penalty when they are fixed in a unique binding conformation. In short, our Goldbodies behave like natural proteins, and the restricted flexibility of the CDR loops on AuNPs makes it possible to put the right groups/atoms in the right places at the right time.
Given the apparent merits (strong affinity and stability, straightforward design) of Goldbodies, our strategy could be used to create other Goldbodies for various applications. Different from other approaches that select functional peptides from a large random library, the design of Goldbodies is rational, simple, and straightforward, namely just choosing the CDR peptides from known natural antibodies and covalently linking both their terminals to AuNPs. Our strategy is certainly not restricted to antibody–antigen interactions but might be applicable to various protein interactions, including ligands–receptors. The grafting peptide fragments could be chosen either from known protein–protein complex structures or information identified by biochemical experiments, or even by bioinformatics methods. Additionally, the NP platforms make it easy to create multispecific Goldbodies. For an antigen, there are many different natural antibodies. We can envisage that for many natural antibodies, there would be a corresponding artificial substitution—Goldbody.
Materials and Methods
Materials.
Chloroauric acid, sodium tetrahydroborate, and trisodium citrate dehydrate were purchased from Sinopharm Chemical Reagent. HEWL, RNase A, and BSA were obtained from Sigma-Aldrich. sEGFR and EGF were obtained from Creative BioMart. Cetuximab was obtained from Shanghai TheraMabs Biotech. Mouse anti-EGFR/Cy5 was obtained from Bioss Antibodies. Tri-N-acetyl-d-glucosamine was obtained from Aladdin Reagent. FITC-PEG [polyethylene glycol; molecular weight (MW) 5K]-SH was obtained from Qian-Bi Bio-Tech (Shanghai). Peptides were synthesized by GL Biochem. N-hydroxysuccinimide, N-ethyl-N-(3-diethylaminopropyl) carbodiimide hydrochloride, ethanolamine⋅HCl (1 M solution, pH 8.5), HBS-EP buffer [10 mM Hepes, 150 mM NaCl, 3 mM EDTA, 0.05% (vol/vol) surfactant P20, pH 7.4], and series S CM5 chips were obtained from GE Healthcare. Ultrapure Millipore water was used. All other chemicals were of analytical grade.
Synthesis of AuNPs.
All glassware was washed with aqua regia and rinsed with plenty of ultrapure water. To synthesize AuNPs with a diameter of ∼3.6 nm, 1 mL 25 mM chloroauric acid was diluted with 98.36 mL of water, and then 0.65 mL 39.47 mM sodium citrate was added. After the mixture was stirred for 1 min, 0.5 mL 0.075 wt % freshly prepared sodium tetrahydroborate in 39.47 mM sodium citrate was added while stirring. The reaction mixture was stirred for 5 min to complete the reaction. The as-prepared AuNPs were stored at room temperature in the dark. The size of the AuNPs was calculated from Aspr/A450 nm of the UV/vis spectrum (24). The concentration of the as-prepared 3.6-nm AuNPs was typically around 75 nM, as calculated from A450 nm/ε450 nm using an ε450 nm of 2.768 × 106 M−1⋅cm−1.
To synthesize AuNPs with a diameter of ∼6.9 nm, 150 mL 2.2 mM sodium citrate aqueous solution was heated in an oil bath at 160 °C, and then 0.936 mL 25 mM chloroauric acid aqueous solution was added quickly under vigorous stirring. The mixture was kept at 160 °C until its color became a stable wine red. Finally, the solution was cooled in ice water to obtain AuNPs. The concentration of the as-prepared 6.9-nm AuNPs was typically around 9.4 nM, as calculated from A450 nm/ε450 nm using an ε450 nm of 2.03 × 107 M−1⋅cm−1.
AuNPs with a diameter of ∼15.0 nm were also prepared by the reduction of chloroauric acid with trisodium citrate. An aqueous solution of trisodium citrate (150 mL, 2.2 mM) was heated at 120 °C with a reflux apparatus, and then 0.936 mL 25 mM chloroauric acid was quickly added under stirring. The reaction was kept at 120 °C until the color of the solution became a wine red. The solution was cooled in ice water to obtain the AuNPs. The concentration of the as-prepared 15.0-nm AuNPs was typically around 0.7 nM, as calculated from A450 nm/ε450 nm using an ε450 nm of 2.18 × 108 M−1⋅cm−1.
Preparation of AuNP–Pep.
The concentration of peptides was calculated using their UV/vis spectra according to their calculated extinction coefficient at 280 nm (web.expasy.org/protparam/). The pH of the AuNP solution was adjusted to 7.4 using 0.2 M trisodium citrate. The peptide dissolved in phosphate buffer (PB) solution (0.01 M, pH 7.4) was added dropwise to the AuNP solution and stirred for 1 h at room temperature. The synthesized AuNP–Pep was purified and concentrated by centrifugal filtration (Millipore Amicon Ultra-15 filter, MW cutoff 10K, 3,000 × g, 20 min, 4 °C). The conjugation of the peptide with Cys to the citrate-passivated AuNPs was efficient within the concentration range in our experiments. SI Appendix, Fig. S3 shows the UV absorbance and fluorescence spectra of the total Pep1 in the reaction solution and the Pep1 in the filtrate of the reaction solution by centrifugal filtration (Millipore Amicon Ultra-15 filter, MW cutoff 10K, 3,000 × g, 30 min, 4 °C). It is clear that free Pep1 is undetectable after the reaction, demonstrating the high efficiency of this conjugation reaction. Therefore, the average number of peptides on one AuNP can be simply adjusted by adjusting the concentration of the reactants of AuNPs and peptides. For example, the preparation of AuNP–5Pep1, AuNP–10Pep1, AuNP–15Pep1, AuNP–20Pep1, AuNP–40Pep1, AuNP–60Pep1, AuNP–80Pep1, and AuNP–100Pep1 was carried out simply by adding dropwise 1 mL 2.25, 4.5, 6.75, 9, 18, 27, 36, and 45 μM Pep1, respectively, to 6 mL 75 nM AuNPs. Similarly, the preparation of AuNP–20Pep2, AuNP–40Pep2, AuNP–60Pep2, AuNP–80Pep2, and AuNP–100Pep2 was carried out simply by adding dropwise 1 mL 9, 18, 27, 36, and 45 μM Pep2, respectively, to 6 mL 75 nM AuNPs.
Tuning the Conformation of Pep1 on AuNPs by Adjusting the Surface Peptide Density.
To reliably quantify the effect of surface peptide density on the activities of the Goldbodies (which reflect the conformation of the active peptide), we kept the number of active Pep1 per AuNP (3.6 nm) at 20 for all comparing species, and tuned the conformation of Pep1 (and hence its activity) by adding different numbers of inactive Pep1s. Therefore, AuNP–20Pep1, AuNP–20Pep1–10Pep1s, AuNP–20Pep1–20Pep1s, AuNP–20Pep1–30Pep1s, AuNP–20Pep1–40Pep1s, AuNP–20Pep1–50Pep1s, AuNP–20Pep1–60Pep1s, AuNP–20Pep1–70Pep1s, and AuNP–20Pep1–80Pep1s were synthesized by adding dropwise 1 mL of peptide solution containing 9 μM Pep1 as well as 0, 4.5, 9, 13.5, 18, 22.5, 27, 31.5, and 36 μM Pep1s, respectively, to 6 mL 75 nM AuNPs, following the above procedure. For comparison, a different AuNP–20Pep1–40Pep1s species was synthesized by adding dropwise 0.5 mL 18 μM Pep1 to 6 mL 75 nM AuNPs and, 1 h later, adding dropwise 0.5 mL 36 μM Pep1s to the above mixture.
Fluorescence Labeling of the Anti-EGFR Goldbody: Synthesis of 30FITC–PEG–AuNP–40Pep2.
To introduce fluorescence to the anti-EGFR Goldbody, AuNPs were conjugated with a mixture of 40 Pep2 and 30 FITC–PEG (MW 5K)-SH, following the above procedure.
Characterization of AuNPs and AuNP–Pep.
The size and surface morphology of AuNPs and AuNP–Pep were characterized by high-resolution (HR)TEM (JEM-2100F; JEOL) and UV/vis spectra (U-3010; Hitachi). The hydrodynamic diameters of nonfunctionalized AuNPs (40 nM in 0.01 M PB, pH 7.4), Goldbodies (40 nM in 0.01 M PB, pH 7.4), and Goldbodies in the presence of their corresponding antigens or control proteins were also measured with a nanosizer (DLS; Malvern ZS90) at 25 °C.
Enzymatic Activity Assay of Lysozyme.
The enzymatic activity of HEWL was determined by the dynamic absorbance at 450 nm of Micrococcus lysodeikticus digested by HEWL. The enzymatic process was recorded using a UV/vis spectrophotometer (U-3010; Hitachi) immediately after mixing HEWL with M. lysodeikticus for 3 min, and the slope of the curve of absorbance versus time represents the activity of HEWL. The relative activities of HEWL in the presence of various inhibitors are presented as the ratio of the corresponding slopes to the slope of free HEWL, and the inhibition rate was calculated as the percentage of relative activity loss. The M. lysodeikticus suspension was freshly prepared by adding 10 mg of dried M. lysodeikticus to 30 mL of PB buffer (0.1 M, pH 6.2). HEWL (1.5 × 10−7 M) was dissolved in PB buffer (0.01 M, pH 7.4). In a typical assay, 0.5 mL of the above HEWL solution was added to 1 mL 60 nM AuNP–Pep and mixed well for 1 min. Then 1 mL of the above M. lysodeikticus suspension was added to the mixture. After vigorous shaking, the mixture was quickly transferred to a cuvette for absorbance measurement. All samples were kept and assays conducted at 25 °C.
IC50 Determination.
The IC50 of AuNP–60Pep1 to inhibit HEWL was determined by measuring the enzymatic activity of 30 nM (final concentration) HEWL in the presence of 0, 0.3, 0.6, 1.2, 1.8, 2.4, 3.0, 4.0, 8.0, 12, 16, 20, 24, or 44 nM (final concentration) AuNP–60Pep1, following the above assay procedure. The IC50 of Tri-NAG to inhibit HEWL was determined by measuring the enzymatic activity of 30 nM (final concentration) HEWL in the presence of 0, 0.2, 0.4, 1.0, 2.0, 4.0, 8.0, 12, 20, 50, or 60 mM (final concentration) Tri-NAG, following the above procedure. The obtained data were plotted with the activity vs. the logarithm of inhibitor concentration, and IC50 is the concentration where the activity (or inhibition) is 50%.
SPR Experiments.
All SPR experiments were carried out at 25 °C on a Biacore T200 instrument (GE Healthcare). HBS EP buffer was selected as the running buffer. HEWL, sEGFR, RNase A, BSA, and EGF were coupled to different channels of series S CM5 sensor chips by a standard amine coupling procedure.
To investigate the binding specificity of the anti-lysozyme Goldbody, 6 nM AuNP–60Pep1, 6 nM AuNP–60Pep1s, 6 nM AuNP–60Pep1m, or 360 nM free Pep1 in running buffer was injected into the HEWL-, RNase A-, or BSA-immobilized channels, respectively, at a flow rate of 30 μL/min. To investigate the binding specificity of the anti-EGFR Goldbody, 6 nM AuNP–60Pep2, 6 nM AuNP–60Pep2s, 6 nM AuNP–60Pep2m, or 360 nM free Pep2 in running buffer was injected into the EGFR-, EGF-, or BSA-immobilized channels, respectively, at a flow rate of 30 μL/min.
To investigate the competitive binding to HEWL between AuNP–60Pep1 and Tri-NAG, 6 nM AuNP–Pep1 or AuNP–Pep1s in the presence of different concentrations of Tri-NAG was injected into the HEWL-immobilized CM5 chip channel at a flow rate of 30 μL/min. To investigate the competitive binding to EGFR between AuNP–60Pep2 and EGF, 200 nM EGFR in the absence and presence of 10 nM AuNP–60Pep2, 10 nM AuNP–60Pep2s, 600 nM free Pep2, or 600 nM free Pep2s was injected into the EGF-immobilized channel at a flow rate of 30 μL/min.
For the kinetic study, HEWL or sEGFR was immobilized onto CM5 chips at a low level (about 30% of the recommended amount for the kinetic study), so that the binding kinetics could be fitted with a simple 1:1 model. AuNP–Pep1, AuNP–Pep2, EGF, or cetuximab with different concentrations in running buffer was injected into the HEWL or sEGFR channel at a flow rate of 30 μL/min. For the anti-HEWL Goldbody, glycine⋅HCl (pH 1.5) was used as the regeneration buffer, while for the anti-EGFR Goldbody, two regeneration steps were used with glycine⋅HCl (pH 1.5) and NaOH (20 mM), respectively. The data were fitted with different binding models, and the 1:1 model gave the best fitting for all our kinetic studies.
Cell Culture.
HeLa cells (human epithelial cervical cancer) were obtained from the Cell Bank of the Chinese Academy of Sciences. HeLa cells were grown in high-glucose DMEM (4.5 g/L glucose) supplemented with 10% FBS (PAN-Biotech) and 1% penicillin/streptomycin. Cells were cultured at 37 °C in a humidified atmosphere with 5% CO2 and 95% air.
Flow Cytometry Analysis of Binding of the Anti-EGFR Goldbody to HeLa Cells.
HeLa cells (2.8 × 105 cells per well) were seeded in six-well plates and incubated for 24 h. Then cells were incubated in culture medium (FBS free) containing 25 nM AuNP–Pep2 or 25 nM AuNP–Pep2s for 1.5 h. After that, the cells were washed twice with ice-cold D-Hanks’ solution (containing 8.0 mg/mL of NaCl, 0.4 mg/mL of KCl, 0.134 mg/mL of Na2HPO4⋅12H2O, 0.06 mg/mL of KH2PO4, and 0.35 mg/mL of Na2HCO3, pH 7.4), trypsinized, suspended in medium (FBS free), and analyzed by flow cytometry to obtain the percentage of cells incorporating AuNP–Pep2 and AuNP–Pep2s. The attachment of AuNP–Pep2 or AuNP–Pep2s to cells induces the increase of the granularity of cells, which can be reflected by increased SSC intensity. There was no significant difference between the control cells and cells treated with AuNP–Pep2s, but cells treated with AuNP–Pep2 showed very significantly increased SSC, indicating the binding of AuNP–Pep2 to HeLa cells.
TEM and Energy-Dispersive Spectroscopy Analysis of Binding of the Anti-EGFR Goldbody to HeLa Cells.
TEM was used to investigate the binding site of the anti-EGFR Goldbody on HeLa cells. After incubation of HeLa cells in the presence of the anti-EGFR Goldbody, thin sections of cells were prepared and investigated under TEM. Briefly, HeLa cells were seeded in 25-cm2 culture flasks and incubated for 24 h. Anti-EGFR Goldbody (AuNP–Pep2) at a concentration of 25 nM was introduced to the culture media (FBS free). After a 1.5-h exposure, the cells were washed three times with ice-cold D-Hanks’ solution and scraped off. After centrifugation (335 × g, 3 min), cells were collected, prefixed with 2.5% glutaraldehyde, postfixed in 1% osmium tetroxide, dehydrated in a graded alcohol series, embedded in epoxy resin, and cut with an ultramicrotome. Thin sections poststained with toluidine blue and lead citrate were imaged by biology TEM (Tecnai G2 Spirit BioTWIN; FEI), and the same sample was inspected by energy-dispersive spectroscopy on an HRTEM instrument (Talos F200X; FEI) to detect AuNPs.
Confocal Fluorescence Imaging of the Colocalization of the Anti-EGFR Goldbody and Natural Anti-EGFR Antibody to HeLa Cells.
HeLa cells (6 × 104 cells per well) were seeded in four-well Lab-Tek chambers (Thermo Fisher Scientific) and incubated for 24 h in a humidified incubator at 37 °C. The cells were first incubated in culture medium (FBS free) containing 40 nM FITC-labeled anti-EGFR Goldbody (30FITC–PEG–AuNP–40Pep2) for 1.5 h. Then, the cells were incubated with 80 nM Cy5-labeled natural anti-EGFR antibody for 15 min. After that, the supernatant was discarded and 500 μL of cold D-Hanks’ buffer was added to each well. The chambers were investigated by confocal microscopy (Olympus; FM 1000), excited with 488-nm (for FITC) and 635-nm (for Cy5) lasers.
Inhibition of EGF-Induced Cell Proliferation by the Anti-EGFR Goldbody.
HeLa cells were seeded in 12-well plates (3.5 × 104 cells per well). One day later, 0.5 mL fresh serum-free culture media or 0.5 mL serum-free culture media containing AuNP–Pep2 or AuNP–Pep2s was added. One hour later, 0.5 mL fresh culture media (20% FBS) containing EGF (20 nM) was added. The cells without exposure to EGF or AuNPs were taken as the control by culturing in 1.0 mL fresh culture media (10% FBS). After culture for 72 h, cells (control, EGF control, AuNP–Pep2, AuNP–Pep2s, AuNP–Pep2 plus EGF, and AuNP–Pep2s plus EGF) were collected and counted on a blood cell counting plate under an optical microscope. Each well was counted three times. Parallel triplicate samples were tested.
Thermal Stability Assay.
AuNP–Pep1 was heated at different temperatures (60, 80, and 100 °C) in a water bath for 1 h and then cooled down to room temperature. After that, the HEWL activity assay was carried out in the presence of preheated AuNP–Pep1. The thermal stability of the Goldbodies was also analyzed by the SPR binding assay after being preheated at 100 °C for 1 h and then cooling down to room temperature.
Statistical Analysis.
All mean values were calculated from at least three independent experiments, and the SDs are shown as error bars in the corresponding figures. Statistically significant differences were determined using the Student’s t test, and the differences were considered significant if P < 0.05.
Supplementary Material
Acknowledgments
We thank our former group members Zhengwei Cai, Xiaojun Ji, and Yu Jiang for their early work on this project. We thank Prof. Yuliang Zhao at the National Center for Nanoscience and Technology, China, for his valuable comments on the manuscript. This work was supported by National Key Research and Development Plan of China Grant 2016YFA0201600 and China Natural Science Foundation Grants 31571024, 21371117, and 31771105.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. N.A.K. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1713526115/-/DCSupplemental.
References
- 1.Peer D, et al. Nanocarriers as an emerging platform for cancer therapy. Nat Nanotechnol. 2007;2:751–760. doi: 10.1038/nnano.2007.387. [DOI] [PubMed] [Google Scholar]
- 2.Zhao Y. Sensing system for mimicking cancer cell–drug interaction. Sci Bull. 2015;60:1218–1219. [Google Scholar]
- 3.Mulvaney P, Weiss PS. Have nanoscience and nanotechnology delivered? ACS Nano. 2016;10:7225–7226. doi: 10.1021/acsnano.6b05344. [DOI] [PubMed] [Google Scholar]
- 4.Pan D, Turner JL, Wooley KL. Folic acid-conjugated nanostructured materials designed for cancer cell targeting. Chem Commun (Camb) 2003;19:2400–2401. doi: 10.1039/b307878g. [DOI] [PubMed] [Google Scholar]
- 5.Farokhzad OC, et al. Targeted nanoparticle-aptamer bioconjugates for cancer chemotherapy in vivo. Proc Natl Acad Sci USA. 2006;103:6315–6320. doi: 10.1073/pnas.0601755103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.El-Sayed IH, Huang X, El-Sayed MA. Selective laser photo-thermal therapy of epithelial carcinoma using anti-EGFR antibody conjugated gold nanoparticles. Cancer Lett. 2006;239:129–135. doi: 10.1016/j.canlet.2005.07.035. [DOI] [PubMed] [Google Scholar]
- 7.Weissleder R, Kelly K, Sun EY, Shtatland T, Josephson L. Cell-specific targeting of nanoparticles by multivalent attachment of small molecules. Nat Biotechnol. 2005;23:1418–1423. doi: 10.1038/nbt1159. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Tian S, Petros RA, Napier ME, Desimone JM. The complex role of multivalency in nanoparticles targeting the transferrin receptor for cancer therapies. J Am Chem Soc. 2010;132:11306–11313. doi: 10.1021/ja1043177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martinez-Veracoechea FJ, Frenkel D. Designing super selectivity in multivalent nano-particle binding. Proc Natl Acad Sci USA. 2011;108:10963–10968. doi: 10.1073/pnas.1105351108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kotov NA. Inorganic nanoparticles as protein mimics. Science. 2010;330:188–189. doi: 10.1126/science.1190094. [DOI] [PubMed] [Google Scholar]
- 11.Huang P-S, Boyken SE, Baker D. The coming of age of de novo protein design. Nature. 2016;537:320–327. doi: 10.1038/nature19946. [DOI] [PubMed] [Google Scholar]
- 12.Marcos E, et al. Principles for designing proteins with cavities formed by curved β sheets. Science. 2017;355:201–206. doi: 10.1126/science.aah7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Boyken SE, et al. De novo design of protein homo-oligomers with modular hydrogen-bond network-mediated specificity. Science. 2016;352:680–687. doi: 10.1126/science.aad8865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bhardwaj G, et al. Accurate de novo design of hyperstable constrained peptides. Nature. 2016;538:329–335. doi: 10.1038/nature19791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clackson T, Wells JA. A hot spot of binding energy in a hormone-receptor interface. Science. 1995;267:383–386. doi: 10.1126/science.7529940. [DOI] [PubMed] [Google Scholar]
- 16.Bogan AA, Thorn KS. Anatomy of hot spots in protein interfaces. J Mol Biol. 1998;280:1–9. doi: 10.1006/jmbi.1998.1843. [DOI] [PubMed] [Google Scholar]
- 17.Wells JA. Systematic mutational analyses of protein-protein interfaces. Methods Enzymol. 1991;202:390–411. doi: 10.1016/0076-6879(91)02020-a. [DOI] [PubMed] [Google Scholar]
- 18.Lafont V, Schaefer M, Stote RH, Altschuh D, Dejaegere A. Protein-protein recognition and interaction hot spots in an antigen-antibody complex: Free energy decomposition identifies “efficient amino acids.”. Proteins. 2007;67:418–434. doi: 10.1002/prot.21259. [DOI] [PubMed] [Google Scholar]
- 19.Harper JW, Vallee BL. A covalent angiogenin/ribonuclease hybrid with a fourth disulfide bond generated by regional mutagenesis. Biochemistry. 1989;28:1875–1884. doi: 10.1021/bi00430a067. [DOI] [PubMed] [Google Scholar]
- 20.Raines RT, Toscano MP, Nierengarten DM, Ha JH, Auerbach R. Replacing a surface loop endows ribonuclease A with angiogenic activity. J Biol Chem. 1995;270:17180–17184. doi: 10.1074/jbc.270.29.17180. [DOI] [PubMed] [Google Scholar]
- 21.Nicaise M, Valerio-Lepiniec M, Minard P, Desmadril M. Affinity transfer by CDR grafting on a nonimmunoglobulin scaffold. Protein Sci. 2004;13:1882–1891. doi: 10.1110/ps.03540504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kiss C, et al. Antibody binding loop insertions as diversity elements. Nucleic Acids Res. 2006;34:e132. doi: 10.1093/nar/gkl681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu S, et al. Nonnatural protein–protein interaction-pair design by key residues grafting. Proc Natl Acad Sci USA. 2007;104:5330–5335. doi: 10.1073/pnas.0606198104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Haiss W, Thanh NTK, Aveyard J, Fernig DG. Determination of size and concentration of gold nanoparticles from UV-vis spectra. Anal Chem. 2007;79:4215–4221. doi: 10.1021/ac0702084. [DOI] [PubMed] [Google Scholar]
- 25.Hamers-Casterman C, et al. Naturally occurring antibodies devoid of light chains. Nature. 1993;363:446–448. doi: 10.1038/363446a0. [DOI] [PubMed] [Google Scholar]
- 26.Desmyter A, et al. Crystal structure of a camel single-domain VH antibody fragment in complex with lysozyme. Nat Struct Biol. 1996;3:803–811. doi: 10.1038/nsb0996-803. [DOI] [PubMed] [Google Scholar]
- 27.Kelly PM, et al. Mapping protein binding sites on the biomolecular corona of nanoparticles. Nat Nanotechnol. 2015;10:472–479. doi: 10.1038/nnano.2015.47. [DOI] [PubMed] [Google Scholar]
- 28.Cedervall T, et al. Understanding the nanoparticle-protein corona using methods to quantify exchange rates and affinities of proteins for nanoparticles. Proc Natl Acad Sci USA. 2007;104:2050–2055. doi: 10.1073/pnas.0608582104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nel AE, et al. Understanding biophysicochemical interactions at the nano-bio interface. Nat Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 30.Yang S-T, Liu Y, Wang Y-W, Cao A. Biosafety and bioapplication of nanomaterials by designing protein-nanoparticle interactions. Small. 2013;9:1635–1653. doi: 10.1002/smll.201201492. [DOI] [PubMed] [Google Scholar]
- 31.Kühnle A, Linderoth TR, Hammer B, Besenbacher F. Chiral recognition in dimerization of adsorbed cysteine observed by scanning tunnelling microscopy. Nature. 2002;415:891–893. doi: 10.1038/415891a. [DOI] [PubMed] [Google Scholar]
- 32.Yu M, et al. True nature of an archetypal self-assembly system: Mobile Au-thiolate species on Au(111) Phys Rev Lett. 2006;97:166102. doi: 10.1103/PhysRevLett.97.166102. [DOI] [PubMed] [Google Scholar]
- 33.Biener MM, Biener J, Friend CM. Sulfur-induced mobilization of Au surface atoms on Au(111) studied by real-time STM. Surf Sci. 2007;601:1659–1667. [Google Scholar]
- 34.Maksymovych P, Voznyy O, Dougherty DB, Sorescu DC, Yates JT., Jr Gold adatom as a key structural component in self-assembled monolayers of organosulfur molecules on Au(111) Prog Surf Sci. 2010;85:206–240. [Google Scholar]
- 35.Cossaro A, et al. X-ray diffraction and computation yield the structure of alkanethiols on gold(111) Science. 2008;321:943–946. doi: 10.1126/science.1158532. [DOI] [PubMed] [Google Scholar]
- 36.Transue TR, De Genst E, Ghahroudi MA, Wyns L, Muyldermans S. Camel single-domain antibody inhibits enzyme by mimicking carbohydrate substrate. Proteins. 1998;32:515–522. doi: 10.1002/(sici)1097-0134(19980901)32:4<515::aid-prot9>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 37.Rich RL, Myszka DG. Survey of the year 2007 commercial optical biosensor literature. J Mol Recognit. 2008;21:355–400. doi: 10.1002/jmr.928. [DOI] [PubMed] [Google Scholar]
- 38.Cao A, Sha Y, Lin D, Lai L, Tang Y. Influence of long-range interactions on α-helix formation. Protein Pept Lett. 1998;5:53–56. [Google Scholar]
- 39.Chipman DM, Grisaro V, Sharon N. The binding of oligosaccharides containing N-acetylglucosamine and N-acetylmuramic acid to lysozyme. The specificity of binding subsites. J Biol Chem. 1967;242:4388–4394. [PubMed] [Google Scholar]
- 40.Jia Y, et al. Overcoming EGFR(T790M) and EGFR(C797S) resistance with mutant-selective allosteric inhibitors. Nature. 2016;534:129–132. doi: 10.1038/nature17960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Roovers RC, et al. A biparatopic anti-EGFR nanobody efficiently inhibits solid tumour growth. Int J Cancer. 2011;129:2013–2024. doi: 10.1002/ijc.26145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schmitz KR, Bagchi A, Roovers RC, van Bergen en Henegouwen PM, Ferguson KM. Structural evaluation of EGFR inhibition mechanisms for nanobodies/VHH domains. Structure. 2013;21:1214–1224. doi: 10.1016/j.str.2013.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li S, et al. Structural basis for inhibition of the epidermal growth factor receptor by cetuximab. Cancer Cell. 2005;7:301–311. doi: 10.1016/j.ccr.2005.03.003. [DOI] [PubMed] [Google Scholar]
- 44.Blank CU, Haanen JB, Ribas A, Schumacher TN. The “cancer immunogram.”. Science. 2016;352:658–660. doi: 10.1126/science.aaf2834. [DOI] [PubMed] [Google Scholar]
- 45.Scott AM, Wolchok JD, Old LJ. Antibody therapy of cancer. Nat Rev Cancer. 2012;12:278–287. doi: 10.1038/nrc3236. [DOI] [PubMed] [Google Scholar]
- 46.Baker M. Reproducibility crisis: Blame it on the antibodies. Nature. 2015;521:274–276. doi: 10.1038/521274a. [DOI] [PubMed] [Google Scholar]
- 47.Reardon S. US government issues historic $3.5-million fine over animal welfare. Nature. May 20, 2016 doi: 10.1038/nature.2016.19958. [DOI] [Google Scholar]
- 48.Šácha P, et al. iBodies: Modular synthetic antibody mimetics based on hydrophilic polymers decorated with functional moieties. Angew Chem Int Ed Engl. 2016;55:2356–2360. doi: 10.1002/anie.201508642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liao W-S, et al. Subtractive patterning via chemical lift-off lithography. Science. 2012;337:1517–1521. doi: 10.1126/science.1221774. [DOI] [PubMed] [Google Scholar]
- 50.Claridge SA, et al. From the bottom up: Dimensional control and characterization in molecular monolayers. Chem Soc Rev. 2013;42:2725–2745. doi: 10.1039/c2cs35365b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Love JC, Estroff LA, Kriebel JK, Nuzzo RG, Whitesides GM. Self-assembled monolayers of thiolates on metals as a form of nanotechnology. Chem Rev. 2005;105:1103–1169. doi: 10.1021/cr0300789. [DOI] [PubMed] [Google Scholar]
- 52.Anfinsen CB. Principles that govern the folding of protein chains. Science. 1973;181:223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- 53.Milburn PJ, Scheraga HA. Local interactions favor the native 8-residue disulfide loop in the oxidation of a fragment corresponding to the sequence Ser-50-Met-79 derived from bovine pancreatic ribonuclease A. J Protein Chem. 1988;7:377–398. doi: 10.1007/BF01024887. [DOI] [PubMed] [Google Scholar]
- 54.Yang Y, et al. An individually coated near-infrared fluorescent protein as a safe and robust nanoprobe for in vivo imaging. Nanoscale. 2013;5:10345–10352. doi: 10.1039/c3nr02508j. [DOI] [PubMed] [Google Scholar]
- 55.Yang X, et al. In situ synthesis of porous silica nanoparticles for covalent immobilization of enzymes. Nanoscale. 2012;4:414–416. doi: 10.1039/c1nr11153a. [DOI] [PubMed] [Google Scholar]
- 56.Cai Z, et al. Encapsulated enhanced green fluorescence protein in silica nanoparticle for cellular imaging. Nanoscale. 2011;3:1974–1976. doi: 10.1039/c0nr00956c. [DOI] [PubMed] [Google Scholar]
- 57.Simons KT, Bonneau R, Ruczinski I, Baker D. Ab initio protein structure prediction of CASP III targets using ROSETTA. Proteins. 1999;(Suppl 3):171–176. doi: 10.1002/(sici)1097-0134(1999)37:3+<171::aid-prot21>3.3.co;2-q. [DOI] [PubMed] [Google Scholar]
- 58.Zhou H-X. Loops, linkages, rings, catenanes, cages, and crowders: Entropy-based strategies for stabilizing proteins. Acc Chem Res. 2004;37:123–130. doi: 10.1021/ar0302282. [DOI] [PubMed] [Google Scholar]
- 59.Goldberg RJ. A theory of antibody-antigen reactions. I. Theory for reactions of multivalent antigen with bivalent and univalent antibody. J Am Chem Soc. 1952;74:5715–5725. [Google Scholar]
- 60.Pauling L, Campbell DH, Pressman D. The nature of forces between antigen and antibody and of the precipitation reaction. Physiol Rev. 1943;23:203–219. [Google Scholar]
- 61.Davies DR, Cohen GH. Interactions of protein antigens with antibodies. Proc Natl Acad Sci USA. 1996;93:7–12. doi: 10.1073/pnas.93.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.