Significance
Phosphite (HPO32−) is the most energetically favorable biological electron donor known, but only one organism capable of growing by phosphite oxidation has been previously identified. Here, we describe a phosphite-oxidizing bacterium that can grow with CO2 as its sole electron acceptor, and we propose a metabolic model in which inorganic carbon is assimilated via the reductive glycine pathway. Although the reductive glycine pathway has previously been identified as a “synthetic” carbon fixation pathway, this study provides evidence that it may actually function as a natural autotrophic pathway. Our results suggest that phosphite may serve as a driver of microbial growth and carbon fixation in energy-limited environments, particularly in aphotic environments lacking alternative terminal electron acceptors.
Keywords: phosphite oxidation, carbon fixation, metagenomics, formatotrophic, reductive glycine pathway
Abstract
Dissimilatory phosphite oxidation (DPO), a microbial metabolism by which phosphite (HPO32−) is oxidized to phosphate (PO43−), is the most energetically favorable chemotrophic electron-donating process known. Only one DPO organism has been described to date, and little is known about the environmental relevance of this metabolism. In this study, we used 16S rRNA gene community analysis and genome-resolved metagenomics to characterize anaerobic wastewater treatment sludge enrichments performing DPO coupled to CO2 reduction. We identified an uncultivated DPO bacterium, Candidatus Phosphitivorax (Ca. P.) anaerolimi strain Phox-21, that belongs to candidate order GW-28 within the Deltaproteobacteria, which has no known cultured isolates. Genes for phosphite oxidation and for CO2 reduction to formate were found in the genome of Ca. P. anaerolimi, but it appears to lack any of the known natural carbon fixation pathways. These observations led us to propose a metabolic model for autotrophic growth by Ca. P. anaerolimi whereby DPO drives CO2 reduction to formate, which is then assimilated into biomass via the reductive glycine pathway.
Although phosphorus has long been recognized as an essential nutrient for life, the role of phosphorus redox chemistry in biology remains poorly understood. Most of the phosphorus on extant Earth exists as phosphate (PO43−), but recent work has detected the reduced inorganic phosphorus compound phosphite (HPO32−) in many environments, including rivers, lakes, swamps, geothermal pools, and wastewater treatment plants (1–4). Phosphite appears to be a key intermediate in the phosphorus redox cycle, accounting for up to 30% of the total dissolved phosphorus in some ecosystems (1). It can be produced naturally through geothermal reactions, volcanic eruptions, lightning discharges, and metal corrosion and can also be present in some industrial products, such as fertilizers and fungicides (3–7). A variety of microorganisms have evolved the ability to utilize phosphite as a phosphorus source by oxidizing phosphite to phosphate, a process known as assimilatory phosphite oxidation (APO) (8–17).
Phosphite is also the most energetically favorable chemotrophic electron donor known due to the extremely low redox potential of the phosphate/phosphite couple (Eo′ = −650 mV) (18). This property, together with its high solubility and chemical stability, allows it to drive cellular growth in a process known as dissimilatory phosphite oxidation (DPO) since the phosphate produced is excreted from the cells as an end product of energy metabolism rather than being incorporated into biomass (13, 19). However, the only dissimilatory phosphite-oxidizing microorganism (DPOM) described to date is Desulfotignum phosphitoxidans FiPS-3, which grows by coupling phosphite oxidation to either sulfate or CO2 reduction (19, 20). Its ability to oxidize phosphite is conferred by the ptx-ptd gene cluster. Two of these genes (ptxED) can also be found in APO bacteria (13, 14, 17, 21). PtxD is a phosphite dehydrogenase that catalyzes the NAD-dependent oxidation of phosphite to phosphate, and PtxE is a transcriptional regulator. The remaining five genes (ptdFCGHI) have so far been found only in FiPS-3 (13, 22, 23). PtdC is an inner membrane transporter that facilitates phosphite uptake, possibly by functioning as a phosphite/phosphate antiporter (22, 23). PtdFGHI are likely involved in energy conservation during DPO, but their functions have yet to be experimentally confirmed (5). Whether this gene cluster is conserved in all DPOM is uncertain.
Here, we expand our understanding of DPOM through 16S ribosomal RNA (rRNA) gene community analysis and genome-resolved metagenomics of phosphite-oxidizing enrichments from anaerobic wastewater treatment sludge grown with CO2 as the sole electron acceptor. We reveal the presence in our enrichments of an uncultivated DPOM possessing a ptx-ptd gene cluster and propose a metabolic model for phosphite oxidation coupled to CO2 reduction and carbon fixation in this organism based on genomic evidence. Our results suggest that inorganic carbon is assimilated via the reductive glycine pathway. Although this pathway has been previously proposed as a viable “synthetic” route for carbon fixation (24), this study provides evidence that it may actually function in a natural system as an autotrophic pathway.
Results
Enrichment for Dissimilatory Phosphite-Oxidizing Microorganisms from Wastewater Treatment Sludge.
To enrich for DPOM, phosphite and CO2 were added as the sole electron donor and electron acceptor, respectively, to defined anaerobic media inoculated with wastewater treatment sludge. Killed controls (autoclaved inoculum with phosphite added) and no-phosphite controls (live inoculum with no phosphite added) were also prepared. The live phosphite-fed enrichment cultures completely oxidized 10 mM phosphite after 29 wk of incubation (Fig. S1). The rate of phosphite oxidation was 50 ± 1 μM/d and was accompanied by a stoichiometric increase in the phosphate concentration in the media. No phosphite oxidation or phosphate accumulation was seen in the killed controls over the same time period (Fig. S1). DNA samples were collected at 0, 20, and 29 wk after inoculation from both the live phosphite-fed cultures and the no-phosphite controls for microbial community composition analysis. Three operational taxonomic units (OTUs) were prevalent (≥1% of the community) only in the phosphite-fed cultures: OTU 21 (Proteobacteria, 4.4 ± 2.4%), OTU 28 (Thermovirga, 1.3 ± 0.7%), and OTU 33 (Desulfomonile, 1.0 ± 1.0%) (Fig. S2).
Given the slow overall rate of phosphite oxidation in our enrichments and the presence of an OTU belonging to the Desulfomonile (OTU 33) among phosphite-enriched taxa, we decided to test variables known to enhance or inhibit growth of this organism in pure culture. The first of these was the addition of cow rumen fluid (RF) to determine if RF would stimulate the growth and rate of phosphite oxidation in our cultures. RF is a complex substrate containing various cofactors and nutrients and has been previously shown to stimulate growth of some fastidious anaerobic microbes, including Desulfomonile tiedjei DCB-1 (25–27). When we added 5% sterilized RF to the media, a substantial increase in phosphite oxidation rates was observed after several transfers. Cultures containing RF oxidized phosphite at a significantly higher rate of 480 ± 10 μM/d compared with 79 ± 36 μM/d in cultures without RF (Fig. 1A) (t test, P < 0.0001).
Fig. 1.
(A) Phosphite oxidation by cultures derived from enrichments amended with rumen fluid. Phosphite (closed symbols with solid lines) and phosphate (open symbols with dashed lines) concentrations in cultures containing 9 mM phosphite and 5% rumen fluid (blue circles); 5% rumen fluid only (red squares); 9 mM phosphite only (green triangles); 9 mM phosphite, 5% rumen fluid, and 5 mM molybdate (orange inverted triangles). Data points represent the average of triplicate cultures, with error bars denoting one SD. DNA samples for community analysis were obtained for all data points. (B–E) Taxonomic composition of microbial communities in cultures containing 9 mM phosphite and 5% rumen fluid (B); 5% rumen fluid only (C); 9 mM phosphite only (D); 9 mM phosphite, 5% rumen fluid, and 5 mM molybdate (E). Each OTU with an average normalized abundance of ≥1% of the community under any treatment is labeled according to the lowest taxonomic rank assigned to it: c, class; d, domain; f, family; g, genus; p, phylum. Each dataset represents the average of independently sequenced triplicate cultures.
In contrast to testing phosphite oxidation enhancement by RF addition, we also investigated the potential for phosphite oxidation inhibition by molybdate. Previous studies have shown that growth of D. tiedjei is inhibited by molybdate concentrations as low as 2 mM (27). When 5 mM molybdate was added to our enrichment cultures containing RF, the average phosphite oxidation rate dropped to 35 ± 12 μM/d (Fig. 1A), indicating a significant inhibitory effect compared with the cultures with no molybdate (t test, P < 0.0001). Stable RF-amended enrichments were maintained for the rest of the study. Furthermore, because CO2 was the sole potential electron acceptor in the growth media, we surmised that phosphite oxidation was coupled to CO2 reduction. Indeed, cultures grown in the presence of CO2 oxidized 8.33 ± 0.84 mM phosphite within 21 d whereas no phosphite oxidation was observed during the same period in cultures without CO2 (Fig. S3). We also confirmed that phosphite oxidation was driving growth in our enrichments; significantly higher growth rates were observed in cultures containing phosphite (0.0093 ± 0.0012 ODU/d) compared with control cultures with no phosphite (0.0030 ± 0.0015 ODU/d) (t test, P = 0.0012) (Fig. S4).
RF-amended enrichments were characterized by 16S rRNA gene community analysis of DNA samples collected at 0, 5, 11, 16, and 19 d. Just 11 OTUs accounted for 96% to 98% of the total community under all treatments (Fig. 1 B–E). Unexpectedly, OTUs 33 and 28 were not detected in any of the RF-amended cultures even though these taxa had been enriched on phosphite in the original cultures. Instead, the cultures at day 0 were dominated by two taxa: OTU 21 (Proteobacteria, 39.1 ± 2.5%) and OTU 38 (Tepidanaerobacter, 33.7 ± 2.2%) (Fig. 1 B–E). OTU 21, which was enriched on phosphite in the original cultures, was the taxon with the largest positive fold difference in final abundance (4.29 ± 0.53) on phosphite and RF compared with RF only (Fig. 1 B and C and Fig. S5). In contrast, OTU 38 had a lower relative abundance in the presence of phosphite than in the RF-only cultures.
Candidatus Phosphitivorax Anaerolimi Phox-21 Is a DPOM from an Uncultivated Deltaproteobacteria Lineage.
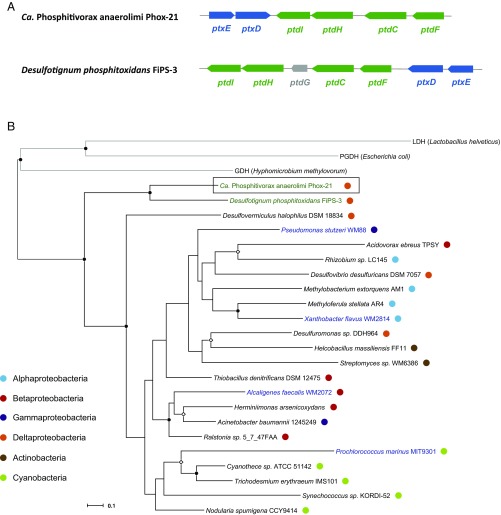
The community dynamics discussed above strongly suggested that OTU 21 (henceforth referred to as Candidatus Phosphitivorax (Ca. P.) anaerolimi strain Phox-21) was the taxon responsible for phosphite oxidation in our enrichments, so we focused our efforts on characterizing this organism. We obtained its full 16S rRNA gene sequence via a clone library and performed qPCR using species-specific 16S ribosomal DNA (rDNA) primers to assess the absolute abundance of this organism during phosphite-dependent growth. Significantly higher growth rates were observed in the presence of phosphite (1.21 ± 0.13 × 105 16S rDNA copies per milliliter per day) compared with the no-phosphite controls (7.26 ± 2.64 × 102 16S rDNA copies per milliliter per day) (t test, P = 0.0008), and growth was concurrent with phosphite oxidation (Fig. 2). A maximum likelihood phylogenetic tree based on the full 16S rRNA gene sequence placed Ca. P. anaerolimi Phox-21 within the candidate Deltaproteobacteria order GW-28, an uncultivated clade most closely related to the Desulfovibrionales (Fig. 3). Phox-21 shares only 83% sequence identity to the DPOM D. phosphitoxidans FiPS-3. The closest relatives to Ca. P. anaerolimi Phox-21 are uncultured clones from anaerobic wastewater and food waste digesters, estuarine sediments, peat wetlands, and sinkhole biomats (Fig. 3).
Fig. 2.
Growth of Ca. Phosphitivorax anaerolimi Phox-21 in enrichment cultures containing either 8 mM phosphite and 5% rumen fluid (blue circles with solid lines) or 5% rumen fluid only (red squares with solid lines). Blue circles with dashed lines indicate phosphite concentrations in phosphite-containing cultures. Phox-21 16S rDNA copy numbers were determined by qPCR using taxon-specific primers and normalized by the total volume of culture sampled for DNA extraction. Data points for 16S rDNA copy numbers represent the geometric mean of triplicate cultures, with error bars denoting one geometric SD. Data points for phosphite concentrations represent the average of triplicate cultures, with error bars denoting one SD.
Fig. 3.
Phylogenetic tree showing the placement of Ca. Phosphitivorax anaerolimi Phox-21 within the Deltaproteobacteria. Selected 16S rRNA sequences were aligned using the Silva aligner, and a maximum likelihood phylogenetic tree was constructed with 1,000 bootstrap resamplings using RAxML-HPC. Members of the Acidobacteria were included as an outgroup. Taxa represented in the Silva reference database were assigned to known orders within the Deltaproteobacteria based on their Greengenes taxonomic assignments. GenBank accession numbers are provided in parentheses. Taxa for which there is experimental evidence of DPO activity are indicated in bold. Internal nodes with bootstrap support of >70% are indicated by closed circles and those with support of >50% by open diamonds. (Scale bar: 0.1 change per nucleotide.)
All attempts to isolate Ca. P. anaerolimi proved unsuccessful. As such, we obtained its genome directly from the enriched community using genome-resolved metagenomics. Two samples were taken at days 11 and 19 from a culture grown on 10 mM phosphite and 5% RF that displayed a high rate of phosphite oxidation (470 μM/d). Another two samples were taken at days 5 and 19 from a culture grown on 10 mM phosphite, 5% RF, and 5 mM molybdate that exhibited a very low rate of phosphite oxidation (28 μM/d) due to molybdate inhibition. Sequencing reads from all four samples were combined and assembled, yielding 50.95 Mbp of sequence data in 20,733 contigs, with an average contig size of 2,458 bp and an N50 of 16,833 bp (Table S1). More than 98% of reads from each of the samples mapped back to the assembled contigs (Table S1). Contigs ≤1,000 bp long were removed from the dataset, and only the remaining 5,997 contigs (43.85 Mbp) were included in the binning step. A total of 2,531 contigs (42.2% of the contigs analyzed) representing 76.8% (33.66 Mbp) of the total sequence data in the analysis were binned into 13 high-quality genome bins (Table S2). Nine of the genome bins recovered were previously identified as prevalent OTUs based on 16S rRNA gene analysis, including bin 2 (Deltaproteobacteria) (Table S2). The remaining four bins did not correspond to any of the prevalent OTUs previously identified. The correspondence between bin 2 and Ca. P. anaerolimi Phox-21 was confirmed by aligning a partial 16S rRNA gene from bin 2 to the full Phox-21 gene from the clone library. In accordance with the abundance patterns revealed by 16S rRNA gene analysis, bin 2 had the highest sequencing depth (581×) of any genome bin in the highly phosphite-oxidizing sample at day 19 (Fig. S6).
The Ca. P. anaerolimi genome encodes the entire ptx-ptd gene cluster for dissimilatory phosphite oxidation, with the exception of ptdG (Fig. 4A). The key gene for phosphite oxidation, phosphite dehydrogenase (ptxD), was not found in any other binned or assembled sequences. No genes for hypophosphite oxidation or phosphonate degradation were found in Phox-21 or in any other binned or assembled sequences (Table S3). All of the ptx-ptd genes of Phox-21 are most closely related to those of D. phosphitoxidans FiPS-3. The Phox-21 PtxD has 55% amino acid identity to that of FiPS-3, and together they form a separate clade from PtxD proteins involved in APO (Fig. 4B). However, in contrast to the FiPS-3 ptx-ptd cluster, which is flanked by transposases, there is no genomic evidence that the Phox-21 gene cluster was horizontally acquired.
Fig. 4.
(A) Ptx-ptd gene clusters of Ca. Phosphitivorax anaerolimi Phox-21 and Desulfotignum phosphitoxidans FiPS-3. IMG gene annotations: ptdC, major facilitator superfamily transporter; ptdF, nucleoside-diphosphate-sugar epimerase; ptdG, nucleotide-binding universal stress protein; ptdH, radical SAM superfamily enzyme; ptdI, hypothetical protein; ptxD, phosphite dehydrogenase; ptxE, transcriptional regulator. Genes highlighted in blue are present in APO organisms, genes highlighted in green are present only in FiPS-3 and Phox-21, and genes highlighted in gray are present only in FiPS-3. (B) Phylogenetic tree of the phosphite dehydrogenase PtxD. Protein sequences from selected organisms were aligned using Clustal Omega, and a maximum likelihood tree was constructed with 1,000 bootstrap resamplings using RAxML-HPC. Black branches indicate PtxD sequences while gray branches indicate related outgroup sequences belonging to the d-hydroxyacid dehydrogenase protein family (GDH, glycerate dehydrogenase; LDH, lactate dehydrogenase; PGDH, 3-phosphoglycerate dehydrogenase). Organisms for which there is experimental evidence of DPO are highlighted in green while those for which there is experimental evidence of APO are highlighted in blue. Colored circles indicate the taxonomic affiliations of the organisms harboring each PtxD sequence. Internal nodes with bootstrap support of >70% are indicated by closed circles and those with support of >50% by open diamonds. (Scale bar: 0.1 change per amino acid residue.)
Metabolic Overview of Ca. Phosphitivorax Anaerolimi Phox-21.
Ca. P. anaerolimi has an incomplete TCA cycle and appears to completely lack an electron transport chain as genes for complexes I to IV are absent from its genome (Fig. 5 and Table S3). It does have genes for glucose, pyruvate, and ethanol metabolism, as well as for a sodium-translocating ferredoxin:NADH oxidoreductase (Rnf) complex (rnfABCDEG). The genome also encodes a NAD-dependent ferredoxin:NADPH oxidoreductase (nfnAB), an electron-bifurcating complex that catalyzes the reduction of two NADP molecules coupled to the oxidation of reduced ferredoxin and NADH in a reversible manner (28). Both an F-type and V-type ATP synthase are present in the genome, with the F-type ATPase likely functioning as a sodium-translocating complex and the V-type ATPase as a proton-translocating complex, based on the identity of key residues in the c subunits of both complexes (29). Additionally, the genome encodes a quinone-dependent, molybdate-binding formate dehydrogenase (fdoGHI), which is a membrane-bound periplasmic complex typically involved in formate oxidation coupled to nitrate or nitrite reduction and an ammonifying periplasmic nitrite reductase (nrfAH) (30). No genes for hydrogenases, dissimilatory sulfate reduction, dissimilatory nitrate reduction, denitrifying nitrite reduction, or anaerobic ammonium oxidation were identified (Table S3). Phox-21 is predicted to be incapable of synthesizing the amino acids alanine, histidine, and threonine, as well as the cofactors tetrahydrofolate (THF), pyridoxal 5′-phosphate (PLP), and Vitamin B12.
Fig. 5.
Functional overview of high-quality binned genomes. Filled boxes indicate the presence of the metabolic process or bioenergetic complex in the corresponding genome while empty boxes indicate its absence. Only metabolisms and complexes that are present in at least one high-quality binned genome are depicted here. See Table S3 for a list of the key functional genes used to indicate the presence of a metabolism or complex. A metabolism was considered present only if all of the genes encoding at least one full enzymatic pathway capable of carrying out that metabolic process were found in the genome. A bioenergetic complex was considered present only if all of the genes encoding the necessary subunits for a functional protein complex were found in the genome.
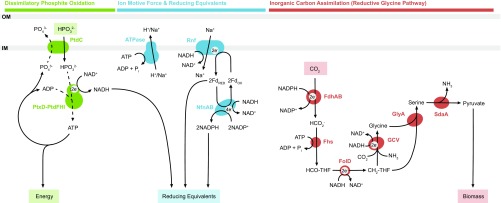
Acetyl-CoA synthase (acsB), a key gene in the autotrophic Wood–Ljungdahl pathway, is present in the Ca. P. anaerolimi genome while key genes for all other known carbon fixation pathways (Calvin–Benson–Bassham cycle, reverse TCA cycle, 3-hydroxypropionate bi-cycle, dicarboxylate/4-hydroxybutyrate cycle, and 3-hydroxypropionate/4-hydroxybutyrate cycle) are absent (Fig. 5 and Table S3). Surprisingly, however, it is missing all of the genes in the carbonyl branch of the Wood–Ljungdahl pathway other than acsB (Fig. S7). These genes are absent from all unbinned sequences as well. On the other hand, Phox-21 does have genes for both the catalytic subunit (fdhA) and the NADP-binding subunit (fdhB) of the CO2-reducing formate dehydrogenase, which catalyzes the first step in the methyl branch of the Wood–Ljungdahl pathway in bacteria (31) (Fig. S7). It also possesses genes for formate:THF ligase (fhs), methenyl-THF cyclohydrolase/methylene-THF dehydrogenase (folD), and methylene-THF reductase (metF), which together constitute the rest of the methyl branch of the Wood–Ljungdahl pathway. Nonetheless, given the absence of key carbonyl branch genes, Phox-21 appears to be incapable of assimilating CO2 via the Wood–Ljungdahl pathway.
We explored the possibility that Phox-21 could reduce CO2 to formate using FdhAB and then assimilate formate using an alternate pathway. The only known formate assimilation pathways are the Wood–Ljungdahl pathway, the serine pathway, and the reductive glycine pathway (32, 33). All three pathways use the enzymes Fhs and FolD (present in Phox-21) to initially reduce formate to methylene-THF, but they have distinct downstream routes for the subsequent conversion of methylene-THF to biomass precursors (32, 33). In addition to having an incomplete Wood–Ljungdahl pathway, Phox-21 lacks genes for the key serine pathway enzymes serine-glyoxylate transaminase and malyl-CoA lyase. However, all of the genes for the reductive glycine pathway (gcvP, glycine dehydrogenase; gcvT, aminomethyltransferase; gcvH, lipoate-binding protein; lpd, dihydrolipoyl dehydrogenase; glyA, serine hydroxymethyltransferase; and sdaA, serine deaminase) are present in the genome of Phox-21, indicating that this is likely a functional route for formate assimilation.
Metabolic Overview of Other Enrichment Community Members.
Like Ca. P. anaerolimi, most of the other enrichment community members have incomplete TCA cycles and lack genes involved in electron transport and respiratory processes (Fig. 5). Only Proteiniphilum sp. EBM-41 (bin 6) has a complete TCA cycle and genes for oxygen respiration (cydAB). There are two methanogens present, Methanoculleus sp. EBM-46 (bin 7) and Methanococcoides sp. EBM-47 (bin 9), and both have the full Wood–Ljungdahl pathway, although only EBM-46 has the uptake hydrogenase necessary for autotrophic growth on H2/CO2 (Fig. 5). Nonetheless, both methanogens appear to be capable of growth on acetate and formate, and EBM-47 has genes for growth on methanol as well. Both Tepidanaerobacter sp. strains, EBM-38 (bin 1) and EBM-49 (bin 13), have all of the genes for the carbonyl branch of the Wood–Ljungdahl pathway but have no CO2-reducing formate dehydrogenase (fdhAB) or uptake hydrogenase, which is consistent with previous genomic analysis of Tepidanaerobacter acetatoxydans and indicates that these organisms are capable of syntrophic acetate oxidation but not of autotrophic growth (34). The remaining seven organisms lack any autotrophic or syntrophic acetate-oxidation pathways and are therefore likely to be obligate fermenters involved in the degradation of RF components (Fig. 5).
Discussion
DPO was discovered over a decade ago in the marine sediment isolate D. phosphitoxidans FiPS-3, but there have been no additional reports of this metabolism in other environments. This study, therefore, represents only the second observation of DPO and provides a description of the community structure and metagenomic context related to this metabolism. Phosphite oxidation in our wastewater sludge enrichments occurred only in the presence of live cells whereas killed controls showed no decrease in phosphite even after 29 wk, consistent with previous reports of phosphite’s chemical stability under anaerobic conditions (6). Moreover, phosphite-oxidizing cultures accumulated phosphate in the media and had significantly higher growth rates than controls lacking phosphite, which points to a dissimilatory process involved in energy metabolism rather than a purely assimilatory process. Genomic analysis of the uncultured bacterium Ca. Phosphitivorax anaerolimi Phox-21, whose abundance was strongly correlated with phosphite oxidation in our enrichments, confirmed the presence of a ptx-ptd gene cluster in its genome. This gene cluster, known to be involved in phosphite oxidation in D. phosphitoxidans, is not present in any other microbial isolate genome currently available in the Integrated Microbial Genomes (IMG) database. Multiple lines of evidence thus indicate that Ca. P. anaerolimi is the organism responsible for DPO in our enrichments.
The CO2 dependence of phosphite oxidation in our enrichments, together with the lack of alternative electron acceptors, implies that Ca. P. anaerolimi couples phosphite oxidation to CO2 reduction, and the presence of a CO2-reducing formate dehydrogenase complex (FdhAB) in its genome provides the putative means by which this metabolism could occur. However, the absence of any of the six known autotrophic pathways in Phox-21 complicates the question of how inorganic carbon might be assimilated by this organism. Phox-21 also appears to lack an electron transport chain, meaning it should be incapable of energy conservation through oxidative phosphorylation.
In light of these observations, we propose that Phox-21 couples phosphite oxidation to CO2 reduction to formate and uses the energy generated by this reaction to assimilate formate via the reductive glycine pathway (Fig. 6). Phosphite can enter the cell via the PtdC transporter, and this process could potentially be coupled to phosphate export, allowing Phox-21 to uptake phosphite in an energy-neutral manner (22, 23). Once inside the cell, phosphite could be oxidized to produce NADH and ATP (Fig. 6). Based on thermodynamic calculations and physiological evidence, Schink et al. (19) have previously proposed that D. phosphitoxidans is able to conserve energy during DPO by directly generating ATP from the oxidation of phosphite. This putative substrate-level phosphorylation step during DPO could potentially be mediated by PtdFHI and would allow for energy conservation in the absence of membrane-associated electron transport, although this has not yet been experimentally confirmed (5). ATP produced in this manner could be used to run proton and sodium translocating ATP synthases in reverse to establish an ion motive force across the cell membrane (Fig. 6). The resulting sodium ion gradient could then drive the Rnf complex to reduce ferredoxin, which could serve as an electron donor for NADPH production by the NfnAB complex (Fig. 6). In this way, it would be possible for Phox-21 to generate all of the energy and reducing equivalents needed for cellular growth. Phox-21 could then reduce CO2 to formate using FdhAB and convert the formate to pyruvate via the reductive glycine pathway (Fig. 6). All of the necessary biomass precursors (acetyl-CoA, oxaloacetate, 2-oxoglutarate, and succinyl-CoA) could be subsequently generated from pyruvate via the partial TCA cycle.
Fig. 6.
Genomics-based metabolic model of dissimilatory phosphite oxidation coupled to CO2 reduction in Ca. Phosphitivorax anaerolimi Phox-21. Dotted lines denote putative mechanisms based on physiological and genomic observations that have yet to be confirmed by direct biochemical evidence. Dissimilatory phosphite oxidation proteins: PtdC, phosphite transporter; PtdF, nucleoside-diphosphate-sugar epimerase; PtdH, radical SAM superfamily enzyme; PtdI, hypothetical protein; PtxD, phosphite dehydrogenase. Ion motive force and reducing equivalents proteins: ATPase, ATP synthase complex; NfnAB, NAD-dependent ferredoxin:NADP oxidoreductase; Rnf, sodium-translocating ferredoxin:NAD oxidoreductase complex. Inorganic carbon assimilation (reductive glycine pathway) proteins: FdhAB, NADP-dependent formate dehydrogenase; Fhs, formate:THF ligase; FolD, methenyl-THF cyclohydrolase/methylene-THF dehydrogenase; GCV, glycine cleavage system (GcvH, lipoate-binding protein; GcvP, glycine dehydrogenase; GcvT, aminomethyltransferase; Lpd, dihydrolipoyl dehydrogenase); GlyA, serine hydroxymethyltransferase; SdaA, serine deaminase.
Reductive glycine pathways without the final deamination to pyruvate have been previously shown to exist in a variety of bacteria (35). However, due to the presence of known carbon fixation pathways in these organisms, Braakman and Smith (35) concluded that the reductive glycine pathway functions as a route for glycine and serine synthesis and not as a primary autotrophic pathway. Furthermore, studies by Bar-Even et al. (32, 33) used computational modeling of known metabolic pathways to identify the full reductive glycine pathway (including deamination to pyruvate) as a potentially efficient and oxygen-tolerant pathway for formatotrophic growth. A subsequent study proposed that the reductive glycine pathway could also function as an autotrophic pathway if a CO2-reducing formate dehydrogenase was present (as it is in Phox-21) (24). Nevertheless, the authors consider this to be a “synthetic” pathway, even though it is composed of natural enzymes, because there has so far been no report of a native reductive glycine pathway capable of supporting formatotrophic or autotrophic growth (30, 31, 34). Given that CO2 is required for DPO in our system and that Phox-21 lacks any of the six known carbon fixation pathways, it appears that Phox-21 is in fact using the reductive glycine pathway as its sole carbon fixation pathway. Further experimental evidence is needed to ascertain the validity of this hypothesis, but, if true, it would mean that the reductive glycine pathway constitutes a seventh natural carbon fixation pathway for autotrophic growth.
It remains unclear why RF stimulated the growth of Phox-21 to such an extent. Its effect could be due to Phox-21’s inability to synthesize certain essential cofactors (e.g., THF and PLP) and amino acids (e.g., alanine, histidine, and threonine), all of which were absent from our original growth media but may be present in RF (25). However, subsequent attempts to grow our enrichments in media lacking RF, but supplemented with THF, PLP, and amino acids, still resulted in substantially lower phosphite oxidation rates than in RF-amended media, suggesting that additional compounds present in RF are needed for robust growth of Phox-21. It is also not clear why molybdate, which is known to be an inhibitor of sulfate reduction, had such a strong inhibitory effect on phosphite oxidation since Phox-21 lacks any sulfate reduction genes. It is possible that molybdate directly targets the phosphite oxidation pathway, but further work is needed to elucidate this point. The presence of other organisms (fermenters, acetate oxidizers, and methanogens) in our enrichment cultures complicates analysis of the physiology of Phox-21 due to the activity of additional metabolic pathways and the possibility of symbiotic interactions between community members. Obtaining a pure culture of Ca. P. anaerolimi would, therefore, greatly facilitate any further investigation of its physiology.
The environmental prevalence and phylogenetic diversity of DPOM is still unclear. Like D. phosphitoxidans FiPS-3, Ca. P. anaerolimi Phox-21 belongs to the Deltaproteobacteria, but the sample size of known DPOM is still far too small to allow us to determine whether this metabolism is restricted to a specific phylogenetic group. There is evidence that the ptx-ptd genes in FiPS-3 were acquired by lateral gene transfer (22), but we do not know how common it is for these genes to be horizontally propagated in the environment or what the phylogenetic range of these events might be. Phox-21 belongs to the GW-28 candidate order, a poorly studied clade with no known cultured representatives. Its closest relatives are all uncultured clones from anaerobic waste sites throughout the world. Other related taxa include uncultured clones from hydrothermal vent environments that could potentially be exposed to phosphite generated by geothermal processes (5). Whether or not the capacity for DPO is a conserved feature of the GW-28 clade remains to be determined.
Our analysis of the Ca. P. anaerolimi genome suggests that it may be possible for organisms to exploit the unique energetic properties of phosphite to grow autotrophically via the reductive glycine pathway with CO2 as the sole electron acceptor. Due to its extremely low redox potential, phosphite is the only known biological electron donor that could theoretically drive chemotrophic CO2 fixation via the reductive glycine pathway (a net consumer of ATP) in the absence of an additional energy source or electron acceptor. Previous work in D. phosphitoxidans has demonstrated that phosphite oxidation can also be coupled to CO2 fixation via the Wood–Ljungdahl pathway (19). Together, these findings suggest that phosphite may be a driver of growth and carbon fixation under energy-limited conditions, like those found in aphotic environments lacking high-potential electron acceptors. As such, it may be advisable to search for DPOM in poorly studied, energy-limited environments, such as deep freshwater and marine sediments, subsurface terrestrial aquifers, and subseafloor basalts. We hope that future research will continue to elucidate the role of dissimilatory phosphite oxidation in microbial systems and its potential impact on global primary production.
Etymology and Description.
We propose the provisional name Candidatus Phosphitivorax anaerolimi for the dissimilatory phosphite-oxidizing bacterium described in this study. The etymology of the genus (nov.) is as follows: Phosphitivorax (“voracious phosphite consumer”); phosphitis (N.L. n.), phosphite; vorax (L. adj.), voracious. The etymology of the species (nov.) is as follows: anaerolimi (“from sludge with no air”); an (Gr. pref.), not; aer (Gr. n.), air; limus (L. n., limi gen. n.), sludge. To summarize, we have Phosphitivorax anaerolimi: phosphite-consuming bacterium from anaerobic sludge.
Description.
The description is as follows: an uncultivated bacterium enriched from anaerobic wastewater treatment sludge at 37 °C and pH ∼7 to 8; belongs to the GW-28 candidate order of the Deltaproteobacteria; capable of growth by dissimilatory phosphite oxidation, leading to phosphate accumulation in the media; genomic evidence suggests that it can grow autotrophically; growth is stimulated by rumen fluid and inhibited by molybdate.
Methods
Sampling, Media Composition, and Growth Conditions.
Sludge samples were obtained from an anaerobic digester operating at 37 °C at the East Bay Municipal Utility District wastewater treatment facility in Oakland, CA. Anaerobic bottles (Bellco) containing 45 mL of basal media were each inoculated with 5 mL of sludge and incubated at 37 °C. The composition of the basal medium (pH = 7) was as follows (per 1 L of distilled water): 5 g/L NaHCO3, 12 g/L Hepes buffer, 1 g/L NH4Cl, 0.5 g/L KCl, 0.5 g/L l-cysteine HCl, 0.001 g/L resazurin, and 10 mL each of vitamins and trace minerals (36). No phosphorus source was added to the basal media. The media were prepared and dispensed into individual bottles under an N2/CO2 (80:20, vol/vol) headspace, sealed with butyl rubber stoppers, and autoclaved. Bicarbonate-free controls were prepared by omitting NaHCO3 from the basal media and degassing it with 100% N2. Na2HPO3 (5 H2O) and Na2MoO4 (2 H2O) (Sigma Aldrich) were added from sterile, anaerobic stocks to the basal media as needed. Rumen fluid from fistulated cows fed a high forage diet was obtained from the University of California (UC), Davis Department of Animal Science (37). Rumen fluid stocks were prepared anaerobically, autoclaved at 121 °C for 20 min, and added to the basal media as needed. Killed controls were autoclaved at 121 °C for 1 h. Samples for ion chromatography (IC) analysis were filtered and stored at 4 °C while samples for DNA extraction were stored at −80 °C.
Measurement of Cell Growth and Ionic Metabolites.
Cell growth was measured as optical density at 600 nm (OD600) using a Cary 50 UV-Vis Spectrophotometer (Agilent Technologies), and growth measurements are given in optical density units (ODUs). Phosphite and phosphate concentrations were measured via ion chromatography (IC) using a Dionex ICS 2100 instrument with an electrical conductivity detector and a Dionex IonPac AS 16 (4 × 250-mm) column (Thermo Fisher Scientific) maintained at 30 °C. Background conductivity was suppressed with a Dionex ASRS 300 suppressor set at 100 mA and operating in recycle mode (38). To improve the separation of the phosphite and phosphate peaks, a gradient elution program was developed in which the mobile phase concentration increased from 12 mM to 35 mM NaOH after 10 min. The total run time of the program was 20 min, with a constant mobile phase flow rate of 1.2 mL/min and a sample injection volume of 25 µL. All growth and metabolite analyses were carried out in triplicate cultures, and the statistical significance of pairwise comparisons was assessed using Student’s t test with an alpha level of 0.05.
16S rRNA Gene Community Analysis.
DNA was isolated from triplicate cultures using the DNeasy PowerSoil Kit (Qiagen). The archaeal and bacterial primer set MiSeq 16S Forward (F) (5′ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCAGCMGCCGCGGTAA 3′) and MiSeq 16S Reverse (R) (5′ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC 3′) was used to amplify a 287-bp segment of the 16S rRNA gene that spans HV region four. These primers were based on primers S-D-Arch-0519-a-S-15 (A519F) and S-D-Bact-0785-a-A-21 (Bakt_805R) (39) but include the necessary Illumina adapters. PCR amplification and library preparation were carried out as described previously (38). Samples were sent to the UC Davis Genome Center for sequencing on an Illumina MiSeq using the MiSeq V2 reagent kit (2 × 250 bp) and software version MiSeq 2.4.1. Illumina amplicon reads were analyzed as described previously (38). Briefly, Mothur v. 1.3.3 (40) was used to cluster reads into OTUs based on a 3% dissimilarity threshold and then generate a table of relative abundances and taxonomic identities for each OTU.
16S rRNA Gene qPCR of Ca. Phosphitivorax Anaerolimi Phox-21.
Full-length 16S rRNA gene sequences were PCR-amplified from genomic DNA extracted from enrichments grown on phosphite and rumen fluid using the universal primers 27F and 1525R (41). A clone library of these sequences was prepared using the TOPO TA Cloning Kit (Thermo Fisher Scientific), and 16S rRNA gene inserts from 20 clones were PCR-amplified using primers M13 F and M13 R. The inserts were Sanger-sequenced at the UC Berkeley DNA Sequencing Facility and compared with the partial 16S rRNA gene of Phox-21 using BLAST to identify a matching full sequence. The full Phox-21 16S rRNA gene was then used to design 16S rDNA qPCR primers specific to Phox-21 using the selective primer design software tool PRISE2 (42). The primer set F4 (5′ ACGTAGGCGGATTGGTAAGT 3′) and R2 (5′ TACTCATCGTTACGGCGTG 3′) was experimentally validated by running a conventional PCR using genomic DNA from phosphite-oxidizing enrichments. Sanger sequencing of PCR products from this test reaction confirmed that only the Phox-21 16S rRNA gene was amplified. Real-time PCR assays were then performed using the Maxima SYBR Green/ROX qPCR kit (Thermo Fisher Scientific). Template DNA (5 μL) was used in a reaction mixture containing 12.5 μL of 2× Master Mix, 0.6 μL of each forward and reverse Phox-21 16S primer, and water to a final volume of 25 μL. Reaction conditions were 95 °C, 15 min; 35 cycles of 95 °C, 15 s; 60 °C, 30 s; and 72 °C, 30 s; and a final extension of 72 °C, 10 min. Standard curves were generated using purified plasmids containing the Phox-21 16S rDNA sequence serially diluted to cover a range of DNA concentrations from 4.5 × 109 copies to 4.5 × 102 copies. All samples and standards were run in duplicate.
Phylogenetic Analysis of Ca. Phosphitivorax Anaerolimi Phox-21.
The full Phox-21 16S rRNA gene sequence from the clone library described above was compared with the National Center for Biotechnology Information (NCBI) nucleotide database using BLAST to identify the top 100 hits among cultured isolates, as well as uncultured clones. Representative sequences from top BLAST hits, as well as other taxa of interest, were chosen for phylogenetic analysis. The Silva aligner (43) was used to align selected 16S rRNA sequences, and taxa represented in the Silva reference database were classified based on their Greengenes taxonomic assignments (44). Selected protein sequences of interest were aligned using Clustal Omega (45). Maximum likelihood phylogenetic trees were constructed from 16S rRNA alignments and protein alignments using RAxML-HPC with 1,000 bootstrap resamplings, and trees were visualized using Dendroscope v3.5.7 (46, 47).
Metagenomic Assembly, Binning, and Annotation.
Metagenomic DNA samples were sent to the QB3 Vincent J. Coates Genomics Sequencing Laboratory at UC Berkeley for sequencing on an Illumina HiSeq 2000 (100-bp paired-end reads). Illumina sequencing reads were trimmed for quality and filtered using Sickle v1.33, with a quality threshold value of 28, and then merged using IDBA-UD v1.0 (48, 49). Merged reads from all samples were combined and assembled using MEGAHIT v1.0.2 with default parameters (50, 51). MEGAHIT is an assembler developed specifically for metagenomic reads that uses succinct de Bruijn graphs and an iterative multiple k-mer size strategy. To assess sequencing depth, reads from each sample were then mapped backed to the combined assembly using BWA-MEM v0.7.10, with default parameters (52). Contigs from the combined assembly were binned into individual genomes using the Anvi’o v1.1.0 platform (53). Contigs were automatically clustered using tetranucleotide frequency and sequencing depth profiles across samples. Bins were then manually refined using the Anvi’o interactive graphical interface, which allows the user to visualize bins and to add or remove contigs based on patterns of sequencing depth, GC content, and taxonomic identity. Taxonomic identities were assigned to contigs based on the most commonly occurring taxonomy assigned to genes within that contig using Rapid Annotations using Subsystems Technology (RAST) (54). Taxonomic classification of bins was determined based on the most abundant taxonomic assignment occurring in at least 25% of the contigs in the bin. Genome bins were assessed for completeness and contamination based on the presence of lineage-specific, conserved, single-copy marker genes using the automated bin evaluation tool CheckM v1.0.1 (55). CheckM calculates “completeness” based on the number of expected marker genes present in a given bin and “contamination” based on the number of marker genes present in multiple copies with less than 90% amino acid identity to each other. High-quality genome bins (>80% completeness and <5% contamination) were submitted to the Integrated Microbial Genomes (IMG) database for gene calling and annotation (56). IMG utilizes Prodigal v2.50 for identification of protein-coding genes, which are then functionally annotated using a custom manually curated pipeline based on BLAST and HMMER searches against multiple protein databases (COG, KEGG, MetaCyc, Pfam, and TIGRfam) (57, 58).
Supplementary Material
Acknowledgments
We thank Carmence Ho and Annette Liao for providing general laboratory support; Martin Musabyimana (East Bay Municipal Utility District) for help in obtaining the wastewater sludge samples used in this study; Kenny Mok (Taga Laboratory, UC Berkeley) for providing rumen fluid samples; Robert Rohde for help developing computational tools for microbial community analysis; and Ke Bi (Computational Genomics Resource Laboratory, UC Berkeley) for aid in troubleshooting computational issues related to the development of a metagenomic analysis pipeline. Funding supporting research on phosphorus redox cycling in the laboratory of J.D.C. is provided by the Energy Biosciences Institute (Berkeley, CA) and the US Department of Agriculture Agricultural Experiment Station program. I.A.F. received funding from the National Science Foundation’s Graduate Research Fellowship Program and a UC Berkeley Chancellor’s Fellowship.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The full 16S rRNA gene sequence of Ca. Phosphitivorax anaerolimi Phox-21 has been deposited in the GenBank (GB) database (accession no. KU898264). MiSeq reads from community 16S rRNA gene amplicon sequencing have been deposited in the National Center for Biotechnology Information Sequence Read Archive (accession no. SRP071909). The combined metagenomic assembly of all four enrichment community samples has been deposited in the Integrated Microbial Genomes (IMG) database (accession no. Ga0100964). Individual genomes recovered from the combined assembly are available in the IMG and GB databases under the following accession nos.: Ca. Phosphitivorax anaerolimi Phox-21, Ga0115057 (IMG), MPOS00000000 (GB); Tepidanaerobacter sp. EBM-38, Ga0115060 (IMG), MPOT00000000 (GB); unclassified bacterium EBM-40, Ga0115061 (IMG), MPOU00000000 (GB); Proteiniphilum sp. EBM-39, Ga0115062 (IMG), MPOV00000000 (GB); Thermotogales bacterium EBM-19, Ga0115064 (IMG), MPOW00000000 (GB); Proteiniphilum sp. EBM-41, Ga0115065 (IMG), MPOX00000000 (GB); Methanoculleus sp. EBM-46, Ga0115067 (IMG), MPOY00000000 (GB); Coprothermobacter sp. EBM-25, Ga0115069 (IMG), MPOZ00000000 (GB); Methanococcoides sp. EBM-47, Ga0115071 (IMG), MPPA00000000 (GB); Spirochaeta sp. EBM-43, Ga0115073 (IMG), MPPB00000000 (GB); Aminobacterium sp. EBM-42, Ga0115070 (IMG), MPPC00000000 (GB); Thermotogales bacterium EBM-38, Ga0115076 (IMG), MPPD00000000 (GB); Tepidanaerobacter sp. EBM-49, Ga0115075 (IMG), MPPE00000000 (GB).
See Commentary on page 7.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1715549114/-/DCSupplemental.
References
- 1.Pasek MA, Sampson JM, Atlas Z. Redox chemistry in the phosphorus biogeochemical cycle. Proc Natl Acad Sci USA. 2014;111:15468–15473. doi: 10.1073/pnas.1408134111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yu X, Geng J, Ren H, Chao H, Qiu H. Determination of phosphite in a full-scale municipal wastewater treatment plant. Environ Sci Process Impacts. 2015;17:441–447. doi: 10.1039/c4em00543k. [DOI] [PubMed] [Google Scholar]
- 3.Pech H, et al. Detection of geothermal phosphite using high-performance liquid chromatography. Environ Sci Technol. 2009;43:7671–7675. doi: 10.1021/es901469t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Han C, et al. Phosphite in sedimentary interstitial water of Lake Taihu, a large eutrophic shallow lake in China. Environ Sci Technol. 2013;47:5679–5685. doi: 10.1021/es305297y. [DOI] [PubMed] [Google Scholar]
- 5.Figueroa IA, Coates JD. Microbial phosphite oxidation and its potential role in the global phosphorus and carbon cycles. Adv Appl Microbiol. 2017;98:93–117. doi: 10.1016/bs.aambs.2016.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Pasek MA. Rethinking early Earth phosphorus geochemistry. Proc Natl Acad Sci USA. 2008;105:853–858. doi: 10.1073/pnas.0708205105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Pasek M, Block K. Lightning-induced reduction of phosphorus oxidation state. Nat Geosci. 2009;2:553–556. [Google Scholar]
- 8.Adams F, Conrad JP. Transition of phosphite to phosphate in soils. Soil Sci. 1953;75:361–371. [Google Scholar]
- 9.Casida LE., Jr Microbial oxidation and utilization of orthophosphite during growth. J Bacteriol. 1960;80:237–241. doi: 10.1128/jb.80.2.237-241.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Malacinski G, Konetzka WA. Bacterial oxidation of orthophosphate. J Bacteriol. 1966;91:578–582. doi: 10.1128/jb.91.2.578-582.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Foster TL, Winans L, Jr, Helms SJ. Anaerobic utilization of phosphite and hypophosphite by Bacillus sp. Appl Environ Microbiol. 1978;35:937–944. doi: 10.1128/aem.35.5.937-944.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martínez A, Osburne MS, Sharma AK, DeLong EF, Chisholm SW. Phosphite utilization by the marine picocyanobacterium Prochlorococcus MIT9301. Environ Microbiol. 2012;14:1363–1377. doi: 10.1111/j.1462-2920.2011.02612.x. [DOI] [PubMed] [Google Scholar]
- 13.White AK, Metcalf WW. Microbial metabolism of reduced phosphorus compounds. Annu Rev Microbiol. 2007;61:379–400. doi: 10.1146/annurev.micro.61.080706.093357. [DOI] [PubMed] [Google Scholar]
- 14.Relyea HA, van der Donk WA. Mechanism and applications of phosphite dehydrogenase. Bioorg Chem. 2005;33:171–189. doi: 10.1016/j.bioorg.2005.01.003. [DOI] [PubMed] [Google Scholar]
- 15.White AK, Metcalf WW. Two C-P lyase operons in Pseudomonas stutzeri and their roles in the oxidation of phosphonates, phosphite, and hypophosphite. J Bacteriol. 2004;186:4730–4739. doi: 10.1128/JB.186.14.4730-4739.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yang K, Metcalf WW. A new activity for an old enzyme: Escherichia coli bacterial alkaline phosphatase is a phosphite-dependent hydrogenase. Proc Natl Acad Sci USA. 2004;101:7919–7924. doi: 10.1073/pnas.0400664101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Costas AM, White AK, Metcalf WW. Purification and characterization of a novel phosphorus-oxidizing enzyme from Pseudomonas stutzeri WM88. J Biol Chem. 2001;276:17429–17436. doi: 10.1074/jbc.M011764200. [DOI] [PubMed] [Google Scholar]
- 18.Roels J, Verstraete W. Biological formation of volatile phosphorus compounds. Bioresour Technol. 2001;79:243–250. doi: 10.1016/s0960-8524(01)00032-3. [DOI] [PubMed] [Google Scholar]
- 19.Schink B, Thiemann V, Laue H, Friedrich MW. Desulfotignum phosphitoxidans sp. nov., a new marine sulfate reducer that oxidizes phosphite to phosphate. Arch Microbiol. 2002;177:381–391. doi: 10.1007/s00203-002-0402-x. [DOI] [PubMed] [Google Scholar]
- 20.Schink B, Friedrich M. Phosphite oxidation by sulphate reduction. Nature. 2000;406:37. doi: 10.1038/35017644. [DOI] [PubMed] [Google Scholar]
- 21.Metcalf WW, Wolfe RS. Molecular genetic analysis of phosphite and hypophosphite oxidation by Pseudomonas stutzeri WM88. J Bacteriol. 1998;180:5547–5558. doi: 10.1128/jb.180.21.5547-5558.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Poehlein A, Daniel R, Schink B, Simeonova DD. Life based on phosphite: A genome-guided analysis of Desulfotignum phosphitoxidans. BMC Genomics. 2013;14:753. doi: 10.1186/1471-2164-14-753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Simeonova DD, Wilson MM, Metcalf WW, Schink B. Identification and heterologous expression of genes involved in anaerobic dissimilatory phosphite oxidation by Desulfotignum phosphitoxidans. J Bacteriol. 2010;192:5237–5244. doi: 10.1128/JB.00541-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cotton CA, Edlich-Muth C, Bar-Even A. Reinforcing carbon fixation: CO2 reduction replacing and supporting carboxylation. Curr Opin Biotechnol. 2017;49:49–56. doi: 10.1016/j.copbio.2017.07.014. [DOI] [PubMed] [Google Scholar]
- 25.Saleem F, et al. The bovine ruminal fluid metabolome. Metabolomics. 2013;9:360–378. [Google Scholar]
- 26.Shelton DR, Tiedje JM. Isolation and partial characterization of bacteria in an anaerobic consortium that mineralizes 3-chlorobenzoic acid. Appl Environ Microbiol. 1984;48:840–848. doi: 10.1128/aem.48.4.840-848.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Deweerd KA, Mandelco L, Tanner RS, Woese CR, Suflita JM. Desulfomonile tiedjei gen. nov. and sp. nov., a novel anaerobic, dehalogenating, sulfate-reducing bacterium. Arch Microbiol. 1990;154:23–30. [Google Scholar]
- 28.Buckel W, Thauer RK. Energy conservation via electron bifurcating ferredoxin reduction and proton/Na(+) translocating ferredoxin oxidation. Biochim Biophys Acta. 2013;1827:94–113. doi: 10.1016/j.bbabio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Mulkidjanian AY, Galperin MY, Makarova KS, Wolf YI, Koonin EV. Evolutionary primacy of sodium bioenergetics. Biol Direct. 2008;3:13. doi: 10.1186/1745-6150-3-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maia LB, Moura JJG, Moura I. Molybdenum and tungsten-dependent formate dehydrogenases. J Biol Inorg Chem. 2015;20:287–309. doi: 10.1007/s00775-014-1218-2. [DOI] [PubMed] [Google Scholar]
- 31.Ragsdale SW, Pierce E. Acetogenesis and the Wood-Ljungdahl pathway of CO(2) fixation. Biochim Biophys Acta. 2008;1784:1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bar-Even A, Noor E, Flamholz A, Milo R. Design and analysis of metabolic pathways supporting formatotrophic growth for electricity-dependent cultivation of microbes. Biochim Biophys Acta. 2013;1827:1039–1047. doi: 10.1016/j.bbabio.2012.10.013. [DOI] [PubMed] [Google Scholar]
- 33.Bar-Even A. Formate assimilation: The metabolic architecture of natural and synthetic pathways. Biochemistry. 2016;55:3851–3863. doi: 10.1021/acs.biochem.6b00495. [DOI] [PubMed] [Google Scholar]
- 34.Müller B, Manzoor S, Niazi A, Bongcam-Rudloff E, Schnürer A. Genome-guided analysis of physiological capacities of Tepidanaerobacter acetatoxydans provides insights into environmental adaptations and syntrophic acetate oxidation. PLoS One. 2015;10:e0121237. doi: 10.1371/journal.pone.0121237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Braakman R, Smith E. The emergence and early evolution of biological carbon-fixation. PLOS Comput Biol. 2012;8:e1002455. doi: 10.1371/journal.pcbi.1002455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Balch WE, Fox GE, Magrum LJ, Woese CR, Wolfe RS. Methanogens: Reevaluation of a unique biological group. Microbiol Rev. 1979;43:260–296. doi: 10.1128/mr.43.2.260-296.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Crofts TS, Men Y, Alvarez-Cohen L, Taga ME. A bioassay for the detection of benzimidazoles reveals their presence in a range of environmental samples. Front Microbiol. 2014;5:592. doi: 10.3389/fmicb.2014.00592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Carlström CI, et al. Characterization of an anaerobic marine microbial community exposed to combined fluxes of perchlorate and salinity. Appl Microbiol Biotechnol. 2016;100:9719–9732. doi: 10.1007/s00253-016-7780-5. [DOI] [PubMed] [Google Scholar]
- 39.Klindworth A, et al. Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res. 2013;41:e1. doi: 10.1093/nar/gks808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schloss PD, et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol. 2009;75:7537–7541. doi: 10.1128/AEM.01541-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weisburg WG, Barns SM, Pelletier DA, Lane DJ. 16S ribosomal DNA amplification for phylogenetic study. J Bacteriol. 1991;173:697–703. doi: 10.1128/jb.173.2.697-703.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang YT, Yang JI, Chrobak M, Borneman J. PRISE2: Software for designing sequence-selective PCR primers and probes. BMC Bioinformatics. 2014;15:317. doi: 10.1186/1471-2105-15-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pruesse E, et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007;35:7188–7196. doi: 10.1093/nar/gkm864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDonald D, et al. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012;6:610–618. doi: 10.1038/ismej.2011.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sievers F, Higgins DG. Clustal omega. Curr Protoc Bioinformatics. 2014;48:3.13.1–3.13.16. doi: 10.1002/0471250953.bi0313s48. [DOI] [PubMed] [Google Scholar]
- 46.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 47.Huson DH, Scornavacca C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 48.Joshi NA, Fass JN. 2011 Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files, Version 1.33. Available at https://github.com/najoshi/sickle. Accessed on September 10, 2015.
- 49.Peng Y, Leung HCM, Yiu SM, Chin FYL. IDBA-UD: A de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28:1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 50.Li D, Liu C-M, Luo R, Sadakane K, Lam T-W. MEGAHIT: An ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph. Bioinformatics. 2015;31:1674–1676. doi: 10.1093/bioinformatics/btv033. [DOI] [PubMed] [Google Scholar]
- 51.Li D, et al. MEGAHIT v1.0: A fast and scalable metagenome assembler driven by advanced methodologies and community practices. Methods. 2016;102:3–11. doi: 10.1016/j.ymeth.2016.02.020. [DOI] [PubMed] [Google Scholar]
- 52.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Eren AM, et al. Anvi’o: An advanced analysis and visualization platform for ‘omics data. PeerJ. 2015;3:e1319. doi: 10.7717/peerj.1319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aziz RK, et al. The RAST server: Rapid annotations using subsystems technology. BMC Genomics. 2008;9:75. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25:1043–1055. doi: 10.1101/gr.186072.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Markowitz VM, et al. IMG: The integrated microbial genomes database and comparative analysis system. Nucleic Acids Res. 2012;40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hyatt D, et al. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huntemann M, et al. The standard operating procedure of the DOE-JGI microbial genome annotation pipeline (MGAP v.4) Stand Genomic Sci. 2015;10:86. doi: 10.1186/s40793-015-0077-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.