Significance
Insulin is a target of CD4 T cells in type 1 diabetes in mice and humans. Why the major epitope in the insulin B chain is presented poorly to the diabetogenic CD4 T cells by the disease-associated major histocompatibility class II (MHCII) alleles has been highly debated. Here we present high-resolution mouse and human MHCII structures and T-cell functional data to show that C-terminal modifications of this epitope are required for binding and presentation in the appropriate position in the MHCII binding groove. These results suggest that pancreas-specific posttranslational modifications of this peptide may play a role in the induction of diabetes and explain how the pathogenic T cells escape deletion in the thymus.
Keywords: autoimmunity, peptide presentation, self-tolerance, posttranslational modification
Abstract
A polymorphism at β57 in some major histocompatibility complex class II (MHCII) alleles of rodents and humans is associated with a high risk for developing type 1 diabetes (T1D). However, a highly diabetogenic insulin B chain epitope within the B:9–23 peptide is presented poorly by these alleles to a variety of mouse and human CD4 T cells isolated from either nonobese diabetic (NOD) mice or humans with T1D. We have shown for both species that mutations at the C-terminal end of this epitope dramatically improve presentation to these T cells. Here we present the crystal structures of these mutated peptides bound to mouse IAg7 and human HLA-DQ8 that show how the mutations function to improve T-cell activation. In both peptide binding grooves, the mutation of B:22R to E in the peptide changes a highly unfavorable side chain for the p9 pocket to an optimal one that is dependent on the β57 polymorphism, accounting for why these peptides bind much better to these MHCIIs. Furthermore, a second mutation of the adjacent B:21 (E to G) removes a side chain from the surface of the complex that is highly unfavorable for a subset of NOD mouse CD4 cells, thereby greatly enhancing their response to the complex. These results point out the similarities between the mouse and human responses to this B chain epitope in T1D and suggest there may be common posttranslational modifications at the C terminus of the peptide in vivo to create the pathogenic epitopes in both species.
The risk of developing type 1 diabetes (T1D) is closely linked to polymorphisms in major histocompatibility complex class II (MHCII) genes (1–4). For example, in humans, the highest T1D genetic risk factor is a single polymorphism in the β chain of HLA-DQ (DQ) within the codon for β57 that lies within the peptide binding groove (5–7). Low-risk DQ alleles and most other MHCII alleles in humans and rodents have a D at this position. Substitutions for this D in the high-risk alleles break a conserved, stabilizing salt bridge to Lys76 in the MHCII α chain (8, 9), creating a strong preference among these alleles for peptides that bind with an acidic amino acid, E or D, at the p9 position, which now can make a salt bridge to the freed-up α76 Lys (9, 10). These findings suggest a priori that the risk associated with these alleles is linked to some aspect of MHCII peptide presentation to CD4 T cells—for example, unique involvement of diabetogenic pancreatic peptides that satisfy the preference for a p9 acidic amino acid. However, despite decades of experimentation, the reasons for this association are still debated.
We have studied a peptide from the insulin B chain, B:9–23 (SHLVEALYLVCGERG), shown to contain an essential epitope(s) for CD4 T cells in the development of spontaneous T1D in the NOD mouse model (11). Many CD4 insulin-recognizing T cell clones from the islets of prediabetic NOD mice target the insulin B:9–23 peptide (12, 13), and CD4 T cells targeting this peptide have been identified in humans as well (14, 15). This peptide can potentially occupy various positions or registers (Rs) in the binding groove of IAg7, the only MHCII molecule in NOD mice (Fig. 1A). For example, R1 places VEALYLVCG in the p1–p9 positions of the groove and R2 places EALYLVCGE in p1–p9. While the binding of this peptide in R1 and R2 has been demonstrated by ourselves and others (13, 16), our data accumulated over the past 8 y have shown that neither of these Rs creates the diabetogenic MHCII–peptide complex for a wide variety of NOD CD4 T-cell clones, which we conclude recognize the peptide in R3, which would place ALYLVCGER in p1–p9 (16–18). This was a highly unexpected result, since in this register the R at the p9 position of this peptide greatly interferes with binding to IAg7. We showed that mutation of p9R to an optimal E dramatically improved binding to IAg7 in R3 by about 100-fold (16). While this increase in binding was sufficient for a strong increase in activation of about half of B:9–23-specific NOD T-cell clones, we showed that a second mutation, p8E to G, was required as well to get strong activation of the other half (17). The two populations defined with these mutated peptides correlated, respectively, with the type A and type B insulin-reactive CD4 T cells, previously described by Unanue and coworkers (13, 19), which they proposed recognized the peptide bound in R2 and R1, respectively. We went on to show that type A T cells were also present in human patients with T1D, revealed by the dramatically improved T-cell response to the B:22R to E mutated peptide bound to HLA-DQ8 (DQ8) (14, 15).
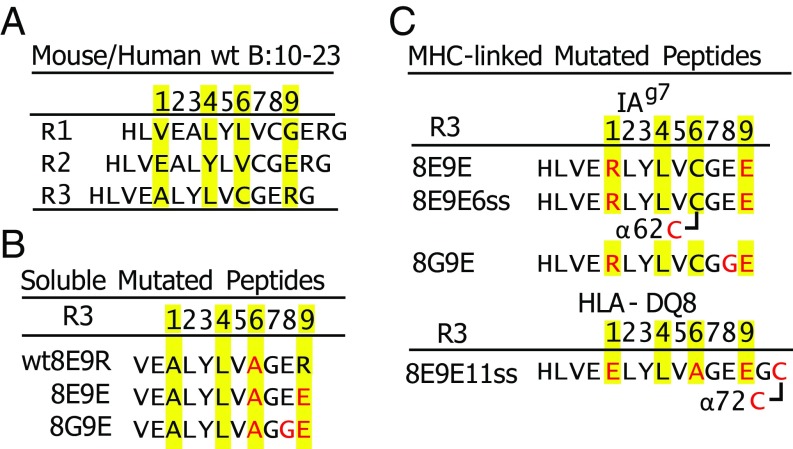
Fig. 1.
Various versions of the insulin B:9–23 peptide used in these studies. (A) Three of the possible Rs (R1–R3) for the binding of the B:10–23 peptide within the IAg7 or DQ8 binding groove. Predicted anchor amino acids at p1, p4, p6, and p9 are highlighted in yellow. (B) Mutated soluble versions of the B:12–22 peptide are shown with the mutations in red. Predicted R3 anchor amino acids are highlighted in yellow. (C) Various versions of the B:10–22 peptide covalently bound to IAg7 or DQ8 via a linker between the peptide C termini and the N termini of the MHCII β chain. Mutations within the peptide, linker, or MHCII α chain are in red. Disulfide bonds between the peptide/linker and MHCII α chain are indicated. See Materials and Methods for details of the constructions.
Thus, rather than promoting the presentation of the diabetic insulin epitope, the MHCII β57 polymorphisms in fact inhibited the presentation of the natural peptide in the proper R. We have proposed that pancreatic posttranslational modifications of this epitope at p9 and sometimes at p8 as well may be required for effective presentation in vivo and could explain how diabetogenic CD4 T cells escape thymic negative selection and find their neo-antigen targets in the pancreas (18, 20). Here we support these ideas with crystallographic structures of the mutated peptides bound to mouse IAg7 and human DQ8. The structures confirm the R3 binding nature of the different versions of the functional MHCII/insulin complexes from both mice and humans. They also elucidate the key feature of p8 in discriminating type A and type B specificities of the T cells in the NOD mouse model. We hypothesize that pancreatic-specific modifications of the C terminus of the B:9–23 peptide in vivo may be needed to create the functional epitopes for CD4 effectors in T1D.
Results
C-Terminal Modifications of an Insulin B Chain Peptide Create IAg7 R3-Bound Superagonists for both Type A and B CD4+ Insulin-Reactive T Cells from NOD Mice.
Our previously published data showed that type A and type B insulin-reactive mouse T cells recognize the B chain 9–23 peptide bound with R3 anchor residues. Both types of T cells required a mutation of the natural R at p9 to E for strong IAg7 binding. In addition, the type A T cells strongly preferred the surface-exposed natural p8E, but the type B T cells preferred the additional p8E to G mutation (14–17). As shown in Fig. 1, to study this phenomenon, we have prepared a variety of insulin peptides modified in this way either as soluble peptides (Fig. 1B) or as peptides tethered in multiple ways to IAg7 or DQ8 (Fig. 1C). We have studied the activity of these peptides with a variety of human and mouse insulin-reactive CD4 T-cell hybridomas or T-cell receptor (TCR)-transduced T-cell avatars whose origins and properties are described in Materials and Methods and Table S1.
To demonstrate the dramatic effects of the 8E9E and 8G9E peptide modifications, we compared their stimulatory activities to that of the wild-type WT8E9R peptide using IL-2 secretion assays with fixed M12.C3-IAg7 B lymphoma cells as antigen-presenting cells (APCs) and eight (four type A and four type B) NOD mouse CD4 T cells as responders (Fig. 2). We used an IAg7 binding peptide from hen egg lysozyme (HEL) as a negative control (21).
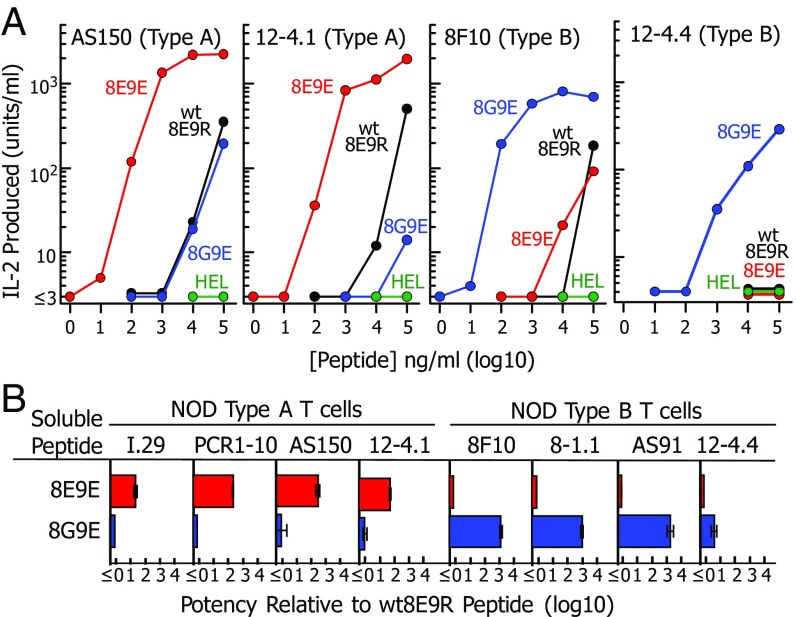
Fig. 2.
Mutations to the B:12–22 peptide create reciprocal superagonists for type A vs. type B insulin-reactive T cells. (A) Various concentrations of soluble WT8E9R (black), 8E9E (red), and 8G9E (blue) peptides (Fig. 1B) were presented by fixed M12.C3-IAg7 cells to two NOD mouse type A and two type B insulin-reactive T cells (Table S1). Results of a single experiment are presented as IL-2 produced after 24 h vs. the concentration of the offered peptide. The HEL peptide was used as a negative control (green). (B) Peptide titration experiments were performed as in A with all eight NOD mouse T cells listed in Table S1. The titration data were fitted with parallel third-order polynominal curves. The peptide potency was defined as the shift in the titration curve relative to that of the WT8E9R peptide. Results are shown as the geometric average and SEM of three separate experiments.
Fig. 2A shows sample peptide titration data for two type A and two type B T cells. None of these cells responded to the HEL control peptide. All responded poorly to the WT8E9R peptide, failing to reach a maximal response with even 100 μg/mL peptide. The 8E9E peptide was about 100× more potent than the WT8E9R with type A T cells but no better than the WT peptide with the type B T cells. Reciprocally, the 8G9E peptide was 100–1,000× more potent with the type B T cells than the WT8E9R peptide but no better than the WT peptide with type A T cells.
These titrations were performed three times with all eight NOD T cells and average potencies of the 8E9E and 8G9E peptides calculated as the shifts in the titration curves compared with that of the WT8E9R peptide (Fig. 2B). In each case, the 8E9E peptide was much more potent than the WT8E9R peptide with the type A, but not type B, T cells, and the 8G9E peptide was much more potent for the type B but not type A T cells. We conclude that the p9R to E mutation is required for strong IAg7 binding and that p8E is required for type A T cells but inhibitory for type B T cells.
Reciprocal Binding of Type A and Type B TCRs to the Modified Insulin Peptides.
As an independent way of showing these reciprocal specificities of the type A and type B T cells, we evaluated the binding kinetics of type A and type B TCRs in vitro with surface plasmon resonance (SPR). We produced soluble TCRs from two type A (I.29 and PCR-1.10) and two type B (8F10 and 8–1.1) T cells as described in Materials and Methods. We also prepared soluble versions of IAg7 tethered to versions of the 8E9E or 8G9E peptide (Fig. 1C) via a flexible linker attached to the IAg7 β chain N terminus (16, 17). These contained the natural C at p6, but the p1 A was mutated to R to further assure stable binding to IAg7 in R3 (21). Also, a second version of the IAg7–8E9E complex was made using IAg7, whose α62N was mutated to C (8E9E6ss), thus allowing a disulfide to form between the peptide p6C and the mutated IAg7 α62C (16, 17). This version was used with the I.29 TCR, since we had shown in a previous study (17) that the fluorescent IAg7–8E9E6ss tetramer bound I.29 better than the 8E9E tetramer. In these experiments, IAg7 bound to the HEL peptide was used as a negative control. These soluble IAg7 constructs were also stabilized with an acid-base leucine zipper and contained a biotinylated peptide tag (17).
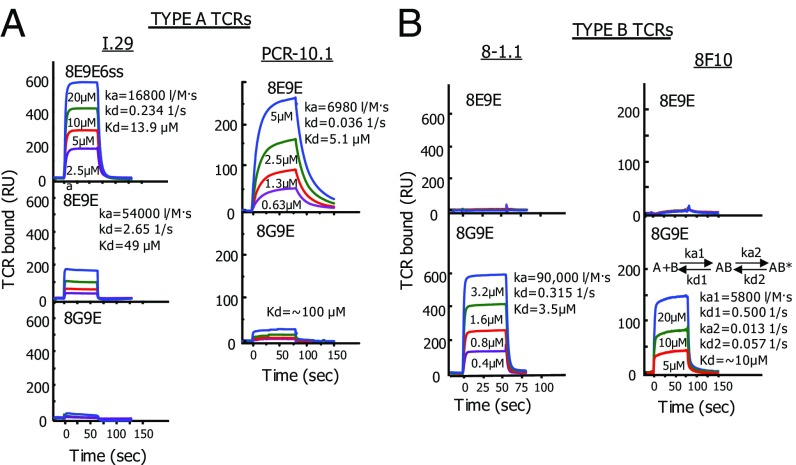
The biotinylated IAg7 complexes were captured in separate flow cells of a BIAcore streptavidin (SA) BIAsensor chip. The various concentrations of soluble type A (Fig. 3A) or type B (Fig. 3B) TCRs were injected through the flow cells while following the binding to and dissociation from the IAg7 complexes via the SPR signals after correction for the fluid phase signal from the flow cell with the control IAg7–HEL complex. As with the stimulation assays in Fig. 2, reciprocal specificities were observed in these TCR binding assays. The type A I.29 TCR bound with first-order kinetics to the IAg7–8E9E6ss ligand, as did the PCR1–10 TCR to IAg7–8E9E, with dissociation constants of 13.9 and 5.1 μM, respectively. The I.29 TCR also bound to the IAg7–8E9E complex that lacked the disulfide but, as expected, with poorer affinity (∼100 uM). These type A TCRs bound very poorly or not at all to the 8G9E complex. The 8–1.1 and 8F10 type B TCRs failed to bind to the 8E9E complex, but both bound strongly to the 8G9E complex. The 8–1.1 TCR bound with first-order kinetics with a dissociation constant of 3.5 μM. The 8F10 TCR showed second-order kinetics, consistent with a two-state reaction, in which the TCR initially bound with rapid kinetics but then bound more strongly after a conformational change, leading to an overall apparent dissociation constant of ∼10 μM. These affinities are typical for conventional CD4 T cells with high affinity for conventional self–MHCII–foreign peptide complexes.
Fig. 3.
Binding of soluble type A and type B TCRs to IAg7–peptide complexes parallels the peptide stimulation activities. (A) The affinities of soluble TCRs from two type A T cells (I.29 and PCR1-10) for IAg7 bearing various mutated insulin B:10–22 peptides were evaluated using SPR. Approximately 2,000 RUs of the biotinylated IAg7 peptides were immobilized in flow cells of a streptavidin containing BIAsensor chip. Biotinylated IAg7-HEL was immobilized in a separate flow cell to correct for the fluid phase SPR signal. The indicated concentrations of the soluble TCRs were injected for ∼75 s and the binding and dissociation of the TCRs followed by the SPR signal (RU). Where there was sufficient binding, kinetics (ka, kd, and Kd) were calculated using standard BIAevalution 4 software. B is similar to A, except the soluble TCRs came from two type B T cells (8-1.1 and 8F10). The 8F10 TCR showed second-order binding kinetics best represented by the equation shown. See Discussion.
High-Resolution Crystal Structures Show How the Peptide Mutations Determine Presentation by IAg7.
Both the T-cell stimulation and SPR experiments above show the superagonist properties of the C-terminally modified peptides. These results are consistent with our previous interpretation of mutational data, concluding that the B:22R to E mutation creates an optimal R3 p9 anchor amino acid that can overcome the B:22R inability to fill the IAg7 p9 pocket and that the adjacent B21E to G mutation removes the R3 p8E side chain that is inhibitory for type B TCRs. However, to offer unequivocal structural evidence for this interpretation, we crystallized the 8E9E, 8E9Ess, and 8G9E complexes with IAg7 after proteolytic removal of the zippers and biotinylation tag from the proteins and solved their structures at 1.8 Å, 2.0 Å, and 2.3 Å, respectively (Fig. 4 and Table S2).
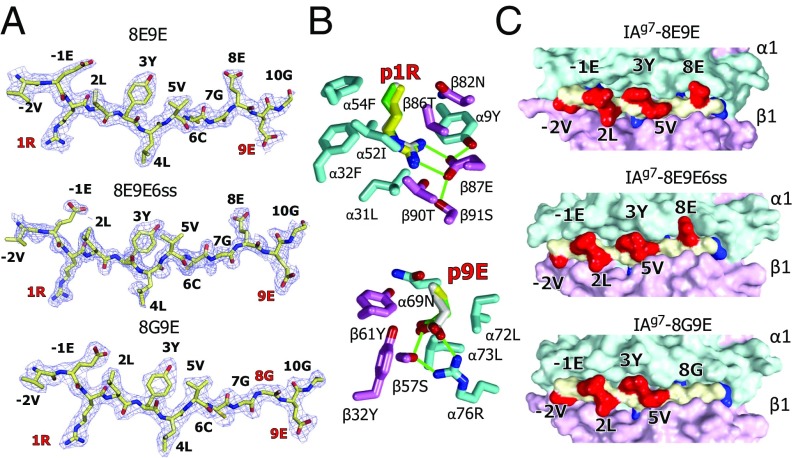
Fig. 4.
Crystal structures show very similar R3 binding of the mutated peptides to IAg7. (A) 2Fo−Fc electron density maps contoured at 1 σ within 1.5 Å of the 8E9E, 8E9E6ss, and 8G9E peptides bound to IAg7 are shown with the amino acid at each position in the MHCII binding groove labeled. The mutated amino acids are labeled in red. (B) Wireframe representations of the p1 pocket (Above) and p9 pocket (Below) of IAg7–8E9E are shown with oxygen, red and nitrogen, blue. Carbons are colored as follows: peptide, white; α chain, cyan; β chain, magenta. The side chains of p1R and p9E from the other two structures are shown superimposed, 8E9E6ss (carbons, yellow) and 8G9E (carbons, green). H bonds and salt bridges are shown with green lines. (C) Water-accessible surfaces of the IAg7 complexes are shown (α chain, cyan; β chain, magenta; peptide backbone, yellow; peptide exposed side chains, red; peptide buried side chains, blue). Exposed amino acids p-2, p-1, p2, p3, p5, and p8 are labeled.
Electron density maps clearly show unambiguous density for the peptides in the three structures (Fig. 4A and Fig. S1). As predicted, all three peptides sit in R3 in the IAg7 binding groove, with peptide anchor amino acid side chains from p1R, p4L, p6C, and p9E pointing into the corresponding IAg7 pockets within the binding groove. The mutated p1 A > R and p9 R > E side chains (Fig. 4B) optimally fill the p1 and p9 pockets, forming numerous salt bridges, H bonds, and van der Waal’s interactions with IAg7 amino acids lining the pockets, confirming our rationale for using these mutations to stabilize R3 binding. As expected, none of the side chains of the anchor residues are exposed on the surface of the complexes, but in all three structures, the side chains of the p-2V, p-1E, p2L, p3Y, and p5V amino acids are similarly positioned on the surface and accessible for potential TCR interaction (Fig. 4C). Most importantly, in the 8E9E and 8E9E6ss structures, the side chain of p8E is well-exposed on the surface, pointing straight up out of the groove, while in the 8G9E structure, the absence of a side chain at the p8 position leaves a stretch of the peptide from p6 to p10 with only the backbone of the peptide exposed on the surface. Surface electrostatic analysis shows that the 8E9E, 8E9E6ss, and 8G9E structures have similar surface potentials around the N-terminal part of the peptide, but the 8G9E structure differs conspicuously from the other two around the p8 position (Fig. S2). Therefore, both exposed surface and charge differences around the p8 position support our previous conclusion that the p8 amino acid is the deciding surface feature in discriminating type A and type B specificities of the T cells.
Comparison of the IAg7–8E9E and IAg7–8E9E6ss structures may also explain why introducing a disulfide between p6C and the α62C in the IAg7–8E9E6ss complex improves I.29 TCR binding and I.29 T-cell activation. For the most part, the TCR-facing surfaces of the 8E9E and 8E9E6ss complexes are very similar (Fig. 4C). However, as shown in Fig. S3, the disulfide introduced between p6C and α62C causes movement of the critical p3Y side chain toward the p5V, bringing the OH of p3Y ∼1.5 Å closer to IAg7 αQ61, which now bridges the OH of p3Y and the backbone O of p6C via 2 H bonds. We suggest that optimal preorienting of these amino acids for I.29 TCR interaction could account for the increased I.29 TCR affinity for the 8E9E6ss complex.
Human DQ8 and Mouse IAg7 Present the Mutated Insulin Peptide Very Similarly.
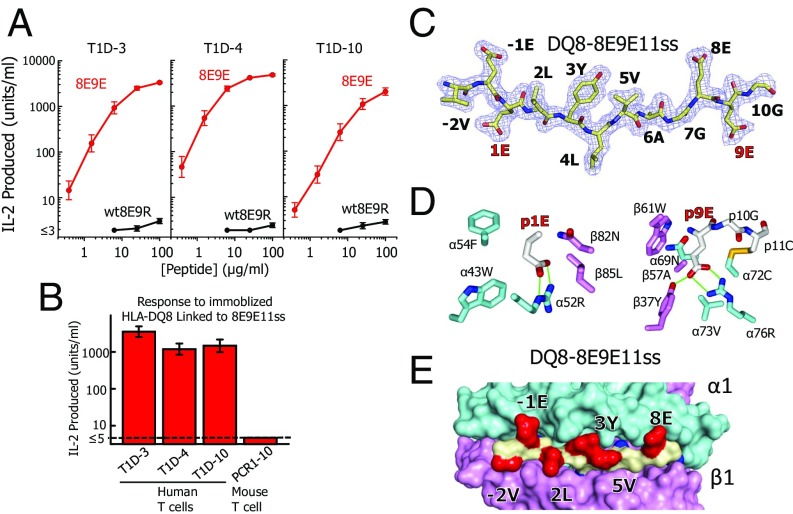
Proinsulin is an important autoantigen in both human T1D and the NOD mouse T1D model. As in mice, a polymorphism in human DQ8 β57 also breaks a salt bridge to α76R, creating a p9 pocket that greatly prefers D or E side chains and rejects K and R (9, 10). Two previous studies have shown that, like the type A mouse insulin-reactive CD4 T cells characterized above, human T1D patients contain CD4 T cells that strongly recognize versions of the B:11–23 peptide (identical between mice and humans) presented by DQ8 only when B:22R is mutated to E (14, 15). We have converted three T-cell clones isolated in one of these studies (14), T1D3, T1D4, and T1D10 (Table S1), to TCR-transduced T-cell avatars. We compared the response of the avatars to the soluble 8E9E vs. WT8E9R peptide presented by a DQ8 APC (Fig. 5A). As with the original T-cell clones, all three avatars responded much more strongly to the 8E9E, than to the WT8E9R, peptide, with the 9R to E mutation improving potency about 1,000×.
Fig. 5.
DQ8 also binds and presents the mutant insulin peptide in R3. (A) Various concentrations of soluble WT8E9R (black) and 8E9E (red) peptides (Fig. 1B) were presented by fixed M12.C3-DQ8 cells to three human type A insulin-reactive T cells (Table S1). Results of a single experiment are presented as IL-2 produced after 24 h vs. the concentration of the offered peptide. Similar results were obtained in a second experiment. (B) A biotinylated soluble version of the DQ8-8E9E11ss was immobilized in culture wells coated with Extravidin (Sigma). The same three insulin-reactive human T cells were added and IL-2 production assayed at 24 h. A mouse T cell (PCR1-10), highly reactive to the same peptide bound to IAg7, was used as the negative control. (C) 2Fo−Fc electron density maps contoured at 1 σ within 1.5 Å of the 8E9E11ss peptide bound to DQ8 are shown with the amino acid at each position in the MHCII binding groove labeled. The mutated amino acids are labeled in red. (D) Wireframe representations of the p1 pocket (Left) and p9 pocket (Right) of DQ8-8E9E11ss complex are shown colored as in Fig. 4C. H bonds and salt bridges are shown as green lines. The disulfide is shown (yellow) between the introduced cysteines at p11 and DQ8 α72. (E) Water-accessible surface of the DQ8-8E9E11ss complex is shown (α chain, cyan; β chain, magenta; peptide backbone, yellow; peptide exposed side chains, red; peptide buried side chains, blue). Exposed amino acids p-2, p-1, p2, p3, p5, and p8 are labeled.
We then prepared a soluble version of the DQ8 complex tethered to the 8E9E peptide with a linker (Fig. 1C). As with IAg7, we stabilized the complex by adding an acid/base leucine zipper to the C termini of the DQ8 β and α chains. Using the same strategy that we used with IAg7 to promote stable peptide binding in R3, we changed the p1 amino acid (B:14) from A to E, an optimal anchor residue for the DQ8 p1 pocket (10). We also added a disulfide bond between cysteines introduced at the first position of the peptide linker (p11) and at α72 of DQ8 to create DQ8–8E9E11ss (see Materials and Methods). Also, as with our IAg7 constructs, we added a biotinylated peptide tag to the C terminus of the acidic half of the leucine zipper. To test the function of this construct, we immobilized the soluble, biotinylated protein in Extravidin-coated culture wells and added each of the three T1D avatars or a fourth negative control T cell, mouse PCR1–10, that responds strongly to this peptide construct when bound to IAg7 (22). IL-2 production was assayed after 24 h (Fig. 5B). The three human T1D avatars responded strongly to DQ8 tethered to 8E9E11ss, but the control PCR1–10 T-cell hybridoma did not respond.
These results predict that the 8E9E insulin peptide binds to DQ8 in R3, very like its binding to IAg7. As confirmation, we crystallized (Table S2) the soluble DQ8–8E9E11ss complex after enzymatically removing the acid/base leucine zipper and biotinylation tag (Materials and Methods). Electron density surrounding the peptide (Fig. 5C and Fig. S1) clearly shows that, as predicted, the 8E9E11ss peptide lies in the DQ8 binding groove in R3 almost identically to how the 8E9E and 8E9E6ss peptides lie in the IAg7 groove (Fig. 4A). The side chains of peptide amino acid mutations at p1 (A to E) and p9 (R to E), introduced to stabilize binding in R3, are firmly held in these pockets by both van der Waals interactions and salt bridges and the disulfide between p11 and DQ8 α72, confirming our strategy for this construction (Fig. 5D). As with the IAg7–8E9E and IAg7–8E9E6ss complexes, the peptide amino acid side chains at p-2, p-1, p2, p3, p5, and p8 are surface-exposed for potential TCR interaction (Fig. 5E).
Discussion
The data that we present here and previously (14–17) argue convincingly that in both mouse and humans, most CD4 T cells reactive to the B:9–23 insulin peptide recognize it bound to IAg7 or DQ8 in R3. The question of R might seem to some a minor issue, but we point out that each shift in R of a peptide in an MHCII binding groove brings an entirely new set of amino acids to the surface. The resulting MHCII–peptide complexes are as different from each other as they are from the same MHCII bound to peptides from other foreign or self-proteins. The structures that we present here indicate that, from the point of view of the responding T cell, when bound to IAg7 in R3, all of the complexes form very similar TCR interaction surfaces that differ from each other primarily at the exposed p8 position. Also, we show that the contribution of the mutated peptide to the surface of DQ8 is virtually identical. Concluding that the epitopes all bind in the same R in IAg7 and DQ8 helps clear the confusion over the conflicting data about this peptide, emphasizes the similarity in how humans and mice recognize this peptide in T1D, and makes possible the design of antigen-specific diagnostic and therapeutic reagents.
Why particular MHCII alleles are strongly associated with risk for various autoimmune diseases in humans and mice has remained a puzzle for several decades. The initial prediction was that certain epitopes from proteins targeted in these diseases would be selected based on their ability to bind to these MHCII alleles. However, more recent data have suggested a very different explanation for the MHCII linkage. One example is in rheumatoid arthritis, where peptidylarginine deiminase conversion of arginine to citrulline in some epitopes from synovial proteins greatly improves their presentation by HLA-DR4 to some pathogenic CD4 T cells (23). Another example is in Celiac disease, where conversion of glutamine to glutamic acid by transglutaminase greatly improves presentation of a number of gliadin epitopes by HLA-DQ2 and -DQ8 to CD4 T cells (24). Since these modifications occur in the target tissue, these results offered a reasonable explanation for how the relevant pathogenic T cells escape negative selection in the thymus but eventually find their targets in the tissue to initiate disease in individuals with the appropriate MHCII alleles. Thus, the high-risk MHCII alleles are unable to present the epitopes in the thymus but effectively present the modified epitope in the target organs.
Our results suggest that a parallel mechanism may be at play in T1D in the CD4 T-cell response to the insulin B:9–23 peptide in mice and humans. The relevant wild-type epitope within this peptide is poorly presented in the relevant R (R3). Our finding that mutations at p8-B:21E and/or p9-B:22R solve this problem raises the question of how these substitutions might occur in vivo in the pancreas. There is no simple enzymatic way to achieve these transitions in situ in the protein. However, we have proposed a solution (20) based on recent evidence showing that a natural proteolytic process, transpeptidation, can carry out these kinds of modifications (reviewed in refs. 25 and 26). Transpeptidation is a form of reverse proteolysis in which the transient covalent bond that is formed between the active-site serine, threonine, or cysteine of proteases and the new carboxylate at a cleavage site, instead of being broken by water to complete the cleavage reaction, is attacked by the N terminus of a nearby peptide to reestablish a peptide bond, thus creating a hybrid peptide. Transpeptidation, first studied in the 1930s, surfaced as a mechanism for generating T-cell epitopes in the 2000s in a series of papers showing how this protease-driven mechanism in the proteasome could account for the creation of MHCI-presented neoantigens in cancer (27, 28). Most recently, Liepe et al. (29) have provided evidence that naturally processed peptides eluted from the human MHCI allele, HLA-A2, contain a very high proportion of chimeric peptides formed in the proteasome by the fusion between fragments from the same or different self-proteins.
We have proposed transpeptidation as a means of creating superagonists in T1D for CD4 T cells specific for insulin and other proteins (20), and recently several examples of fused peptide agonists have been identified in a mouse primary insulinoma tumor for T cells specific for chromogranin A and islet amyloid polypeptide (30). An examination of the sequences of mouse and human proinsulin reveals many potential donors within C peptide that could be fused by internal transpeptidation deletion to an insulin B chain acceptor cleaved at B:20 or B:21 to generate peptides similar or identical to the mutated peptides used in our experiments. The transpeptidation reaction is favored in situations where proteins are digested by the appropriate proteases in a confined space with limited water and a high local concentration of the donor peptide, often achieved by donors that are within the same protein as the acceptor. In the case of MHCI epitopes, these conditions are achieved within the proteasome. In the case of MHCII, the lysosome may be a more likely site for the reaction, either in the beta cell itself or in islet-resident APCs (B cells, dendritic cells, etc.). Within the beta cell, two stress-related events could trigger the process within proinsulin. The first is crinophagy, where the level of beta cell secretory granules is regulated by fusion to lysosomes during normal and stress-induced catabolism of granules (31). The second is autophagy, where ER stress leads to enhanced misfolding of proinsulin, which is eliminated from the ER in vesicles that are destroyed in part by fusion with lysosomes (32).
Materials and Methods
T-Cell Hybridomas and T-Cell Transfectomas.
For APCs, we used two versions of the M12.C3 B-cell lymphoma, one expressing IAg7 (M12.C3.g7) (33) and the other expressing DQ8 (M12.C3-DQ8-8) (34). The 12–4.1, 12–4.4, PCR-1–10, and -8–1.1 hybridomas or avatar transductomas were produced at the Barbara Davis Center (11, 35, 36). The 12–4.1 T cell was transduced with high-affinity mutant human CD4 (37) to improve its antigen sensitivity. The AS150 and AS91 T-cell hybridomas were obtained from Emil Unanue at Washington University, St. Louis. The original I.29 T-cell hybridoma was obtained from Matteo Levisetti, Washington University, St. Louis. In this paper, to avoid confusion, we are using the original type A vs. type B nomenclature for the T cells instead of our original type II vs. type I nomenclature. E. Unanue also provided us with the CDR3 sequences of the 8F10 TCR, which we used to construct a 5KC-transduced avatar. The T1D3, T1D4, and T1D10 human T-cell clones were produced at the Benaroya Institute (14). We cloned the TCR genes from these T cells and constructed 5KC-transduced T-cell avatars as above, coexpressing unmutated human CD4. All cells were cultured in enriched MEM containing 10% FCS as previously described (38).
Antigen Presentation Assays and Soluble Peptides.
T-cell hybridomas or TCR-transduced avatars (105 cells) were mixed with 105 paraformaldehyde fixed (17) M12.C3.g7 or M12.C3.DQ8-8 APCs and cultured overnight with various concentrations of peptide in a volume of 250 µL. Secreted IL-2 was assayed with either a functional assay, following the growth and survival of the HT-2 IL-2–dependent cell line (38), or with an ELISA-based assay for IL-2 (39). For presentation of soluble DQ8-p8E9E11ss, a biotin-tagged version (22) was captured in Extravidin (Sigma-Aldrich)-coated culture wells. Hybridomas or avatars were added, and IL-2 production assessed as above with HT-2. The potencies of mutant peptides compared with the WT8E9R peptide were calculated by fitting a set of parallel third-order polynominal curves to the titration data and determining the shift of the mutant and chimeric peptide data on the log10 x axis relative to the WT8E9R data, which were assigned a potency value of 1.
All soluble versions of insulin B chain containing peptides had the natural C at B:19 (p6) changed to A to avoid peptide dimerization in solution. This mutation had been previously shown to have no detrimental effect on peptide binding to IAg7 or T-cell activation (16). Soluble peptides (>95% pure) were obtained from either CHI Scientific or Schafer-N.
Protein Expression and Purification.
As previously described (17, 20), acid-base leucine zipper stabilized, soluble IAg7 and human HLA-DQ8 molecules with covalently attached peptides were produced in baculovirus-infected insect cells and purified by immunoaffinity chromatography. A biotinylation peptide tag was attached to the C terminus of the acidic half of the zipper, which was enzymatically biotinylated with BirA, produced in our own laboratory, after purification of the molecule. For crystallization, the zippers and biotinylation tag were removed with papain.
Two expression systems were used for producing soluble TCRs for biophysical studies. Soluble I.29, PCR1–10, and PCR8–1.1 TCRs were produced in baculovirus-infected insect cells as previously described (40), and the V domains of 8F10 were expressed in a bacterial vector to produce a soluble scFV tagged with 6His (41, 42), which was purified from the transformed bacteria periplasm using immobilized Ni++ chromatography.
Surface Plasmon Resonance Measurements.
Approximately 2,000 RUs (resonance units) of biotinylated IAg7 bound to the 8E9E, 8E9E6ss, 8G9E, or HEL peptides were captured in flow cells of a BIAcore streptavidin BIAsensor chip. Various concentrations of soluble refolded TCRs were injected and the association and dissociation kinetics recorded and then corrected for the fluid phase SPR signal using the data from the IAg7-HEL flow cell as baseline. Kinetics was analyzed with BIAcore BIAEval 4 software.
Protein Crystallization and X-Ray Data Collection, Data Processing, and Structural Analysis.
The MHCII complexes were crystalized using the hanging-drop vapor-diffusion method. X-ray diffraction data were collected at the Advanced Photon Source, Argonne National Laboratory. Details are contained within SI Methods and Table S2.
Supplementary Material
Acknowledgments
We acknowledge the contribution of Dr. Brian Stadinski during the early stages of this project. This work was supported by NIH Grants 5T32-AI-074491 (to Y.W.), ES-025797 (to S.D.), AI-18785 (to P.M.), DK-032083 (to T.S.), and AI-118688 (to J.W.K.) as well as JDRF Grant 1-PNF-2015-126-A-R, University of Colorado CCTSI Grant KL2 TR001080, and a Boettcher Foundation Investigator award (to S.D.). The synchrotron data were collected at Advanced Photon Source Argonne National Laboratory, Beamline 24 ID-C, whose work is based upon research conducted at the Northeastern Collaborative Access Team beamlines, which are funded by the National Institute of General Medical Sciences from National Institutes of Health Grant P41 GM103403. The Pilatus 6M detector on 24-ID-C beamline is funded by NIH-ORIP HEI Grant S10 RR029205.
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.rcsb.org [PDB ID codes (IAg7p8E9E) 6BLQ, (IAg7p8E9E6ss) 6BLR, (IAg7p8G9E) 6BLX, and (DQ8p8E9E11ss) 5UJT].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1716527115/-/DCSupplemental.
References
- 1.Aly TA, et al. Genetic prediction of autoimmunity: Initial oligogenic prediction of anti-islet autoimmunity amongst DR3/DR4-DQ8 relatives of patients with type 1A diabetes. J Autoimmun. 2005;25(Suppl):40–45. doi: 10.1016/j.jaut.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 2.Stadinski B, Kappler J, Eisenbarth GS. Molecular targeting of islet autoantigens. Immunity. 2010;32:446–456. doi: 10.1016/j.immuni.2010.04.008. [DOI] [PubMed] [Google Scholar]
- 3.Mordes JP, Bortell R, Blankenhorn EP, Rossini AA, Greiner DL. Rat models of type 1 diabetes: Genetics, environment, and autoimmunity. ILAR J. 2004;45:278–291. doi: 10.1093/ilar.45.3.278. [DOI] [PubMed] [Google Scholar]
- 4.Concannon P, Rich SS, Nepom GT. Genetics of type 1A diabetes. N Engl J Med. 2009;360:1646–1654. doi: 10.1056/NEJMra0808284. [DOI] [PubMed] [Google Scholar]
- 5.Hu X, et al. Additive and interaction effects at three amino acid positions in HLA-DQ and HLA-DR molecules drive type 1 diabetes risk. Nat Genet. 2015;47:898–905. doi: 10.1038/ng.3353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Morel PA, Dorman JS, Todd JA, McDevitt HO, Trucco M. Aspartic acid at position 57 of the HLA-DQ beta chain protects against type I diabetes: A family study. Proc Natl Acad Sci USA. 1988;85:8111–8115. doi: 10.1073/pnas.85.21.8111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Quartey-Papafio R, et al. Aspartate at position 57 of nonobese diabetic I-Ag7 beta-chain diminishes the spontaneous incidence of insulin-dependent diabetes mellitus. J Immunol. 1995;154:5567–5575. [PubMed] [Google Scholar]
- 8.Corper AL, et al. A structural framework for deciphering the link between I-Ag7 and autoimmune diabetes. Science. 2000;288:505–511. doi: 10.1126/science.288.5465.505. [DOI] [PubMed] [Google Scholar]
- 9.Lee KH, Wucherpfennig KW, Wiley DC. Structure of a human insulin peptide-HLA-DQ8 complex and susceptibility to type 1 diabetes. Nat Immunol. 2001;2:501–507. doi: 10.1038/88694. [DOI] [PubMed] [Google Scholar]
- 10.Suri A, Walters JJ, Gross ML, Unanue ER. Natural peptides selected by diabetogenic DQ8 and murine I-A(g7) molecules show common sequence specificity. J Clin Invest. 2005;115:2268–2276. doi: 10.1172/JCI25350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakayama M, et al. Germline TRAV5D-4 T-cell receptor sequence targets a primary insulin peptide of NOD mice. Diabetes. 2012;61:857–865. doi: 10.2337/db11-1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wegmann DR, Gill RG, Norbury-Glaser M, Schloot N, Daniel D. Analysis of the spontaneous T cell response to insulin in NOD mice. J Autoimmun. 1994;7:833–843. doi: 10.1006/jaut.1994.1066. [DOI] [PubMed] [Google Scholar]
- 13.Levisetti MG, Suri A, Petzold SJ, Unanue ER. The insulin-specific T cells of nonobese diabetic mice recognize a weak MHC-binding segment in more than one form. J Immunol. 2007;178:6051–6057. doi: 10.4049/jimmunol.178.10.6051. [DOI] [PubMed] [Google Scholar]
- 14.Yang J, et al. Autoreactive T cells specific for insulin B:11-23 recognize a low-affinity peptide register in human subjects with autoimmune diabetes. Proc Natl Acad Sci USA. 2014;111:14840–14845. doi: 10.1073/pnas.1416864111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakayama M, et al. Regulatory vs. inflammatory cytokine T-cell responses to mutated insulin peptides in healthy and type 1 diabetic subjects. Proc Natl Acad Sci USA. 2015;112:4429–4434. doi: 10.1073/pnas.1502967112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stadinski BD, et al. Diabetogenic T cells recognize insulin bound to IAg7 in an unexpected, weakly binding register. Proc Natl Acad Sci USA. 2010;107:10978–10983. doi: 10.1073/pnas.1006545107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crawford F, et al. Specificity and detection of insulin-reactive CD4+ T cells in type 1 diabetes in the nonobese diabetic (NOD) mouse. Proc Natl Acad Sci USA. 2011;108:16729–16734. doi: 10.1073/pnas.1113954108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Marrack P, Kappler JW. Do MHCII-presented neoantigens drive type 1 diabetes and other autoimmune diseases? Cold Spring Harb Perspect Med. 2012;2:a007765. doi: 10.1101/cshperspect.a007765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mohan JF, Petzold SJ, Unanue ER. Register shifting of an insulin peptide-MHC complex allows diabetogenic T cells to escape thymic deletion. J Exp Med. 2011;208:2375–2383. doi: 10.1084/jem.20111502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jin N, et al. N-terminal additions to the WE14 peptide of chromogranin A create strong autoantigen agonists in type 1 diabetes. Proc Natl Acad Sci USA. 2015;112:13318–13323. doi: 10.1073/pnas.1517862112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Latek RR, et al. Structural basis of peptide binding and presentation by the type I diabetes-associated MHC class II molecule of NOD mice. Immunity. 2000;12:699–710. doi: 10.1016/s1074-7613(00)80220-4. [DOI] [PubMed] [Google Scholar]
- 22.Crawford F, Kozono H, White J, Marrack P, Kappler J. Detection of antigen-specific T cells with multivalent soluble class II MHC covalent peptide complexes. Immunity. 1998;8:675–682. doi: 10.1016/s1074-7613(00)80572-5. [DOI] [PubMed] [Google Scholar]
- 23.Wegner N, et al. Autoimmunity to specific citrullinated proteins gives the first clues to the etiology of rheumatoid arthritis. Immunol Rev. 2010;233:34–54. doi: 10.1111/j.0105-2896.2009.00850.x. [DOI] [PubMed] [Google Scholar]
- 24.Sollid LM, Jabri B. Celiac disease and transglutaminase 2: A model for posttranslational modification of antigens and HLA association in the pathogenesis of autoimmune disorders. Curr Opin Immunol. 2011;23:732–738. doi: 10.1016/j.coi.2011.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Berkers CR, de Jong A, Ovaa H, Rodenko B. Transpeptidation and reverse proteolysis and their consequences for immunity. Int J Biochem Cell Biol. 2009;41:66–71. doi: 10.1016/j.biocel.2008.08.036. [DOI] [PubMed] [Google Scholar]
- 26.Cresswell P. Cell biology. Cutting and pasting antigenic peptides. Science. 2004;304:525–527. doi: 10.1126/science.1097508. [DOI] [PubMed] [Google Scholar]
- 27.Hanada K, Yewdell JW, Yang JC. Immune recognition of a human renal cancer antigen through post-translational protein splicing. Nature. 2004;427:252–256. doi: 10.1038/nature02240. [DOI] [PubMed] [Google Scholar]
- 28.Vigneron N, et al. An antigenic peptide produced by peptide splicing in the proteasome. Science. 2004;304:587–590. doi: 10.1126/science.1095522. [DOI] [PubMed] [Google Scholar]
- 29.Liepe J, et al. A large fraction of HLA class I ligands are proteasome-generated spliced peptides. Science. 2016;354:354–358. doi: 10.1126/science.aaf4384. [DOI] [PubMed] [Google Scholar]
- 30.Delong T, et al. Pathogenic CD4 T cells in type 1 diabetes recognize epitopes formed by peptide fusion. Science. 2016;351:711–714. doi: 10.1126/science.aad2791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sandberg M, Borg LA. Intracellular degradation of insulin and crinophagy are maintained by nitric oxide and cyclo-oxygenase 2 activity in isolated pancreatic islets. Biol Cell. 2006;98:307–315. doi: 10.1042/BC20050085. [DOI] [PubMed] [Google Scholar]
- 32.Sun J, et al. Proinsulin misfolding and endoplasmic reticulum stress during the development and progression of diabetes. Mol Aspects Med. 2015;42:105–118. doi: 10.1016/j.mam.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Abiru N, et al. Peptide and major histocompatibility complex-specific breaking of humoral tolerance to native insulin with the B9-23 peptide in diabetes-prone and normal mice. Diabetes. 2001;50:1274–1281. doi: 10.2337/diabetes.50.6.1274. [DOI] [PubMed] [Google Scholar]
- 34.Michels AW, et al. Islet-derived CD4 T-cells targeting proinsulin in human autoimmune diabetes. Diabetes. 2016;66:722–734. doi: 10.2337/db16-1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Daniel D, Gill RG, Schloot N, Wegmann D. Epitope specificity, cytokine production profile and diabetogenic activity of insulin-specific T cell clones isolated from NOD mice. Eur J Immunol. 1995;25:1056–1062. doi: 10.1002/eji.1830250430. [DOI] [PubMed] [Google Scholar]
- 36.Simone E, et al. T cell receptor restriction of diabetogenic autoimmune NOD T cells. Proc Natl Acad Sci USA. 1997;94:2518–2521. doi: 10.1073/pnas.94.6.2518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang XX, et al. Affinity maturation of human CD4 by yeast surface display and crystal structure of a CD4-HLA-DR1 complex. Proc Natl Acad Sci USA. 2011;108:15960–15965. doi: 10.1073/pnas.1109438108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.White J, Kappler J, Marrack P. Production and characterization of T cell hybridomas. Methods Mol Biol. 2000;134:185–193. doi: 10.1385/1-59259-682-7:185. [DOI] [PubMed] [Google Scholar]
- 39.Allicotti G, Borras E, Pinilla C. A time-resolved fluorescence immunoassay (DELFIA) increases the sensitivity of antigen-driven cytokine detection. J Immunoassay Immunochem. 2003;24:345–358. doi: 10.1081/IAS-120025772. [DOI] [PubMed] [Google Scholar]
- 40.Kappler J, White J, Kozono H, Clements J, Marrack P. Binding of a soluble alpha beta T-cell receptor to superantigen/major histocompatibility complex ligands. Proc Natl Acad Sci USA. 1994;91:8462–8466. doi: 10.1073/pnas.91.18.8462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Feng D, Bond CJ, Ely LK, Maynard J, Garcia KC. Structural evidence for a germline-encoded T cell receptor-major histocompatibility complex interaction ‘codon’. Nat Immunol. 2007;8:975–983. doi: 10.1038/ni1502. [DOI] [PubMed] [Google Scholar]
- 42.Yin L, Crawford F, Marrack P, Kappler JW, Dai S. T-cell receptor (TCR) interaction with peptides that mimic nickel offers insight into nickel contact allergy. Proc Natl Acad Sci USA. 2012;109:18517–18522. doi: 10.1073/pnas.1215928109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.