Significance
More than 71 million people are infected with HCV, and eradication of this pandemic will likely require a vaccine. Induction of broadly neutralizing monoclonal antibodies (bNAbs) is a goal of vaccine development, but no single bNAb neutralizes all strains of HCV. Here, we measured neutralizing activity of 35 combinations of neutralizing monoclonal antibodies (NAbs), showing that some NAbs form combinations with greater neutralizing breadth than any individual bNAb. One combination was also exceptionally potent because it blocks virus binding to three different HCV receptors. These data suggest that full-length envelope protein might have an advantage as a vaccine antigen relative to truncated protein or single-epitope scaffolds, since it might induce combinations of NAbs that are synergistic, with complementary neutralizing breadth.
Keywords: neutralizing antibody, hepatitis C virus, synergy, antagonism, neutralizing breadth
Abstract
There is an urgent need for a vaccine to combat the hepatitis C virus (HCV) pandemic, and induction of broadly neutralizing monoclonal antibodies (bNAbs) against HCV is a major goal of vaccine development. Even within HCV genotype 1, no single bNAb effectively neutralizes all viral strains, so induction of multiple neutralizing monoclonal antibodies (NAbs) targeting distinct epitopes may be necessary for protective immunity. Therefore, identification of optimal NAb combinations and characterization of NAb interactions can guide vaccine development. We analyzed neutralization profiles of 12 human NAbs across diverse HCV strains, assigning the NAbs to two functionally distinct clusters. We then measured neutralizing breadth of 35 NAb combinations against genotype 1 isolates, with each combination including one NAb from each neutralization cluster. Many NAbs displayed complementary neutralizing breadth, forming combinations with greater neutralization across diverse strains than any individual bNAb. Remarkably, one of the most broadly neutralizing combinations of two NAbs, designated HEPC74/HEPC98, also displayed enhanced potency, with interactions matching the Bliss independence model, suggesting that these NAbs inhibit HCV infection through independent mechanisms. Subsequent experiments showed that HEPC74 primarily blocks HCV envelope protein binding to CD81, while HEPC98 primarily blocks binding to scavenger receptor B1 and heparan sulfate. Together, these data identify a critical vulnerability resulting from the reliance of HCV on multiple cell surface receptors, suggesting that vaccine induction of multiple NAbs with distinct neutralization profiles is likely to enhance the breadth and potency of the humoral immune response against HCV.
Despite the development of highly effective direct-acting antivirals (DAAs) for treatment of hepatitis C virus (HCV) infection, a vaccine is still needed to combat the HCV pandemic. Most infected individuals are unaware of their status and may continue to expose others (1). Most infected persons do not have access to DAAs, and currently available treatments do not provide protection against reinfection after cure (2–4).
One objective of HCV vaccine development is the induction of broadly neutralizing monoclonal antibodies (bNAbs) against the virus. Dozens of bNAbs have been isolated from infected humans. These bNAbs target overlapping but distinct epitopes on the HCV envelope proteins (E1 and E2) and neutralize diverse HCV strains (5–15). Combinations of bNAbs are protective against HCV challenge in animal models (9, 10, 16, 17), and spontaneous clearance of HCV without treatment in humans is associated with early development of bNAbs (18–20), suggesting that bNAbs may play a key role in immune-mediated control of human HCV infection.
While some human bNAbs show impressive neutralizing breadth (5, 9, 20–22), HCV is an extraordinarily diverse virus, so no single bNAb neutralizes all viral strains (21–25). This resistance makes it less likely that vaccine induction of any single bNAb would provide adequate protection against a diverse HCV quasispecies challenge. Therefore, induction of multiple neutralizing monoclonal antibodies (NAbs) targeting distinct epitopes may be necessary. However, several studies have suggested that some antibodies against HCV are antagonistic (26–28), and the most advantageous NAb combinations have not yet been identified. New studies are needed to better define NAb interactions, and to identify optimal NAb combinations.
We and others have shown that human NAbs have distinct patterns of relative neutralizing potency across diverse HCV strains, also called neutralization profiles (20–22). Two NAbs could have enhanced neutralizing breadth in combination if their neutralization profiles across diverse HCV strains are complementary, so that viral strains resistant to one NAb are sensitive to the other. The efficacy of NAb combinations is also influenced by potency against viral strains recognized by both NAbs. Synergy or antagonism between NAbs can be defined relative to either the Loewe additivity model (29, 30), which assumes that two inhibitors have similar mechanisms or compete for the same binding site, or the Bliss independence model (31), which assumes that inhibitors have independent binding sites and independent mechanisms.
To identify optimal NAb combinations, we analyzed neutralization profiles of 12 NAbs, assigning them to two functionally distinct neutralization clusters. We then measured neutralization of 11 genetically and antigenically diverse genotype 1 HCV pseudoparticles (HCVpp) by 35 NAb combinations, with each combination including one NAb from each neutralization cluster. For a subset of combinations, we compared experimental neutralization to neutralization predicted by the Loewe additivity and Bliss independence models, to identify synergy, additivity, antagonism, or independence between NAbs.
Results
Selection of NAb Combinations for Analysis.
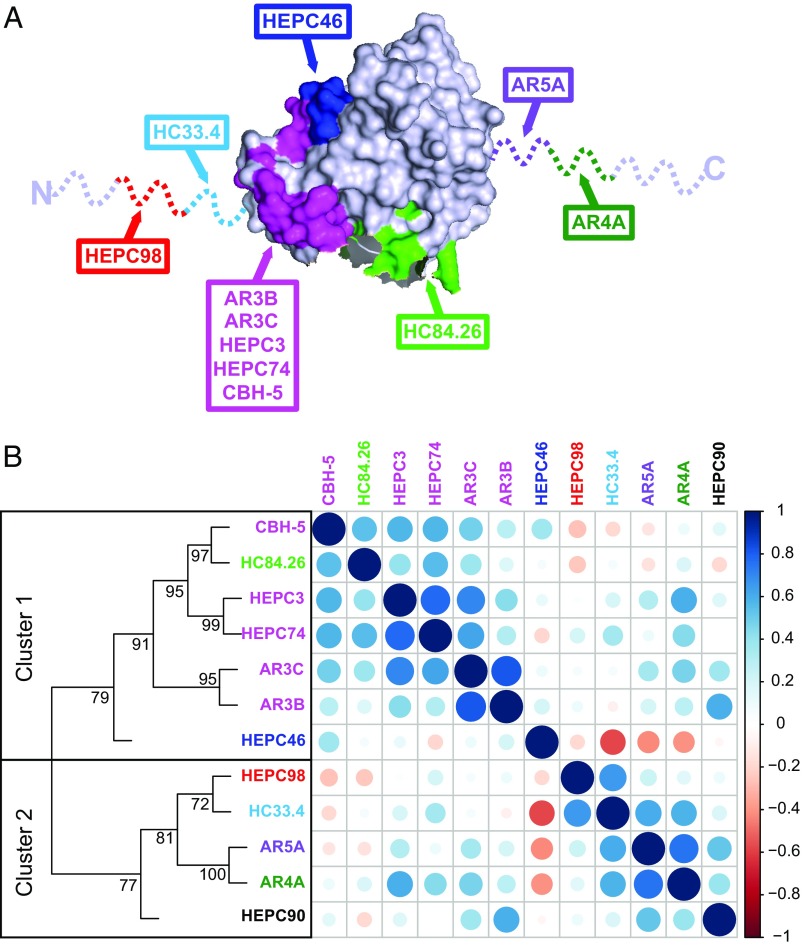
We selected a panel of 12 human NAbs targeting distinct epitopes on HCV E2. This panel includes some of the most broadly neutralizing anti-HCV antibodies described to date, as well as less broadly neutralizing mAbs, with binding epitopes that overlap to varying degrees (Fig. 1A and Table S1). We analyzed the neutralizing breadth of each of these NAbs in prior studies using a diverse panel of 19 genotype 1 HCVpp (20, 22–24). We previously showed that these quantitative neutralization data across diverse strains comprise a neutralization profile for each NAb, which can be compared among NAbs to understand functional relationships (22). We calculated pairwise Spearman correlations between neutralization profiles of these 12 NAbs and performed hierarchical clustering analysis using these pairwise correlations (Fig. 1B). As we have seen in prior analyses, these NAbs segregated into two distinct neutralization clusters. We tested all possible combinations comprising one NAb from each neutralization cluster (35 combinations), since we hypothesized that these combinations would have greatest potential for complementarity.
Fig. 1.
NAb binding epitopes and hierarchical clustering. (A) The crystallized structure of the HCV E2 protein, strain H77, from Kong et al. (49) acquired from Protein Data Bank, accession 4MWF, with colors modified in PyMOL. The E2 structure is in gray, with previously defined NAb binding residues identified by alanine scanning highlighted in various colors. NAbs targeting the magenta region bind to largely overlapping but distinct epitopes. Dashed lines indicate regions missing from the crystal structure. The binding epitope of HEPC90 is not known. (B) Hierarchical clustering of NAbs based on neutralization profiling. NAbs are colored-coded according to the scheme in A. For each NAb, relative neutralization of each of 19 HCVpp was measured to generate a neutralization profile. Pairwise Spearman correlations (r) between NAb neutralization profiles were then calculated to identify functional similarities among NAbs. Circles at each intersection of the heat map were scaled by the magnitude of the correlation between the indicated NAbs. R values approaching +1 (dark blue) and −1 (dark red) indicate positive or negative correlations, respectively. Hierarchical clustering analysis using these pairwise correlations is depicted as a tree. Numbers at tree nodes are approximately unbiased (AU) test values (51), which indicate strength of support for a particular cluster.
Selection of a Diverse and Representative Genotype 1 HCVpp Panel.
We previously developed a panel of 19 genotype 1 HCVpp that was selected to maximize genetic diversity among functional E1E2 genes (18, 22). To date, we have used this panel of 19 HCVpp to measure neutralizing breadth of 32 mAbs with binding epitopes across E1E2, including the mAbs in the current study (20, 22). Using these data, we identified a subset panel of 11 HCVpp that recapitulates the full range of neutralization sensitivity of the larger panel of 19 HCVpp (Fig. S1A). Neutralizing breadth of each of 32 mAbs measured using either the full 19 HCVpp panel or the 11 HCVpp subset are very similar. The mean breadth of these mAbs (percentage of HCVpp in the panel that were neutralized >50% at a mAb concentration of 10 µg/mL) was 38% (range 0 to 95%) using the 19 HCVpp panel and 36% (range 0 to 91%) using the 11 HCVpp panel (P = not significant by paired t test). Strains in the 11 HCVpp panel differ at an average of 74 amino acids (13%) [range 26 (5%) to 121 (22%)], and isolates are distributed across a phylogenetic tree including 634 genotype 1a and 1b reference sequences from GenBank (32) (Fig. S1B). Variability in 20 amino acid windows across E1E2 is nearly identical for the 11 HCVpp panel, the 19 HCVpp panel, and in the reference set of 634 genotype 1 sequences (Fig. S1C). The panel of 11 HCVpp also contains 96% of E1E2 amino acid polymorphisms present with at least 10% frequency in the 634 genotype 1 reference sequences (Fig. S1D). Since it appears to be representative of genotype 1 E1E2 diversity at the level of conformational B cell epitopes, 20 amino acid windows, and the majority of single amino acid polymorphisms, we elected to use this 11 HCVpp panel for neutralization breadth testing of NAb combinations.
Combinations Show Greater Neutralization Across Diverse Strains than Individual NAbs.
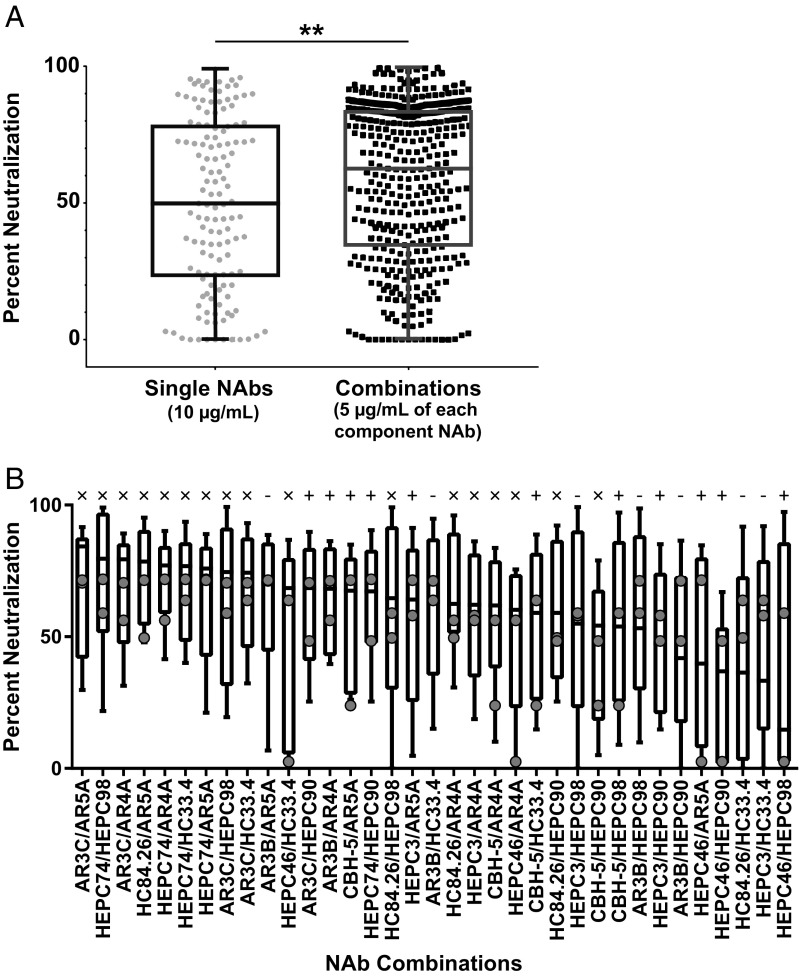
We measured neutralization of the 11 HCVpp panel by 35 NAb combinations or by their individual component NAbs. Each NAb combination was tested at a total concentration of 10 µg/mL, consisting of 5 µg/mL of each NAb, and each individual NAb was tested separately at 10 µg/mL against the same panel. We have shown previously that percentage neutralization of HCVpp by NAbs at 10 µg/mL can be used to make reliable quantitative comparisons of both the number of HCVpp recognized by each NAb (breadth) and the quantitative level of neutralization of each HCVpp (potency) (16, 20, 22–25). We first compared neutralization by all individual NAbs to neutralization by all NAb combinations. The median percentage neutralization of HCVpp in the panel by individual NAbs at 10 µg/mL was 50%, while the median percentage neutralization by NAb combinations at the same total concentration was significantly greater at 62% (P < 0.01, unpaired two-tailed t test) (Fig. 2A).
Fig. 2.
Combinations show greater neutralization across diverse strains than individual NAbs. Percent neutralization of 11 HCVpp by individual NAbs at 10 µg/mL or NAb combinations at 10 µg/mL total concentration (5 µg/mL of each NAb). (A) Percent neutralization of 11 HCVpp by all individual NAbs (n = 132; 12 NAbs × 11 HCVpp) and all NAb combinations (n = 385; 35 NAb combinations × 11 HCVpp). Each symbol indicates mean percent neutralization of an individual HCVpp, measured in duplicate. **P < 0.01 by unpaired, two-sided t test. (B) Neutralization of 11 HCVpp by each NAb combination, with combinations arranged from highest to lowest median percentage neutralization. Horizontal lines indicate medians, bars indicate 25th to 75th percentile, and whiskers indicate range. Gray circles indicate the median percent neutralization value of each component NAb in the combination when tested individually at 10 µg/mL (x, combinations with median percent neutralization greater than that of either component NAb; +, combinations with median percent neutralization between that of component NAbs; −, combinations with median percent neutralization less than either component NAb).
We then evaluated neutralization by each combination individually (Fig. 2B). Median percentage neutralization of HCVpp in the panel by NAb combinations ranged from 84% (AR3C/AR5A) down to 15% (HEPC46/HEPC98), while median percentage neutralization by individual NAbs ranged from 72% (HEPC74) down to 2% (HEPC46). Notably, 17 of 35 NAb combinations had a higher median percentage neutralization of HCVpp in the panel than either of their component NAbs tested individually. Eleven combinations produced median percentage neutralization between that of their two component NAbs. Only 7 of 35 combinations showed lower median percentage neutralization than either component NAb tested individually. Overall, these results suggest that many NAb combinations show greater neutralization across diverse strains than their individual component NAbs tested at the same total antibody concentration, and relatively few combinations are disadvantageous.
Combinations Have Complementary Neutralizing Breadth.
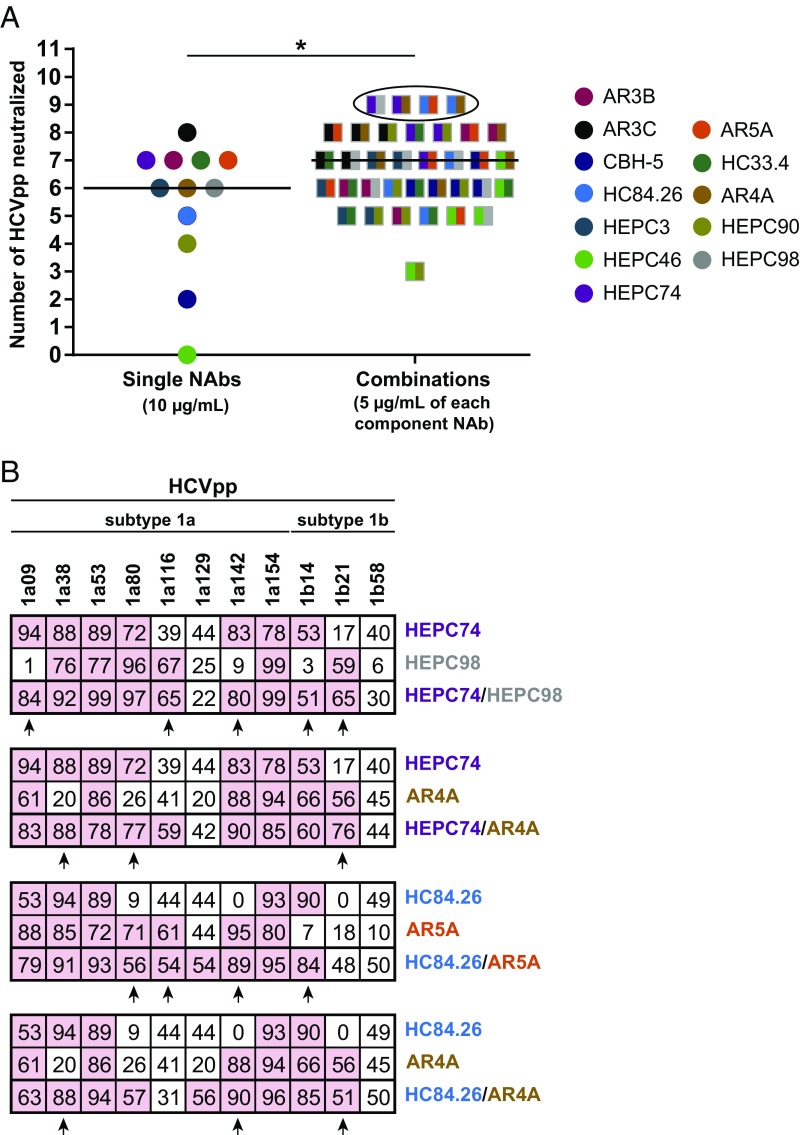
Next, we compared neutralizing breadth of individual NAbs and combinations, with breadth defined as the number of HCVpp neutralized by greater than 50%. Individual NAbs neutralized a median of 6 of the 11 HCVpp (55%; range 0 to 8 HCVpp), while NAb combinations neutralized a median of 7 HCVpp (64%; range 3 to 9 HCVpp) at the same total antibody concentration (P < 0.05, unpaired two-tailed t test) (Fig. 3A). Four combinations, HEPC74/HEPC98, HEPC74/AR4A, HC84.26/AR5A, and HC84.26/AR4A, neutralized 9 of the 11 HCVpp (81%), exceeding the neutralizing breadth of any individual bNAb tested. By examining the neutralization profiles of these NAbs individually or in combination, we observed clear evidence of complementation, as all HCVpp that were neutralized by at least one of the two NAbs in a combination also were neutralized by the combination (Fig. 3B). Complementation was also observed between most other NAb combinations (Fig. S2). Taken together, these results suggest that NAbs from distinct neutralization clusters have enhanced neutralizing breadth in combination due to complementation, with some combinations displaying greater breadth than any individual bNAb.
Fig. 3.
NAbs have complementary neutralizing breadth. (A) Neutralization of 11 HCVpp by individual NAbs at 10 µg/mL or NAb combinations at 10 µg/mL total concentration (5 µg/mL of each NAb). HCVpp were counted as neutralized if infectivity was reduced by more than 50%. Colors indicate the two NAbs in each combination. Horizontal lines indicated medians. Neutralization profiles of circled combinations are shown in B. *P < 0.05 by unpaired, two-sided t test. (B) Neutralization profiles of the NAb combinations with greatest neutralizing breadth. Values are mean percent neutralization by 10 µg/mL of each individual NAb or 10 µg/mL total concentration of NAb combinations, measured in duplicate. Pink indicates NAb/HCVpp tests with percent neutralization of >50%. Arrows indicate complementary neutralization by the two NAbs in each combination.
Of these 35 combinations tested, we selected three for more detailed analyses: one with greater percentage neutralization of HCVpp in the panel than either of its component NAbs (HEPC74/HEPC98), one with median percentage neutralization between that of its two component NAbs (HEPC3/HEPC90), and one with lower median percentage neutralization than either of its component NAbs (HC84.26/HC33.4). As a positive control for Loewe additivity, we also performed all subsequent analyses with a combination of two cluster 1 NAbs (HEPC3/HEPC74) with largely overlapping epitopes.
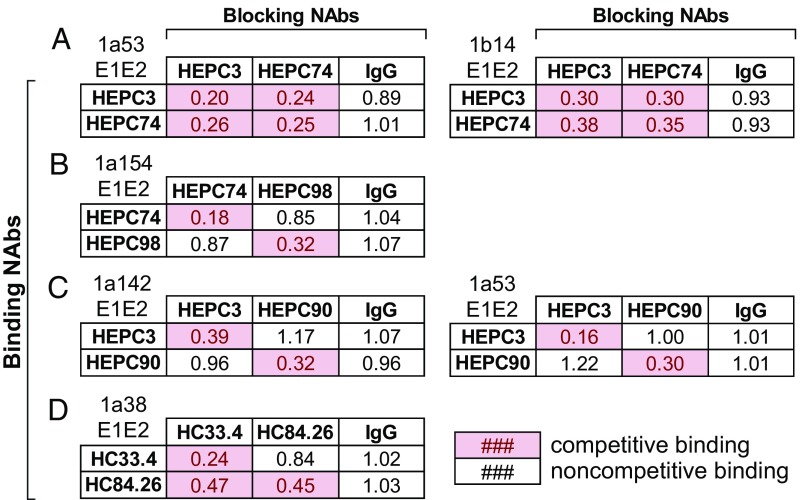
Binding Competition.
E1E2-binding competition ELISAs were performed with the four NAb combinations of interest. For each NAb combination, we selected one to two E1E2 strains from HCVpp that were neutralized potently by each NAb in the combination. Competition was defined as a 50% or greater reduction in binding of a biotinylated NAb in the presence of a blocking NAb. Competitive binding with self was used as a positive control, and noncompetitive binding with nonspecific human IgG served as a negative control. HEPC3 and HEPC74, the component NAbs in the control combination, bound competitively to both 1a53 E1E2 and 1b14 E1E2 (Fig. 4A), which was expected given their largely overlapping binding epitopes (Fig. 1A). In contrast, HEPC74 and HEPC98 bound to 1a154 E1E2 noncompetitively (Fig. 4B), which also was expected given their nonoverlapping binding epitopes (Fig. 1A). HEPC3 and HEPC90 bound noncompetitively to both 1a142 E1E2 and 1a53 E1E2 (Fig. 4C). Interestingly, HC33.4 and HC84.26, which have adjacent but nonoverlapping binding epitopes, showed unidirectional binding competition. HC84.26 reduced binding by HC33.4 by 53%, while HC33.4 only reduced the binding of HC84.26 by 16% (Fig. 4D). This unidirectional binding competition between these NAbs was also observed in a prior study (13).
Fig. 4.
Competition binding between NAbs. Binding of 2 µg/mL of biotinylated NAbs on the y axis (“Binding NAbs”) to the indicated E1E2 proteins was measured in the presence or absence of the NAbs on the x axis (“Blocking NAbs”) at a concentration of 20 µg/mL (HEPC3, HEPC74, HEPC98, HEPC90/1a53 E1E2) or 50 µg/mL (HC84.26, HEPC90/1a142 E1E2). Values shown are binding of the biotinylated NAb in the presence of blocking NAb, relative to binding in the absence of blocking NAb, averaged from duplicate measurements. Pink boxes indicate competition, which was defined as a relative binding value of <0.50. Competition between HEPC3 and HEPC74 (A), HEPC74 and HEPC98 (B), HEPC3 and HEPC90 (C), and HC33.4 and HC84.26 (D).
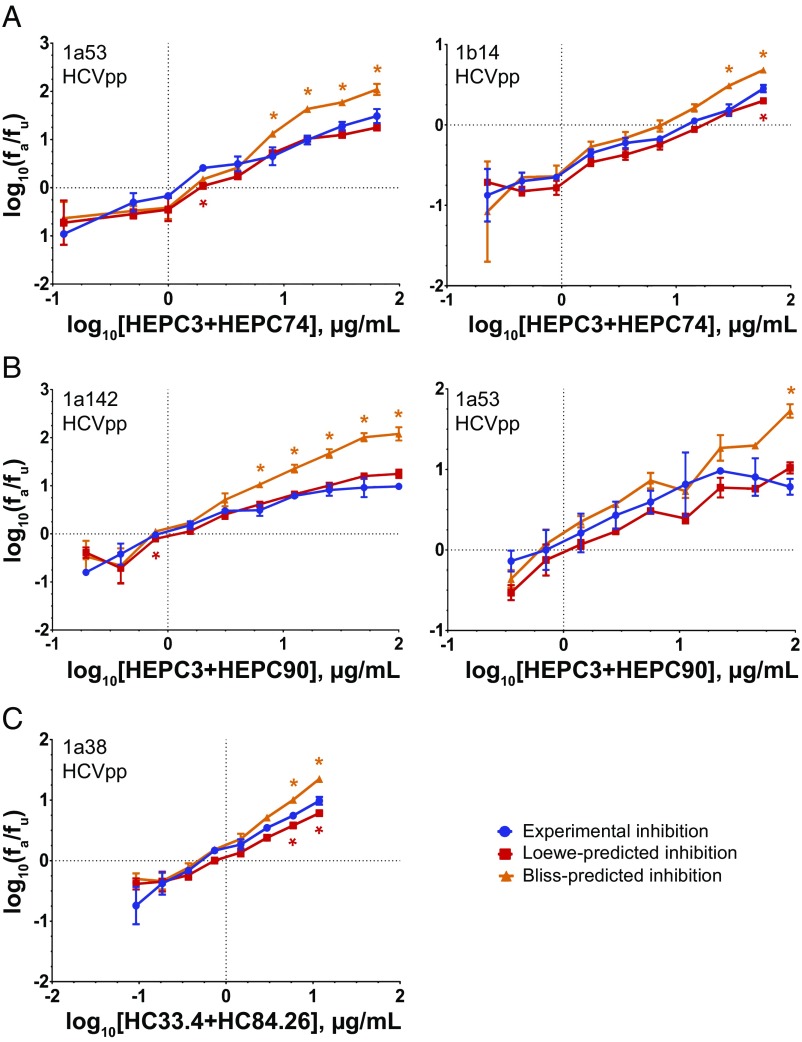
HEPC3/HEPC74, HEPC3/HEPC90, and HC84.26/HC33.4 Neutralization Follows a Pattern Consistent with Loewe Additivity.
To better understand interactions between these NAb pairs, we compared neutralization by serial dilutions of NAb combinations to neutralization predicted by the Loewe additivity or Bliss independence models, which can be calculated from neutralization curves of individual component NAbs. We developed a script in Python to perform these calculations (see Materials and Methods). The Loewe additivity model assumes that two inhibitors have similar mechanisms or compete for the same binding site. Experimental neutralization by a combination matching the Loewe prediction indicates additive effects of the component NAbs, while neutralization greater than or less than the Loewe prediction indicates synergy or antagonism, respectively (30, 33). Experimental neutralization by a combination matching the Bliss independence prediction suggests that the component NAbs have both independent binding sites and independent mechanisms of inhibition (30, 33). We analyzed combination effects relative to the Loewe additivity model, since many NAbs have overlapping binding sites and common inhibitory mechanisms. However, mechanisms of neutralization for many NAbs are not known, and we hypothesized that some might have independent mechanisms, since HCV entry requires multiple cell surface receptors, including CD81, scavenger receptor-B1 (SR-B1), claudin, and occludin (34). We therefore analyzed combination effects relative to the Bliss independence model as well.
As expected, neutralization of strain 1a53 HCVpp by the control combination HEPC3/HEPC74 followed a Loewe additivity pattern (Fig. 5A). Experimental neutralization differed from that predicted by the Loewe model at only one of nine antibody concentrations, while experimental neutralization differed significantly from the Bliss prediction at four of nine concentrations (P < 0.05 for each point, paired two-tailed t test/Holm−Sidak correction for multiple comparisons). These results were confirmed with three additional independent experiments (Fig. S3A). HEPC3/HEPC74 neutralization of a second HCVpp, 1b14, also primarily followed Loewe additivity (Fig. 5A). Experimental neutralization differed from neutralization predicted by the Loewe model at only one of nine concentrations, and differed significantly from the Bliss prediction at two of nine concentrations (P < 0.05 for each), with a trend toward difference from Bliss at two additional points. These results also were confirmed with three additional independent experiments (Fig. S3B). Additive effects between these NAbs were expected, since they bind to largely overlapping epitopes and compete for binding to E1E2 (Fig. 4A).
Fig. 5.
Assessment of NAb combinations for neutralizing synergy, additivity, antagonism, or independence. Median effect plots comparing experimental inhibition (blue) to the inhibition predicted by either the Loewe additivity (red) or Bliss independence (orange) models. Statistically significant differences between the experimental inhibition and Loewe or Bliss predictions are indicated with red and orange asterisks, respectively. Experimental neutralization values are the means of duplicate measurements. Loewe and Bliss values are the means of four individual predictions made at each antibody concentration using data from individual NAb neutralization, which was measured in duplicate. Error bars indicate SDs. Statistical significance was determined by paired t tests, corrected for multiple comparisons using the Holm−Sidak method. (A) HEPC3/HEPC74 tested against 1a53 HCVpp (Left) or 1b14 HCVpp (Right). (B) HEPC3/HEPC90 tested against 1a142 HCVpp (Left) and 1a53 HCVpp (Right). (C) HC84.26/HC33.4 tested against 1a38 HCVpp.
Neutralization by HEPC3/HEPC90 also matched the Loewe additivity prediction (Fig. 5B). Experimental neutralization of strain 1a142 HCVpp by this combination differed from the Loewe prediction at only 1 of 10 concentrations, and differed significantly from the Bliss prediction at 5 of 10 concentrations (P < 0.05 for each). This result was confirmed with two additional independent experiments (Fig. S3C). Experimental neutralization of a second strain, 1a53 HCVpp, differed from Loewe-predicted neutralization at zero of nine concentrations, and differed significantly from the Bliss prediction at one of nine concentrations (P < 0.05 for each), with a trend toward difference from Bliss at two additional points. These results also were confirmed in a second independent experiment (Fig. S3D). Since HEPC3 and HEPC90 do not compete for E1E2 binding, neutralization matching Loewe additivity suggests that these NAbs may act through common or dependent inhibitory mechanisms.
HC84.26/HC33.4 neutralization of 1a38 HCVpp primarily followed Loewe additivity as well, except that at the highest antibody concentrations the combination demonstrated slight synergy relative to the Loewe prediction. At these two concentrations, experimental neutralization was significantly greater than the Loewe prediction, but significantly lower than the Bliss prediction (P < 0.05 for each) (Fig. 5C). This result was confirmed in a second independent experiment (Fig. S3E), and these results agree with analysis of this combination in a prior study (13).
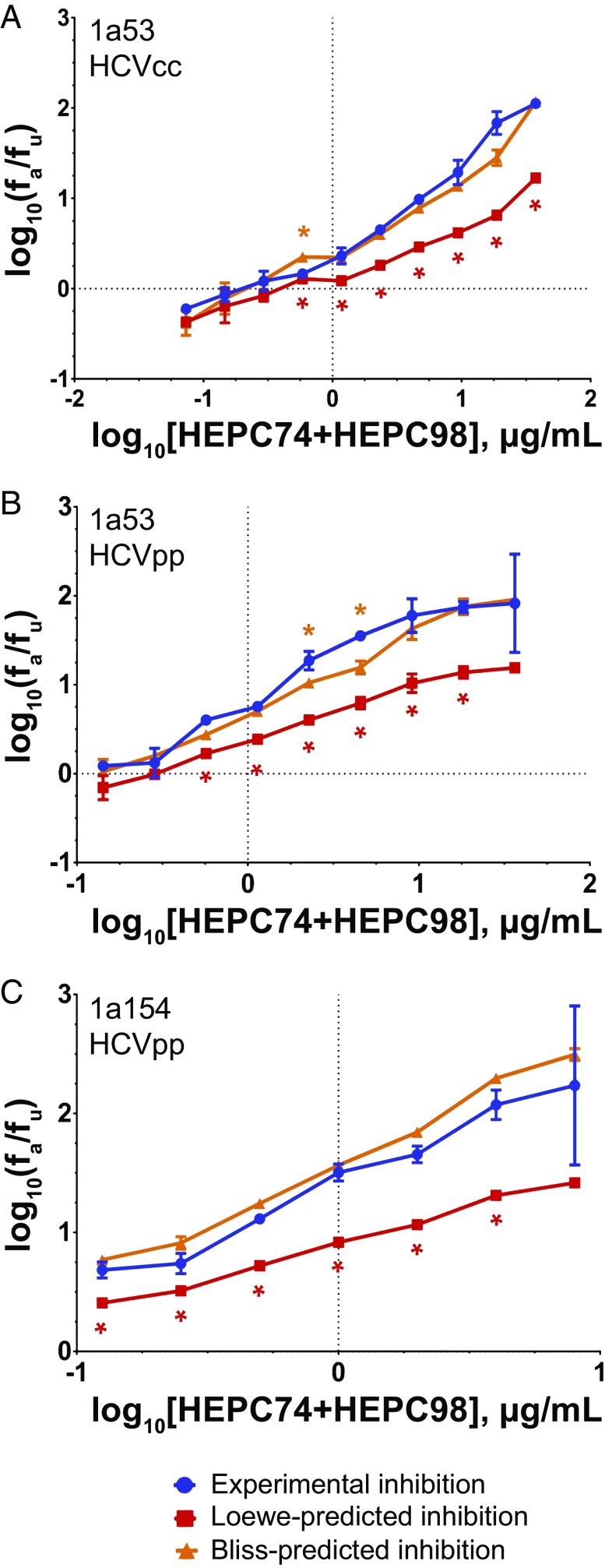
HEPC74/HEPC98 Neutralization Is Synergistic Relative to the Loewe Additivity Model and Follows Bliss Independence.
Neutralization by serial dilutions of HEPC74/HEPC98 was tested using both replication competent cell culture virus (HCVcc) and HCVpp. Remarkably, HEPC74/HEPC98 not only demonstrated strong synergy relative to the Loewe model prediction, but neutralization by the combination also matched the Bliss independence prediction very closely (Fig. 6). Experimental neutralization of 1a53 HCVcc exceeded Loewe-predicted neutralization at 7 of 10 concentrations (P < 0.05 for each), while it differed from Bliss-predicted neutralization at only 1 of 10 concentrations. Notably, neutralization of strain 1a53 HCVpp closely reproduced results obtained with 1a53 HCVcc, in agreement with our prior studies showing that neutralization of HCVpp accurately represents neutralization of E1E2-matched replication competent virus (22, 24). Experimental neutralization of strain 1a53 HCVpp significantly exceeded the neutralization predicted by the Loewe additivity model at six of nine concentrations (P < 0.05 for each), while it differed significantly from the Bliss prediction at only two of nine concentrations. This result was supported by three additional independent experiments (Fig. S3F).
Fig. 6.
HEPC74/HEPC98 neutralization follows Bliss independence. Median effect plots comparing experimental inhibition (blue) to the inhibition predicted by either the Loewe additivity (red) or Bliss independence (orange) models. Statistically significant differences between the experimental inhibition and Loewe or Bliss predictions are indicated with red and orange asterisks, respectively. Experimental neutralization values are the means of duplicate measurements. Loewe and Bliss values are the means of four individual predictions made at each antibody concentration using data from individual NAb neutralization, which was measured in duplicate. Error bars indicate SDs. Statistical significance was determined by paired t tests, corrected for multiple comparisons using the Holm−Sidak method. (A) HEPC74/HEPC98 tested against 1a53 HCVcc. (B) HEPC74/HEPC98 tested against 1a53 HCVpp. (C) HEPC74/HEPC98 tested against 1a154 HCVpp.
HEPC74/HEPC98 neutralization of a second strain, 1a154 (H77) HCVpp, was also synergistic relative to the Loewe model and also followed Bliss independence. Experimental neutralization significantly exceeded Loewe-predicted neutralization at six of seven concentrations (P < 0.05 for each), and differed from Bliss-predicted neutralization at zero of seven concentrations. This result also was supported by two additional independent experiments (Fig. S3G). Taken together, these results suggest that HEPC74 and HEPC98 are potent in combination because the component NAbs have independent inhibitory mechanisms.
HEPC74 and HEPC98 Have Distinct Mechanisms of Inhibition.
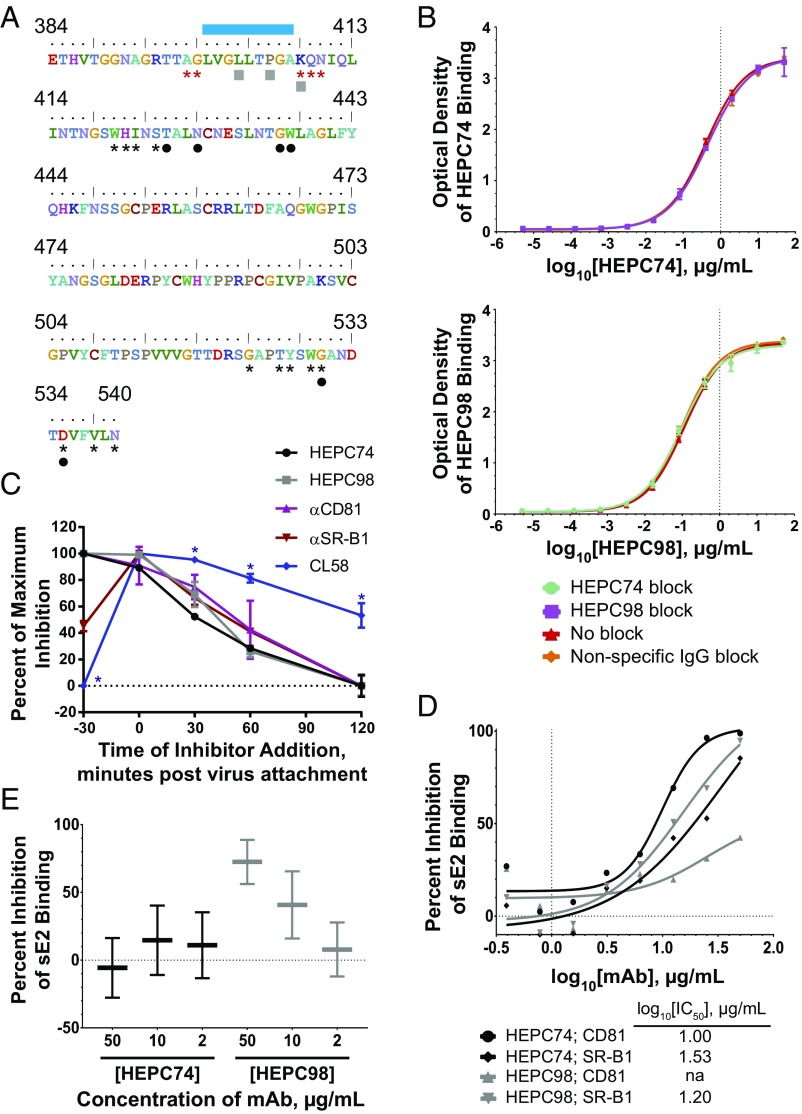
We assessed the binding sites of HEPC74 and HEPC98 on E2 relative to previously identified sites of E2 binding to two primary HCV receptors, CD81 (35) and SR-B1 (36), and an accessory receptor, heparan sulfate (36) (Fig. 7A). The binding site of HEPC74 overlaps with the E2−CD81 binding site, while the binding site of HEPC98 overlaps with the E2−SR-B1 and E2−heparan sulfate binding sites.
Fig. 7.
HEPC74 and HEPC98 have distinct mechanisms of neutralization. (A) Amino acids 384 to 540 of strain 1a154 (H77) E2. Binding residues of HEPC74 and HEPC98 are indicated with black circles and gray squares, respectively. SR-B1 binding residues and the heparan sulfate binding domains determined in a prior study (36) are marked with red asterisks and a blue bar, respectively. CD81 binding residues identified in a prior study (35) are marked with black asterisks. (B) ELISA binding of HEPC74 (Top) or HEPC98 (Bottom) to strain 1a154 E1E2 alone, or in the presence serial dilutions of nonspecific human IgG, HEPC74, or HEPC98. Values are the means of duplicate measurements, and error bars indicate SDs. (C) Timing of inhibition of strain 1a154 HCVpp entry by HEPC74, HEPC98, anti-CD81, anti−SR-B1, or CL58 peptide (claudin inhibitor). HCVpp were incubated with cells at 4 °C for 4 h to allow attachment without entry, then cells were washed and shifted to 37 °C (Time 0) to allow subsequent entry steps. Inhibitors were added 30 min before HCVpp addition to cells (T-30), immediately after attachment (T0), or 30, 60, or 120 min after HCVpp attachment. Maximal activity of each inhibitor was adjusted to 100% to facilitate comparison. Values are the means of duplicates, and error bars indicate SEM. *P < 0.05 by paired t tests between CL58 and all other inhibitors, corrected for multiple comparisons using the Holm−Sidak method. One experiment representative of two independent experiments is shown. (D) Inhibition of strain 1a154 sE2 binding to CHO cells expressing either CD81 or SR-B1 on their surface. The sE2 binding was detected by flow cytometry, and each point was calculated from 10e4 events. Background binding to wild-type CHO cells was subtracted from mean fluorescence intensity (MFI) values. One experiment representative of three independent experiments is shown. (E) Inhibition of strain 1a154 sE2 binding to heparan sulfate. ELISA wells were coated with heparan sulfate, then sE2 that had been preincubated with serial dilutions of HEPC74 or HEPC98 was added. Background binding of sE2 to wells without heparan sulfate was subtracted, and percent inhibition of binding was calculated relative to binding in the presence of nonspecific human IgG. Horizontal lines are means of duplicate wells, and whiskers represent range.
Given data showing that hypervariable region 1 (HVR1), the binding site of HEPC98, can mask the CD81 binding site (37), which is also the binding site of HEPC74, we surmised that binding of HEPC98 might enhance or inhibit binding of HEP74 by exposing or occluding the HEPC74 epitope. To rule this out, we quantitated E1E2 binding of serial dilutions of HEPC74 in the presence or absence of HEPC98, and binding of serial dilutions of HEPC98 in the presence or absence of HEPC74, confirming that binding of one antibody does not enhance or inhibit binding of the other (Fig. 7B).
To further assess mechanisms of neutralization, we measured timing of activity of each NAb relative to timing of HCVpp cell surface attachment. For comparison, we also measured timing of activity of blocking antibodies against CD81 and SR-B1, and of CL58, a peptide inhibitor of claudin, which is a cell surface molecule required for entry after virus binding to CD81 and SR-B1 (38) (Fig. 7C). All inhibitors showed maximal or near-maximal activity when added immediately after binding of HCVpp to hepatoma cells (T0). Interestingly, CL58 and anti−SR-B1 showed reduced inhibitory activity when incubated with cells and then removed from medium before HCVpp attachment (T-30), which could be due to low-affinity binding or inaccessibility of binding sites before virus attachment. HEPC74, HEPC98, anti-CD81, and anti−SR-B1 all showed progressively declining activity relative to T0 when added 30, 60, or 120 min after HCVpp attachment, while CL58 maintained significantly more activity than the other inhibitors at these later time points. Together, these data suggest that HEPC74 and HEPC98 block HCV entry at an early postattachment step, upstream of claudin engagement, with timing of maximal activity similar to anti-CD81 and anti−SR-B1 mAbs.
We then quantified for each NAb inhibition of strain 1a154 (H77) soluble E2 (sE2) binding to CD81 or SR-B1 on the surface of Chinese hamster ovary (CHO) cells (Fig. 7D). We first confirmed that each NAb bound sE2 in ELISA, and also serially diluted sE2 to define the linear range of sE2 binding to CD81−CHO and SR-B1−CHO cells (Fig. S4). A concentration of sE2 within this linear binding range was preincubated with serial dilutions of either HEPC74, HEPC98, or nonspecific human IgG, then used to stain CHO cells. HEPC74 inhibited binding to CD81 (IC50 log10[1.00] µg/mL) and inhibited binding to SR-B1 somewhat less potently (IC50 log10[1.53] µg/mL). As expected given its binding epitope, HEPC98 only showed significant inhibition of binding to SR-B1 (IC50 log10[1.20] µg/mL).
Finally, we measured inhibition of sE2 binding to heparan sulfate, an accessory HCV receptor. As predicted by their E2 binding epitopes, HEPC98 inhibited sE2 binding to heparan, while HEPC74 had no effect. Overall, investigations of HEPC74 and HEPC98 inhibitory mechanisms confirm that HEPC74 binds at the E2−CD81 binding site and inhibits E2−CD81 binding more potently than E2−SR-B1 binding, while HEPC98 binds at the E2−SR-B1 and E2−heparan binding sites and primarily inhibits E2−SR-B1 binding as well as E2−heparan binding. These notable differences between mechanisms of neutralization may explain the observation that neutralization by this combination follows Bliss independence.
Discussion
In this study, we assigned 12 human NAbs to two distinct functional clusters and tested 35 NAb combinations for neutralization of a panel of 11 genetically and antigenically diverse HCVpp. Many NAbs displayed complementary neutralizing breadth, forming combinations with greater neutralization across the HCVpp panel than any individual bNAb. The pairing of HEPC74/HEPC98 showed both enhanced neutralizing breadth and enhanced potency relative to its component NAbs. Remarkably, neutralization with this combination matched neutralization predicted by the Bliss independence model, suggesting that these NAbs act through independent mechanisms. Our investigations of these inhibitory mechanisms suggest that HEPC74 primarily blocks E2−CD81 binding, while HEPC98 primarily blocks E2−SR-B1 and E2−heparan binding.
The discovery of Bliss independence between HEPC74 and HEPC98 is remarkable, since both NAbs bind to the E2 protein. In contrast, many inhibitor pairs that follow Bliss independence, such as drugs used in combination for HIV or HCV treatment, target distinct viral proteins with different functions (33). These data identify a critical vulnerability resulting from the reliance of HCV on multiple cell surface receptors, suggesting that vaccine induction of both HEPC74-like and HEPC98-like antibodies could be particularly effective.
A recent study found antagonism between a murine mAb, H77.16, which binds to HVR1 of E2, and a set of human bNAbs, including a bNAb designated HC-11. These data led to the conclusion that vaccine induction of NAbs against HVR1 might be undesirable (26). In contrast, in our study, the combination of HEPC74 and a human NAb HEPC98, which also targets HVR1, was synergistic and broadly neutralizing, showing that human antibodies targeting HVR1 can be advantageous. The contrast between the findings of these two studies is interesting, since H77.16 and HEPC98 bind to overlapping epitopes, as do HC-11 and HEPC74. These contrasting results may be the result of greater spatial separation between E2-bound HEPC98 and HEPC74 relative to E2-bound H77.16 and HC-11. This model is plausible, since recent studies have suggested that this region of E2 is highly flexible (39, 40), so NAbs with overlapping epitopes might bind to different epitope conformations or bind with different angles of approach. The structural relationship between these adjacent neutralizing epitopes certainly warrants further investigation.
Several recent publications have discussed vaccination strategies designed to favor induction of NAbs against specific E2 epitopes, and limit formation of antibodies against other epitopes. These include molecular scaffold approaches that present single epitopes (41), or truncation of E2 to eliminate variable regions and better expose conserved epitopes (26, 42). In contrast, data presented here suggest that vaccination with full-length E1E2 could also be advantageous, since it might induce combinations of NAbs with distinct mechanisms of neutralization, leading to neutralizing synergy and complementary neutralizing breadth. Alternatively, epitope scaffolds might be designed to present both HEPC74-like and HEPC98-like epitopes.
Several studies have previously evaluated synergy/antagonism between anti-HCV mAbs using models based on Loewe additivity. Carlsen et al. (21) observed synergy between HC84.26 and AR4A, a NAb combination also included in our study. We observed complementation of neutralizing breadth between these NAbs, further suggesting that this pairing may be advantageous. Keck et al. (13) previously evaluated another combination, HC84.26/HC33.4. In agreement with the Keck study, we observed unidirectional competition for E2 binding between the mAbs, as well as additive neutralization at most antibody concentrations, with slight synergy relative to Loewe at the highest antibody concentrations. Interestingly, however, this was one of the less broadly neutralizing combinations in our study (Fig. 3A). This result may be explained, in part, by the presence in the HCVpp panel of four strains with leucine or isoleucine polymorphisms at the 442 locus in E2, which confer resistance to HC84.26 (16, 22). HC84.26 resistance may be slightly exaggerated by this panel, since isoleucine (I) or leucine (L) polymorphisms at position 442 were present in 36% of isolates in this panel of 11 HCVpp, while they are present in only 17% of isolates in the reference panel of 634 genotype 1a and 1b sequences from GenBank. Importantly, however, HC33.4 also showed only marginal potency against some of these same strains, and thus did not fully complement this deficiency in HC84.26 breadth. Together, these results highlight the importance of complementarity of neutralizing breadth in addition to synergy/antagonism when identifying ideal NAb combinations.
A limitation of this study is the use of only genotype 1 HCVpp or HCVcc to measure neutralizing breadth and synergy. We have chosen to focus on this genotype since it is most prevalent worldwide, and no bNAb isolated to date effectively neutralizes all genotype 1 HCV strains (9, 22, 23, 43). While we have selected a panel of genotype 1 HCVpp that is genetically and phenotypically diverse, future studies are needed to confirm and extend these findings using larger panels of genotype 1 HCVpp, as well as HCV from other genotypes. This approach will be particularly important for further evaluation of combinations including HEPC98, since it binds to a highly variable region of E2.
Overall, our evaluation of 35 NAb combinations suggests that enhanced neutralizing breadth by combinations of NAbs from distinct functional neutralization clusters is common. We also discovered a potent and broadly neutralizing combination of NAbs with interactions following Bliss independence, suggesting that NAbs binding to distinct epitopes on E2 may inhibit HCV infection through independent mechanisms. Together, these data define superior NAb combinations, identify a critical vulnerability resulting from the reliance of HCV on multiple cell surface receptors, and suggest that vaccine induction of multiple bNAbs with distinct neutralization profiles is likely to enhance the breadth and potency of the humoral immune response against HCV.
Materials and Methods
See SI Materials and Methods for detailed experimental procedures.
Cell Lines.
HEK293T cells and Hep3B cells were obtained from American Type Culture Collection. CHO cells expressing recombinant human CD81 or SR-B1 were a gift from Matthew Evans, Icahn School of Medicine, Mount Sinai, New York.
Antibodies.
MAbs CBH-5 (11), HC84.26 (5), and HC33.4 (13) were a gift of Steven Foung, Stanford University School of Medicine, Stanford, CA. MAbs AR3B, AR3C (10), AR4A, and AR5A (9) were a gift of Mansun Law, Scripps Research Institute, La Jolla, CA. All other antibodies were isolated in the laboratory of J.E.C. (20).
Hierarchical Clustering of NAbs.
NAbs were grouped into functionally related clusters based upon their neutralization profiles as previously described (20, 22).
E1E2 Sequence Analysis.
Evolutionary analyses were conducted in Molecular Evolutionary Genetics Analysis version 7 (MEGA7) (44). Diversity plots were generated using VarPlot v1.2 (45) (https://sray.med.som.jhmi.edu/SCRoftware/VarPlot/) with 1aa steps and 20 aa windows. Inclusion of reference panel polymorphisms in the 11 and 19 HCVpp panels was calculated using a previously described reference alignment from GenBank (32) and the package “seqinr” in R, as previously described (22).
HCVpp Production and Neutralization.
HCVpp were produced by lipofectamine-mediated transfection of HCV E1E2, pNL4-3.Luc.R-E-, and pAdVantage (Promega) plasmids into HEK293T cells as previously described (22, 46, 47). Neutralization assays were performed as described previously (48).
HCVcc Production and Neutralization.
Generation of an HCVcc chimera expressing the 1a53 E1E2 proteins was previously described (24, 25). HCVcc neutralization assays were performed in triplicate as previously described (24, 25).
Identification of Synergy/Additivity/Antagonism or Independence Using the Loewe Additivity and Bliss Independence Models.
Briefly, for four selected NAb combinations, two HCVpp were selected for each NAb combination that were at least partially neutralized by 10 µg/mL of each NAb in that combination [fraction unaffected (fu) < 0.3, except HEPC3/1b14 (0.63), HEPC90/1a53 (0.65)]. The combination of HEPC74/HEPC98 was also tested using 1a53 strain HCVcc. Neutralization by serial twofold dilutions of NAbs combined in a fixed ratio adjusted for their relative IC50s was measured (30). Neutralization by serial dilutions of individual component NAbs was simultaneously measured, using the same antibody concentrations tested in the NAb combination neutralization curve. The fu and fraction affected (fa) values from the individual NAb neutralization curves were used to calculate neutralization for each NAb combination predicted by the Loewe additivity and the Bliss independence models.
Quality Control for Loewe/Bliss Analysis.
Only individual NAb curves with linear relationships on median effect plot with R squared of >0.85 were used to calculate Loewe additivity and Bliss independence curves (Fig. S5). To confirm that NAbs were combined in optimal ratios to allow discrimination between Loewe additivity and Bliss independence, predicted Loewe and Bliss curves for each combination were confirmed to be statistically different from each other for at least three antibody concentrations (Fig. S5).
Binding Competition Assays.
MAb binding to E1E2 was quantitated using ELISA as previously described (7).† Blocking NAbs were added at either 50 µg/mL or 20 µg/mL, and biotinylated binding NAbs were added at their EC75.
Timecourse Inhibition Experiments.
HCVpp were incubated with cells at 4 °C for 4 h to allow attachment without entry, then cells were washed and shifted to 37 °C (Time 0) to allow subsequent entry steps. Inhibitors HEPC74, HEPC98, CL58 peptide (38), anti-CD81 mAb (BD 555675), anti−SR-B1 mAb (BD 610882), or isotype control mAb (BD 560550) were added 30 min before HCVpp addition to cells (T-30), immediately after attachment (T0), or 30, 60, or 120 min after HCVpp attachment.
Expression of sE2.
A truncated, soluble form of strain 1a154 (H77) E2 ectodomain (sE2), encompassing residues 384 to 645, as previously described (49), was cloned into a mammalian expression vector (phCMV3_Ig Kappa_HIS, a gift of Leopold Kong, The Scripps Research Institute, La Jolla, CA) and expressed by transfection into HEK293T or HEK293-6E cells.
sE2 Binding to CHO Cells.
CHO−CD81 and CHO−SR-B1 binding experiments were carried out as previously described (23, 50).
sE2 Binding to Heparan Sulfate.
Heparan binding experiments were carried out essentially as previously described (36).
Statistical Analysis.
All statistical analyses were carried out in Prism v7.02 (Graphpad Software). Neutralizing breadth and neutralizing potency were compared by unpaired, two-sided t test. Experimental neutralization values were compared with Loewe-predicted and Bliss-predicted values by multiple t tests, corrected for multiple comparisons by the Holm−Sidak method, without assumption of consistent SD between points.
Study Approval.
All protocols were approved by the Institutional Review Board of the Johns Hopkins Hospital, and informed consent was obtained from all study participants.
Supplementary Material
Acknowledgments
We thank Dr. Robert Siliciano for useful discussions, Michelle Colbert for technical support, James Notaro for Python guidance, and Dr. Mansun Law and Dr. Steven Foung for the generous gift of monoclonal antibodies. We thank Tiffany Luong and the Caltech Protein Expression Center for help with sE2 expression. A.I.F. is a Cancer Research Institute Irvington Fellow supported by the Cancer Research Institute. This project received support from the US National Institutes of Health Grants K08 AI102761, U19 AI088791, and R01 AI127469.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
†Netski DM, et al., 11th International Symposium on Hepatitis C Virus and Related Viruses, October 3−7, 2004, Heidelberg, Germany.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1718441115/-/DCSupplemental.
References
- 1.Armstrong GL, et al. The prevalence of hepatitis C virus infection in the United States, 1999 through 2002. Ann Intern Med. 2006;144:705–714. doi: 10.7326/0003-4819-144-10-200605160-00004. [DOI] [PubMed] [Google Scholar]
- 2.Simmons B, Saleem J, Hill A, Riley RD, Cooke GS. Risk of late relapse or reinfection with hepatitis C virus after achieving a sustained virological response: A systematic review and meta-analysis. Clin Infect Dis. 2016;62:683–694. doi: 10.1093/cid/civ948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cox AL. MEDICINE. Global control of hepatitis C virus. Science. 2015;349:790–791. doi: 10.1126/science.aad1302. [DOI] [PubMed] [Google Scholar]
- 4.Falade-Nwulia O, Sulkowski M. The HCV care continuum does not end with cure: A call to arms for the prevention of reinfection. J Hepatol. 2017;66:267–269. doi: 10.1016/j.jhep.2016.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Keck ZY, et al. Human monoclonal antibodies to a novel cluster of conformational epitopes on HCV E2 with resistance to neutralization escape in a genotype 2a isolate. PLoS Pathog. 2012;8:e1002653. doi: 10.1371/journal.ppat.1002653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Keck ZY, et al. Mapping a region of hepatitis C virus E2 that is responsible for escape from neutralizing antibodies and a core CD81-binding region that does not tolerate neutralization escape mutations. J Virol. 2011;85:10451–10463. doi: 10.1128/JVI.05259-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keck ZY, et al. Mutations in hepatitis C virus E2 located outside the CD81 binding sites lead to escape from broadly neutralizing antibodies but compromise virus infectivity. J Virol. 2009;83:6149–6160. doi: 10.1128/JVI.00248-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kong L, et al. Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1. Proc Natl Acad Sci USA. 2012;109:9499–9504. doi: 10.1073/pnas.1202924109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Giang E, et al. Human broadly neutralizing antibodies to the envelope glycoprotein complex of hepatitis C virus. Proc Natl Acad Sci USA. 2012;109:6205–6210. doi: 10.1073/pnas.1114927109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Law M, et al. Broadly neutralizing antibodies protect against hepatitis C virus quasispecies challenge. Nat Med. 2008;14:25–27. doi: 10.1038/nm1698. [DOI] [PubMed] [Google Scholar]
- 11.Hadlock KG, et al. Human monoclonal antibodies that inhibit binding of hepatitis C virus E2 protein to CD81 and recognize conserved conformational epitopes. J Virol. 2000;74:10407–10416. doi: 10.1128/jvi.74.22.10407-10416.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keck ZY, et al. A point mutation leading to hepatitis C virus escape from neutralization by a monoclonal antibody to a conserved conformational epitope. J Virol. 2008;82:6067–6072. doi: 10.1128/JVI.00252-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keck Z, et al. Cooperativity in virus neutralization by human monoclonal antibodies to two adjacent regions located at the amino terminus of hepatitis C virus E2 glycoprotein. J Virol. 2013;87:37–51. doi: 10.1128/JVI.01941-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krey T, et al. Structural basis of HCV neutralization by human monoclonal antibodies resistant to viral neutralization escape. PLoS Pathog. 2013;9:e1003364. doi: 10.1371/journal.ppat.1003364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansson DX, et al. Human combinatorial libraries yield rare antibodies that broadly neutralize hepatitis C virus. Proc Natl Acad Sci USA. 2007;104:16269–16274. doi: 10.1073/pnas.0705522104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keck ZY, et al. Affinity maturation of a broadly neutralizing human monoclonal antibody that prevents acute hepatitis C virus infection in mice. Hepatology. 2016;64:1922–1933. doi: 10.1002/hep.28850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.de Jong YP, et al. Broadly neutralizing antibodies abrogate established hepatitis C virus infection. Sci Transl Med. 2014;6:254ra129. doi: 10.1126/scitranslmed.3009512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Osburn WO, et al. Clearance of hepatitis C infection is associated with the early appearance of broad neutralizing antibody responses. Hepatology. 2014;59:2140–2151. doi: 10.1002/hep.27013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pestka JM, et al. Rapid induction of virus-neutralizing antibodies and viral clearance in a single-source outbreak of hepatitis C. Proc Natl Acad Sci USA. 2007;104:6025–6030. doi: 10.1073/pnas.0607026104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bailey JR, et al. Broadly neutralizing antibodies with few somatic mutations and hepatitis C virus clearance. JCI Insight. 2017;2:92872. doi: 10.1172/jci.insight.92872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlsen TH, et al. Breadth of neutralization and synergy of clinically relevant human monoclonal antibodies against HCV genotypes 1a, 1b, 2a, 2b, 2c, and 3a. Hepatology. 2014;60:1551–1562. doi: 10.1002/hep.27298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bailey JR, et al. Naturally selected hepatitis C virus polymorphisms confer broad neutralizing antibody resistance. J Clin Invest. 2015;125:437–447. doi: 10.1172/JCI78794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El-Diwany R, et al. Extra-epitopic hepatitis C virus polymorphisms confer resistance to broadly neutralizing antibodies by modulating binding to scavenger receptor B1. PLoS Pathog. 2017;13:e1006235. doi: 10.1371/journal.ppat.1006235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wasilewski LN, Ray SC, Bailey JR. Hepatitis C virus resistance to broadly neutralizing antibodies measured using replication-competent virus and pseudoparticles. J Gen Virol. 2016;97:2883–2893. doi: 10.1099/jgv.0.000608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wasilewski LN, et al. A hepatitis C virus envelope polymorphism confers resistance to neutralization by polyclonal sera and broadly neutralizing monoclonal antibodies. J Virol. 2016;90:3773–3782. doi: 10.1128/JVI.02837-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Keck ZY, et al. Antibody response to hypervariable region 1 interferes with broadly neutralizing antibodies to hepatitis C virus. J Virol. 2016;90:3112–3122. doi: 10.1128/JVI.02458-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang P, et al. Depletion of interfering antibodies in chronic hepatitis C patients and vaccinated chimpanzees reveals broad cross-genotype neutralizing activity. Proc Natl Acad Sci USA. 2009;106:7537–7541. doi: 10.1073/pnas.0902749106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kachko A, et al. Antibodies to an interfering epitope in hepatitis C virus E2 can mask vaccine-induced neutralizing activity. Hepatology. 2015;62:1670–1682. doi: 10.1002/hep.28108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loewe S, Muischnek H. Combinated effects I announcement–Implements to the problem. Naunyn Schmiedebergs Arch Exp Pathol Pharmakol. 1926;114:313–326. [Google Scholar]
- 30.Chou TC. Theoretical basis, experimental design, and computerized simulation of synergism and antagonism in drug combination studies. Pharmacol Rev. 2006;58:621–681. doi: 10.1124/pr.58.3.10. [DOI] [PubMed] [Google Scholar]
- 31.Bliss CI. The toxicity of poisons applied jointly. Ann Appl Biol. 1939;26:585–615. [Google Scholar]
- 32.Munshaw S, et al. Computational reconstruction of Bole1a, a representative synthetic hepatitis C virus subtype 1a genome. J Virol. 2012;86:5915–5921. doi: 10.1128/JVI.05959-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jilek BL, et al. A quantitative basis for antiretroviral therapy for HIV-1 infection. Nat Med. 2012;18:446–451. doi: 10.1038/nm.2649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zeisel MB, Fofana I, Fafi-Kremer S, Baumert TF. Hepatitis C virus entry into hepatocytes: Molecular mechanisms and targets for antiviral therapies. J Hepatol. 2011;54:566–576. doi: 10.1016/j.jhep.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 35.Owsianka AM, et al. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J Virol. 2006;80:8695–8704. doi: 10.1128/JVI.00271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guan M, et al. Three different functional microdomains in the hepatitis C virus hypervariable region 1 (HVR1) mediate entry and immune evasion. J Biol Chem. 2012;287:35631–35645. doi: 10.1074/jbc.M112.382341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bankwitz D, et al. Hepatitis C virus hypervariable region 1 modulates receptor interactions, conceals the CD81 binding site, and protects conserved neutralizing epitopes. J Virol. 2010;84:5751–5763. doi: 10.1128/JVI.02200-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Si Y, et al. A human claudin-1-derived peptide inhibits hepatitis C virus entry. Hepatology. 2012;56:507–515. doi: 10.1002/hep.25685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kong L, et al. Structural flexibility at a major conserved antibody target on hepatitis C virus E2 antigen. Proc Natl Acad Sci USA. 2016;113:12768–12773. doi: 10.1073/pnas.1609780113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Vasiliauskaite I, et al. Conformational flexibility in the immunoglobulin-like domain of the hepatitis C virus glycoprotein E2. MBio. 2017;8:e00382-17. doi: 10.1128/mBio.00382-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pierce BG, et al. Structure-based design of hepatitis C virus vaccines that elicit neutralizing antibody responses to a conserved epitope. J Virol. 2017;91:e01032-17. doi: 10.1128/JVI.01032-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prentoe J, Velázquez-Moctezuma R, Foung SK, Law M, Bukh J. Hypervariable region 1 shielding of hepatitis C virus is a main contributor to genotypic differences in neutralization sensitivity. Hepatology. 2016;64:1881–1892. doi: 10.1002/hep.28705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keck ZY, et al. Affinity maturation of a broadly neutralizing human monoclonal antibody that prevents acute hepatitis C virus infection in mice. Hepatology. 2016;64:1922–1933. doi: 10.1002/hep.28850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ray SC, et al. Acute hepatitis C virus structural gene sequences as predictors of persistent viremia: Hypervariable region 1 as a decoy. J Virol. 1999;73:2938–2946. doi: 10.1128/jvi.73.4.2938-2946.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hsu M, et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc Natl Acad Sci USA. 2003;100:7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Logvinoff C, et al. Neutralizing antibody response during acute and chronic hepatitis C virus infection. Proc Natl Acad Sci USA. 2004;101:10149–10154. doi: 10.1073/pnas.0403519101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Dowd KA, Netski DM, Wang XH, Cox AL, Ray SC. Selection pressure from neutralizing antibodies drives sequence evolution during acute infection with hepatitis C virus. Gastroenterology. 2009;136:2377–2386. doi: 10.1053/j.gastro.2009.02.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kong L, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–1094. doi: 10.1126/science.1243876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sabo MC, et al. Neutralizing monoclonal antibodies against hepatitis C virus E2 protein bind discontinuous epitopes and inhibit infection at a postattachment step. J Virol. 2011;85:7005–7019. doi: 10.1128/JVI.00586-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Suzuki R, Shimodaira H. Pvclust: An R package for assessing the uncertainty in hierarchical clustering. Bioinformatics. 2006;22:1540–1542. doi: 10.1093/bioinformatics/btl117. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.