Abstract
The present study aimed to develop a rational therapy based on the genetic epidemiology, molecular mechanism evaluation and in vitro antibiotic combinations activity in multidrug-resistant Acinetobacter baumannii (MDRAB). MDRAB was screened by the Kirby-Bauer method. The random amplified polymorphic DNA technique was used to establish genetic fingerprinting, and a series of resistance genes were detected by polymerase chain reaction. Antimicrobial agents including amikacin (AK), cefoperazone/sulbactam (SCF I/II), meropenem (MEM), minocycline (MINO) and ciprofloxacin (CIP) were used to determine the minimum inhibitory concentrations (MICs) and interactions between antibiotics by the broth microdilution method and chequerboard assays. In total, 34 MDRAB strains were isolated and classified into 8 phenotypes A-H, according to their general drug susceptibilities. A total of 4 major genotypes (I–IV) were clustered at 60% a genotypic similarity threshold. High positive rates of β-lactamase TEM-1, topoisomerase IV, oxacillinase (OXA)-23, AdeB family multidrug efflux RND transporter adeB, β-lactamase AmpC, class 1 integrons (Int-1), 16S rRNA methylase rmtA, phosphotransferase aph(3), 16S rRNA methyltransferase armA were presented to exceed 90%, acetylyltransferase aac(3)-I, aac(6′-I, ant(3″)-I, 16S rRNA methylase rmtB, oxacillinase OXA-24 and metallo-β-lactamase IMP-5 genes demonstrated positive rates of 29.4–85.29%, while adeRS two-component system was not observed in any strain. MEM+SCF I or SCF II primarily exhibited synergistic effects. AK+SCF I, AK+SCF II, MINO+SCF I, MINO+SCF II, MINO+CIP and MINO+MEM primarily presented additive effects. AK+CIP demonstrated 70.59% antagonism. The antibacterial activity of SCF I was superior compared with that of SCF II. The results indicated the polyclonal genetic epidemiological trend of MDRAB in the Second Xiangya Hospital, and verified the complexity of genetic resistance. In addition, combinations suggested to be efficacious were MEM+SCF I and MEM+SCF II, which were more effective compared with other combinations for the management of MDRAB infection.
Keywords: multidrug-resistant Acinetobacter baumannii, resistance genes, in vitro antimicrobial activity, antibiotic interaction
Introduction
Acinetobacter baumannii, a leading nosocomial pathogen, has been identified to induce serious infections and high mortality rates in intensive care units (ICUs) (1,2). Multidrug-resistant Acinetobacter baumannii (A. baumannii) (MDRAB), with resistance to at least three classes of antibiotics among cephalosporins, carbapenems, β-lactamases, aminoglycosides and quinolones (3), is only susceptible to certain agents such as tigecycline, and polymyxins due to intrinsic and acquired resistance (4,5). In addition, several pandrug-resistant (PDR) A. baumannii strains with resistance to almost all available antibiotics have been identified in previous decades (6,7).
Abuse of broad-spectrum antibiotics has been demonstrated to be a major cause for the development of drug resistance of A. baumannii. At present, a number of genes responsible for drug resistance have been identified though long-term studies on the mechanisms of bacterial resistance (7–10). Production of β-lactamase is suggested to be associated with the bacterial resistance to penicillin, cephalosporins and carbapenems (9). At present, four major categories have been available for the β-lactamase protein-encoding genes, including narrow-spectrum β-lactamase, extended-spectrum β-lactamase (ESBLs), metallo-β-lactamases (MBLs) and oxacillinase (OXA)-type carbapenemases (10). Bacterial resistance to aminoglycoside usually results in the production of aminoglycoside-modifying enzymes. Aminoglycoside resistance genes, including acetyltransferase (aac), phosphotransferase (aph) and adenylyltransferase (aad), have been frequently identified in MDRAB (11). For example, MDRAB has been demonstrated to acquire antimicrobial resistance genes via class 1 integrons (Int-1), which contain single or multiple gene cassettes (12). Carbapenemase and aminoglycoside resistance genes were localized within Int-1 (13). In addition, 16S rRNA methylases may confer resistance to aminoglycosides (14). The increased production of fluoroquinolone-resistant A. baumannii was demonstrated to be markedly associated with spontaneous mutations in the quinolone resistance-determining regions (QR-DRs) in DNA gyrase (gyrA) or topoisomerase IV (parC) (15).
MDRAB remains a challenge for the clinical management of life-threatening infections, including bacteremia, pneumonia and wound infections. At present, MDRAB is a lethal threat to public health due to the lack of effective antimicrobial agents available. Currently, combination therapy has been considered to be a promising method for the management of infection. Previous studies revealed that tigecycline and polymyxins were active against MDRAB (4,5), but their application is inhibited due to high toxicity and low commercial availability in China. Consequently, the effective combinations of clinical drugs may be an improved choice for treating MDRAB infection. In the present study, the genotypes and encoding resistance genes of MDRAB were determined. Based on the phenotypic analysis, the gene structure and molecular determinants that confer alternative MDRAB phenotypes were investigated. Furthermore, five drugs were used to evaluate the in vitro activity of various antibiotic combinations against MDRAB, in order to provide reliable data to support novel clinical combination therapies.
Materials and methods
Bacterial isolates
A total of 34 consecutive and non-repetitive MDRAB strains were identified using the MicroScan WalkAway-96 system (Siemens AG, Munich, Germany) from the Second Xiangya Hospital (Changsha, China) between February 2011 and May 2011. Kirby-Bauer antibiotic susceptibility testing (K-B test) was utilized to determine the susceptibility to several clinically significant antimicrobial agents. MDRAB was defined as the presence of resistance to at least three classes of antibiotics, including: Cephalosporins, carbapenems, β-lactamases, aminoglycosides and quinolones. In the present study, a total of 34 strains were collected (Table I), 30 of which were identified from sputum samples from patients with nosocomial pneumonia, referring to criteria for radiologically-confirmed pneumonia occurring ≥48 h after hospitalization in non-intubated patients (16), 3 were isolated from wound secretion and one was isolated from fluid drainage. The samples were primarily collected from the ICU Respiratory and Cardiothoracic Surgery departments. The interpretation of susceptibility test and breakpoints was performed according to the Clinical and Laboratory Standard Institute (CLSI) criteria (17). The present study was approved by the Ethics Committee of The Second Xiangya Hospital, Central South University (Changsha, China). Written informed consent was obtained from all patients.
Table I.
Characteristics of study isolates.
| Clinical antimicrobial susceptibility | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NOx | PT | Date of strain separated | Ward | TZP | CAZ | SCF | ATM | IPM | AK | FEP | MEM | TIM | CIP | SXT | CTX | LEV | CN | SAM |
| 1AI | 51M | 3/9 | 3 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| 2AI | 13M | 3/9 | 4 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| 3BI | 46M | 3/23 | 1 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 4AI | 64M | 3/22 | 10 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| 5AI | 34M | 3/11 | 8 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| 6BI | 1M | 3/17 | 3 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 7CI | 65M | 3/12 | 1 | R | R | R | R | R | R | R | R | R | R | R | R | I | R | R |
| 8AI | 7M | 5/14 | 4 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| 9DI | 88F | 3/31 | 6 | R | R | S | R | R | R | R | R | R | R | R | R | R | R | R |
| 10BI | 33F | 3/28 | 3 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 11BII | 25F | 3/30 | 3 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 12BII | 48F | 3/28 | 1 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 13BII | 30F | 3/28 | 1 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 14BII | 47F | 4/2 | 2 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 15BII | 55M | 4/21 | 3 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 16BIV | 25M | 4/21 | 1 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 17AII | 63M | 3/20 | 1 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| 18BI | 72F | 3/23 | 1 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 19CI | 57M | 3/12 | 1 | R | R | R | R | R | R | R | R | R | R | R | R | I | R | R |
| 20AI | 78M | 3/18 | 1 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| 21AII | 41M | 3/10 | 4 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| 22EII | 80F | 3/30 | 1 | R | R | I | R | R | R | R | R | R | R | R | R | R | I | R |
| 23FI | 46M | 4/5 | 4 | R | R | I | R | R | R | I | R | R | R | R | R | R | R | R |
| 24BII | 43M | 4/3 | 9 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 25AII | 55M | 4/5 | 5 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| 26AII | 55M | 4/6 | 5 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| 27BII | 57F | 4/7 | 2 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 28AI | 70F | 4/7 | 2 | R | R | R | R | R | R | R | R | R | R | R | R | R | R | R |
| 29DII | 69M | 4/5 | 2 | R | R | S | R | R | R | R | R | R | R | R | R | R | R | R |
| 30GII | 87F | 2/28 | 1 | R | S | R | R | R | R | R | R | R | R | R | R | R | S | S |
| 31BIII | 56M | 2/28 | 1 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 32BI | 6F | 2/28 | 7 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 33BI | 27M | 3/4 | 1 | R | R | I | R | R | R | R | R | R | R | R | R | R | R | R |
| 34HI | 79M | 2/28 | 1 | R | S | I | R | R | R | R | R | R | R | R | R | R | R | R |
PT, patient age and sex. Wards: 1, Intensive care unit; 2, Department of Respiratory; 3, Cardiothoracic surgery; 4, Orthopedics; 5, Neurology; 6, Geriatric Ward; 7, Department of Minimally Invasive Surgery; 8, Neurosurgery; 9, Urology; 10, Digestive system department. Antimicrobials: TZP, piperacillin/tazobactam; CAZ, ceftazidime; SCF, cefoperazone/sulbactam; ATM, aztreonam; IPM, imipenem; AK, amikacin; FEP, cefepime; MEM, meropenem; TIM, ticarcillin/clavulanic acid; CIP, ciprofloxacin; SXT, trimethoprim/sulfamethoxazole; CTX, cefotaxime; LEV, levofloxacin; CN, gentamicin; SAM, ampicillin/sulbactam. NOx: The isolates were classified into 8 phenotypes (A-H) according to their susceptibility to the tested clinical antimicrobials; A, resistant to all the aforementioned drugs; B, only intermediate to SCF; C, only intermediate to LEV; D, only susceptible to SCF; E, intermediate to SCF and CN; F, intermediate to SCF and FEP; G, susceptible to CAZ, CN and SAM; H, susceptible to CAZ and intermediate to SCF; R, resistance; I, intermediary; S, sensitivity.
Random amplified polymorphic DNA (RAPD) genotyping and detection of drug resistance genes by polymerase chain reaction (PCR)
Bacterial DNA was extracted and purified by using Tiangen UltraClean Microbial DNA Isolation kit (Tiangen Biotech Co., Ltd., Beijing, China) according to the manufacturer's instructions. MDRAB genotyping was conducted using RAPD (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA) with an AP2 primer (5-GTTTCGCTCC-3) designed by Primers Express software (version 2.0; Applied Biosystems; Thermo Fisher Scientific, Inc.) and synthesized by Sangon Biotech Co., Ltd. (Shanghai, China), as previously described (18). The genes encoding resistance, including β-lactamase TEM-1, AmpC, metallo-β-lactamase IMP-5, oxacillinase (OXA)-23, OXA-24, acetylyltransferase aac(3)-I, aac(6′)-I, ant(3″)-I, 16S rRNA methyltransferase armA, 16S rRNA methylase rmtA, rmtB, phosphotransferase aph-(3), AdeB family multidrug efflux RND transporter adeB, adeRS two-component system, Int-1 and ParC genes, were detected by PCR. All primers were designed based on the sequences in GenBank (19) using Primer Express software v2.0 (Applied Biosystems; Thermo Fisher Scientific, Inc., Foster City, CA, USA) (Table II). The PCR total reaction volume of 20 µl, containing 10 µl 2X TaqMan PCR master mix, 1 µl forward and reverse primers (10 µmol/l), 7 µl ddH2O and 1 µl DNA template. The amplification conditions of the target genes were presented in Table III. Following amplification, 3 µl of products were electrophoresed on a 1.5% agarose gel (Oxoid; Thermo Fisher Scientific, Inc.) and visualized using ethidium bromide (Sigma-Aldrich; Merck KGaA, Darmstadt, Germany) for 20 min at a voltage of 150 V.
Table II.
Primers of resistance genes.
| Primer sequences (5′-3′) | |||
|---|---|---|---|
| Target genes | Forward | Reverse | Size, bp |
| TEM-1 | TTCGTGTCGCCCTTATTC | ACGCTCGTCGTTTGGTAT | 512 |
| IMP-5 | CTACCGCAGCAGAGTCTTTG | AACCAGTTTTGCCTTACCAT | 587 |
| OXA-23 | TGTCATAGTATTCGTCGTT | TTCCCAAGCGGTAAA | 453 |
| OXA-24 | TTTGCCGATGACCTT | TAGCTTGCTCCACCC | 175 |
| AmpC | CGACAGCAGGTGGAT | GGTTAAGGTTGGCATG | 510 |
| aac(3)-I | ACCTACTCCCAACATCAGCC | ATATAGATCTCACTACGCGC | 158 |
| aac(6′)-I | TATGAGTGGCTAAATCGA | CCCGCTTTCTCGTAGCA | 395 |
| ant(3″)-I | TGATTTGCTGGTTACGGTGAC | CGCTATGTTCTCTTGCTTTTG | 284 |
| armA | GGGGTCTTACTATTCTG | TTCCCTTCTCCTTTC | 503 |
| rmtA | CCTAGCGTCCATCCTTTCCTC | AGCGATATCCAACACACGATGG | 315 |
| rmtB | ATGAACATCAACGATGCCCTC | TTATCCATTCTTTTTTATCAAGTATAT | 756 |
| aph(3) | ATACAGAGACCACCATACAGT | GGACAATCAATAATAGCAAT | 234 |
| adeB | GTATGAATTGATGCTGC | CACTCGTAGCCAATACC | 1,000 |
| adeRS | CTCAGACTCCCGTGATCATGTTG | CGTAAGTCTTCGACTAAGTGAGA | 1,115 |
| Int-1 | GCACCGCCAACTTTC | CCTTGATGTTACCCGAGA | 433 |
| ParC | CTGAACAGGCTTACTTGAA | AAGTTATCTTGCCATTCG | 200 |
Table III.
Polymerase chain reaction conditions of target genes.
| Amplification | ||||||
|---|---|---|---|---|---|---|
| Target genes | Initialdenaturation (°C, min) | Denaturation (°C, sec) | Annealing (°C, sec) | Extension (°C, sec) | Cycles (n) | Final extension (°C, min) |
| AP2 | 95, 6 | 95, 45 | 33, 45 | 72, 120 | 45 | 72, 10 |
| TEM-1 | 94, 5 | 94, 60 | 55, 60 | 72, 50 | 30 | 72, 7 |
| IMP | 94, 5 | 94, 60 | 55, 60 | 72, 50 | 30 | 72, 7 |
| OXA-23 | 94, 5 | 94, 30 | 48, 30 | 72, 35 | 30 | 72, 10 |
| OXA-24 | 94, 5 | 94, 30 | 48, 30 | 72, 35 | 30 | 72, 10 |
| AmpC | 94, 5 | 94, 30 | 50, 30 | 72, 50 | 30 | 72, 10 |
| aac(3)-I | 94, 5 | 94, 30 | 55, 30 | 72, 30 | 35 | 72, 10 |
| aac(6′)-I | 94, 5 | 94, 30 | 55, 30 | 72, 30 | 35 | 72, 10 |
| ant(3″)-I | 94, 5 | 94, 30 | 55, 30 | 72, 30 | 35 | 72, 10 |
| rmtA | 93, 2 | 93, 20 | 55, 30 | 72, 30 | 30 | 72, 5 |
| aph (3) | 93, 2 | 93, 20 | 55, 30 | 72, 30 | 30 | 72, 5 |
| armA | 94, 5 | 94, 30 | 47, 30 | 72, 35 | 30 | 72, 10 |
| rmtB | 94, 5 | 94, 30 | 55, 30 | 72, 60 | 30 | 72, 10 |
| adeB | 95, 5 | 95, 30 | 53, 60 | 72, 60 | 30 | 72, 7 |
| adeRS | 95, 5 | 95, 30 | 53, 40 | 72, 60 | 30 | 72, 7 |
| Int-1 | 94, 5 | 94, 30 | 53, 30 | 72, 60 | 30 | 72, 10 |
| ParC | 94, 4 | 94, 30 | 53, 30 | 72, 40 | 30 | 72, 7 |
Antimicrobial agents and minimum inhibitory concentration (MIC) assays
Antimicrobial agents used in the present study were amikacin (AK), cefoperazone/sulbactam [SCF, SCFI 1:1 (cefoperazone:sulbactam) and SCFII 2:1 (cefoperazone:sulbactam)], meropenem (MEM), minocycline (MINO) and ciprofloxacin (CIP). MIC assays were performed in 96-well microtiter plates by the broth microdilution method, according to the CLIS protocol (17). Bacteria were cultured in 10% horse blood agar (Oxoid; Thermo Fisher Scientific, Inc.) for 20–24 h until cells reached the exponential phase. The inoculums were adjusted with fresh Cationic adjustment of Mueller-Hinton Broth [Oxoid; Thermo Fisher Scientific, Inc.; CAMHB, containing Ca2+ (10–25 mg/l) and Mg2+ (10–12.5 mg/l)] to produce solutions with ~5×105 colony forming units (CFUs)/ml in a final volume of 100 µl. Subsequently, the bacteria were cultured using various concentrations of drugs: AK and SCF, 256, 128, 64, 32, 16, 8 µg/ml; MEM and MINO, 64, 32, 16, 8, 4, 2 µg/ml; CIP, 16, 8, 4, 2, 1, 0.5 µg/ml, for 18–20 h at 37°C. The average MIC (MICG), the concentration that inhibited 50% of growth (MIC50) or 90% of strains (MIC90) were calculated. All tests were performed in triplicate, and growth and sterility controls were conducted simultaneously.
Chequerboard assay
A chequerboard assay was used to determine the potential interactions between antibiotics. In each assay, a combination of two antibiotics randomly chosen from the total five was used, and the range of drug concentration was identical to the MIC assays. The drugs in the 96-well plates were diluted with CAMHB by checkerboard method as previously described (20). The broth microdilution plates were inoculated with each MDRAB isolate (initial concentration of bacteria was 0.5 McFarland) for 18–24 h at 37°C, to yield ~5×105 CFU/ml in a 100 µl volume. The effect of the combinations was analyzed through measuring the fractional inhibitory concentration index (FIC) with the following formula: FICA + FICB, where FICA was the ratio of MIC of drug A in combination compared with that of drug A used alone, and FICB was the ratio of MIC of drug B in combination compared with that of drug B used alone. The interaction was defined as synergy (FIC ≤0.5), addition (0.5< FIC ≤1), indifference (1< FIC ≤2) or antagonism (FIC >2), respectively.
Results
Antimicrobial susceptibility
The MDRAB phenotype was determined according to the susceptibility results. In total, 34 strains were isolated, among which 11 strains (29.41%) were PDR A. baumannii with resistance to almost all clinically significant agents. All the strains were resistant to piperacillin/tazobactam, imipenem, meropenem, amikacin, cefotaxime, ticarcillin/clavulanic acid and ciprofloxacin, while only 14 strains (41.18%) were resistant to cefoperazone/sulbactam. The isolates were also resistant to other tested drugs including ceftazidime (97.06%), aztreonam (97.06%), trimethoprim/sulfamethoxazole (97.06%), cefepime (94.12%), gentamicin (94.12%), ampicillin/sulbactam (94.12%) and levofloxacin (94.12%). The strains were classified into 8 phenotypes (A-H) based on the resistance to the aforementioned primary clinical drugs (Table I).
Genotypic diversity and molecular determinants for MDR
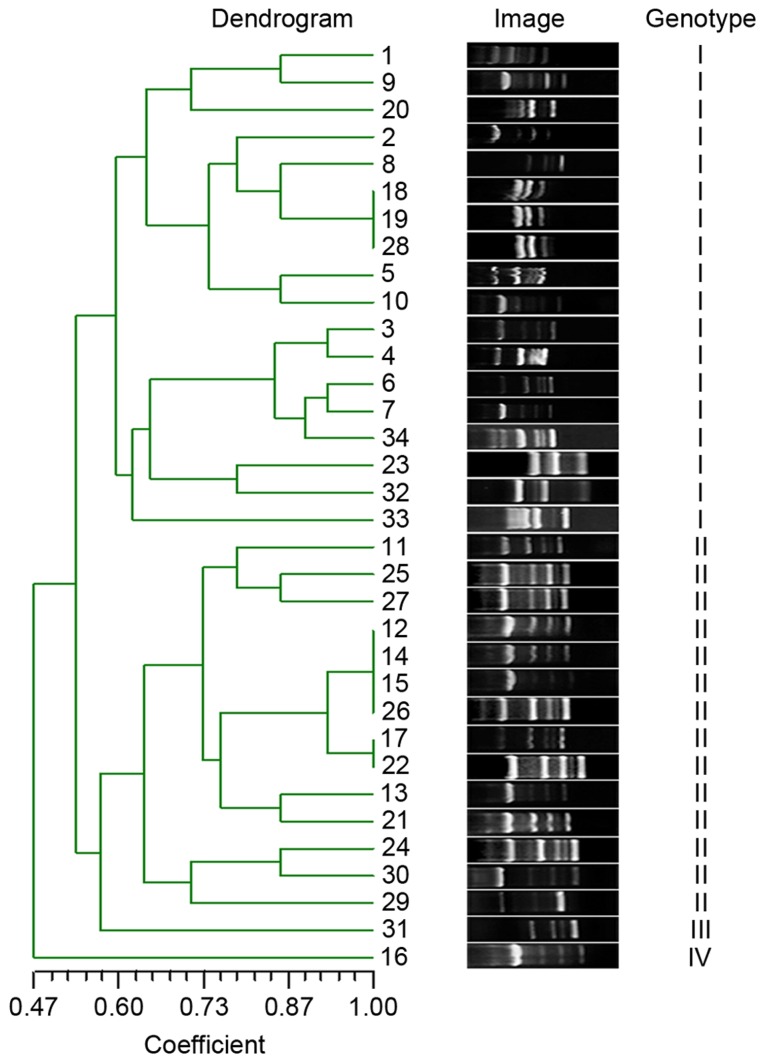
To detect the extent of genotypic diversity of the tested MDRAB, RAPD was performed and the fingerprint images were analyzed by NTsys 2.10e (Exeter Software; Exeter Publishing Ltd., East Setauket, NY, USA) using dice similarity index for cluster analysis and unweighted pair group average for dendrogram construction. The isolates were clustered into four major genotypes (I–IV) at 60% genotypic similarity threshold (Fig. 1).
Figure 1.
RAPD fingerprinting of MDRAB strains. The gel image displayed diversity RAPD genotyping pattern of each MDRAB isolates representing various phenotypes from different wards. The genotypic similarities were calculated by NTsys 2.10e using dice similarity index and UPGMA, presented in the left of the glue image with coefficients and lines. Four genotypes (I–IV) were formed at a 60% similarity level. RAPD, randomly amplified polymorphic DNA; MDRAB, multidrug resistant Acinetobacter baumannii; UPGMA, unweighted pair group average.
Table IV summarizes the distribution of resistance genes in the strains. TEM-1 and ParC were identified in all strains. A total of seven genes demonstrated high positive rates, including OXA-23 (97.06%), AdeB (97.06%), AmpC (94.12%), Int-1 (94.12%), rmtA (94.12%), aph(3) (91.18%) and armA (91.18%). The other genes, including aac(3)-I, aac(6′-I, ant(3″)-I, rmtB, OXA-24 and IMP-5, demonstrated positive rates of 29.41, 32.35, 76.47, 41.18, 85.29 and 64.71%, respectively.
Table IV.
Distribution of positivity of various resistance genes.
| No. | aac(3)-I | aac(6′)-I | ant(3”)-I | OXA-23 | OXA-24 | TEM-1 | IMP-5 | AmpC | ArmA | RmtA | RmtB | Aph(3) | AdeRS | AdeB | Int1 | ParC |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | N | N | P | P | P | P | N | P | P | P | N | P | N | P | P | P |
| 2 | N | N | P | P | P | P | P | P | N | P | P | P | N | P | P | P |
| 3 | N | N | P | P | P | P | P | P | P | P | N | P | N | P | P | P |
| 4 | N | N | P | P | P | P | P | P | P | P | N | P | N | P | P | P |
| 5 | N | N | P | P | P | P | P | P | P | P | N | P | N | P | P | P |
| 6 | N | N | P | P | P | P | P | P | P | N | N | P | N | P | P | P |
| 7 | N | N | P | P | P | P | P | P | N | P | N | P | N | P | P | P |
| 8 | N | N | P | P | P | P | P | P | P | P | P | P | N | P | P | P |
| 9 | N | N | P | P | P | P | P | P | P | P | P | P | N | P | P | P |
| 10 | N | N | P | P | P | P | N | P | P | P | N | N | N | P | P | P |
| 11 | N | N | P | P | P | P | P | P | P | P | N | P | N | P | P | P |
| 12 | N | N | P | P | P | P | P | P | P | P | N | P | N | P | P | P |
| 13 | P | N | P | P | P | P | P | P | P | P | N | P | N | P | P | P |
| 14 | N | N | P | P | P | P | P | P | P | P | P | P | N | P | P | P |
| 15 | N | N | P | P | P | P | P | P | P | P | N | P | N | P | P | P |
| 16 | N | N | P | P | P | P | P | P | P | P | P | P | N | P | P | P |
| 17 | N | N | P | P | P | P | P | P | P | P | N | P | N | P | P | P |
| 18 | N | N | P | P | P | P | P | P | P | P | N | P | N | P | P | P |
| 19 | N | N | P | P | N | P | P | P | P | P | N | P | N | P | P | P |
| 20 | N | P | N | P | P | P | P | P | P | P | P | P | N | P | P | P |
| 21 | N | N | P | P | P | P | P | P | P | P | N | P | N | P | P | P |
| 22 | N | N | P | P | P | P | P | P | P | P | P | P | N | P | P | P |
| 23 | P | P | N | P | N | P | N | P | P | P | N | P | N | P | P | P |
| 24 | P | P | N | P | N | P | N | P | P | P | N | P | N | P | P | P |
| 25 | N | N | N | N | N | P | N | P | N | P | P | P | N | P | P | P |
| 26 | P | N | N | P | P | P | N | N | P | P | P | P | N | P | N | P |
| 27 | P | P | P | P | P | P | N | P | P | P | N | P | N | P | P | P |
| 28 | P | P | N | P | P | P | N | P | P | P | P | P | N | P | N | P |
| 29 | P | P | P | P | P | P | N | P | P | P | P | P | N | P | P | P |
| 30 | N | P | N | P | N | P | N | N | P | P | P | N | N | P | P | P |
| 31 | P | P | P | P | P | P | P | P | P | P | P | P | N | P | P | P |
| 32 | P | P | P | P | P | P | P | P | P | P | P | P | N | P | P | P |
| 33 | N | P | N | P | P | P | N | P | P | P | N | P | N | P | P | P |
| 34 | P | P | P | P | P | P | N | P | P | P | N | N | N | N | P | P |
| P (%) | 29.41 | 32.35 | 76.47 | 97.06 | 85.29 | 100.00 | 64.71 | 94.12 | 91.18 | 97.06 | 41.18 | 91.18 | 0.00 | 97.06 | 94.12 | 100.00 |
P (%), percentage of strains positive for each gene; P, positive; N, negative.
MIC and the interaction of drug combinations
The antibiotic susceptibility levels, expressed as MIC of AK, SCF I, SCF II, MEM, MINO and CIP, were preliminarily determined for the 34 MDRAB isolates. The distribution of MIC50, MIC90 and MICG are presented in Table V. The majority of isolates were resistant to CIP (91.18%), SCF II (91.18%), amikacin (85.29%), SCF I (82.35%) and MEM (73.53%), while less isolates (5.88%) were resistant to MINO.
Table V.
MIC parameters of drugs alone use or in combination.
| Antibiotics usage | MIC50 (µg/ml) | MIC90 (µg/ml) | MICG (µg/ml) |
|---|---|---|---|
| AK alone | >256.00 | >256.00 | >222.24 |
| SCF I alone | 64.00 | >256.00 | >90.83 |
| SCF II alone | 128.00 | >256.00 | >130.35 |
| MEM alone | 33.60 | >64.00 | >46.39 |
| MINO alone | 3.50 | >25.00 | >5.50 |
| CIP alone | >16.00 | >16.00 | >14.13 |
| AK/SCF I | |||
| AK | 8 | 8 | 8 |
| SCF I | 32 | 256 | 29.88 |
| AK/SCF II | |||
| AK | 8 | >256 | >24.24 |
| SCF II | 64 | >256 | >57.41 |
| MEM/SCF I | |||
| MEM | 2 | 32 | 2.82 |
| SCF I | 8 | 128 | 17.18 |
| MEM/SCF II | |||
| MEM | 2 | >64 | >6.14 |
| SCF II | 8 | >256 | >26.12 |
| CIP/SCF I | |||
| CIP | 0.5 | >16 | >0.96 |
| SCF I | 32 | >256 | >48.24 |
| CIP/SCF II | |||
| CIP | 0.5 | >16 | >1.85 |
| SCF II | 64 | >256 | >87.76 |
| MINO/SCF I | |||
| MINO | 2 | 2 | 2 |
| SCF I | 8 | 8 | 8 |
| MINO/SCF II | |||
| MINO | 2 | 4 | 3.24 |
| SCF II | 8 | 8 | 8 |
| AK/MEM | |||
| AK | 8 | 8 | >37.18 |
| MEM | 16 | >64 | >89.82 |
| AK/CIP | |||
| AK | >256 | >256 | >176.99 |
| CIP | >16 | >16 | >12.25 |
| CIP/MEM | |||
| CIP | >16 | >16 | >14.40 |
| MEM | 16 | >64 | >24.65 |
| CIP/MINO | |||
| CIP | 0.5 | 1 | 0.53 |
| MINO | 2 | 4 | 2.47 |
| MEM/MINO | |||
| MEM | 2 | 2 | 2 |
| MINO | 2 | 2 | 2 |
MIC, minimum inhibitory concentration; MIC50, the concentration that inhibits the growing of 50% of strains; MIC90, the concentration that inhibits the growing of 90% of strains; MICG, the average MIC; AK, amikacin; SCF, SCFI 1:1 and SCFII 2:1, cefoperazone/sulbactam; MEM, meropenem; MINO, minocycline; CIP, ciprofloxacin; FIC, fractional inhibitory concentration index.
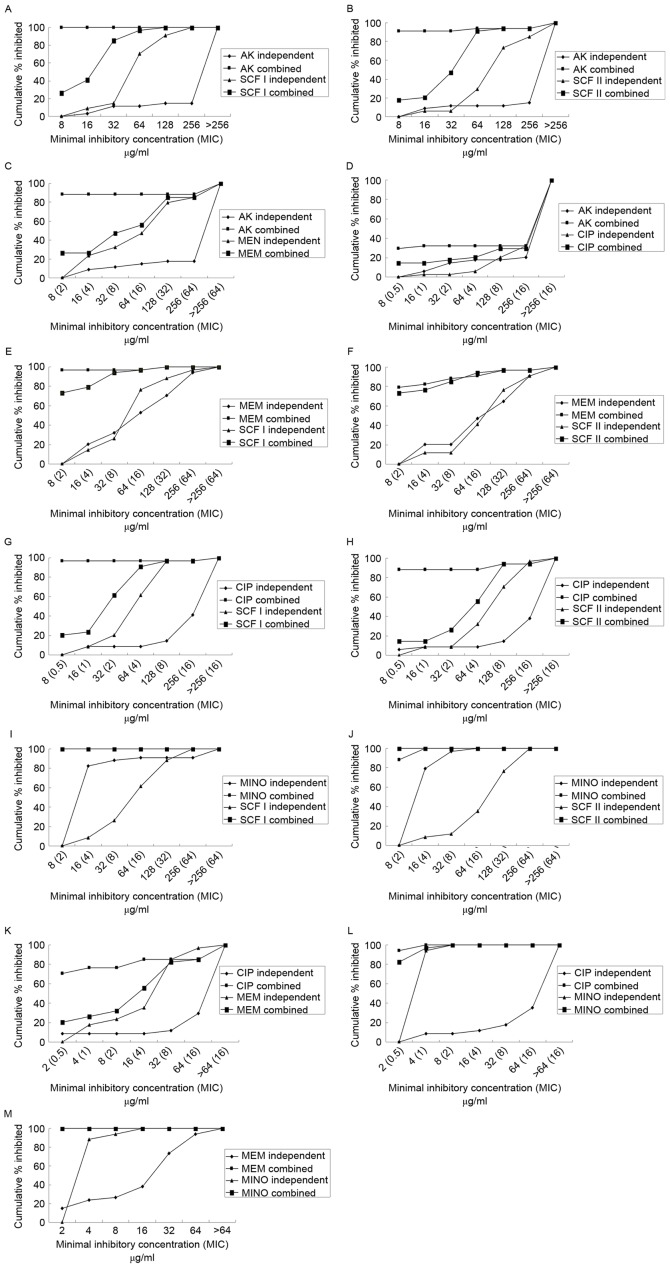
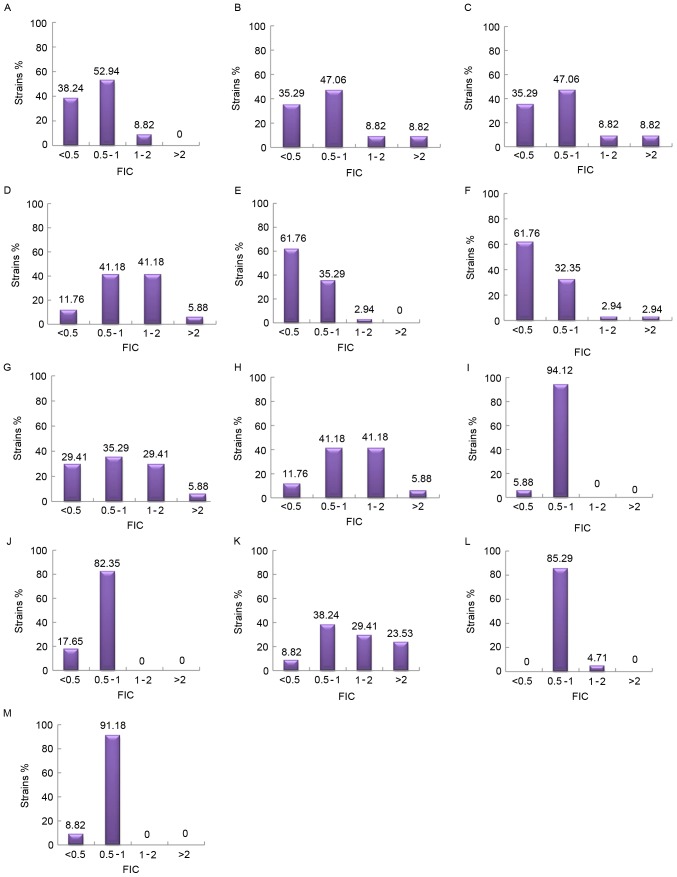
A chequerboard assay was performed with random combinations of two drugs (Table V). Fig. 2 demonstrates the percentage of isolates inhibited at various MIC of antibiotics when use alone or in combination. The majority of the drug combinations exhibited superior inhibitory effects compared with each used alone. All the combinations demonstrated synergism, with the exception of CIP with MINO. MEM in combination with SCF I or SCF II primarily demonstrated synergy, while the combination of AK+SCF I, AK+SCF II, MINO+SCF I, MINO+SCF II, MINO+CIP, and MINO+MEM primarily exhibited additive effects. Concurrently, the combination of AK+CIP demonstrated evidence of antagonism (Fig. 3).
Figure 2.
Cumulative percentages of MDRAB strains that were inhibited by increasing concentrations of various types of drugs used alone or combination. (A) AK+SCFI; (B) AK+SCFII; (C) AK+MEM; (D) AK+CIP; (E) MEM+SCFI; (F) MEM+SCFII; (G) CIP+SCFI; (H) CIP+SCFII; (I) MINO+SCFI; (J) MINO+SCFII; (K) CIP+MEM; (L) CIP+MINO; and (M) MEM+MINO. There were two cumulative percentage lines overlapped (to 100% bacteriostasis) in parts I and M of MINO combining with SCFI and MEM. AK, amikacin; SCF, SCFI 1:1 and SCFII 2:1, cefoperazone/sulbactam; MEM, meropenem; MINO, minocycline; CIP, ciprofloxacin.
Figure 3.
The distribution of FIC of various combinations of antimicrobial drugs. (A) AK+SCFI; (B) AK+SCFII; (C) AK+MEM; (D) AK+CIP; (E) MEM+SCFI; (F) MEM+SCFII; (G) CIP+SCFI; (H) CIP+SCFII; (I) MINO+SCFI; (J) MINO+SCFII; (K) CIP+MEM; (L) CIP+MINO; and (M) MEM+MINO. Synergy, FIC ≤0.5; addition, 0.5< FIC ≤1; indifference, 1< FIC ≤2; antagonism, FIC >2. AK, amikacin; SCF, SCFI 1:1 and SCFII 2:1, cefoperazone/sulbactam; MEM, meropenem; MINO, minocycline; CIP, ciprofloxacin; FIC, fractional inhibitory concentration index.
Discussion
Antibiotic resistance patterns observed in A. baumannii exhibits the capacity to cause an epidemic globally. MDRAB has been demonstrated to lead to serious hospital-acquired infection, as limited therapeutic options are available (20). In the present study, the features of 34 MDRAB strains isolated from the Second Xiangya Hospital were investigated, including antimicrobial susceptibility, genotypes, screening of antibiotic resistance genes, MIC assay and antibiotic interactions.
In the antimicrobial susceptibility study, eight phenotypes (A-H) were classified using the K-B test. The results of the drug susceptibility were in accordance with previous studies (21,22) that highlighted the efficiency of cefoperazone/sulbactam against MDRAB. However, the expression of characteristic bacterial phenotypes may be easily affected by various environmental factors. Therefore, only determining the phenotype was not sufficient for the complete epidemiological typing of various strains. Organism identification based on the genotype is considered to be more reliable, as the genotype of each organism is unique and invariable. In the present study, a AP2 primer known to be efficient in RAPD genotyping was used to amplify the genomic DNA of MDRAB strains (23). In total, four genotypes (I–IV) were formed at a 60% similarity level. The genotypic distribution was polyclonal, which was opposite to a previous study (24). In general, the ‘classic’ outbreaks of MDRAB may be more frequently induced by a single clone spread among people, whereas for the prevalence of polyclones in the Second Xiangya Hospital, it may be associated the existence of mobile genetic elements, for example integrons. The variety of patient wards and isolate origin was hypothesized to be responsible for the transmission of different subtypes. When comparing the phenotypes with diverse fingerprinting profiles, it is noteworthy to select isolates of the same phenotype with different genotypes, as it manifested that the environment may affect the discrepancy between genotype and phenotype.
To additionally investigate the resistant mechanisms of MDRAB, the expression of the genes associated with drug resistance was detected among the 34 isolates. Differences were observed in the genetic characteristics of β-lactam, aminoglycoside and quinolones resistance. Previously, the carbapenem resistance associated with class D β-lactamase genes had been suggested to cause serious therapeutic problems in clinical practice (25,26). The high positive rate of OXA-23 (33/34) in the present study was consistent with a previous study (27), while the proportion of OXA-24-positive strains (29/34) was increased compared with a previous study (28). The production of OXA-23 and OXA-24 β-lactamase may be the major cause for the selected MDRAB representing 100% resistance to imipenem and meropenem. Other β-lactamase genes, including TEM-1 (class A), IMP-5 (class B) and AmpC (class C) were also identified in the present study, which may be associated with the resistance of MDRAB to various types of β-lactams including aztreonam, ceftazidime, cefepime, and cefoperazone/sulbactam. At present, aminoglycoside-modifying enzymes and 16S rRNA methylases have been suggested to be the most important mechanism for bacterial resistance against aminoglycosides (14). In the present study, the co-existence of aph(3) (91.18%), ant(3″)-I (76.47%), aac(3)-I (29.41%), aac(6′)-I (32.35%), armA (91.18%), rmtA (94.12%) and rmtB (41.18%) resistance genes confers the high resistance to amikacin and gentamicin in MDRAB. Mutations in ParC were observed in all isolates, which may assist in explaining the 100% resistance to ciprofloxacin. The result was similar to a previous study (29). Among the isolates, the positive rates of Int1 and adeB were 94.1 and 97.1%, respectively, while the adeRS was completely negative. Integron and efflux pump genes are non-specific resistance genes. Concurrently, integrons are widely present in MDRAB, particularly Int-1, which provides A. baumannii with a gene capture system adapted to circumvent the challenges of multiple-antibiotic treatment regimens (30). The results of the present study demonstrated that Int1 serves a crucial role in multiple drug resistance. In addition, the identification of the co-existence of Int-1 and the majority of the β-lactamase and aminoglycoside genes was notable, which is in accordance with a previous study (31). This highlighted the importance of the roles of Int-1 in the horizontal spreading of antibiotic resistance genes, which may finally result in the polyclonal prevalence of MDRAB in the Second Xiangya Hospital. AdeB is a vital component of the AdeABC efflux pump, and the expression of adeABC is regulated by the adeRS two-component regulatory system (32). The sophisticated feedback between them may account for the high positive ratio of adeB and absolute absence of adeRS observed in the present study.
The results of the present study confirmed that several genes were associated with MDR. Abuse of antibiotic chemotherapeutics affords major challenges in treating MDRAB infections. At present, traditional therapy regimens are not efficient to manage these life-threatening infections. Tigecycline and polymyxins have been considered as the last resort for treating MDRAB infections (4,5). However, they are still widely used across mainland China due to the lack of any other qualified commercial products. On this basis, the present study aimed to identify an effective regimen to manage MDRAB infections through a combination of antibiotics that are frequently used in clinical practice. In the present study, 6 drugs (iAK, SCF I, SCF II, MEM, MINO and CIP) were selected to study the in vitro activity of drug combinations against MDRAB. The lower MIC of SCFI compared with SCF II demonstrated a comparatively increased rate of in vitro activity of SCF I against MDRAB, which was incompatible with previous studies (33,34). This discrepancy may arise from the geographical and biological evolutional differences. In addition, a high percentage (76.47%) of MDRAB was susceptible to minocycline, indicating the potential of this drug for the treatment of this fatal infection (35). The chequerboard assay indicated synergism for all the tested combinations, particularly the combination of MEM+SCF I, and MEM+SCF II, with the exception of CIP+MINO. Conversely, the combinations of MINO+SCF I, MINO+SCF II, CIP+MINO and MEM+MINO demonstrated additive effects. In summary, the combination of cefoperazone-sulbactam or meropenem-minocycline has been indicated to be more active compared with ciprofloxacin-amikacin, which is similar to the recent surveillance data (16,21). The present study also demonstrated that meropenem and cefoperazone-sulbactam were generally active in MDRAB. In a previous study, the combined utilization of meropenem and sulbactam was considered a therapeutic option for A. baumannii infection (36). However, only a small number of studies have been focused on the study of the efficiency of a MEM+SCF combination. The present study attempted to investigate the in vitro activity of two types of SCF (SCF I 1:1; SCF II 2:1) combined with MEM against 34 strains of MDRAB. The results indicated a marked synergistic interaction in the majority tested isolates. Although no significant differences were observed in the activity of cefoperazone-sulbactam combined with meropenem, it revealed a novel potential option for clinical combination therapy. Meropenem belongs to the family of β-lactam antibiotics, while cefoperazone-sulbactam is a type of the third-generation cephalosporin and β-lactamase inhibitor. When meropenem is combined with cefoperazone-sulbactam, they may bind to different types of penicillin bonding proteins, executing their bactericidal effects. Concurrently, sulbactam may irreversibly inhibit β-lactamase activity. This may be the most probable explanation for the synergism observed. MINO was active against MDRAB whenever it is used alone or combination. Although it is only a second-line antibiotic for the majority of clinical bacterial infections, its potential antibacterial activity against MDRAB should not be neglected. In the present study, the combination of AK+CIP produced an antagonism of 70.59%. Therefore, the combined use of these drugs should be avoided in clinical practice.
In conclusion, the identification of fingerprinting diversity highlights the issues with the polyclonal and horizontal spread of MDRAB in the Second Xiangya Hospital. Although the co-occurrence of numerous resistance-encoding genes presented a completely threaten for the active therapy, the determination of efficacious combinations among minocycline, meropenem and cefoperazone-sulbactam, particularly MEM+SCFI and MEM+SCFII, provides improved choices for the rational clinical combination therapy for MDRAB infections. Additionally, MINO may be the alternative choice to overcome the critical resistance of A. baumannii. The present study failed to depict the pharmacokinetics and pharmacodynamics of these drug combinations. Future studies should focus on updating these data and proceed to additionally identify the clinical effects of combination therapy.
Acknowledgements
The present study was supported by the China National Natural Scientific Foundation (grant no. 81470133), and the Science and Technology Planning Project of Hunan Province of China (grant no. 2015JC3035).
References
- 1.Visca P, Seifert H, Towner KJ. Acinetobacter infection - an emerging threat to human health. IUBMB Life. 2011;63:1048–1054. doi: 10.1002/iub.600. [DOI] [PubMed] [Google Scholar]
- 2.Munoz-Price LS, Weinstein RA. Acinetobacter infection. N Engl J Med. 2008;358:1271–1281. doi: 10.1056/NEJMra070741. [DOI] [PubMed] [Google Scholar]
- 3.Dent LL, Marshall DR, Pratap S, Hulette RB. Multidrug resistant Acinetobacter baumannii: A descriptive study in a city hospital. BMC Infect Dis. 2010;10:196. doi: 10.1186/1471-2334-10-196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Principe L, D'Arezzo S, Capone A, Petrosillo N, Visca P. In vitro activity of tigecycline in combination with various antimicrobials against multidrug resistant Acinetobacter baumannii. Ann Clin Microbiol Antimicrob. 2009;8:18. doi: 10.1186/1476-0711-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lim TP, Tan TY, Lee W, Sasikala S, Tan TT, Hsu LY, Kwa AL. In-vitro activity of polymyxin B, rifampicin, tigecycline alone and in combination against carbapenem-resistant Acinetobacter baumannii in Singapore. PLoS One. 2011;6:e18485. doi: 10.1371/journal.pone.0018485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Weng XB, Mi ZH. Phylogenetic analysis on pandrug-resistant Acinetobacter baumannii strains. Zhonghua Liu Xing Bing Xue Za Zhi. 2011;32:948–949. (In Chinese) [PubMed] [Google Scholar]
- 7.Zhao WS, Liu GY, Mi ZH, Zhang F. Coexistence of blaOXA-23 with armA and novel gyrA mutation in a pandrug-resistant Acinetobacter baumannii isolate from the blood of a patient with haematological disease in China. J Hosp Infect. 2011;77:278–279. doi: 10.1016/j.jhin.2010.11.006. [DOI] [PubMed] [Google Scholar]
- 8.Wen Y, Ouyang Z, Yu Y, Zhou X, Pei Y, Devreese B, Higgins PG, Zheng F. Mechanistic insight into how multidrug resistant Acinetobacter baumannii response regulator AdeR recognizes an intercistronic region. Nucleic Acids Res. 2017;45:9773–9787. doi: 10.1093/nar/gkx624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang H, Huang L, Barnie PA, Su Z, Mi Z, Chen J, Aparna V, Kumar D, Xu H. Characterization and distribution of drug resistance associated β-lactamase, membrane porin and efflux pump genes in MDR A. Baumannii isolated from Zhenjiang, China. Int J Clin Exp Med. 2015;8:15393–15402. [PMC free article] [PubMed] [Google Scholar]
- 10.Jiang W, Liu H, Zhong M, Yang YC, Xiao DW, Huang WF. Study on the resistant genes to carbapenems and epidemiological characterization of multidrug-resistant Acinetobacter baumannii isolates. Microb Drug Resist. 2013;19:117–123. doi: 10.1089/mdr.2012.0049. [DOI] [PubMed] [Google Scholar]
- 11.Ramirez MS, Tolmasky ME. Aminoglycoside modifying enzymes. Drug Resist Updat. 2010;13:151–171. doi: 10.1016/j.drup.2010.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Petersen A, Guardabassi L, Dalsgaard A, Olsen JE. Class I integrons containing a dhfrl trimethoprim resistance gene cassette in aquatic Acinetobacter spp. FEMS Microbiol Lett. 2000;182:73–76. doi: 10.1111/j.1574-6968.2000.tb08876.x. [DOI] [PubMed] [Google Scholar]
- 13.Azizi O, Shakibaie MR, Badmasti F, Modarresi F, Ramazanzadeh R, Mansouri S, Shahcheraghi F. Class 1 integrons in non-clonal multidrug-resistant Acinetobacter baumannii from Iran, description of the new blaIMP-55 allele in In1243. J Med Microbiol. 2016;65:928–936. doi: 10.1099/jmm.0.000315. [DOI] [PubMed] [Google Scholar]
- 14.Tada T, Miyoshi-Akiyama T, Kato Y, Ohmagari N, Takeshita N, Hung NV, Phuong DM, Thu TA, Binh NG, Anh NQ, et al. Emergence of 16S rRNA methylase-producing Acinetobacter baumannii and Pseudomonas aeruginosa isolates in hospitals in Vietnam. BMC Infect Dis. 2013;13:251. doi: 10.1186/1471-2334-13-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JK, Lee YS, Park YK, Kim BS. Mutations in the gyrA and parC genes in ciprofloxacin-resistant clinical isolates of Acinetobacter baumannii in Korea. Microbiol Immunol. 2005;49:647–653. doi: 10.1111/j.1348-0421.2005.tb03643.x. [DOI] [PubMed] [Google Scholar]
- 16.American Thoracic Society, corp-author. Infectious Diseases Society of America: Guidelines for the management of adults with hospital-acquired, ventilator-associated, and healthcare-associated pneumonia. Am J Respir Crit Care Med. 2005;171:388–416. doi: 10.1164/rccm.200405-644ST. [DOI] [PubMed] [Google Scholar]
- 17.Clinical and Laboratory Standards Institute: Performance standards for antimicrobial susceptibility testing: Twenty-second informational supplement M100-S21. CLSI. 2011;31:64–65. [Google Scholar]
- 18.Zhang T, Wang M, Xie Y, Li X, Dong Z, Liu Y, Wang L, Yang M, Song H, Cao H, Cao W. Active efflux pump adeB is involved in multidrug resistance of Acinetobacter baumannii induced by antibacterial agents. Exp Ther Med. 2017;13:1538–1546. doi: 10.3892/etm.2017.4141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Wheeler DL. GenBank. Nucleic Acids Res. 2005;33:D34–D38. doi: 10.1093/nar/gki063. (Database Issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daoud Z, Mansour N, Masri K. Synergistic combination of carbapenems and colistin against P. aeruginosa and A. baumannii. Open J Med Microbiol. 2013;3:253–258. doi: 10.4236/ojmm.2013.34038. [DOI] [Google Scholar]
- 21.Huang J, Chen EZ, Qu HP, Mao EQ, Zhu ZG, Ni YX, Han LZ, Tang YQ. Sources of multidrug-resistant Acinetobacter baumannii and its role in respiratory tract colonization and nosocomial pneumonia in intensive care unit patients. Chin Med J (Engl) 2013;126:1826–1831. [PubMed] [Google Scholar]
- 22.Xu T, Xia W, Rong G, Pan S, Huang P, Gu B. A 4-year surveillance of antimicrobial resistance patterns of Acinetobacter baumanni in a university-affiliated hospital in China. J Thorac Dis. 2013;5:506–512. doi: 10.3978/j.issn.2072-1439.2013.08.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koeleman JG, Stoof J, Biesmans DJ, Savelkoul PH, Vandenbroucke-Grauls CM. Comparison of amplified ribosomal DNA restriction analysis, random amplified polymorphic DNA analysis, and amplified fragment length polymorphism fingerprinting for identification of Acinetobacter genomic species and typing of Acinetobacter baumannii. J Clin Microbiol. 1998;36:2522–2529. doi: 10.1128/jcm.36.9.2522-2529.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fontana C, Favaro M, Minelli S, Bossa MC, Testore GP, Leonardis F, Natoli S, Favalli C. Acinetobacter baumannii in intensive care unit: A novel system to study clonal relationship among the isolates. BMC Infect Dis. 2008;8:79. doi: 10.1186/1471-2334-8-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Niranjan DK, Singh NP, Manchanda V, Rai S, Kaur IR. Multiple carbapenem hydrolyzing genes in clinical isolates of Acinetobacter baumannii. Indian J Med Microbiol. 2013;31:237–241. doi: 10.4103/0255-0857.115626. [DOI] [PubMed] [Google Scholar]
- 26.Feizabadi MM, Fathollahzadeh B, Taherikalani M, Rasoolinejad M, Sadeghifard N, Aligholi M, Soroush S, Mohammadi-Yegane S. Antimicrobial susceptibility patterns and distribution of blaOXA genes among Acinetobacter spp. Isolated from patients at Tehran hospitals. Jpn J Infect Dis. 2008;61:274–278. [PubMed] [Google Scholar]
- 27.Thapa B, Tribuddharat C, Srifuengfung S, Dhiraputra C. High prevalence of bla(OXA)-23 in oligoclonal carbapenem-resistant Acinetobacter baumannii from Siriraj Hospital, Mahidol University, Bangkok, Thailand. Southeast Asian J Trop Med Public Health. 2010;41:625–635. [PubMed] [Google Scholar]
- 28.Chen Z, Liu W, Zhang Y, Li Y, Jian Z, Deng H, Zou M, Liu Y. Molecular epidemiology of carbapenem-resistant Acinetobacter spp. from XiangYa Hospital, in Hunan Province, China. J Basic Microbiol. 2013;53:121–127. doi: 10.1002/jobm.201100420. [DOI] [PubMed] [Google Scholar]
- 29.Sung JY, Kwon KC, Cho HH, Koo SH. Antimicrobial resistance determinants in imipenem-nonsusceptible Acinetobacter calcoaceticus-baumannii complex isolated in Daejeon, Korea. Korean J Lab Med. 2011;31:265–270. doi: 10.3343/kjlm.2011.31.4.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Peymani A, Farajnia S, Nahaei MR, Sohrabi N, Abbasi L, Ansarin K, Azhari F. Prevalence of class 1 integron among multidrug-resistant Acinetobacter baumannii in Tabriz, northwest of Iran. Pol J Microbiol. 2012;61:57–60. [PubMed] [Google Scholar]
- 31.Farajnia S, Azhari F, Alikhani MY, Hosseini MK, Peymani A, Sohrabi N. Prevalence of PER and VEB type extended spectrum betalactamases among multidrug resistant acinetobacter baumannii Isolates in North-West of Iran. Iran J Basic Med Sci. 2013;16:751–755. [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon EJ, Courvalin P, Grillot-Courvalin C. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: Major role for AdeABC overexpression and AdeRS mutations. Antimicrob Agents Chemother. 2013;57:2989–2995. doi: 10.1128/AAC.02556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jones RN, Barry AL, Packer RR, Gregory WW, Thornsberry C. In vitro antimicrobial spectrum, occurrence of synergy, and recommendations for dilution susceptibility testing concentrations of the cefoperazone-sulbactam combination. J Clin Microbiol. 1987;25:1725–1729. doi: 10.1128/jcm.25.9.1725-1729.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barry AL, Jones RN. Criteria for disk susceptibility tests and quality control guidelines for the cefoperazone-sulbactam combination. J Clin Microbiol. 1988;26:13–17. doi: 10.1128/jcm.26.1.13-17.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pei G, Mao Y, Sun Y. In vitro activity of minocycline alone and in combination with cefoperazone-sulbactam against carbapenem-resistant Acinetobacter baumannii. Microb Drug Resist. 2012;18:574–577. doi: 10.1089/mdr.2012.0076. [DOI] [PubMed] [Google Scholar]
- 36.Deveci A, Coban AY, Acicbe O, Tanyel E, Yaman G, Durupinar B. In vitro effects of sulbactam combinations with different antibiotic groups against clinical Acinetobacter baumannii isolates. J Chemother. 2012;24:247–252. doi: 10.1179/1973947812Y.0000000029. [DOI] [PubMed] [Google Scholar]