Abstract
Secreted protein acidic and rich in cysteine-like 1 (SPARCL1), a member of extracelluar matrix glycoprotein, has been reported to be associated with various tumor types. The present study aimed to evaluate the prognostic value of SPARCL1 in patients with colorectal cancer. Tissue microarray blocks were constructed based on 79 patients who underwent radical surgery at the Kunshan First People's Hospital between 2008 and 2010. Thirty pairs of fresh-frozen tissues were also obtained for total protein extraction. The expression of SPARCL1 protein was analyzed using immunohistochemistry and western blotting analyses, and the association between overexpressed SPARCL1 and clinicopathological factors was evaluated. Survival analysis with Kaplan Meier curves and Cox regression analysis was used to analyze the prognostic value of SPARCL1. According to western blot analyses, SPARCL1 protein expression in colorectal tumors was significantly lower compared with corresponding normal tissues. The expression of SPARCL1 was markedly decreased from differentiation I to III, and the negative rate of SPARCL1 was higher at Duke's stage C compared with B. Though without any difference between ‘positive’ and ‘negative’ in overall survival, significantly higher survival in patients with positive SPARCL1 expression at Duke's stage B was detected in the present study. These results indicated that SPARCL1 may be a potential tumor suppressor gene and associated with good prognosis.
Keywords: secreted protein acidic and rich in cysteine-like 1, colorectal cancer, survival, prognosis, immunohistochemistry, western blotting
Introduction
Colorectal cancer (CRC), associated with high morbidity and mortality rates, has become one of the most common types of cancer (1). Even with the improvement in diagnostic and therapeutic approaches, the prognosis of colorectal cancer remains poor primarily due to local recurrence, and distal metastases (2). Therefore, the identification of novel reliable prognostic markers to predict the prognosis, and provide better and more suitable therapy for patients with colorectal cancer is warranted.
Secreted protein acidic and rich in cysteine-like 1 (SPARCL1), also known as SC1, Hevin and Mast 9, belongs to the SPARC-associated family of matricellular proteins (3,4). The SPARCL1 gene is localized at chromosome 4q22 and is expressed in various normal tissues, including the brain, heart, lung and lymphoid tissues (5,6). However, SPARCL1 is not expressed or expressed at a low level in various cancer types, except in hepatocellular carcinoma (5,7).
Recent studies have suggested that the most important function of SPARCL1 is modulating high endothelial cell adhesion to the basement membrane (3). In addition, SPARCL1 is able to suppress tumor growth by prolonging the G1 phase and inducing cell differentiation (8,9).
The prognostic role of SPARCL1 in patients with colorectal cancer appears to be controversial. Certain studies have suggested that elevated expression of SPARCL1 is associated with a better prognosis compared with low SPARCL1 expression, and with reduced odds ratios (ORs) of lymph node involvement and distant organ metastasis (9,10). Other studies have demonstrated that SPARCL1 is a negative regulator in the progression of CRC (11). Notably, the expression of SPARCL1 has been observed to be increased from Dukes' stages A to B, but then decreased from Dukes' stages B to C and D (11). The aim of the present study was to clarify the association between SPARCL1 expression and CRC progression and explore its prognostic value in patients with colorectal cancer.
Materials and methods
Tissue samples
The clinical and pathological data of 79 patients who were diagnosed with colorectal cancer at Dukes' stage B or C, and underwent radical surgery at Kunshan First People's Hospital (Kunshan, China) between January 2008 and December 2010 were reviewed. The mean age was 59.5 years, with 38 males and 41 females. None of the patients had received radiotherapy or chemotherapy prior to surgery. Thirty pairs of fresh-frozen colorectal tumors and matched normal tissues were also collected for total protein extraction and stored at −80°C. Written informed consent was obtained from all patients and the study was approved by the ethical approval of Kunshan First People's Hospital Ethics Committee. Follow-up data were available for all patients and the duration ranged between 3 and 60 months with a mean of 40.29±2.42 months.
Tissue microarray (TMA) construction
In each case, three representative tumor regions were selected, from which tissue cylinders with diameters of 0.6 mm were arrayed into a recipient block using a tissue chip microarrayer (Beecher Instruments, Inc., Silver Spring, MD, USA). Subsequently, the recipient block was cut into 5-µm thick sections on slides pretreated with adhesion agent (Jiangsu Haimen Shitai Experimental Equipment Co. Ltd., Haimen, China) to support adhesion of the tissue samples.
Protein extraction and western blotting
Thirty pairs of fresh-frozen CRC specimens and corresponding adjacent normal tissues were used for western blotting. Total protein of each tissue was extracted using RIPA lysis buffer (Beyotime Institute of Biotechnology, Haimen, China) for 10–15 min at 0°C, and then the supernatant was collected, in which the concentration of protein was measured using a BCA protein assay kit (Pierce; Thermo Fisher Scientific, Inc., Waltham, MA, USA). Supernatants from each sample were all mixed with 5X SDS-PAGE Sample Loading Buffer (Beyotime Institute of Biotechnology, Haimen, China) and boiled for 8–10 min at 96°C. A total of 15 mg sample was resolved per lane of 6–12% SDS-PAGE, transferred to polyvinylidene difluoride membranes (EMD Millipore, Billerica, MA, USA), and then blocked in 5% non-fat dry milk with TBS-Tween 20 for 1–2 h at room temperature (depending on seasonal changes in room temperature; 1 h in summer, 2 h in winter). Subsequently, the primary antibodies, mouse anti-human SPARCL1 polyclonal antibody (1:500; cat. no. ab107533, Abcam, Cambridge, UK) and mouse β-actin monoclonal antibody (1:500; cat. no. AA128, Beyotime Institute of Biotechnology), were used to incubate the membranes in 4°C overnight. Accordingly, horseradish peroxidase (HRP)-conjugated goat anti-mouse IgG secondary antibody (1:1,000; cat. no. A0216, Beyotime Institute of Biotechnology) were used to incubate the membranes for 1 h at 37°C. Membranes were then detected using an Enhanced Chemiluminescence Detection system (Beyotime Institute of Biotechnology). The relative densities of proteins were quantified using ImageJ software (version 1.8.0; National Institutes of Health, Bethesda, MD, USA). The formula using to calculate the relative SPARCL1 expression was as follows: Gray value (SPARCL1)/gray value (β-actin).
Immunohistochemistry (IHC)
A mouse anti-human SPARCL1 polyclonal antibody (cat. no. ab107533; 1:200; Abcam, UK) was used as the primary antibody. A Streptavidin-HRP kit (CW2069A; CWBio, Beijing, China) was used for IHC according to the manufacturer's protocol. The slides were deparaffinized, rehydrated and heated in a microwave for 10–15 min in 10 mmol/l citrate buffer (Sigma-Aldrich; Merck KGaA, Darmstadt Germany). After the slides were cooled to room temperature, the slides were treated with reagent1 and reagent2 to block non-specific staining, and then treated with the primary antibody for 12 h. Slides were then incubated with reagent3 and reagent4 (anti-rabbit/mouse IgG) for 5 min, then treated with DAB and hematoxylin for visualization.
Evaluation of immunohistochemical staining
A stained TMA slide was scanned using electron microscopy and analyzed with ImageScope software (version 11; Leica Microsystems GmbH, Wetzlar, Germany). Protein expression was assessed in a semi-quantitative manner by two pathologists (Dr Xiao-jiao Gao and Dr Fang Chen; Department of Pathology, Kunshan First People's Hospital Affiliated to Jiangsu University), who were unaware of any patient information. The percentage of tumor cells with staining of the cytoplasm was evaluated as follows: 0%, 0; 1–10%, 1; 11–50%, 2; 51–80%, 3; 81–100%, 4. The staining intensity was also evaluated as follows: No expression, 0; weak, 1; moderate, 2; or strong, 3. The values were multiplied, resulting in an immunoreactivity score (IRS) ranging between 0 and 12. Mean scores calculated from three TMAs were the final numeric values. Samples were divided into negative/under-expressed (IRS<3) and positive/over-expressed (IRS>2) levels according to SPARCL1 expression. Any disagreement was resolved by discussion.
Statistical analysis
Data are presented as the means ± standard deviation. The SPSS software (version 20.0; IBM Corp., Armonk, NY, USA) was used for statistical analyses and P<0.05 was considered to indicate a statistically significant difference. Paired Student's t-tests were used to compare SPARCL1 protein expression in tumors with normal tissues. Pearson's Chi squared and Fisher's tests were used to analyze the associations of SPARCL1 expression with clinicopathological characteristics. In addition, the Cox univariate and multivariate regression analyses, and Kaplan-Meier curves with log rank test were also used to analyze overall survival (OS). Hazard ratios (HRs) with 95% confidence intervals (CI) were used as the main indicate for OS.
Results
SPARCL1 protein expression in tumors and normal tissues
SPARCL1 protein expression in thirty paired CRC and normal tissues was visualized by western blot analyses. The data of four representative samples derived from a total of thirty paired specimens are presented in Fig. 1A. The comparison of average relative SPARCL1 expression rates between total thirty tumors and normal tissues are summarized in Fig. 1B (the relative SPARCL1 expression of tumors vs. normal tissues, was 1.370±0.185 vs. 0.901±0.136; P<0.0001), which indicated a significantly lower expression pattern of SPARCL1 protein in colorectal tumors compared with corresponding normal tissues.
Figure 1.
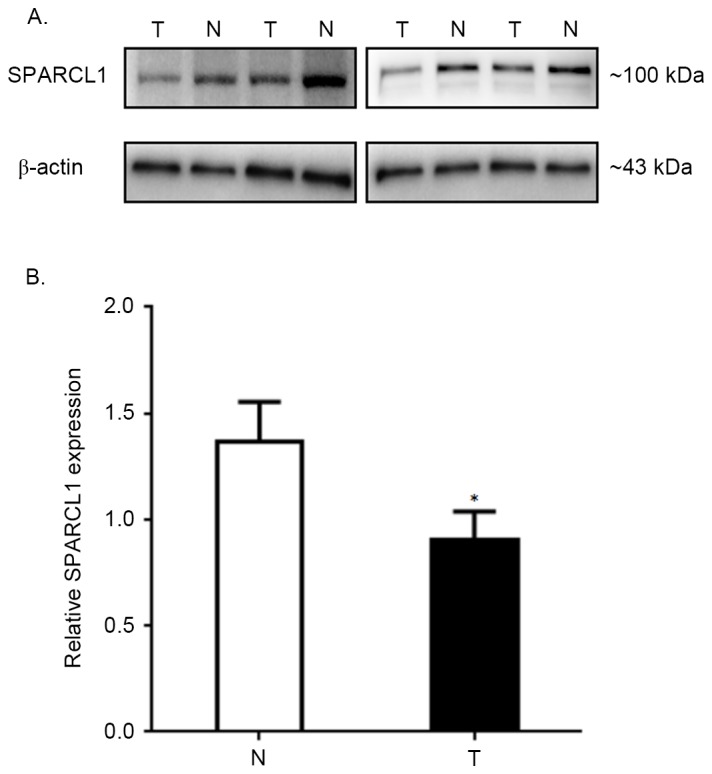
SPARCL1 expression in 30 pairs of tissues investigated by western blotting. (A) Patterns of four representative pairs are demonstrated; the molecular weight of SPARCL1 was ~100 kDa, while that of β-actin was ~43 kDa. (B) The mean relative SPARCL1 expression rates of 30 pairs are presented. The expression level of SPARCL1 in tumors was significantly lower compared with that in normal tissues. *P<0.0001. T, colorectal tumors; N, adjacent normal tissues; SPARCL1, secreted protein acidic and rich in cysteine-like 1.
Association between clinicopathological factors and SPARCL1 expression
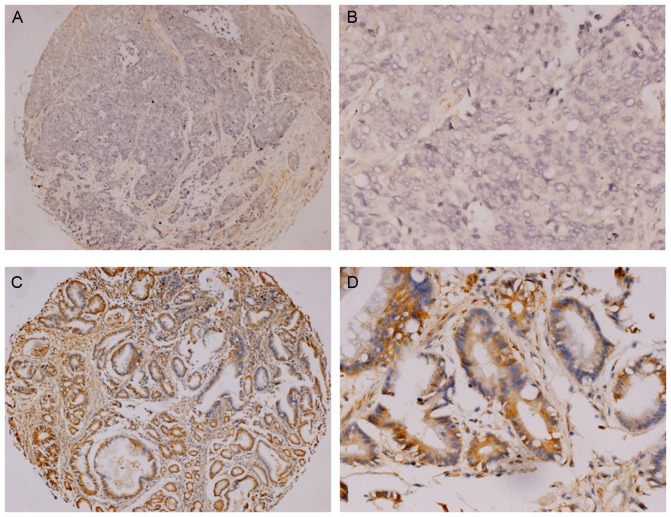
A total of 79 patients with radically resected Duke's stage B or C tumors, were included in the present study. Of the 79 patients, 48 cases (60.76%) were negative expression for SPARCL1 (Fig. 2A and B) and the other 31 (39.24%) were positive (Fig. 2C and D). Significant differences were identified in the SPARCL1 expression status in relation to differentiation and Duke's stages (P<0.05; Table I), but no significant differences were identified among other parameters. The associations between clinicopathological features of patients with colorectal cancer and SPARCL1 expression are presented in Table I.
Figure 2.
Expression of SPARCL1 protein in the primary colorectal cancer. (A and B) Negative staining and (C and D) positive staining for SPARCL1 are presented. Original magnification, ×100 for (A) and (C) and ×400 for (B) and (D). SPARCL1, secreted protein acidic and rich in cysteine-like 1.
Table I.
Association between SPARCL1 expression and clinicopathological variables.
| SPARCL1 expression (%) | |||||
|---|---|---|---|---|---|
| Variable | N | Positive | Negative | χ2 value | P-value |
| Total | 79 | 31 (39.24) | 48 (60.76) | ||
| Age, years | |||||
| ≤60 | 44 | 21 (47.73) | 23 (52.27) | 3.00 | >0.05 |
| >60 | 35 | 10 (28.57) | 25 (71.43) | ||
| Gender | |||||
| Male | 38 | 18 (47.37) | 20 (52.63) | 2.03 | >0.05 |
| Female | 41 | 13 (31.71) | 28 (68.29) | ||
| Tumor size, cm | |||||
| ≤2 | 12 | 3 (25.00) | 9 (75.00) | 1.20 | >0.05 |
| 2–5 | 43 | 18 (41.86) | 25 (58.14) | ||
| >5 | 24 | 10 (41.67) | 14 (58.33) | ||
| Location | |||||
| Right colon | 37 | 13 (35.14) | 24 (64.86) | −4.69 | >0.05 |
| Left colon | 18 | 9 (50.00) | 9 (50.00) | ||
| Rectum | 24 | 9 (37.50) | 15 (62.50) | ||
| Differentiation | |||||
| I | 13 | 7 (53.85) | 6 (46.15) | 8.54 | <0.05 |
| II | 44 | 21 (47.73) | 23 (52.27) | ||
| III | 22 | 3 (13.64) | 19 (86.36) | ||
| Dukes stage | |||||
| B | 44 | 19 (43.18) | 25 (56.82) | 4.81 | <0.05 |
| C | 35 | 12 (34.29) | 23 (65.71) | ||
| Serum CEA, ng/ml | |||||
| <5 | 32 | 16 (50.00) | 16 (50.00) | 2.61 | >0.05 |
| ≥5 | 47 | 15 (31.91) | 32 (68.09) | ||
| Serum CA19-9, U/ml | |||||
| <37 | 68 | 26 (38.24) | 42 (61.76) | 0.01 | >0.05 |
| ≥37 | 11 | 5 (45.45) | 6 (54.55) | ||
| Serum CA12-5, U/ml | |||||
| <35 | 52 | 21 (40.38) | 31 (59.62) | 0.08 | >0.05 |
| ≥35 | 27 | 10 (37.04) | 17 (62.96) | ||
| Complicationa | |||||
| Yes | 23 | 7 (30.43) | 16 (69.57) | 1.06 | >0.05 |
| No | 56 | 24 (42.86) | 32 (57.14) | ||
| Survival | |||||
| Yes | 37 | 17 (45.95) | 20 (54.05) | 1.31 | >0.05 |
| No | 42 | 14 (33.33) | 28 (66.67) | ||
Complications include obstruction, fistula, hematoma and peritonitis. CEA, carcinoembryonic antigen; CA, cancer antigen; SPARCL1, secreted protein acidic and rich in cysteine-like 1.
Survival analysis
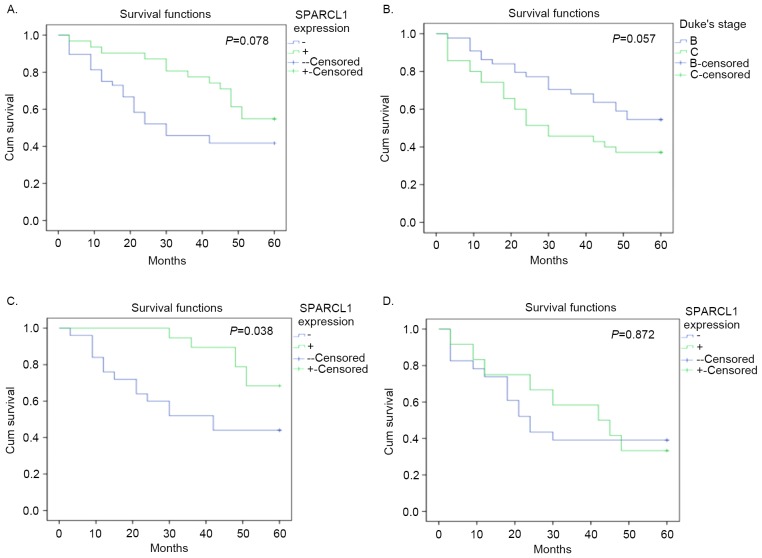
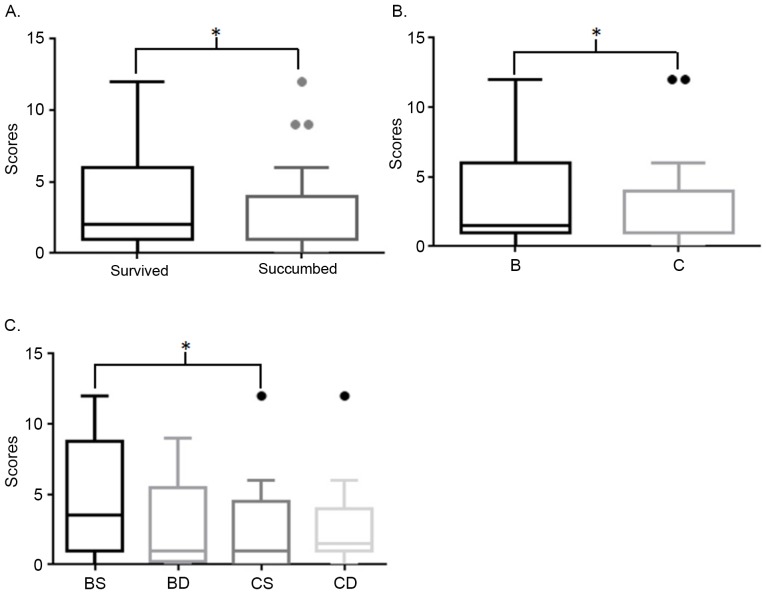
Although no difference between positive and negative expression status was identified in overall survival analysis, and no significance in SPARCL1 expression was detected in the Cox regression analysis (P>0.05; Tables I and II; Fig. 4A), immunohistochemical staining scores of patients who survived were significantly higher compared with those who succumbed (Fig. 3A). In addition, scores of patients at Duke's stage B who survived were significantly higher compared with patients with stage C (Fig. 3). Furthermore, the following Kaplan Meier curves demonstrated that positive expression of SPARCL1 was a potentially good indicator of prognosis at Duke's stage B, but not at stage C (P=0.038 and P=0.872, respectively; Fig. 4). The HR of overall survival calculated by log rank test, was 0.5755 (95% CI, 0.3108–1.045; P=0.078). The HR of patients at stage B was 0.3850 (95% CI, 0.1603–0.9277; P=0.038). The HR of patients at stage C was 0.9337 (95% CI, 0.3928–2.198; P=0.872).
Table II.
Univariate and multivariate analyses of factors in terms of overall survival.
| Univariate analysis | Multivariate analysis | |||||||
|---|---|---|---|---|---|---|---|---|
| 95% CI | 95% CI | |||||||
| Factors | HR | Lower | Upper | P-value | HR | Lower | Upper | P-value |
| SPARCL1 (+ vs. -) | 0.572 | 0.301 | 1.090 | 0.090 | 0.613 | 0.313 | 1.203 | 0.155 |
| Differentiation (I vs. II&III) | 0.378 | 0.122 | 1.173 | 0.092 | 0.529 | 0.158 | 1.774 | 0.303 |
| Differentiation (I&II vs. III) | 0.937 | 0.472 | 1.860 | 0.851 | 1.084 | 0.532 | 2.212 | 0.824 |
| Duke's stage (B vs. C) | 0.567 | 0.309 | 1.041 | 0.067 | 1.385 | 0.725 | 2.646 | 0.324 |
HR, hazard ratio; CI, confidence interval; SPARCL1, secreted protein acidic and rich in cysteine-like 1.
Figure 4.
Overall survival of patients with colorectal cancer. (A) The difference between positive and negative SPARCL1 expression was not significant. (B) The difference between Duke's stage B and C was not significant. (C) The difference between positive and negative SPARCL1 expression in Duke's stage B was significant. (D) The difference between positive and negative SPARCL1 expression in Duke's stage C was not significant. SPARCL1, secreted protein acidic and rich in cysteine-like 1; cum, cumulative.
Figure 3.
Comparison of immunoreactivity scores. (A) Survived vs. succumbed. (B) B vs. C; (C). BS vs. BD vs. CS vs. CD. *P<0.05. B, Duke's stage B; C, Duke's stage C; S, survived; D, succumbed.
Discussion
In the present study, the expression of SPARCL1 protein was investigated in colorectal carcinoma tissues through western blot analysis and detected a significantly lower expression in colorectal tumors compared with in adjacent normal tissues. The associations between SPARCL1 expression, clinicopathological factors and overall survival in patients with colorectal cancer were also analyzed. Since it was markedly decreased from differentiation I to III, it was hypothesized that the downregulation of SPARCL1 serves an essential role during the development and progression of colorectal cancer. Due to significantly higher survival in patients with positive SPARCL1 expression at Duke's stage B, SPARCL1 may be used as a potential biomarker for the prognosis of colorectal cancer. Despite a recent study demonstrated that SPARCL1 was a possible biomarker for the diagnosis of colorectal cancer, Zhang et al (11) reported that upregulated SPARCL1 was associated with poor survival. This conclusion was different the results of the present study, whereby SPARCL1 was an indicator of good prognosis at stage B, but not at C. In addition, another study reported that high SPARCL1 expression was associated with better prognosis in patients with rectal cancer with radiotherapy, but not in patients without radiotherapy (12). Therefore, high SPARCL1 expression may indicate better prognosis in patients with colorectal cancer at early stage or with neoadjuvant therapy, implying that SPARCL1 is a valuable target, and a significant marker of diagnosis. In previous studies, downregulated SPARCL1 expression was demonstrated to be significantly associated with lymphatic metastasis and poor grade of breast cancer, gastric adenocarcinoma, prostate cancer, and lung adenocarcinoma (13–16). In hepatocellular carcinoma, the expression of SPARCL1 was elevated in tissues, but overexpression of Hevin and SPARC significantly delayed tumor growth in vivo (7).
Gene mutations, loss of heterozygosity (LOH), and aberrant promoter methylation were three common possible mechanisms of gene inactivation (17). To the best of our knowledge, SPARCL1 gene was mapped to chromosome 4, which involves numerous hotspots for LOH in various tumor types, and a previous report, which included 10 paired tumor tissues with matched normal tissues, demonstrated that LOH may be responsible for the downregulation of SPARCL1 (8,18). However, following the transfection of SPARCL1 into colorectal cancer cells, including RKO and SW620 cells, overexpressed SPARCL1 protein significantly inhibited the growth, migration, and invasion of cancer cells (9). The aforementioned studies support the results of the current study, indicating that SPARCL1 may be an indicator of good prognosis. However, the molecular signaling pathway underlying SPARCL1 interaction remains unclear.
SPARCL1, also known as ‘SPARC-like 1’, exhibits functions similar or opposite to SPARC to a certain extent (3,4). SPARCL1 is putatively counter-adhesive to dermal fibroblasts in a similar way to that of SPARC; but compared with SPARC, SPARCL1 does not significantly inhibit the proliferation of cells (19). Likewise, high expression of SPARC and SPARCL1 may be two independent indicators of good prognosis in colorectal, and hepatocellular carcinomas (7,9,20); however, in other tumor types, their prognostic value appeared to be the opposite (18,21–23). In addition, the SLF (SPARC-like fragment), split from SPARCL1, similar to SPARC, may antagonize SPARCL1 and regulate synaptogenic activity (19). However, no previous study reported the influence of SPARC expression on prognosis of SPARCL1. In the present study, the prognosis of SPARCL1 overexpression was discussed, but the value of SPARC was not evaluated. In spite of the result that elevated SPARCL1 expression in patients at Duke's stage B predicted a better prognosis, the level of SPARC expression may also influence the outcome.
In the present study, though with no significant value in Cox regression analysis, it was demonstrated that the level of SPARCL1 expression was associated with the overall survival of patients with Duke's stage B. Overexpression of SPARCL1 indicated the patients at Duke's stage B with a longer survival. This result further suggested that SPARCL1 is an ‘early oncoprotein’ in colorectal carcinoma. However, in view of the sample size with 79 patients, which was small, further studies should be performed to reveal the potential values of SPARCL1.
In conclusion, elevated SPARCL1 expression in patients with Duke's stage B colorectal cancer, presented an association with better prognosis. Herein, upregulated SPARCL1 may be a candidate biomarker of good prognosis for early colorectal carcinoma.
Acknowledgements
The present study was supported by Jiangsu General University Program of Practical Innovation of Professional Postgraduate Degree (grant no. SJLX16_0450), Jiangsu University Program of Scientific Research (grant no. 15A358), Suzhou Youth Science and Technology Program of ‘Science and Education’ (grant no. KJXW2015053), Kunshan Science and Technology Program of Social Development (grant no. KS1654) and Jiangsu University Science and Technology Program of Clinical Medicine (grant no. JLY20160040).
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 2.Ng L, Poon RT, Pang R. Biomarkers for predicting future metastasis of human gastrointestinal tumors. Cell Mol Life Sci. 2013;70:3631–3656. doi: 10.1007/s00018-013-1266-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Girard JP, Springer TA. Cloning from purified high endothelial venule cells of hevin,a close relative of the antiadhesive extracellular-matrix protein SPARC. Immunity. 1995;2:113–123. doi: 10.1016/1074-7613(95)90083-7. [DOI] [PubMed] [Google Scholar]
- 4.Hambrock HO, Nitsche DP, Hansen U, Bruckner P, Paulsson M, Maurer P, Hartmann U. SC1/hevin. An extracellular calcium-modulated protein that binds collagen I. J Biol Chem. 2003;278:11351–11358. doi: 10.1074/jbc.M212291200. [DOI] [PubMed] [Google Scholar]
- 5.Isler SG, Schenk S, Bendik I, Schraml P, Novotna H, Moch H, Sauter G, Ludwig CU. Genomic organization and chromosomal mapping of SPARC-like 1, a gene down regulated in cancers. Int J Oncol. 2001;18:521–526. doi: 10.3892/ijo.18.3.521. [DOI] [PubMed] [Google Scholar]
- 6.Bendik I, Schraml P, Ludwig CU. Characterization of MAST9/Hevin, a SPARC-like protein, that is down-regulated in non-small cell lung cancer. Cancer Res. 1998;58:626–629. [PubMed] [Google Scholar]
- 7.Lau CP, Poon RT, Cheung ST, Yu WC, Fan ST. Sparc and hevin expression correlate with tumour angiogenesis in hepatocellular carcinoma. J Pathol. 2006;210:459–468. doi: 10.1002/path.2068. [DOI] [PubMed] [Google Scholar]
- 8.Claeskens A, Ongenae N, Neefs JM, Cheyns P, Kaijen P, Cools M, Kutoh E. Hevin is down-regulated in many cancers and is a negative regulator of cell growth and proliferation. Br J Cancer. 2000;82:1123–1130. doi: 10.1054/bjoc.1999.1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu H, Zhang H, Ge W, et al. Secreted protein acidic and rich in cysteines-like 1 suppresses aggressiveness and predicts better survival in colorectal cancers. Clin Cancer Res. 2012;18:5438–5448. doi: 10.1158/1078-0432.CCR-12-0124. [DOI] [PubMed] [Google Scholar]
- 10.Yu SJ, Yu JK, Ge WT, Hu HG, Yuan Y, Zheng S. SPARC1, Shp2, MSH2, E-cadherin, p53, ADCY-2, and MAPK are prognosis-related in colorectal cancer. World J Gastroenterol. 2011;17:2028–2036. doi: 10.3748/wjg.v17.i15.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang H, Widegren E, Wang DW, Sun XF. SPARCL1: A potential molecule associated with tumor diagnosis, progression and prognosis of colorectal cancer. Tumour Biol. 2011;32:1225–1231. doi: 10.1007/s13277-011-0226-x. [DOI] [PubMed] [Google Scholar]
- 12.Kotti A, Holmqvist A, Albertsson M, Sun XF. SPARCL1 expression increases with preoperative radiation therapy and predicts better survival in rectal cancer patients. Int J Radiat Oncol Biol Phys. 2014;88:1196–1202. doi: 10.1016/j.ijrobp.2013.12.041. [DOI] [PubMed] [Google Scholar]
- 13.Cao F, Wang K, Zhu R, Hu YW, Fang WZ, Ding HZ. Clinicopathological significance of reduced SPARCL1 expression in human breast cancer. Asian Pac J Cancer Prev. 2013;14:195–200. doi: 10.7314/APJCP.2013.14.1.195. [DOI] [PubMed] [Google Scholar]
- 14.Jakharia A, Borkakoty B, Singh S. Expression of SPARC like protein 1 (SPARCL1), extracellular matrix-associated protein is down regulated in gastric adenocarcinoma. J Gastrointest Oncol. 2016;7:278–283. doi: 10.3978/j.issn.2078-6891.2015.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiang Y, Qiu Q, Jiang M, Jin R, Lehmann BD, Strand DW, Jovanovic B, DeGraff DJ, Zheng Y, Yousif DA, et al. SPARCL1 suppresses metastasis in prostate cancer. Mol Oncol. 2013;7:1019–1030. doi: 10.1016/j.molonc.2013.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isler SG, Ludwig CU, Chiquet-Ehrismann R, Schenk S. Evidence for transcriptional repression of SPARC-like 1, a gene downregulated in human lung tumors. Int J Oncol. 2004;25:1073–1079. [PubMed] [Google Scholar]
- 17.Lerebours F, Olschwang S, Thuille B, Schmitz A, Fouchet P, Buecher B, Martinet N, Galateau F, Thomas G. Fine deletion mapping of chromosome 8p in non-small-cell lung carcinoma. Int J Cancer. 1999;81:854–858. doi: 10.1002/(SICI)1097-0215(19990611)81:6<854::AID-IJC3>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 18.Li P, Qian JX, Yu GZ, Chen Y, Liu K, Li J, Wang J. Down-regulated SPARCL1 is associated with clinical significance in human gastric cancer. J Surg Oncol. 2012;105:31–37. doi: 10.1002/jso.22025. [DOI] [PubMed] [Google Scholar]
- 19.Bradshaw AD. Diverse biological functions of the SPARC family of proteins. Int J Biochem Cell Biol. 2012;44:480–488. doi: 10.1016/j.biocel.2011.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu QZ, Gao XH, Chang WJ, Wang HT, Wang H, Cao GW, Fu CG. Secreted protein acidic and rich in cysteine expression in human colorectal cancer predicts postoperative prognosis. Eur Rev Med Pharmacol Sci. 2015;19:1803–1811. [PubMed] [Google Scholar]
- 21.Wang Z, Hao B, Yang Y, Wang R, Li Y, Wu Q. Prognostic role of SPARC expression in gastric cancer: A meta-analysis. Arch Med Sci. 2014;10:863–869. doi: 10.5114/aoms.2014.46207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Esposito I, Kayed H, Keleg S, Giese T, Sage EH, Schirmacher P, Friess H, Kleeff J. Tumor-suppressor function of SPARC-like protein 1/Hevin in pancreatic cancer. Neoplasia. 2007;9:8–17. doi: 10.1593/neo.06646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han W, Cao F, Chen MB, Lu RZ, Wang HB, Yu M, Shi CT, Ding HZ. Prognostic value of SPARC in patients with pancreatic cancer: A systematic review and meta-analysis. Plos One. 2016;11:e0145803. doi: 10.1371/journal.pone.0145803. [DOI] [PMC free article] [PubMed] [Google Scholar]