Abstract
High-risk human papillomavirus (HPV) is the primary cause of cervical carcinoma (CC). Viral integration into the host chromosomes is associated with neoplastic progression, and epigenetic changes may occur as a result. The objective of the present study was to analyze HPV L1 gene methylation and to compare the use of quantitative polymerase chain reaction (qPCR), in situ hybridization (ISH) and L1 methylation analysis as methods for detecting HPV integration. Cervical scrapes or biopsy samples positive for HPV 16 or 18, from 187 female patients with CC, squamous intraepithelial lesions (SILs) or no intraepithelial lesion (non-IL) were analyzed. Methylation of the L1 gene was determined using bisulfite modification followed by PCR, and HPV integration was subsequently analyzed. HPV 16 L1 gene methylation was revealed to increase with histological grade, with statistically significant differences observed as follows: Low-grade SIL vs. CC, P<0.0001 and non-IL vs. CC, P<0.0001. HPV 18 L1 gene methylation also increased according to histological grade, however, no statistically significant differences were observed. Methylation at CpG site 5608 of the HPV 16 L1 gene was associated with all grades of cervical lesions, whereas methylation at CpG site 5617 demonstrated the strongest association with CC (odds ratio, 42.5; 95% confidence interval, 4.7–1861; P<0.0001). The concordance rates between the various methods for the detection of the physical status of HPV 16 and HPV 18 were 96.1% for qPCR and ISH, 76.7% for qPCR and L1 gene methylation, and 84.8% for ISH and L1 gene methylation. In conclusion, methylation of the HPV 16 L1 gene increases significantly according to the grade of the cervical lesion, and methylation at CpG sites 5608 and 5617 of this gene may be used as prognostic biomarkers. ISH and L1 gene methylation have good concordance with qPCR with regards to the detection of HPV integration. Therefore, these are useful methods in determining the physical state of HPV.
Keywords: DNA methylation, human papillomavirus 16, human papillomavirus 18, biomarkers, cervical cancer, L1 gene, squamous intraepithelial lesion, integration
Introduction
The primary cause of cervical carcinoma (CC) is persistent infection with high-risk human papillomavirus (HR-HPV) (1). The infecting HPV type, viral persistence, viral integration status and viral load contribute to the pathogenesis of CC (1). Among the HR-HPVs, HPV 16 and 18 are the most common in cases of CC globally (2).
CC was the fourth most common type of cancer among women globally in 2012 (3). In Mexico, it is the second most frequent cancer type amongst women and the most common female cancer type in women aged 15–44 years, with ~9.4% of women in the general population estimated to harbor cervical HPV infection, and 70.2% of all CC cases being attributed to HPV 16 or 18 (4). In the state of Guerrero in Southern Mexico, CC is the most common type of cancer in women. This state also has the fifth highest mortality rate from CC in the country (5). HPV 16 is the most common genotype in CC, followed by HPV 18 (6).
Cytological screening of Papanicolaou smears has reduced the incidence and mortality rates of CC. However, the low sensitivity and low reproducibility of cytological tests, despite the availability of HPV DNA testing as an adjunct procedure, has resulted in a lack of discrimination between transient and persistent infections (7).
HPV genomes replicate as episomal DNA during productive infections, and the integration of HR-HPV genomes into the host chromosome has been associated with neoplastic progression (8). The E2 HPV gene has been revealed to be broken more frequently compared with other sites (9–11). This break results in a loss of the negative-feedback control for the transcription of oncogenes E6 and E7 (12), enabling tumor development and progression (13,14). Therefore, HPV integration into the human genome is considered to be a notable event in cervical carcinogenesis. Although certain studies suggest that integration is a late event (14,15), others have proposed that integration may be an early event in carcinogenesis (16–20).
Based on this information, a quantitative polymerase chain reaction (qPCR) analysis method that uses the PCR amplification of the E2 and E6 open reading frames (ORFs) and a calculation of E2/E6 ratios in order to measure integration was developed (20). Identification of the integrated HPV has been proposed as a useful biomarker for lesion progression. Nevertheless, qPCR is a technically difficult method for use in clinical practice.
HPV integration may render viral and cellular genes susceptible to epigenetic changes, which may in turn regulate their expression (21). The L1 gene in HPV 16 and 18 has been observed to be frequently hypermethylated in cancer, compared with the infrequent methylation observed in premalignant lesions (18,22–26). It has been reported that the methylation of the HPV 16 L1 gene indicates integration and is associated with the grade of premalignant lesions and cancer types, as well as being a potential clinical marker of CC progression (18,27–29).
HPV integration and the grade of cervical neoplasia may also be identified using a biotinyl tyramide-based in situ hybridization (ISH) method. Punctate signals within the nucleus are consistent with the integration of HPV into host cell chromosomes, whereas diffuse signal patterns throughout the nucleus are consistent with episomal DNA (30–33).
The use of novel tools alongside the cytological diagnosis of premalignant lesions may improve the sensitivity of this method, increasing its effectiveness, which would reduce the number of misdiagnosed cases and prevent unnecessary invasive treatments in women diagnosed with premalignant lesions. As HPV integration is a viral marker for lesion progression, it is important to use a technically simple and affordable method for the detection of HPV integration in clinical practice.
The aim of the present study was to analyze L1 gene methylation in cervical lesions of various grades, and to perform a comparative analysis between L1 methylation, ISH and RT-qPCR as methods to determine the physical status of HPV 16 and 18. Additionally, the ease of use of these methods was compared.
Materials and methods
Clinical specimens
A total of 187 female patients between 28–51 years of age were enrolled in the present study, selected from the 1,115 female residents of the state of Guerrero in Southern Mexico. The patients were admitted to the Unit for Early Diagnosis of Cervical Cancer and HPV at the Autonomous University of Guerrero (Chilpancingo, Mexico) and the Cancer Institute of the State of Guerrero (Acapulco, Mexico) between November 2013 and August 2015. Cervical scrapes or biopsies were collected for cytomorphological examination using liquid cytology or histological examination. Papanicolau smears were reviewed by a certified cytopathologist and biopsies by a certified histopathologist. DNA was extracted from the cervical scrapes or biopsies using the phenol-chloroform isoamyl alcohol method (34) for HPV detection and genotyping with an INNO-LiPA genotyping kit (Fujirebio Europe, Gent, Belgium) (35), without amendment and according the manufacturer's protocol.
All patients included in the current study presented with HPV 16 (n=154) or HPV 18 (n=33), and CC (n=33), high-grade squamous intraepithelial lesion (HSIL; n=13), low-grade squamous intraepithelial lesion (LSIL; n=102) or no intraepithelial lesion (non-IL; n=39). Cytological diagnosis was performed according to the Bethesda System (36), and histological diagnosis was performed according to the classification system of the International Federation of Gynecology and Obstetrics (37). Clinical data from the patients was obtained from the database of the Molecular Biomedicine and Cytopathology Laboratories at the School of Chemistry and Biology, Autonomous University of Guerrero.
The present study was approved by the Bioethics Committee of the Autonomous University of Guerrero. Written informed consent was obtained from all patients prior to the study.
The samples were studied to analyze L1 gene methylation in the different grades of cervical lesion and to compare L1 methylation, ISH and qPCR as methods for determining the physical status of HPV 16 and 18.
Analysis of the methylation of the HPV 16 and HPV 18 L1 gene
Methylation of the L1 gene from HPV 16 and 18 was analyzed using a bisulfite conversion reaction, wherein unmethylated cytosine is converted into uracil, followed by PCR amplification. DNA (400–1,200 ng) from cervical samples was treated with bisulfite for 150 min at 64°C using an EZ DNA Methylation-Gold™ kit (Zymo Research Corp., Irvine, CA, USA), according to the manufacturer's protocol.
The 5′ and middle regions of the HPV 16 L1 gene, and the 5′ and 3′ regions of the HPV 18 L1 gene were amplified using PCR. Amplifications were performed using 100–150 ng bisulfite-treated genomic DNA, 6 µl Amplitaq Gold® 360 Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc., Waltham, MA, USA), 1 µmol sense primer and 1 µmol antisense primer in a 15-µl final reaction volume. The following primers were used for amplification: HPV 16 L1 5′ region forward, 5′-TTGTTGATGTAGGTGATTTTTATTTATATTTTAGTTCCA-3′ and reverse, 5′-CTAATACCCACACCTAATAACTAACC-3′ as previously described by Oka et al (28) at 59°C, HPV 16 L1 middle region forward, 5′-ATTTAGATTATATTAAAATGGTGTTAGAAT-3′ and reverse, 5′-AAATAATTAATTACCCCAACAAATACC-3′ as previously described by Brandsma et al (27) at 54°C. HPV 18 L1 5′ region forward, 5′-GGTTAGTTTTTAGGTGTTGGT-3′ and reverse, 5′-CACACAACTACCAAATAAAACA-3′ (Tm, 54°C); and HPV 18 L1 3′ region forward, 5′-TTATTAGTTTGGTGGATATATATTG-3′ and reverse, 5′-AAAACATACAAACACAACAATAAATA-3′ as previously described by Badal et al (38), at 59°C. The thermocycling conditions were as follows: 10 min at 96°C, followed by 40 cycles for 30 sec at 96°C, 30 sec at 54°C or 59°C, depending on the primer used, and 7 min at 72°C. The PCR-amplified fragments were sequenced using the Big Dye Terminator version 3.1 Cycle Sequencing in an ABI PRISM 310 sequencer (Applied Biosystems; Thermo Fisher Scientific, Inc.). The methylation of 12 (5602 to 6580) and 18 (6015 to 7123) CpG dinucleotides in the HPV 16 and HPV 18 L1 gene, respectively, was analyzed using LALIGN online platform (http://embnet.vital-it.ch/software/LALIGN_form.html). For the analysis of the L1 gene methylation variation in the samples, the L1 gene methylation ratio (L1MR) was calculated using the formula L1MR (%)=(number of methylated CpGs in the analyzed region of the L1 gene)/(number of all CpGs in the analyzed region of the L1 gene) ×100. Oka et al (28) suggested that the L1MR is associated with the grade of cervical intraepithelial neoplasia (CIN) and indicates the HPV genome status in the cell, as follows: A high L1MR value indicates HPV genome integration, whereas a low L1MR indicates episomal HPV genomes. Complete methylation (>80%) is considered to indicate HPV integration, intermediate methylation (20–80%) is considered to indicate a mixed status and low methylation (<20%) is considered to indicate an episomal HPV genome (39).
Analysis of HPV 16 and 18 integrations using qPCR
The physical state of HPV 16 was analyzed using qPCR, amplifying two regions of the E2 gene and a region of the E6 gene using the following primers and TaqMan probes (Applied Biosystems; Thermo Fisher Scientific, Inc.): E2A region (transactivation domain) forward, 5′-TGGATATACAGTGGAAGTGCAGTTTG-3′ and reverse, 5′-CCACAGTTACTGATGCTTCTTCACA-3′ primers, and 5′FAM-ATGGAGACATATGCAATACAAT-NFQ probe; E2B region (hinge) forward, 5′-CAGCAACGAAGTATCCTCTCCT-3′ and reverse, 5′-GCGACGGCTTTGGTATGG-3′ primers, and 5′FAM-AAGTGCTGCCTAATAATTTC-NFQ probe; and E6 region forward, 5′-ACCGTTGTGTGATTTGTTAATTAGGTGTA-3′ and reverse, 5′-GCTTTTTGTCCAGATGTCTTTGCTT-3′ primers, and 5′FAM-AAAGCCACTGTGTCCTG-NFQ probe.
Similarly, the physical state of HPV 18 was analyzed by amplifying two regions of the E2 gene and a region of the E6 gene using the following primers and TaqMan probes (Applied Biosystems; Thermo Fisher Scientific, Inc.): E2N1 region (transactivation domain) forward, 5′-GCAACTAATACGTTGGGAAAATGCA-3′ and reverse, 5′-GCTGGCACCACCTGGT-3′ primers, and 5′FAM-CTGTATGCCATGTTCCC-NFQ probe; E2H1 region forward, 5′-GATTGTAATGACTCTATGTGCAGTACCA-3′ and reverse, 5′-CGGTGCCCACGGACA-3′ primers, and FAM-ACGACACGGTATCCGC-NFQ probe; and E6 region forward, 5′-AAAACTAACTAACACTGGGTTATACAATT-3′ and reverse, 5′-CTGCTGGATTCAACGGTTTCTG primers, and 5′FAM-CACCGCAGGCACCTTA-NFQ probe.
To calculate the number of cells, a region of the RNAse P single copy gene was amplified using forward, 5′-AGATTTGGACCTGCGAGCG-3′ and reverse, 5′-GAGCGGCTGTCTCCACAAGT-3′ primers, and a VIC-TTCTGACCTGAAGGCTCTGCGCG-TAMRA RNAse P probe (cat no. 4403328 Applied Biosystems; Thermo Fisher Scientific, Inc.).
In all cases, PCR was performed in a final volume of 10 µl containing 50 ng DNA, 5 µl Universal MasterMix II (Applied Biosystems; Thermo Fisher Scientific, Inc.) and 0.5 µl of a mix containing specific primers and probes for each gene and region. DNA amplification by qPCR was performed in an ABI-PRISM 7500 (Applied Biosystems; Thermo Fisher Scientific, Inc.) using the following thermocycler conditions: 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 sec and 60°C for 1 min as previously described by Zhang et al (40). All reactions were run in duplicate, and the results were averaged for analysis. The positive control consisted of DNA from the SiHa cell line which contains 1–2 integrated copies of the HPV 16 genome as previously described by Flores et al (41) and Saunier et al (42), and the HeLa cell line, which contains integrated HPV 18. The negative controls consisted of the reaction components without DNA.
A standard curve for the quantification of the RNAse P gene was generated using the commercial kit Taqman Copy Number Reference Assay RNAse P (Applied Biosystems; Thermo Fisher Scientific) with genomic DNA obtained from 10,000, 5,000, 2,500, 1,250 and 625 cells. Standard curves for the E2 and E6 genes were generated by determining the quantification cycle (Cq) (43) value of detection with concentrations of 108, 107, 106, 105, 104 and 103 copies/µl of plasmids containing the HPV 16 or HPV 18 genome as was performed by Zhang et al (40). All assays were performed in duplicate. The number of copies of each gene (x) in the sample was calculated with the formula x=exp[(Y-b)/m] where Y is the Cq value, and b and m are constant values obtained from the equation graph for the standard curve of each gene (44).
The physical state of HPV 16 was determined by calculating the ratios of E2A/E6 and E2B/E6. To ensure the validity of the method, these ratios were determined using the standard curves obtained from various dilutions of pUC8 and DNA from SiHa cells; pUC8 contains the complete HPV 16 genome, representing the episomal form, and SiHa cells were used as the integrated form (44). Viral genome integration was defined by the absence of E2 or an E2/E6 ratio of <0.0010. An E2A/E6 ratio of 0.0010–0.9270 was defined as a mixture of episomal and integrated forms. A ratio of >0.9270 was defined as the episomal form. An E2B/E6 ratio of 0.0010–0.9116 was defined as mixed form, and an E2B/E6 ratio of >0.9116 was defined as the episomal form (40).
The physical state of HPV 18 was determined by calculating the ratios of E2N1/E6 and E2H1/E6. To ensure the validity of this method, these ratios were determined using standard curves obtained from various dilutions of a plasmid containing the HPV 18 genome, representing the episomal form. Viral genome integration was defined by the absence of E2 or an E2/E6 ratio of <0.0010. An E2N1/E6 ratio of 0.0010–0.9151 was defined as the mixed form, and >0.9151 was defined as the episomal form. An E2H1/E6 ratio of 0.0010–0.9206 was defined as the mixed form, and an E2H1/E6 ratio >0.9206 was defined as the episomal form (40).
Analysis of HPV 16 and 18 integration using ISH
Detection of HR-HPV genome integration was performed using ISH with a GenPoint Tyramide Signal Amplification system (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA). Cytological samples were placed in a solution of acetone for 5 min at room temperature. The monolayer smears were digested for 1 min at 37°C with proteinase K (Invitrogen; Thermo Fisher Scientific, Inc.; 1:1,000). Biotinylated viral DNA with probes for 13 HR-HPV genotypes (types 16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59 and 68) and individual probes (Dako; Agilent Technologies, Inc.) directed at HPV 6 and HPV 11 were subsequently applied. The slides were denatured for 10 min at 95°C, hybridized with a biotinylated HPV DNA probe (Dako; Agilent Technologies, Inc.) for 20 h at 37°C and then placed in an astringent solution (1:50) at 55°C for 20 min. Primary streptavidin peroxidase (1:100) was added for 40 min at room temperature, followed by biotinyl-tyramide for 20 min at 37°C, and horseradish peroxidase conjugated streptavidin (cat no. N100; Thermo Fisher Scientific, Inc.) undiluted, as the secondary antibody, was subsequently added for 20 min at 37°C. The chromogen 3,3′-diaminobenzidene (Dako; Agilent Technologies, Inc.) was then added and Mayer's hematoxylin (Merck KGaA, Darmstadt, Germany) at 37°C for 10 sec was used as a contrast dye. Positive reactions were visualized as brown inside of the nucleus in a LEICA DM 1000 light microscope and, according to the type of signal, were classified as diffuse (episomal state), punctate (integrated state) or diffuse and punctate (mixed state) (30,31,33). SiHa cell lines (containing HPV 16) were used as positive controls to demonstrate an integrated state; the same cell lines without probes were used as negative controls.
Statistical analysis
L1MR was compared between groups using one-way analysis of variance and Tukey's multiple comparison test as a post-hoc test using GraphPad Prism V5-03 software (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 was considered to indicate a statistically significant difference. Odds ratios (ORs) were calculated to determine the risk of lesion progression due to the methylation of specific CpG sites using STATA version 11 software (StataCorp LLC, College Station, TX, USA; http://www.stata.com/). Concordance between the various methods for the detection of viral integration was also calculated (45).
Results
Methylation of the HPV 16 and HPV 18 L1 gene in cervical specimens from women with CC, SIL and non-IL
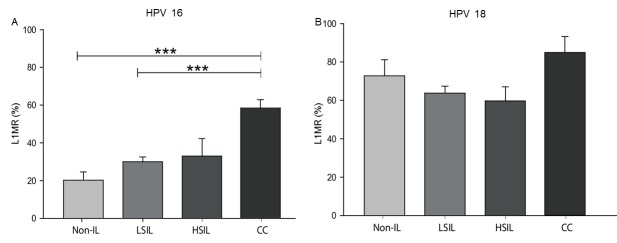
The methylation status of the HPV 16 and HPV 18 L1 gene was evaluated in 187 cervical specimens from women with CC, HSIL, LSIL and non-IL. The median HPV 16 L1MRs were 20.2% for non-IL, 30.0% for LSIL, 33.0% for HSIL and 58.6% for CC. The HPV 16 L1MR was associated with histological grade, as statistically significant differences were identified in the following comparisons: LSIL vs. CC (P<0.0001) and non-IL vs. CC (P<0.0001; Fig. 1A). For HPV 18, the median L1MRs were 73.3% for non-IL, 64.1% for LSIL, 69.7% for HSIL and 85.7% for CC; however, these differences between histological grades were not significant (Fig. 1B).
Figure 1.
L1 gene methylation rate in four distinct tissue types. (A) HPV 16 and (B) HPV 18 L1 gene methylation rate (L1MR, %) in non-IL, LSIL, HSIL and CC tissues. ***P<0.0001 with comparisons indicated by lines. HPV, human papillomavirus; L1MR, L1 methylation ratio; non-IL, non-intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion; CC, cervical carcinoma.
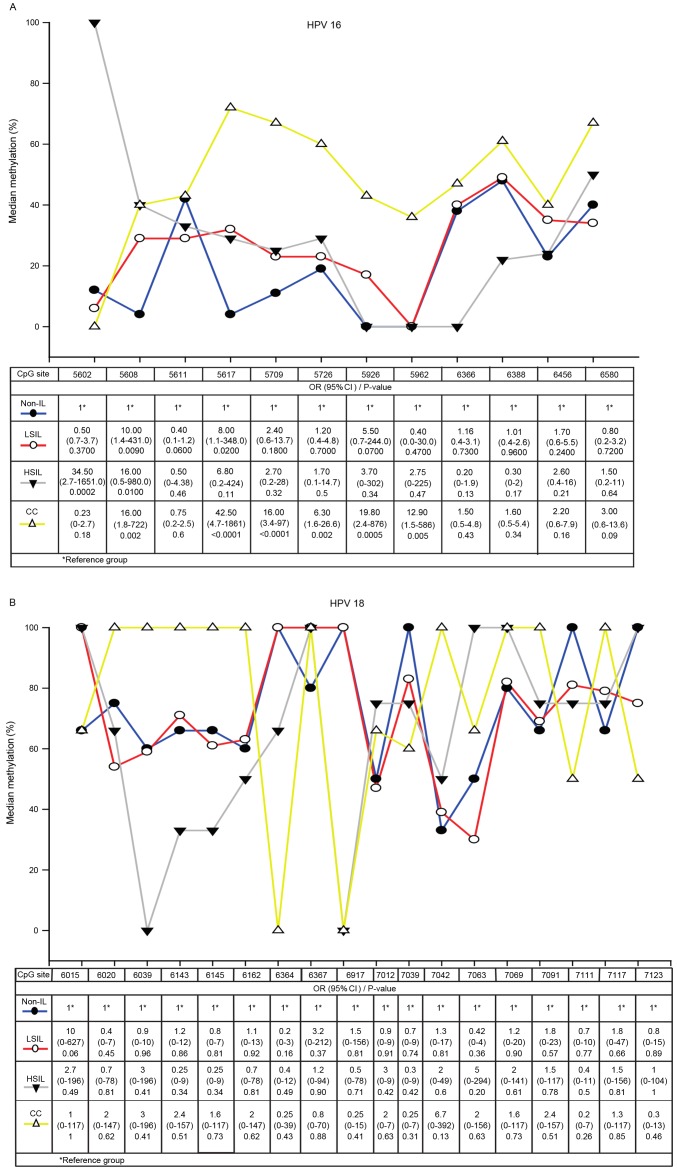
A total of 12 CpG dinucleotides (from 5602 to 6580) in the HPV 16 L1 gene and 18 CpG sites (from 6015 to 7123) in the HPV 18 L1 gene were analyzed in specimens from patients with non-IL, LSIL, HSIL and CC. In HPV 16, the methylation of CpG sites 5608 and 5617 were associated with LSIL, the methylation of CpG sites 5602 and 5608 were associated with HSIL, and CpG sites 5608, 5617, 5709, 5726, 5926, 5962 were associated with CC. The strongest association with CC was CpG site 5617 (OR, 42.5; 95% confidence interval, 4.7–1861; P<0.0001), followed by CpG sites 5926, 5608, 5709 and 5962. Only CpG site 5608 was associated with all grades of cervical lesions. In CC samples, the highest L1MR was at CpG site 5617 (71.4%) (Fig. 2A).
Figure 2.
Methylation in the L1 gene at various CpG sites in (A) HPV 16 and (B) HPV 18 in non-IL, LSIL, HSIL and CC. The x-axis indicates each individual CpG site by the nucleotide position in the L1 gene. HPV, human papillomavirus; non-IL, non-intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion; CC, cervical carcinoma; OR, odds ratio; CI, confidence interval.
High L1MRs at CpG sites in HPV 18 were detected in all grades of cervical lesions. 100% methylation was detected at 10 CpG sites in CC, 5 CpG sites in HSIL, 5 CpG sites in LSIL and 5 CpG sites in non-IL samples. However, the methylation differences in CC, HSIL, LSIL compared with non-IL were not significant (Fig. 2B).
Analysis of HPV 16 and HPV 18 integration using qPCR, ISH and L1 methylation
The physical statuses of HPV 16 and HPV 18 were evaluated using qPCR, ISH and L1 methylation in cervical specimens from CC, HSIL, LSIL and non-IL. HPV 18 was more frequently integrated compared with HPV 16; HPV 18 was integrated in 50% (qPCR), 100% (ISH) and 67% (L1 methylation) of CC specimens, whilst HPV 16 was integrated in 52% (qPCR), 92% (ISH) and 16% (L1 methylation) of CC specimens. Non-IL and LSIL demonstrated high rates of integration of HPV 16, as follows: 50 and 39% by qPCR, 33 and 45% by ISH, and only 6 and 7% by L1 methylation, respectively. HPV 18 was not integrated in non-IL, according to the qPCR and ISH analyses, but did exhibit integration in LSIL (10% by qPCR, 53% by ISH and 25% by L1 methylation; Table I).
Table I.
Physical status of HPV 16 and HPV 18 determined by qPCR, ISH and L1 methylation.
| A, HPV 16 physical status, n (%) | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| qPCR (n=100) | ISH (n=140) | L1 methylation (n=154) | |||||||
| Diagnosis | Episomal (n=2) | Mixed (n=52) | Integrated (n=46) | Episomal (n=4) | Mixed (n=66) | Integrated (n=70) | Episomal (n=51) | Mixed (n=91) | Integrated (n=12) |
| Non-IL | 1 (4) | 12 (46) | 13 (50) | 3 (10) | 17 (57) | 10 (33) | 17 (52) | 16 (48) | 0 (6) |
| LSIL | 1 (2) | 27 (59) | 18 (39) | 1 (1) | 43 (54) | 36 (45) | 28 (34) | 48 (59) | 6 (7) |
| HSIL | 0 (0) | 1 (33) | 2 (67) | 0 (0) | 4 (67) | 2 (33) | 4 (45) | 4 (45) | 1 (10) |
| CC | 0 (0) | 12 (48) | 13 (52) | 0 (0) | 2 (8) | 22 (92) | 2 (7) | 23 (77) | 5 (16) |
| B, HPV 18 physical status, n (%) | |||||||||
| qPCR (n=19) | ISH (n=30) | L1 methylation (n=33) | |||||||
| Diagnosis | Episomal (n=5) | Mixed (n=12) | Integrated (n=2) | Episomal (n=1) | Mixed (n=14) | Integrated (n=15) | Episomal (n=0) | Mixed (n=23) | Integrated (n=10) |
| Non-IL | 2 (67) | 1 (33) | 0 (0) | 1 (17) | 5 (83) | 0 (0) | 0 (0) | 4 (67) | 2 (33) |
| LSIL | 1 (10) | 8 (80) | 1 (10) | 0 (0) | 9 (47) | 10 (53) | 0 (0) | 15 (75) | 5 (25) |
| HSIL | 2 (50) | 2 (50) | 0 (0) | 0 (0) | 0 (0) | 2 (100) | 0 (0) | 3 (75) | 1 (25) |
| CC | 0 (0) | 1 (50) | 1 (50) | 0 (0) | 0 (0) | 3 (100) | 0 (0) | 1 (33) | 2 (67) |
HPV, human papillomavirus; qPCR, quantitative polymerase chain reaction; ISH, in situ hybridization; non-IL, non-intraepithelial lesion; LSIL, low-grade squamous intraepithelial lesion; HSIL, high-grade squamous intraepithelial lesion; CC, cervical carcinoma.
The concordance rate between qPCR and ISH for the detection of the physical status of HPV 16 and HPV 18 was 96.1%. The concordance rates between qPCR and L1 methylation, and between ISH and L1 methylation were 76.7 and 84.8%, respectively (Table II).
Table II.
Concordance rates of qPCR, ISH and HPV L1 methylation in the detection of the physical status of HPV 16 and HPV 18 in clinical samples.
| A, qPCR/ISH (n=107) | |||
|---|---|---|---|
| qPCR | |||
| Physical status | Mixed/integrated | Episomal | Positive concordance, %a |
| ISH | 96.1 | ||
| Mixed/integrated | 98 | 2 | |
| Episomal | 6 | 1 | |
| B, qPCR/L1 methylation (n=120) | |||
| qPCR | |||
| Physical status | Mixed/integrated | Episomal | Positive concordance, %a |
| L1 methylation | 76.7 | ||
| Mixed/integrated | 74 | 7 | |
| Episomal | 38 | 0 | |
| C, ISH/L1 methylation (n=172) | |||
| ISH | |||
| Physical status | Mixed/integrated | Episomal | Positive concordance, %a |
| L1 methylation | 84.8 | ||
| Mixed/integrated | 123 | 2 | |
| Episomal | 42 | 3 | |
Values of 60–80% were considered to indicate good concordance between the techniques and values >80% were considered to indicate very good concordance. HPV, human papillomavirus; qPCR, quantitative PCR; ISH, in situ hybridization.
Discussion
The purpose of the present study was to investigate L1 gene methylation between various grades of cervical lesions positive for HPV 16 or HPV 18. Additionally, L1 methylation, ISH and qPCR were compared as methods with which to determine the integration of HPV 16 and HPV 18 in cervical specimens, including CC, premalignant lesions and non-cervical lesions.
The results indicated that the HPV 16 L1 gene undergoes a significant progressive increase in methylation rate according to the increase of the grade of the cervical lesion, in concordance with previous studies (23,28,46–48). However, higher methylation rates in non-IL and LSIL specimens were observed in the present study compared with those in a previous study (28). Methylation of L1 increases progressively through the four pathological categories (non-IL, LSIL, HSIL and CC). However, methylation is also present in the absence of a cervical lesion in tissue positive for HPV 16 (L1MR, 20.2% in non-IL).
A progressive increase in the methylation of the HPV 18 L1 gene was detected in non-IL, LSIL, HSIL and CC samples, with no significant differences observed between the different grades of cervical lesion. There are certain differences from the results obtained from various previous studies. Badal et al (38) reported the presence of L1 methylation in HPV 18-positive samples from CC and asymptomatic patients, whereas Turan et al (25) reported unmethylated HPV 18 L1 genes in samples from asymptomatic infections and infections with premalignant lesions, but strong methylation in all CCs. Kalantari et al (47) reported that methylation of HPV 16 and HPV 18 is lowest in cases of asymptomatic infection and increases progressively during the progression into cancer.
Kalantari et al (47) hypothesized that highly methylated LSIL and HSIL samples are those that have undergone certain molecular changes that predispose them to develop into cancer. The present study concurs with this hypothesis, however, the level of methylation should be taken into account even in patients without cervical lesions, as this may indicate the possibility of lesion progression.
The usefulness of specific sites of methylation in the HPV genome in identifying women who are at an increased risk of developing CC has been suggested previously (24,48–50).
In the present study, 12 methylation CpG sites of the HPV 16 L1 gene were analyzed in samples from non-IL, LSIL, HSIL and CC tissues. Methylation of the 5608 CpG site was associated with the development of LSIL, HSIL and CC, whilst the methylation of the 5617 CpG site was associated with LSIL and CC, and the methylation of the 5709, 5726, 5926 and 5962 CpG sites was associated exclusively with CC. Mirabello et al (48) identified a strong association between methylation at the 6457 CpG site and CIN2 (HSIL), and in a subsequent study a strong association between methylation at the 5602, 5608, 5611 and 5617 CpG sites and cervical precancerous tissue was revealed (50). Sun et al (29) revealed increased methylation of the CpG sites in HPV 16 L1 associated with CIN1, CIN2 and CIN3 (LSIL/HSIL). Lorincz et al (24) reported a higher methylation of the 6367 and 6389 L1 CpG sites in women with CIN2/3. Brandsma et al (51) reported that the frequency of methylation was the highest for CIN3, followed by CIN2 and CIN1 at the 5602, 5608, 5709, 5726, 6367, 6389, 6581 (referred to as 5600, 5606, 5707, 5724, 6365, 6387 and 6579) L1 CpG sites. The results of the present study coincide with the aforementioned studies, and the observed differences in what the sites of CpG methylation in the HPV 16 L1 gene have been identified to be may be due to the circulating HPV 16 variants in each geographical region. A previous study performed in the state of Guerrero in Southern Mexico revealed 27 variants of HPV 16 that belong to European and Asian-American sub-lineages (52). Nucleotide differences between variants may determine changes in HPV 16 methylation.
There is increasing evidence that the elevated methylation of specific HPV 16 CpG sites in the L1 gene ORF may be a biomarker of progression from premalignant lesions to CC. The present study concurs with Mirabello et al (48) in that a methylation assay in one or a small number of CpG sites would be easier to perform, minimize costs and could potentially serve as a prognostic biomarker to predict the development of premalignant lesions and CC.
In HPV 18, analysis of 18 CpG sites was performed in all grades of cervical lesions, revealing that none of the sites were significantly associated with the development of CC or premalignant lesions. However, high L1MRs in a number of CpG sites were detected in non-IL, LSIL, HSIL and CC, which may indicate that potentially certain changes have occurred that predispose the tissue to develop into cancer, as proposed by Kalantari et al (46), considering that viral methylation appears to occur years prior to the detection of premalignant lesions (48).
The integration of HR-HPV genomes is a key event in cervical carcinogenesis, as it contributes to neoplastic transformation. The integration of viral genomes is a consequence of chromosomal instability induced by deregulated E6 and E7 oncogene expression (53). There are several methods that may be used in order to detect HPV integration in the human genome, including qPCR (20), ISH (30,31) and L1 methylation (18). However, few studies have compared such methods by which to determine the physical state of HPV. Fujii et al (54) compared ISH with qPCR using SIL and CC samples, reporting an 86% concordance rate between the two methods. Biesaga et al (55) additionally compared ISH with qPCR to assess the physical viral genome state in CC samples and revealed no significant difference between these methods, concluding that ISH may be used for the assessment of viral genome status.
To the best of our knowledge, no previous studies comparing the use of qPCR, ISH and L1 gene methylation assay methods have been performed in order to evaluate HPV 16 integration. In the present study, these three methods were compared. Good concordance was identified between them, and each method possessed certain advantages. qPCR is a useful technique for detecting integrated and episomal forms of HPV. This method has the advantage of additionally calculating the viral copy numbers for episomal and integrated HPV, making it a prognostic tool for determining the risk of CC (20). L1 methylation is a frequent result of integration. Therefore, it has been proposed that L1 gene methylation indicates integration, and may be used as a potential biomarker of cancer progression (18,56). ISH with signal amplification by biotinyl tyramide to demonstrate HPV DNA has the advantage of preserving the histological context of a lesion and allows the detection of low-copy DNA (57). It is generally accepted that a diffuse signal indicates an episomal status and that punctate signals indicate integrated HPV (31). The ISH assay with signal amplification by biotinyl tyramide is convenient for clinical purposes (58). According to the results of the present study, it has a high concordance rate with qPCR (96.1%) and also with the HPV L1 gene methylation assay (84.8%).
In conclusion, methylation of the HPV 16 L1 gene increases significantly according to the grade of cervical lesion. CpG site 5608 is associated with premalignant lesions and CC, whereas CpG site 5617 had the strongest association with CC; therefore, these two sites may be useful as prognostic biomarkers. Furthermore, ISH and L1 methylation analyses have good concordance with qPCR and are, therefore, useful methods for determining the physical state of HPV. However, the combination of these techniques may provide more reliable results.
Acknowledgements
The present study was supported by the National Council of Science and Technology (grant no. 201579). The authors would like to thank Dr Marco Antonio Jiménez-López of the Cancer Institute of the State of Guerrero ‘Dr. Arturo Beltrán Ortega’ for the histopathological diagnosis, and the technicians of the Cytopathology and Histochemistry Laboratory (School of Chemical and Biological Sciences, Autonomous University of Guerrero) for their assistance.
References
- 1.zur Hausen H. Papillomaviruses and cancer: From basic studies to clinical application. Nat Rev Cancer. 2002;2:342–350. doi: 10.1038/nrc798. [DOI] [PubMed] [Google Scholar]
- 2.Muñoz N, Bosch FX, de Sanjosé S, Herrero R, Castellsagué X, Shah KV, Snijders PJ, Meijer CJ. International Agency for Research on Cancer Multicenter Cervical Cancer Study Group: Epidemiologic classification of human papillomavirus types associated with cervical cancer. N Engl J Med. 2003;348:518–527. doi: 10.1056/NEJMoa021641. [DOI] [PubMed] [Google Scholar]
- 3.Ferlay J, Soerjomataram I, Ervik M, Dikshit R, Eser S, Mathers C, Rebelo M, Parkin DM, Forman D, Bray F. GLOBOCAN 2012 v1.0, Cancer Incidence and Mortality Worldwide: IARC Cancer Base No. 11[Internet] Lyon, France: International Agency for Research on Cancer; 2013. [Sep 17;2017 ]. [Google Scholar]
- 4.Bruni LB-R L, Serrano B, Brotons M, Cosano R, Munoz J, Bosch FX, de Sanjosé S, Castellsagué X. ICO Information centre on HPV and cancer (HPV Information Centre). Human Papillomavirus and Related Diseases in México. Summary Reports. 2014 [Google Scholar]
- 5.de Secretaría Salud. Información estadística. https://www.gob.mx/cms/uploads/attachment/file/12951/MortalidadCaCu2000a2013.pdf. [May 30;2016 ];Estadísticas de cáncer cervicouterino. [Google Scholar]
- 6.Illades-Aguiar B, Alarcón-Romero Ldel C, Antonio-Véjar V, Zamudio-López N, Sales-Linares N, Flores-Alfaro E, Fernández-Tilapa G, Vences-Velázquez A, Muñoz-Valle JF, Leyva-Vázquez MA. Prevalence and distribution of human papillomavirus types in cervical cancer, squamous intraepithelial lesions, and with no intraepithelial lesions in women from Southern Mexico. Gynecol Oncol. 2010;117:291–296. doi: 10.1016/j.ygyno.2010.01.036. [DOI] [PubMed] [Google Scholar]
- 7.Pinto AP, Degen M, Villa LL, Cibas ES. Immunomarkers in gynecologic cytology: The search for the ideal ‘biomolecular Papanicolaou test’. Acta Cytol. 2012;56:109–121. doi: 10.1159/000335065. [DOI] [PubMed] [Google Scholar]
- 8.Pett M, Coleman N. Integration of high-risk human papillomavirus: A key event in cervical carcinogenesis? J Pathol. 2007;212:356–367. doi: 10.1002/path.2192. [DOI] [PubMed] [Google Scholar]
- 9.Badaracco G, Venuti A, Sedati A, Marcante ML. HPV16 and HPV18 in genital tumors: Significantly different levels of viral integration and correlation to tumor invasiveness. J Med Virol. 2002;67:574–582. doi: 10.1002/jmv.10141. [DOI] [PubMed] [Google Scholar]
- 10.Kalantari M, Karlsen F, Kristensen G, Holm R, Hagmar B, Johansson B. Disruption of the E1 and E2 reading frames of HPV 16 in cervical carcinoma is associated with poor prognosis. Int J Gynecol Pathol. 1998;17:146–153. doi: 10.1097/00004347-199804000-00009. [DOI] [PubMed] [Google Scholar]
- 11.Romanczuk H, Howley PM. Disruption of either the E1 or the E2 regulatory gene of human papillomavirus type 16 increases viral immortalization capacity. Proc Natl Acad Sci USA. 1992;89:3159–3163. doi: 10.1073/pnas.89.7.3159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jeon S, Lambert PF. Integration of human papillomavirus type 16 DNA into the human genome leads to increased stability of E6 and E7 mRNAs: Implications for cervical carcinogenesis. Proc Natl Acad Sci USA. 1995;92:1654–1658. doi: 10.1073/pnas.92.5.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kalantari M, Blennow E, Hagmar B, Johansson B. Physical state of HPV16 and chromosomal mapping of the integrated form in cervical carcinomas. Diagn Mol Pathol. 2001;10:46–54. doi: 10.1097/00019606-200103000-00008. [DOI] [PubMed] [Google Scholar]
- 14.Kalantari M, Villa LL, Calleja-Macias IE, Bernard HU. Human papillomavirus-16 and −18 in penile carcinomas: DNA methylation, chromosomal recombination and genomic variation. Int J Cancer. 2008;123:1832–1840. doi: 10.1002/ijc.23707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.zur Hausen H. Papillomaviruses causing cancer: Evasion from host-cell control in early events in carcinogenesis. J Natl Cancer Inst. 2000;92:690–698. doi: 10.1093/jnci/92.9.690. [DOI] [PubMed] [Google Scholar]
- 16.Gallo G, Bibbo M, Bagella L, Zamparelli A, Sanseverino F, Giovagnoli MR, Vecchione A, Giordano A. Study of viral integration of HPV-16 in young patients with LSIL. J Clin Pathol. 2003;56:532–536. doi: 10.1136/jcp.56.7.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang LW, Chao SL, Lee BH. Integration of human papillomavirus type-16 and type-18 is a very early event in cervical carcinogenesis. J Clin Pathol. 2008;61:627–631. doi: 10.1136/jcp.2007.052027. [DOI] [PubMed] [Google Scholar]
- 18.Kalantari M, Chase DM, Tewari KS, Bernard HU. Recombination of human papillomavirus-16 and host DNA in exfoliated cervical cells: A pilot study of L1 gene methylation and chromosomal integration as biomarkers of carcinogenic progression. J Med Virol. 2010;82:311–320. doi: 10.1002/jmv.21676. [DOI] [PubMed] [Google Scholar]
- 19.Kulmala SM, Syrjänen SM, Gyllensten UB, Shabalova IP, Petrovichev N, Tosi P, Syrjänen KJ, Johansson BC. Early integration of high copy HPV16 detectable in women with normal and low grade cervical cytology and histology. J Clin Pathol. 2006;59:513–517. doi: 10.1136/jcp.2004.024570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peitsaro P, Johansson B, Syrjänen S. Integrated human papillomavirus type 16 is frequently found in cervical cancer precursors as demonstrated by a novel quantitative real-time PCR technique. J Clin Microbiol. 2002;40:886–891. doi: 10.1128/JCM.40.3.886-891.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Das P, Thomas A, Mahantshetty U, Shrivastava SK, Deodhar K, Mulherkar R. HPV genotyping and site of viral integration in cervical cancers in Indian women. PLoS One. 2012;7:e41012. doi: 10.1371/journal.pone.0041012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002;16:6–21. doi: 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- 23.Kalantari M, Calleja-Macias IE, Tewari D, Hagmar B, Lie K, Barrera-Saldana HA, Wiley DJ, Bernard HU. Conserved methylation patterns of human papillomavirus type 16 DNA in asymptomatic infection and cervical neoplasia. J Virol. 2004;78:12762–12772. doi: 10.1128/JVI.78.23.12762-12772.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lorincz AT, Brentnall AR, Vasiljević N, Scibior-Bentkowska D, Castanon A, Fiander A, Powell N, Tristram A, Cuzick J, Sasieni P. HPV16 L1 and L2 DNA methylation predicts high-grade cervical intraepithelial neoplasia in women with mildly abnormal cervical cytology. Int J Cancer. 2013;133:637–644. doi: 10.1002/ijc.28050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Turan T, Kalantari M, Calleja-Macias IE, Cubie HA, Cuschieri K, Villa LL, Skomedal H, Barrera-Saldaña HA, Bernard HU. Methylation of the human papillomavirus-18 L1 gene: A biomarker of neoplastic progression? Virology. 2006;349:175–183. doi: 10.1016/j.virol.2005.12.033. [DOI] [PubMed] [Google Scholar]
- 26.Turan T, Kalantari M, Cuschieri K, Cubie HA, Skomedal H, Bernard HU. High-throughput detection of human papillomavirus-18 L1 gene methylation, a candidate biomarker for the progression of cervical neoplasia. Virology. 2007;361:185–193. doi: 10.1016/j.virol.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brandsma JL, Sun Y, Lizardi PM, Tuck DP, Zelterman D, Haines GK, III, Martel M, Harigopal M, Schofield K, Neapolitano M. Distinct human papillomavirus type 16 methylomes in cervical cells at different stages of premalignancy. Virology. 2009;389:100–107. doi: 10.1016/j.virol.2009.03.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Oka N, Kajita M, Nishimura R, Ohbayashi C, Sudo T. L1 gene methylation in high-risk human papillomaviruses for the prognosis of cervical intraepithelial neoplasia. Int J Gynecol Cancer. 2013;23:235–243. doi: 10.1097/IGC.0b013e31827da1f6. [DOI] [PubMed] [Google Scholar]
- 29.Sun C, Reimers LL, Burk RD. Methylation of HPV16 genome CpG sites is associated with cervix precancer and cancer. Gynecol Oncol. 2011;121:59–63. doi: 10.1016/j.ygyno.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Evans MF, Aliesky HA, Cooper K. Optimization of biotinyl-tyramide-based in situ hybridization for sensitive background-free applications on formalin-fixed, paraffin-embedded tissue specimens. BMC Clin Pathol. 2003;3:2. doi: 10.1186/1472-6890-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Evans MF, Mount SL, Beatty BG, Cooper K. Biotinyl-tyramide-based in situ hybridization signal patterns distinguish human papillomavirus type and grade of cervical intraepithelial neoplasia. Mod Pathol. 2002;15:1339–1347. doi: 10.1038/modpathol.3880698. [DOI] [PubMed] [Google Scholar]
- 32.Vega-Peña A, Illades-Aguiar B, Flores-Alfaro E, López-Bayghen E, Leyva-Vázquez MA, Castañeda-Saucedo E, Alarcón-Romero Ldel C. Risk of progression of early cervical lesions is associated with integration and persistence of HPV-16 and expression of E6, Ki-67, and telomerase. J Cytol. 2013;30:226–232. doi: 10.4103/0970-9371.126644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zubillaga-Guerrero MI, Illades-Aguiar B, Leyva-Vazquez MA, Flores-Alfaro E, Castañeda-Saucedo E, Muñoz-Valle JF, Alarcón-Romero LC. The integration of HR-HPV increases the expression of cyclins A and E in cytologies with and without low-grade lesions. J Cytol. 2013;30:1–7. doi: 10.4103/0970-9371.107504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ausubel F, Brent R, Kingston R, Moore D, Seidman JG, Smith JA, Struhl K. Short Protocols in Molecular Biology. 1995:2–3. [Google Scholar]
- 35.Kleter B, van Doorn LJ, Schrauwen L, Molijn A, Sastrowijoto S, Schegget Ter J, Lindeman J, Harmsel Ter B, Burger M, Quint W. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J Clin Microbiol. 1999;37:2508–2517. doi: 10.1128/jcm.37.8.2508-2517.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Solomon D, Davey D, Kurman R, Moriarty A, O'Connor D, Prey M, Raab S, Sherman M, Wilbur D, Wright T, Jr, et al. The 2001 Bethesda system: Terminology for reporting results of cervical cytology. JAMA. 2002;287:2114–2119. doi: 10.1001/jama.287.16.2114. [DOI] [PubMed] [Google Scholar]
- 37.Benedet JL, Bender H, Jones H, III, Ngan HY, Pecorelli S. FIGO staging classifications and clinical practice guidelines in the management of gynecologic cancers. FIGO Committee on Gynecologic Oncology. Int J Gynaecol Obstet. 2000;70:209–262. doi: 10.1016/S0020-7292(00)90001-8. [DOI] [PubMed] [Google Scholar]
- 38.Badal S, Badal V, Calleja-Macias IE, Kalantari M, Chuang LS, Li BF, Bernard HU. The human papillomavirus-18 genome is efficiently targeted by cellular DNA methylation. Virology. 2004;324:483–492. doi: 10.1016/j.virol.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 39.Reuschenbach M, Huebbers CU, Prigge ES, Bermejo JL, Kalteis MS, Preuss SF, Seuthe IM, Kolligs J, Speel EJ, Olthof N, et al. Methylation status of HPV16 E2-binding sites classifies subtypes of HPV-associated oropharyngeal cancers. Cancer. 2015;121:1966–1976. doi: 10.1002/cncr.29315. [DOI] [PubMed] [Google Scholar]
- 40.Zhang D, Zhang Q, Zhou L, Huo L, Zhang Y, Shen Z, Zhu Y. Comparison of prevalence, viral load, physical status and expression of human papillomavirus-16, −18 and −58 in esophageal and cervical cancer: A case-control study. BMC Cancer. 2010;10:650. doi: 10.1186/1471-2407-10-650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Flores R, Papenfuss M, Klimecki WT, Giuliano AR. Cross-sectional analysis of oncogenic HPV viral load and cervical intraepithelial neoplasia. Int J Cancer. 2006;118:1187–1193. doi: 10.1002/ijc.21477. [DOI] [PubMed] [Google Scholar]
- 42.Saunier M, Monnier-Benoit S, Mauny F, Dalstein V, Briolat J, Riethmuller D, Kantelip B, Schwarz E, Mougin C, Prétet JL. Analysis of human papillomavirus type 16 (HPV16) DNA load and physical state for identification of HPV16-infected women with high-grade lesions or cervical carcinoma. J Clin Microbiol. 2008;46:3678–3685. doi: 10.1128/JCM.01212-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 44.Chang L, He X, Yu G, Wu Y. Effectiveness of HPV 16 viral load and the E2/E6 ratio for the prediction of cervical cancer risk among Chinese women. J Med Virol. 2013;85:646–654. doi: 10.1002/jmv.23490. [DOI] [PubMed] [Google Scholar]
- 45.Kwiecien R, Kopp-Schneider A, Blettner M. Concordance analysis: Part 16 of a series on evaluation of scientific publications. Dtsch Arztebl Int. 2011;108:515–521. doi: 10.3238/arztebl.2011.0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fernandez AF, Rosales C, Lopez-Nieva P, Graña O, Ballestar E, Ropero S, Espada J, Melo SA, Lujambio A, Fraga MF, et al. The dynamic DNA methylomes of double-stranded DNA viruses associated with human cancer. Genome Res. 2009;19:438–451. doi: 10.1101/gr.083550.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kalantari M, Osann K, Calleja-Macias IE, Kim S, Yan B, Jordan S, Chase DM, Tewari KS, Bernard HU. Methylation of human papillomavirus 16, 18, 31, and 45 L2 and L1 genes and the cellular DAPK gene: Considerations for use as biomarkers of the progression of cervical neoplasia. Virology. 2014;448:314–321. doi: 10.1016/j.virol.2013.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mirabello L, Schiffman M, Ghosh A, Rodriguez AC, Vasiljevic N, Wentzensen N, Herrero R, Hildesheim A, Wacholder S, Scibior-Bentkowska D, et al. Elevated methylation of HPV16 DNA is associated with the development of high grade cervical intraepithelial neoplasia. Int J Cancer. 2013;132:1412–1422. doi: 10.1002/ijc.27750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Clarke MA, Wentzensen N, Mirabello L, Ghosh A, Wacholder S, Harari A, Lorincz A, Schiffman M, Burk RD. Human papillomavirus DNA methylation as a potential biomarker for cervical cancer. Cancer Epidemiol Biomarkers Prev. 2012;21:2125–2137. doi: 10.1158/1055-9965.EPI-12-0905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mirabello L, Frimer M, Harari A, McAndrew T, Smith B, Chen Z, Wentzensen N, Wacholder S, Castle PE, Raine-Bennett T, et al. HPV16 methyl-haplotypes determined by a novel next-generation sequencing method are associated with cervical precancer. Int J Cancer. 2015;136:E146–E153. doi: 10.1002/ijc.29119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brandsma JL, Harigopal M, Kiviat NB, Sun Y, Deng Y, Zelterman D, Lizardi PM, Shabanova VS, Levi A, Yaping T, et al. Methylation of twelve CpGs in human papillomavirus type 16 (HPV16) as an informative biomarker for the triage of women positive for HPV16 infection. Cancer Prev Res (Phila) 2014;7:526–533. doi: 10.1158/1940-6207.CAPR-13-0354. [DOI] [PubMed] [Google Scholar]
- 52.Ortiz-Ortiz J, Alarcón-Romero Ldel C, Jiménez-Lopez MA, Garzón-Barrientos VH, Calleja-Macías I, Barrera-Saldaña HA, Leyva-Vázquez MA, Illades-Aguiar B. Association of human papillomavirus 16 E6 variants with cervical carcinoma and precursor lesions in women from Southern Mexico. Virol J. 2015;12:29. doi: 10.1186/s12985-015-0242-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vinokurova S, Wentzensen N, Kraus I, Klaes R, Driesch C, Melsheimer P, Kisseljov F, Dürst M, Schneider A, von Knebel Doeberitz M. Type-dependent integration frequency of human papillomavirus genomes in cervical lesions. Cancer Res. 2008;68:307–313. doi: 10.1158/0008-5472.CAN-07-2754. [DOI] [PubMed] [Google Scholar]
- 54.Fujii T, Masumoto N, Saito M, Hirao N, Niimi S, Mukai M, Ono A, Hayashi S, Kubushiro K, Sakai E, et al. Comparison between in situ hybridization and real-time PCR technique as a means of detecting the integrated form of human papillomavirus 16 in cervical neoplasia. Diagn Mol Pathol. 2005;14:103–108. doi: 10.1097/01.pas.0000162755.84026.9f. [DOI] [PubMed] [Google Scholar]
- 55.Biesaga B, Szostek S, Klimek M, Jakubowicz J, Wysocka J. Comparison of the sensitivity and specificity of real-time PCR and in situ hybridization in HPV16 and 18 detection in archival cervical cancer specimens. Folia Histochem Cytobiol. 2012;50:239–247. doi: 10.5603/FHC.2012.0033. [DOI] [PubMed] [Google Scholar]
- 56.Bryant D, Tristram A, Liloglou T, Hibbitts S, Fiander A, Powell N. Quantitative measurement of Human Papillomavirus type 16 L1/L2 DNA methylation correlates with cervical disease grade. J Clin Virol. 2014;59:24–29. doi: 10.1016/j.jcv.2013.10.029. [DOI] [PubMed] [Google Scholar]
- 57.Bryan JT, Taddeo F, Skulsky D, Jansen KU, Frain BM, Qadadri B, Brown DR. Detection of specific human papillomavirus types in paraffin-embedded sections of cervical carcinomas. J Med Virol. 2006;78:117–124. doi: 10.1002/jmv.20512. [DOI] [PubMed] [Google Scholar]
- 58.De Marchi Triglia R, Metze K, Zeferino LC, De Angelo Andrade Lucci LA. HPV in situ hybridization signal patterns as a marker for cervical intraepithelial neoplasia progression. Gynecol Oncol. 2009;112:114–118. doi: 10.1016/j.ygyno.2008.09.047. [DOI] [PubMed] [Google Scholar]