Abstract
The lack of early diagnostic markers and novel therapeutic targets for clear cell renal cell carcinoma (ccRCC) negatively affects patient prognosis. Cancer metabolism is an attractive area for the understanding of the molecular mechanism of carcinogenesis. The present study attempted to identify metabolic changes from the view of the expression of metabolism-associated genes between control samples and those of ccRCC at different disease stages. Data concerning ccRCC gene expression obtained by RNA-sequencing was obtained from The Cancer Genome Atlas and data on metabolism-associated genes were extracted using the Recon2 model. Following analysis of differential gene expression, multiple differentially expressed metabolic genes at each tumor-node-metastasis disease stage were identified, compared with control non-disease samples: Metabolic genes (305) were differentially expressed in stage I disease, 323 in stage II disease, 355 in stage III disease and 363 in stage IV disease. Following enrichment analysis for differential metabolic genes, 22 metabolic pathways were identified to be dysregulated in multiple stages of ccRCC. Abnormalities in hormone, vitamin, glucose and lipid metabolism were present in the early stages of the disease, with dysregulation to reactive oxygen species detoxification and amino acid metabolism pathways occurring with advanced disease stages, particularly to valine, leucine, and isoleucine metabolism, which was substantially dysregulated in stage IV disease. The xenobiotic metabolism pathway, associated with multiple cytochrome P450 family genes, was dysregulated in each stage of the disease. This pathway is worthy of substantial attention since it may aid understanding of drug resistance in ccRCC. The results of the present study offer information to aid further research into early diagnostic biomarkers and therapeutic targets of ccRCC.
Keywords: clear cell renal cell carcinoma, metabolism pathway, diagnostic biomarkers, drug resistance
Introduction
Renal cell carcinoma (RCC) is the most common form of kidney cancer to affect adult populations in Western countries in 2013 (1). There are three common forms of RCC: Clear cell renal cell carcinoma (ccRCC), papillary renal cell carcinoma (of which there are types 1 and 2) and chromophobe renal cell carcinomas (1,2). According to the World Health Organization (2004), ccRCC accounts for 70–85% of all cases of kidney cancer (1,2). Surgical resection of the primary tumor is often performed in patients with ccRCC; however, it is only effective for patients diagnosed at an earlier stage of ccRCC (3). ccRCC develops and progresses asymptomatically at the early stage of the disease, which results in a low rate of early detection (4). Furthermore, the benefit of a chemotherapeutic approach is limited owing to drug resistance (3). Therefore, the development of novel approaches for early diagnostic biomarkers and improvement of treatment for patients with late stage ccRCC is critical. A promising strategy for the development of anticancer agents is targeting cancer metabolism (5).
Metabolic changes serve a key function in the progression of ccRCC (6–9). For example, Linehan et al (6) considered kidney cancer a metabolic disease rather than a single disease. Shayman et al (7) and others reported that targeting glycosphingolipid metabolism, hormone signaling pathways or vitamin D metabolism may be novel therapeutic approaches for treating RCC (8,9). However, to the best of our knowledge, no previous study has identified the alteration of metabolism-associated genes in ccRCC. A total of 1,603 available metabolism-associated genes involving 99 metabolic pathways from Recon2 model were extracted in the present study. Recon2 is the most comprehensive representation of human metabolism to date (10). The present study investigated changes from the view of metabolism-associated gene expression between control samples and those at different stages of ccRCC. These results will offer information to aid further research into early diagnostic biomarkers and therapeutic targets.
Materials and methods
Data collection
RNA-sequencing gene expression data for cohorts of 72 healthy controls and 532 patients with ccRCC (including clinical data) were extracted from The Cancer Genome Atlas (TCGA; https://cancergenome.nih.gov/) in April 2016 (11). The ccRCC group included 267 stage I, 57 stage II, 124 stage III and 84 stage IV patients based on the tumor-node-metastasis classification system (12). Table I shows the clinical characteristics of patients by stage. As presented in Table I, as ccRCC progressed, the survival rate of patients decreased. In addition, it is noteworthy that the number of male patients with ccRCC was increased compared with that of female patients.
Table I.
Clinical characteristics of patients by disease stage.
| Sex | Ethnicity | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Stages | Patients, n | Age, yearsa | Male | Female | Survival time, monthsb | Asian | African descent | Caucasian | No record |
| Stage I | 267 | 59.6 (25–90) | 162 | 105 | 45.0 | 4 | 40 | 221 | 2 |
| Stage II | 57 | 59.7 (39–86) | 43 | 14 | 49.0 | 1 | 5 | 49 | 2 |
| Stage III | 124 | 63.4 (32–88) | 81 | 43 | 42.1 | 3 | 6 | 112 | 3 |
| Stage IV | 84 | 59.9 (33–84) | 59 | 25 | 34.1 | 0 | 5 | 79 | 0 |
| Total | 532 | 60.6 (26–90) | 345 | 187 | 44.3 | 8 | 56 | 461 | 7 |
Mean (range)
mean.
Overall metabolic gene expression profile
Each sample included 20,501 genes in the data from TCGA. A total of 1,638 metabolic genes were extracted from the Recon2 model. There are 1,603 common genes between the Recon2 model and data from TCGA. A comparison of the expression of these 1,603 metabolic genes expression between control and ccRCC cases with a heat map (generated using the ‘pheatmap’ function in R) (13). All gene expression values were scaled prior to plotting. Clustering was unsupervised.
Analysis of differential metabolism-associated gene expression
Genes with a fold change >2 and a false discovery rate (FDR) <0.05 were considered to be differentially expressed genes. This study was performed using the limma package (14) in R (version 3.2.3). Four comparisons were executed: Control vs. stage I disease samples, control vs. stage II, control vs. stage III and control vs. stage IV. The number of differentially expressed metabolism-associated genes between control and ccRCC samples of multiple stages were represented using a Venn diagram. A total of 4 heat maps were produced to visualize the differentially expressed metabolism-associated genes in each comparison. Gene expression values were scaled prior to plotting. Clustering was unsupervised.
Enrichment analysis of metabolic pathways
In total, there are 99 metabolic pathways in Recon2 model and each pathway includes specific metabolic genes. The present study used the following method of hypergenometric distribution for enrichment analysis of metabolic pathways:
Where N is the total number of metabolic genes, n is the total number of differential metabolic genes, M is the number of genes of a specific pathway, k is the number of differential metabolic genes of a specific pathway.
Results
Metabolism-associated gene expression profiles are separated between control and case
The gene expression profile of metabolism-associated genes was different between ccRCC and control samples (Fig. 1). Approximately 1/3 of the metabolic genes were upregulated and 2/3 were downregulated in ccRCC cases compared with the control samples. These results revealed that almost all metabolic genes were overexpressed or underexpressed in patients with ccRCC, which supports the results of numerous previous studies (6–9), which found that metabolic changes are significantly associated with ccRCC. This result also indicates that further research concerning the function that metabolism-associated genes serve in ccRCC is warranted.
Figure 1.
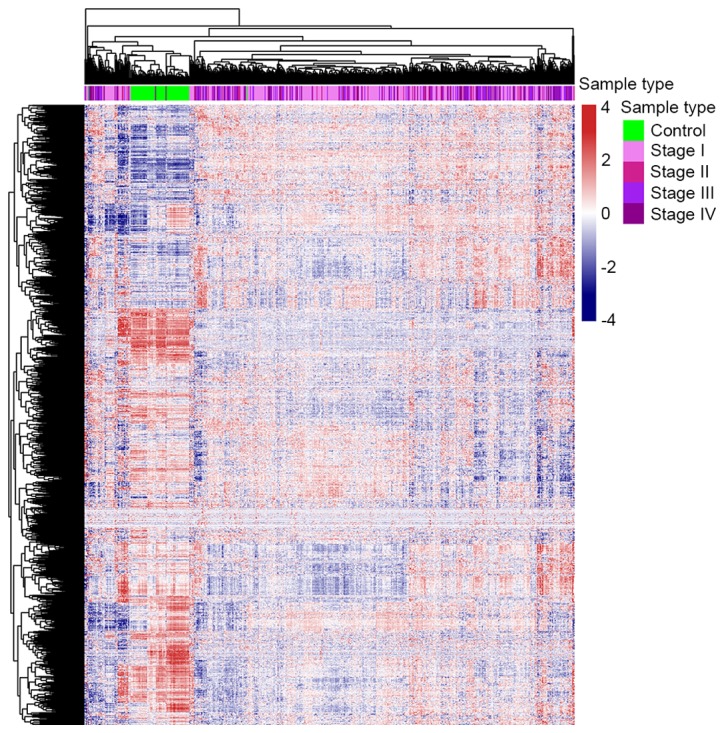
Overview of the expression of 1,603 metabolic genes in control and clear cell renal cell carcinoma samples.
Differential metabolism-associated gene expression
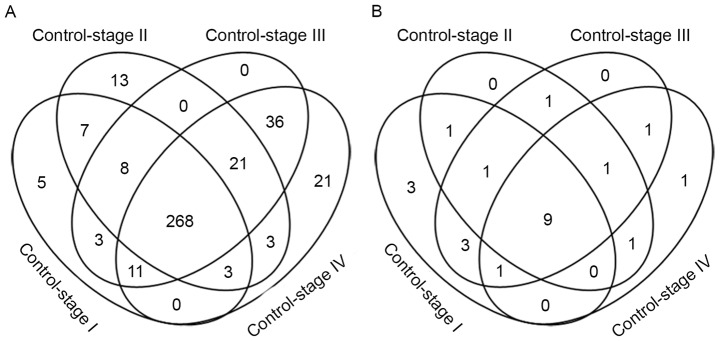
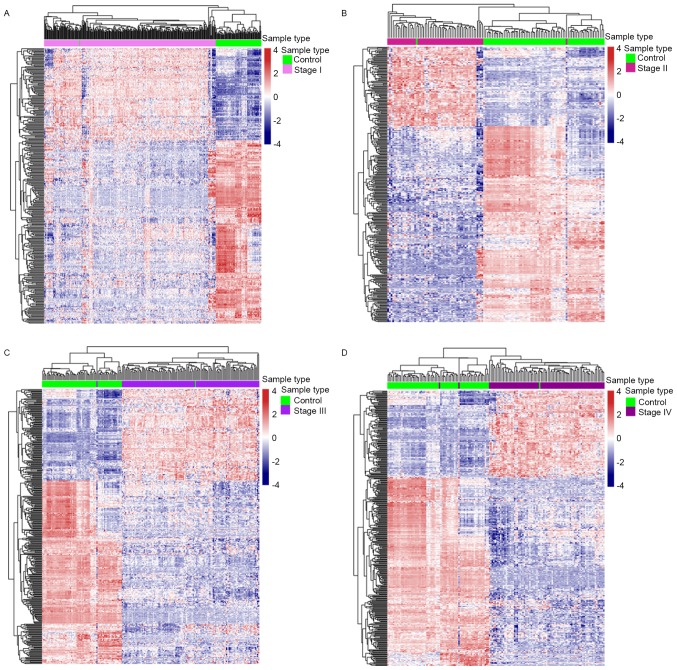
Metabolic genes were differentially expressed (FDR <0.05; fold change >2) in each comparison: 305 metabolic genes were differentially expressed between control and stage I samples, 323 metabolic genes differed between control and stage II samples, 355 metabolic genes differed between control and stage III samples and 363 metabolic genes differed between control and stage IV disease (Fig. 2A). Heat maps of the differentially expressed metabolic genes in each comparison were provided (Fig. 3). There were a total of 407 differential metabolism-associated genes in the 4 comparisons. With more advanced disease stage, there were more differentially expressed metabolic genes between ccRCC and control samples.
Figure 2.
Number of differential metabolic genes and metabolic pathways enriched in different stages of clear cell renal cell carcinoma. (A) Differentially expressed metabolic genes (FDR <0.05, fold change >2) in each comparison between control and disease samples. (B) The metabolic pathways enriched in each comparison. FDR, false discovery rate.
Figure 3.
Heat maps of differential metabolism-associated genes in each comparison. (A) Control vs. stage I. (B) Control vs. stage II. (C) Control vs. stage III. (D) Control vs. stage IV. Gene expression profiles were divided into 2 distinct groups under the condition of fully unsupervised clustering in each comparison.
There were 12 differentially expressed metabolic genes that only appeared at the initial stage of the disease, stage I or stage II, including: β-1,3-N-acetylglucosaminyltransferase 8 (B3GNT8), hydroxysteroid 11-β dehydrogenase 1 (HSD11B1), hyaluronan synthase 2 (HAS2), adenylate cyclase 8 (ADCY8), ADCY2, phosphodiesterase 6G (PDE6G), ectonucleotide pyrophosphatase/phosphodiesterase 1 (ENPP1), 3-hydroxyacyl-CoA dehydratase 1 (PTPLA), solute carrier family 5 member 1 (SLC5A1), SLC18A2, SLC10A2, SLC22A1. Seven of these genes (B3GNT8, HSD11B1, HAS2, ADCY2, ADCY8, PDE6G, ENPP1) have been reported to be associated with cancer. Multiple studies have revealed that increased expression of B3GNT8 and HSD11B1 is associated with different types of cancer, including glioma and laryngeal carcinoma (15–18). Lien et al (19) reported that HAS2 serves a function in producing aggressive phenotypes in primary breast carcinoma. Single nucleotide polymorphisms in ADCY2 and ADCY8 are associated with glioma risk (20). The differential expression of PDE6 G and ENPP1 occurs in breast cancer (19,21). However, the association between the other 5 genes (PTPLA, SLC5A1, SLC18A2, SLC10A2, SLC22A1) and ccRCC has not yet been reported. According to the present study, the solute carrier family genes (SLC5A1, SLC18A2, SLC10A2, SLC22A1) exhibited an increased expression in patients with early ccRCC. The present study demonstrates that abnormalities in the expression of these genes disappear in the late stage of ccRCC.
There were 268 differentially expressed metabolic genes shared by each disease stage. Cytochrome P450 family genes are worthy of substantial attention since 19 of them were differentially expressed in every ccRCC disease stage, including cytochrome P450 family 2 subfamily B member 6 (CYP2B6), CYP4F2, CYP17A1, CYP2J2, CYP24A1, CYP27B1, CYP4F3, CYP1A1, CYP3A5, CYP39A1, CYP1B1, CYP11A1, CYP26B1, CYP21A2, CYP1A2, CYP2C18, CYP4A22, CYP4B1, CYP3A4. The expression of CYP2J2, CYP3A5 and CYP21A2 was downregulated in ccRCC samples; the remaining 16 genes were significantly upregulated, particularly CYP2B6, CYP17A1, CYP4F2, CYP1A1 and CYP39A1, the expression of which was >4 times that in the control. This gene family encodes the proteins that localize to the endoplasmic reticulum and catalyze multiple reactions associated with drug metabolism. The US Food and Drug Association has approved drugs that target 4 of these genes: CYP51A1, CYP19A1, CYP17A1 and CYP11B1 (22).
A total of 89 metabolic genes were expressed differentially at the late stages of the disease (stages III and IV), of which 21 only appeared in stage IV disease, including SLC5A8, pyrroline-5-carboxylate reductase 1 (PYCR1), acyl-CoA synthetase short chain family member 3 (ACSS3), SLC22A2, ATPase Na+/K+ transporting subunit β1 (ATP1B1), B3GNTL1, ST6 N-acetylgalactosaminide α-2,6-sialyltransferase 3 (ST6GALNAC3), ADCY1, procollagen-lysine, 2-oxoglutarate 5-dioxygenase 3 (PLOD3), SLC27A2, PDE7B, thymidine kinase 1 (TK1), aldo-keto reductase family 1 member C1 (AKR1C1), superoxide dismutase 2 (SOD2), glucosylceramidase β3 (gBA3), ADCY10, PLOD1, methylmalonyl-CoA mutase (MUT), asparaginase like 1 (ASRGL1), dimethylglycine dehydrogenase (DMGDH) and SLC6A12. A total of 14 of these genes (PYCR1, ATP1B1, PLOD3, PDE7B, TK1, AKR1C1, SOD2, GBA3, PLOD1, MUT, ASRGL1, DMGDH, ADCY1, ADCY10) have been reported to be associated with cancer (23–30). According to the Recon2 model (10), the majority of these genes participate in nucleic acid and amino acid metabolism: ADCY1, PDE7B, TK1 and ADCY10 participate in nucleic acid metabolism; PYCR1 participates in arginine and proline metabolism; ACSS3 participates in tryptophan metabolism; PLOD3 participates in lysine metabolism; MUT participates in valine, leucine, and isoleucine metabolism; ASRGL1 participates in alanine and aspartate metabolism; and DMGDH participates in glycine, serine, alanine and threonine metabolism.
Enrichment of metabolic pathways
A total of 22 metabolic pathways were enriched by 4 groups of differential metabolic genes (FDR <0.05; Fig. 2B and Table II). Associations between the majority of these pathways and ccRCC have been previously reported (7–9,31–36); however, few of these studies investigated alterations to these metabolic pathways in different stages of the disease. These changes are presented in Table II.
Table II.
Metabolic pathways containing differentially expressed genes enriched in the 4 stages of clear cell renal cell carcinoma compared with control samples.
| Pathway | Control vs. stage I | Control vs. stage II | Control vs. stage III | Control vs. stage IV |
|---|---|---|---|---|
| Androgen and estrogen synthesis and metabolism | X | |||
| Propanoate metabolism | X | |||
| Cytochrome metabolism | X | |||
| Chondroitin synthesis | X | X | ||
| Arachidonic acid metabolism | X | X | ||
| Eicosanoid metabolism | X | X | ||
| Vitamin A metabolism | X | X | ||
| Exchange/demand reaction | X | X | X | |
| Blood group synthesis | X | X | X | |
| D-alanine metabolism | X | X | X | X |
| Transport, extracellular | X | X | X | X |
| Glyoxylate and dicarboxylate metabolism | X | X | X | X |
| Xenobiotics metabolism | X | X | X | X |
| Vitamin E metabolism | X | X | X | X |
| Vitamin D metabolism | X | X | X | X |
| Pyruvate metabolism | X | X | X | X |
| Glycosphingolipid metabolism | X | X | X | X |
| Phenylalanine metabolism | X | X | ||
| Glycine, serine, alanine and threonine metabolism | X | X | ||
| Glycolysis/gluconeogenesis | X | X | X | |
| Reactive oxygen species detoxification | X | X | ||
| Valine, leucine and isoleucine metabolism | X |
X, denotes that the corresponding pathway is enriched by the corresponding comparison (false discovery rate <0.05); blank cells denote FDR >0.05.
The xenobiotic metabolic pathway contains 23 cytochrome P450 family genes, of which 9 were expressed abnormally in each ccRCC disease stage: CYP2B6, CYP2J2, CYP1A1, CYP3A5, CYP3A4, CYP4B1, CYP2C18, CYP1A2 and CYP1B1. As aforementioned, the majority of enzymes that are encoded by these genes are known to metabolize xenobiotics, including the anticancer drugs cyclophosphamide and ifosphamide. Drug resistance induced by dysregulated xenobiotic metabolic pathways may be a major cause of chemotherapy resistance for patients with ccRCC (34,35).
From Table II, pathways that appear disordered in the early ccRCC disease stages are mainly associated with hormone, vitamin, glucose and lipid metabolism. Disorders of reactive oxygen species (ROS) detoxification and amino acid metabolism begin at the late stages of ccRCC. The valine, leucine and isoleucine metabolic pathway contains 35 genes (10) and the number of differentially expressed genes in this pathway increases significantly in the late stages of disease: Seven differentially expressed genes were observed in stage I disease, 6 in stage II, 10 in stage III and 13 in stage IV.
Discussion
Previously, studies of metabolic genes and pathways have attracted great interest in cancer research, to aid understanding of the molecular mechanisms in the process of carcinogenesis (37–39). Monteiro et al (40) revealed the important function that a metabolomic approach could serve in the study of biomarkers in RCC. Kim et al (41) identified potential biomarkers and pathogenic pathways in kidney cancer with urine metabolomic analysis. Shim et al (42) reported that L-2-hydroxyglutarate functions as an epigenetic modifier and putative oncometabolite in renal cancer. Armitage et al (43) analyzed the current trends and future perspectives of metabolomics in cancer biomarker identification. However, owing to the technical challenges in identifying metabolites, the advances in metabolomics have not been as great as those in genomics or transcriptomics (44). The present study used metabolism-associated gene expression to analyze metabolic changes at different disease stages. Although the heterogeneity between the expression of these genes should not be ignored (45), changes to the expression of these genes may not be consistent with changes in metabolite levels for a specific pathway. Previous studies revealed a positive association between mRNA expression and protein levels (46,47) and the metabolic pathway must disorder when a certain proportion of metabolic genes are differentially expressed within a specific pathway. Metabolism-associated gene expression was anomalous in patients with ccRCC.
To solve the issues surrounding early diagnosis and treatment for patients with ccRCC, an analysis of differentiation between control and case with each stage of the disease is necessary. Alterations to metabolic pathways differ at different stages of ccRCC. Abnormalities in hormone, vitamin, glucose and lipid metabolism start with the early stage of the disease and ROS detoxification and amino acid metabolism are aggravating with disease duration, particularly valine, leucine and isoleucine metabolism, which is badly damaged in stage IV. Shayman (7) revealed that eliglustat or associated analogues that inhibit glucosylceramide synthase reversed the disease phenotype and may be a potential treatment strategy for kidney disease. Recently, Czarnecka et al (8) summarized the function that hormonal signaling serves in RCC and hypothesized that inhibitors of this pathway could be used as therapeutics against this cancer. The association between circulating vitamin D and kidney cancer risk has been reported by multiple research teams (9,32). Trachootham et al (36) demonstrated that the increased generation of ROS could be exploited for therapeutic benefits to decrease drug resistance, based on the biochemical properties of cancer cells.
Whether the altered signal molecules or metabolites in stage I or II ccRCC may be biomarkers, permitting the early diagnosis of ccRCC, depends on future research. Drug resistance is a major barrier for treating advanced disease, rendering the xenobiotic metabolism pathway, associated with multiple cytochrome P450 family genes, worthy of substantial attention. Previous studies have revealed that abnormalities in CYP1B1, CYP1A1, CYP3A4 or CYP3A5 expression are associated with ccRCC, particularly drug resistance (34,48–54). The present study demonstrated that the overall disorder of the cytochrome P450 family genes may be the primary cause of marked drug resistance in patients with ccRCC. In addition to CYP1B1, CYP1A1, CYP3A4 and CYP3A5, the importance of other genes in this family requires further study. These genes may serve as therapeutic targets of ccRCC. Further studies on early diagnostic markers may focus on the metabolites in hormone, vitamin, glucose and lipid metabolic pathways.
Acknowledgements
The present study was supported by the National Basic Research Program of China (grant no. 2013CB835100) and the National Natural Science Foundation of China (grant nos. 31401142 and 31401137).
References
- 1.Bhatt JR, Finelli A. Landmarks in the diagnosis and treatment of renal cell carcinoma. Nat Rev Urol. 2014;11:517–525. doi: 10.1038/nrurol.2014.194. [DOI] [PubMed] [Google Scholar]
- 2.Shuch B, Amin A, Armstrong AJ, Eble JN, Ficarra V, Lopez-Beltran A, Martignoni G, Rini BI, Kutikov A. Understanding pathologic variants of renal cell carcinoma: Distilling therapeutic opportunities from biologic complexity. Eur Urol. 2015;67:85–97. doi: 10.1016/j.eururo.2014.04.029. [DOI] [PubMed] [Google Scholar]
- 3.Escudier B, Porta C, Schmidinger M, Algaba F, Patard JJ, Khoo V, Eisen T, Horwich A, ESMO Guidelines Working Group Renal cell carcinoma: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2014;25(Suppl 3):iii49–iii56. doi: 10.1093/annonc/mdu259. [DOI] [PubMed] [Google Scholar]
- 4.Abe H, Kamai T. Recent advances in the treatment of metastatic renal cell carcinoma. Int J Urol. 2013;20:944–955. doi: 10.1111/iju.12187. [DOI] [PubMed] [Google Scholar]
- 5.Rahman M, Hasan MR. Cancer metabolism and drug resistance. Metabolites. 2015;5:571–600. doi: 10.3390/metabo5040571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Linehan WM, Srinivasan R, Schmidt LS. The genetic basis of kidney cancer: A metabolic disease. Nat Rev Urol. 2010;7:277–285. doi: 10.1038/nrurol.2010.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shayman JA. Targeting glycosphingolipid metabolism to treat kidney disease. Nephron. 2016;134:37–42. doi: 10.1159/000444926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Czarnecka AM, Niedzwiedzka M, Porta C, Szczylik C. Hormone signaling pathways as treatment targets in renal cell cancer (Review) Int J Oncol. 2016;48:2221–2235. doi: 10.3892/ijo.2016.3460. [DOI] [PubMed] [Google Scholar]
- 9.Mondul AM, Weinstein SJ, Moy KA, Männistö S, Albanes D. Vitamin D-binding protein, circulating vitamin D and risk of renal cell carcinoma. Int J Cancer. 2014;134:2699–2706. doi: 10.1002/ijc.28596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thiele I, Swainston N, Fleming RM, Hoppe A, Sahoo S, Aurich MK, Haraldsdottir H, Mo ML, Rolfsson O, Stobbe MD, et al. A community-driven global reconstruction of human metabolism. Nat Biotechnol. 2013;31:419–425. doi: 10.1038/nbt.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cancer genome atlas research, corp-author. Comprehensive molecular characterization of clear cell renal cell carcinoma. Nature. 2013;499:43–49. doi: 10.1038/nature12222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wittekind C. 2010 TNM system: On the 7th edition of TNM classification of malignant tumors. Pathologe. 2010;31:331–332. doi: 10.1007/s00292-010-1349-3. (In German) [DOI] [PubMed] [Google Scholar]
- 13.Kolde R. Pheatmap: Pretty Heatmaps. R package version 1.0.8. 2015. https://CRAN.R-project.org/package=pheatmap https://CRAN.R-project.org/package=pheatmap
- 14.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK. limma powers differential expressionanalyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jiang Z, Hu S, Hua D, Ni J, Xu L, Ge Y, Zhou Y, Cheng Z, Wu S. β3GnT8 plays an important role in CD147 signal transduction as an upstream modulator of MMP production in tumor cells. Oncol Rep. 2014;32:1156–1162. doi: 10.3892/or.2014.3280. [DOI] [PubMed] [Google Scholar]
- 16.Shen L, Yu M, Xu X, Gao L, Ni J, Luo Z, Wu S. Knockdown of β3GnT8 reverses 5-fluorouracil resistance in human colorectal cancer cells via inhibition the biosynthesis of polylactosamine-type N-glycans. Int J Oncol. 2014;45:2560–2568. doi: 10.3892/ijo.2014.2672. [DOI] [PubMed] [Google Scholar]
- 17.Liu J, Shen L, Yang L, Hu S, Xu L, Wu S. High expression of β3GnT8 is associated with the metastatic potential of human glioma. Int J Mol Med. 2014;33:1459–1468. doi: 10.3892/ijmm.2014.1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hua D, Qin F, Shen L, Jiang Z, Zou ST, Xu L, Cheng ZH, Wu SL. β3GnT8 regulates laryngeal carcinoma cell proliferation via targeting MMPs/TIMPs and TGF-β1. Asian Pac J Cancer Prev. 2012;13:2087–2093. doi: 10.7314/APJCP.2012.13.5.2087. [DOI] [PubMed] [Google Scholar]
- 19.Lien HC, Lee YH, Jeng YM, Lin CH, Lu YS, Yao YT. Differential expression of hyaluronan synthase 2 in breast carcinoma and its biological significance. Histopathology. 2014;65:328–339. doi: 10.1111/his.12390. [DOI] [PubMed] [Google Scholar]
- 20.Warrington NM, Sun T, Rubin JB. Targeting brain tumor cAMP: The case for sex-specific therapeutics. Front Pharmacol. 2015;6:153. doi: 10.3389/fphar.2015.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dong H, Claffey KP, Brocke S, Epstein PM. Expression of phosphodiesterase 6 (PDE6) in human breast cancer cells. SpringerPlus. 2013;2:680. doi: 10.1186/2193-1801-2-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Law V, Knox C, Djoumbou Y, Jewison T, Guo AC, Liu Y, Maciejewski A, Arndt D, Wilson M, Neveu V, et al. DrugBank 4.0: Shedding new light on drug metabolism. Nucleic Acids Res. 2014;42:D1091–D1097. doi: 10.1093/nar/gkt1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong Y, Kim WJ, Bang CY, Lee JC, Oh YM. Identification of alternative splicing and fusion transcripts in non-small cell lung cancer by RNA sequencing. Tuberc Respir Dis (Seoul) 2016;79:85–90. doi: 10.4046/trd.2016.79.2.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicastri A, Gaspari M, Sacco R, Elia L, Gabriele C, Romano R, Rizzuto A, Cuda G. N-glycoprotein analysis discovers new up-regulated glycoproteins in colorectal cancer tissue. J Proteome Res. 2014;13:4932–4941. doi: 10.1021/pr500647y. [DOI] [PubMed] [Google Scholar]
- 25.Nisman B, Appelbaum L, Yutkin V, Nechushtan H, Hubert A, Uziely B, Pode D, Peretz T. Serum thymidine kinase 1 activity following nephrectomy for renal cell carcinoma and radiofrequency ablation of metastases to lung and liver. Anticancer Res. 2016;36:1791–1797. [PubMed] [Google Scholar]
- 26.Wenners A, Hartmann F, Jochens A, Roemer AM, Alkatout I, Klapper W, van Mackelenbergh M, Mundhenke C, Jonat W, Bauer M. Stromal markers AKR1C1 and AKR1C2 are prognostic factors in primary human breast cancer. Int J Clin Oncol. 2016;21:548–556. doi: 10.1007/s10147-015-0924-2. [DOI] [PubMed] [Google Scholar]
- 27.Wang L, Seino J, Tomotake H, Funakoshi Y, Hirayama H, Suzuki T. Co-Expression of NEU2 and GBA3 causes a drastic reduction in cytosolic sialyl free N-glycans in human MKN45 stomach cancer cells-evidence for the physical interaction of NEU2 and GBA3. Biomolecules. 2015;5:1499–1514. doi: 10.3390/biom5031499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilkes DM, Bajpai S, Wong CC, Chaturvedi P, Hubbi ME, Wirtz D, Semenza GL. Procollagen lysyl hydroxylase 2 is essential for hypoxia-induced breast cancer metastasis. Mol Cancer Res. 2013;11:456–466. doi: 10.1158/1541-7786.MCR-12-0629. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Edqvist PH, Huvila J, Forsström B, Talve L, Carpén O, Salvesen HB, Krakstad C, Grénman S, Johannesson H, Ljungqvist O, et al. Loss of ASRGL1 expression is an independent biomarker for disease-specific survival in endometrioid endometrial carcinoma. Gynecol Oncol. 2015;137:529–537. doi: 10.1016/j.ygyno.2015.03.055. [DOI] [PubMed] [Google Scholar]
- 30.Liu G, Hou G, Li L, Li Y, Zhou W, Liu L. Potential diagnostic and prognostic marker dimethylglycine dehydrogenase (DMGDH) suppresses hepatocellular carcinoma metastasis in vitro and in vivo. Oncotarget. 2016;7:32607–32616. doi: 10.18632/oncotarget.8927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu Z, Yao Y, Song Q, Yang J, Zhao X, Yang P, Kang J. Metabolism-related enzyme alterations identified by proteomic analysis in human renal cell carcinoma. Onco Targets Ther. 2016;9:1327–1337. doi: 10.2147/OTT.S91953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Joh HK, Giovannucci EL, Bertrand KA, Lim S, Cho E. Predicted plasma 25-hydroxyvitamin D and risk of renal cell cancer. J Natl Cancer Inst. 2013;105:726–732. doi: 10.1093/jnci/djt082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shang Y, Yi S, Cui D, Han G, Liu C. Vitamin E Intake and risk of renal cell carcinoma: A meta-analysis of 7 case-control studies. J Ren Nutr. 2015;25:339–344. doi: 10.1053/j.jrn.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 34.Mitsui Y, Chang I, Fukuhara S, Hiraki M, Arichi N, Yasumoto H, Hirata H, Yamamura S, Shahryari V, Deng G, et al. CYP1B1 promotes tumorigenesis via altered expression of CDC20 and DAPK1 genes in renal cell carcinoma. BMC Cancer. 2015;15:942. doi: 10.1186/s12885-015-1951-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Narjoz C, Favre A, McMullen J, Kiehl P, Montemurro M, Figg WD, Beaune P, de Waziers I, Rochat B. Important role of CYP2J2 in protein kinase inhibitor degradation: A possible role in intratumor drug disposition and resistance. PLoS One. 2014;9:e95532. doi: 10.1371/journal.pone.0095532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trachootham D, Alexandre J, Huang P. Targeting cancer cells by ROS-mediated mechanisms: A radical therapeutic approach? Nat Rev Drug Discov. 2009;8:579–591. doi: 10.1038/nrd2803. [DOI] [PubMed] [Google Scholar]
- 37.Rodrigues D, Monteiro M, Jeronimo C, Henrique R, Belo L, Bastos ML, de Pinho P Guedes, Carvalho M. Renal cell carcinoma: A critical analysis of metabolomic biomarkers emerging from current model systems. Transl Res. 2017;180:1–11. doi: 10.1016/j.trsl.2016.07.018. [DOI] [PubMed] [Google Scholar]
- 38.Masui K, Cavenee WK, Mischel PS. Cancer metabolism as a central driving force of glioma pathogenesis. Brain Tumor Pathol. 2016;33:161–168. doi: 10.1007/s10014-016-0265-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johnson C, Warmoes MO, Shen X, Locasale JW. Epigenetics and cancer metabolism. Cancer Lett. 2015;356:309–314. doi: 10.1016/j.canlet.2013.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Monteiro MS, Carvalho M, de Lourdes Bastos M, de Pinho PG. Biomarkers in renal cell carcinoma: A metabolomics approach. Metabolomics. 2014;10:1210–1222. doi: 10.1007/s11306-014-0659-5. [DOI] [Google Scholar]
- 41.Kim K, Taylor SL, Ganti S, Guo L, Osier MV, Weiss RH. Urine metabolomic analysis identifies potential biomarkers and pathogenic pathways in kidney cancer. OMICS. 2011;15:293–303. doi: 10.1089/omi.2010.0094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shim EH, Livi CB, Rakheja D, Tan J, Benson D, Parekh V, Kho EY, Ghosh AP, Kirkman R, Velu S, et al. L-2-Hydroxyglutarate: An epigenetic modifier and putative oncometabolite in renal cancer. Cancer discov. 2014;4:1290–1298. doi: 10.1158/2159-8290.CD-13-0696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Armitage EG, Barbas C. Metabolomics in cancer biomarker discovery: Current trends and future perspectives. J Pharm Biomed Anal. 2014;87:1–11. doi: 10.1016/j.jpba.2013.08.041. [DOI] [PubMed] [Google Scholar]
- 44.You L, Zhang B, Tang YJ. Application of stable isotope-assisted metabolomics for cell metabolism studies. Metabolites. 2014;4:142–165. doi: 10.3390/metabo4020142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hakimi AA, Reznik E, Lee CH, Creighton CJ, Brannon AR, Luna A, Aksoy BA, Liu EM, Shen R, Lee W, et al. An integrated metabolic atlas of clear cell renal cell carcinoma. Cancer Cell. 2016;29:104–116. doi: 10.1016/j.ccell.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwanhausser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, Selbach M. Global quantification of mammalian gene expression control. Nature. 2011;473:337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 47.Nagaraj N, Wisniewski JR, Geiger T, Cox J, Kircher M, Kelso J, Pääbo S, Mann M. Deep proteome and transcriptome mapping of a human cancer cell line. Mol Syst Biol. 2011;7:548. doi: 10.1038/msb.2011.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meng FD, Ma P, Sui CG, Tian X, Jiang YH. Association between cytochrome P450 1A1 (CYP1A1) gene polymorphisms and the risk of renal cell carcinoma: A meta-analysis. Sci Rep. 2015;5:8108. doi: 10.1038/srep08108. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 49.Chang I, Fukuhara S, Wong DK, Gill A, Mitsui Y, Majid S, Saini S, Yamamura S, Chiyomaru T, Hirata H, et al. Cytochrome P450 1B1 polymorphisms and risk of renal cell carcinoma in men. Tumour Biol. 2014;35:10223–10230. doi: 10.1007/s13277-014-2292-3. [DOI] [PubMed] [Google Scholar]
- 50.Quivy A, Daste A, Harbaoui A, Duc S, Bernhard JC, Gross-Goupil M, Ravaud A. Optimal management of renal cell carcinoma in the elderly: A review. Clin Interv Aging. 2013;8:433–442. doi: 10.2147/CIA.S30765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang G, Hou J, Ma L, Xie J, Yin J, Xu D, Chang W, Tan X, Su T, Zhang H, Cao G. Risk factor for clear cell renal cell carcinoma in Chinese population: A case-control study. Cancer epidemiol. 2012;36:177–182. doi: 10.1016/j.canep.2011.09.006. [DOI] [PubMed] [Google Scholar]
- 52.Sverko A, Sobočanec S, Kusic B, Kušić B, Mačak-Šafranko Z, Sarić A, Leniček T, Kraus O, Andrišić L, Korolija M, et al. Superoxide dismutase and cytochrome P450 isoenzymes might be associated with higher risk of renal cell carcinoma in male patients. Int Immunopharmacol. 2011;11:639–645. doi: 10.1016/j.intimp.2010.12.023. [DOI] [PubMed] [Google Scholar]
- 53.Kollmannsberger C, Soulieres D, Wong R, Scalera A, Gaspo R, Bjarnason G. Sunitinib therapy for metastatic renal cell carcinoma: Recommendations for management of side effects. Can Urol Assoc J. 2007;1(Suppl 2):S41–S54. doi: 10.5489/cuaj.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McFadyen MC, Melvin WT, Murray GI. Cytochrome P450 CYP1B1 activity in renal cell carcinoma. Br J Cancer. 2004;91:966–971. doi: 10.1038/sj.bjc.6602053. [DOI] [PMC free article] [PubMed] [Google Scholar]